Abstract
Background
Pyrazinamide (PZA) is among the first-line drugs for the treatment of tuberculosis. In vitro, it kills semidormant mycobacteria only at low pH. The purpose of this study was to compare PZA resistance with pyrazinamidase (PZase) activity and the genotype to better understand the molecular basis of PZA resistance and to expand the profile of pncA mutations worldwide.
Results
Of the 28 tested strains of Mycobacterium tuberculosis, 6 were susceptible to PZA and positive for PZase activity and had no pncA mutations. Twenty-one strains were resistant to PZA and negative for PZase activity and had mutations in the pncA gene, including 15 point mutations, 5 insertions, and 2 deletions. One strain had no mutation in the pncA gene, even though it was resistant to PZA and negative for PZase activity. Three isolates had adenine to guanine point mutations in the -11 upstream region, making this the most common type of pncA mutations in this study, with at least two different RFLP patterns.
Conclusion
These data help in the understanding of the molecular basis of PZA resistance. An adenine to guanine point mutation in the -11 upstream region was the most common type of pncA mutation in our isolates. The results of pncA mutation analyses should be carefully interpreted for epidemiologic purposes.
Background
Pyrazinamide (PZA) is among the first-line drugs used to treat tuberculosis. In vitro, it kills semidormant mycobacteria only at low pH [1]. In vitro susceptibility testing sometimes fails because of the poor growth of mycobacteria at low pH. Therefore, the pyrazinamidase (PZase) test, which was originally used for the differentiation of Mycobacterium tuberculosis from weakly niacin-positive strains of M. bovis, has been used to identify susceptible strains of M. tuberculosis, because PZase converts the prodrug PZA to pyrazinoic acid, the active form of the drug [2]. The pncA gene encodes PZase, and mutations in pncA are associated with resistance to PZA or loss of PZase activity [3]. The purpose of this study was to compare PZase activity with the genotype to better understand the molecular basis of PZA resistance and to expand the profile of pncA mutations worldwide.
Materials and Methods
Bacterial strains, PZA susceptibility and PZase activity
Twenty-eight clinical isolates of M. tuberculosis were included. Twenty-three PZase-negative clinical isolates were provided from Korean Institute of Tuberculosis, and these strains had originally been collected from various sites in this country for the purpose of susceptibility testing. Five PZase-positive isolates were collected randomly among the clinical isolates grown at Pusan National University Hospital (PNUH). The type strain M. tuberculosis H37Rv was included as a PZA-susceptible, and thus PZase-positive, control. All isolates were grown in Löwenstein-Jensen medium at 37°C for 3 to 4 weeks. The PZA susceptibility was tested by using Löwenstein-Jensen medium at pH 5.6 with 100 and 500 μg of PZA per mL [4]. The PZase assay was performed by the method described in the Clinical Microbiology Procedure Handbook [2]. Briefly, 6.5 g of Dubos broth base, 0.1 g of PZA, 2.0 g of sodium pyruvate and 15.0 g of agar were dissolved in 1 L of distilled water and heated to dissolve the components. The solution was dispensed in 5-mL amounts into screw-cap tubes and stored at 2 to 8°C until use after solidification of the agar with the tubes in an upright position. A heavy loopful of growth from an actively growing subculture was inoculated. After incubation at 37°C for 4 or 7 days, 1 mL of freshly prepared 1% ferrous ammonium sulfate was added to each tube. A pink band in the agar indicated a positive test.
Genomic DNA preparation, PCR and DNA sequencing
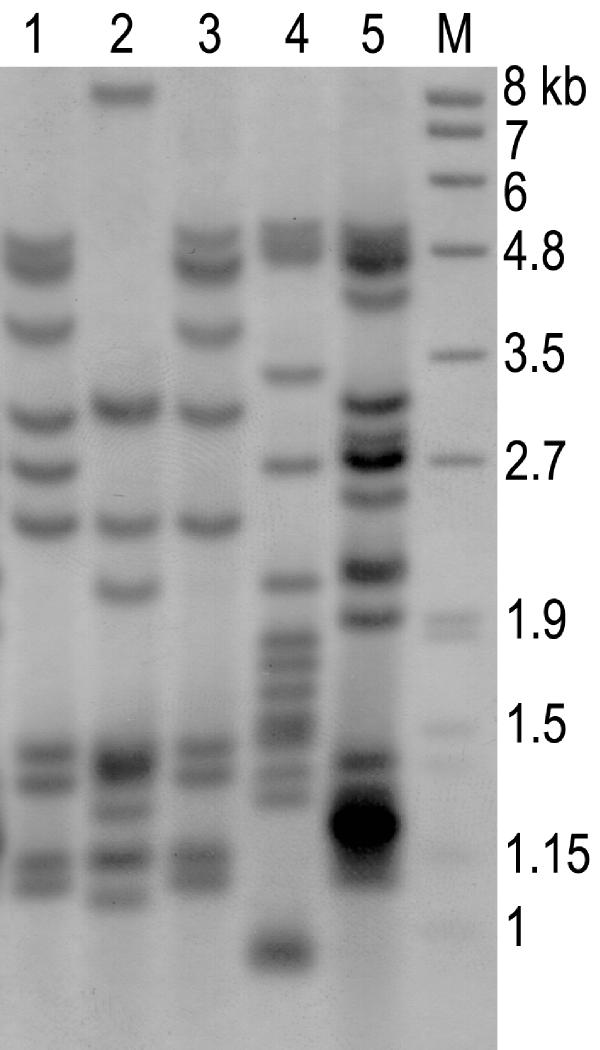
DNA was extracted using an InstaGene matrix kit (Bio-Rad Laboratories Inc., Hercules, CA). A 720-bp segment, including the entire open reading frame, of the pncA gene was amplified by using the conditions and the set of primers P1 and P6 [5]. The PCR products were cut from the gel and purified with the QiaAmp PCR purification kit (QIAGEN GmbH, Germany), according to the manufacturer's instructions. The gel-purified PCR products were quantitated, adjusted to a 200-μmol concentration, and used for direct sequencing by the ABI 377 automatic DNA sequencer (Applied Biosystems Inc., Foster, CA) with 4 pM of each of the above-mentioned primers. The RFLP analysis (Figure 1) was performed by an internationally standardized method for the three clinical isolates showing the same pncA mutations [6], and the hybridized membrane was detected by the colorimetric method using the Roche digoxigenin detection kit (F. Hoffmann-La Roche Ltd., Switzerland). The 245-bp mycobacterial IS probe was amplified by PCR with a DIG DNA labeling kit (Roche) using INS-1 and INS-2 primers.
Figure 1.
RFLP analysis patterns of three strains containing promoter mutations. Lane M, molecular markers; lanes 1-3, tested strains containing -11 upstream adenine to guanine substitutions; lane 4, M. tuberculosis H37Rv; lane 5, clinical isolate as a control strain.
Results and discussion
All five isolates collected from our hospital were PZase-positive and susceptible to PZA with minimal inhibitory concentrations (MIC) of <100 μg/mL, which is consistent with the initial routine tests. These isolates had pncA sequences identical to the published sequences of pncA of M. tuberculosis H37Rv [3]. Among the 23 isolates collected from the Korean Institute of Tuberculosis, one was PZase-positive and susceptible to PZA (MIC <100 μg/mL). It had no pncA mutation in the entire open reading frame, including the upstream region. All of the remaining 22 isolates were negative for PZase activity and resistant to PZA (MIC >500 μg/mL). Among them, 21 organisms (96%) had mutations in the pncA gene. Eleven organisms had twelve point mutations, including two point mutations in one organism, which resulted in one silent mutation, one nonsense mutation, and 10 missense mutations. Of those, six mutations have not previously been described. One strain with a silent mutation actually had another missense-type point mutation. Three isolates had upstream mutations at nucleotide -11, the single most common mutation, resulting in an adenine to guanine change. One had a 3-bp insertion, resulting in a slipped-strand mispairing of PncA. Four had 1- or 2-bp insertions, and two had a 2- or 234-bp deletion, all resulting in frameshift mutations in PncA (Table 1). A clustering tendency was apparent in that 40% (6/15) of the point mutations were located in the region between residues 132 and 142 of the PZase sequence. Among the three strains with the promoter region mutations, one isolate showed a clearly different RFLP pattern from the other two.
Table 1.
pncA nucleotide and amino acid changes in PZase-negative M. tuberculosis clinical isolates from Korea
| Mutation | Nucleotide | Amino acid | No. of |
| site | changes | changes | isolates |
| -11 | A to Gb | Mutation in promoter | 3 |
| 23 | TCG insertion | Slipped-strand mispairing | 1 |
| 41 | G41A | Missense (Cys14 Tyr) | 1 |
| 56 | 234-bp deletion | Frameshift | 1 |
| 172 | T172Cc | Missense (Phe58 Leu) | 1 |
| 190 | T190G | Missense (Tyr64 Asp) | 1 |
| 212 | A212Gd | Missense (His71 Arg) | 1 |
| 227 | C227Te | Missense (Thr76 Ile) | 1 |
| 317 | CT insertion | Frameshift | 1 |
| 382 | AG insertion | Frameshift | 1 |
| 393 | GT insertion | Frameshift | 1 |
| 393 | T insertion | Frameshift | 1 |
| 395 | G395T | Missense (Gly132 Val) | 2 |
| 403 | A403Cd | Missense (Thr135 Pro) | 1 |
| 407 | A407G | Missense (Asp136 Gly) | 1 |
| 421 | C421T | Nonsense (Gln141 Termination) | 1 |
| 425, 180a | C425Tf, C180T | Missense (Thr142 Met), Silent (Gly60 Gly) | 1 |
| 513 | GC deletion | Frameshift | 1 |
The PZase test has been used for the differentiation of M. tuberculosis from M. bovis, M. avium complex from niacin-negative M. bovis, and M. marinum from M. kansasii [2]. The test also has been used for the detection of PZA-resistant M. tuberculosis strains [7]. However, the test has a shortcoming in that if old colonies are used, false-negative results may be obtained. It seems that in stationary- or death-phase colonies, the enzyme activity is reduced below the limits needed to get a positive reaction. This explains why one of the 23 strains that had resulted in a negative PZase reaction turned out to be PZase positive in this study.
Of the 22 PZase-negative strains, 21 (96%) showed pncA mutations in this study. In other reports [5,8,9], pncA mutations were found in 72% to 87% of PZA-resistant strains and in 97% of PZase-negative strains. Sreevatsan et al [9] suggested the possibility of another mechanism of PZA resistance because no mutation in pncA or its upper promoter was found in 28% of PZA-resistant M. tuberculosis strains. In contrast, Hewlett et al [10] demonstrated the low reproducibility of susceptibility to PZA, and Scorpio et al [5] proved false resistance to PZA in susceptible strains. So, if any other resistance mechanism exists, it plays only a minor role in PZA resistance, and more than 95% of PZA-resistant M. tuberculosis strains likely harbor pncA mutations in this study.
Twenty-two mutations included single nucleotide substitutions, resulting in silent, missense, or nonsense mutations, and deletions and insertions of as many as 234 nucleotides. Of those, six single point mutations in seven strains have not been described in previous studies [3, 4, 5, 9, 11, 12, 13, 14, 15, 16]. These mutations are arrayed along virtually the entire length of the gene [9], even though a clustering tendency was apparent in that 40% (6/15) of the single point mutations were located in the region between residues 132 and 142 of the PZase. The clustering tendency in this region was described previously [5, 12]. In addition, however, we think that attention should be paid to the mutations in the promoter region, especially the -11 upstream region. Although no strains having this type of mutation were described in some studies, the results of other studies, including the present one, demonstrate that it is the most common type of pncA mutation [4, 5, 9, 11, 13, 14, 16] (Table 2). Moreover, most of the mutations were substitutions of guanine for adenine. The consensus sequence TATAAT, known as the pribnow box or -10 site, is located approximately 10 bp upstream of the transcription start site of many bacterial genes. So, it seems that mutation of the -11 upstream site blocks binding of RNA polymerase to the promoter site, resulting in inhibition of transcription in the correct place. However, further experiments such as in vitro mutagenesis should be performed to demonstrate the relation between the promoter mutation and loss of PZase activity. Theoretically, the same mutations of the pncA genes would rarely be present in unrelated isolates because mutations occur randomly along the whole pncA gene. Therefore, pncA mutations could be a useful tool for epidemiologic investigations. In fact, Cheng et al reported that 21 strains having the same mutations were found to be highly related by molecular typing, suggesting an outbreak from a single source [4]. However, the same mutations within a region with a clustering tendency, such as the -11 upstream region and the region between residues 132 and 142, do not necessarily mean related strains. In the present study, one of the strains with the promoter mutation showed an RFLP pattern different from those of the other two, suggesting different sources of infection. In fact, the three strains were collected during the years 1985, 1990 and 1993, and the two strains of the same guanine to thymine substitution at nt 395 were collected in 1990 and 1997, suggesting that strains with the same mutations came from different sources. Thus, we think that pncA mutations should be applied carefully for epidemiologic analysis.
Table 2.
Type and frequency of mutations described in the pncA promoter region and the region between residues 132 and 142 of the PncAa
| Mutation site | Nucleotide change | Amino acid change | Frequency | |
| -16∼-11 | AACGTA to GGCAGTT | Mutation in promoter | 1 | |
| -12 | T to G | Mutation in promoter | 1 | |
| -11 | A to C | Mutation in promoter | 1 | |
| -11 | A to G | Mutation in promoter | 13 | |
| -7 | T to C | Mutation in promoter | 1 | |
| 394 | G to A | Gly132 | Ser | 2 |
| 395 | G to A or T | Gly132 | Asp or Val | 4 |
| 398 | T to C | Ile133 | Thr | 1 |
| 401 | C to T | Ala134 | Val | 3 |
| 403 | A to C | Thr135 | Pro | 2 |
| 406 | G to C | Asp136 | His | 1 |
| 407 | A to G | Asp136 | Gly | 1 |
| 410 | A to C or G | His137 | Pro or Arg | 4 |
| 413 | G to A or C | Cys138 | Thr or Ser | 5 |
| 415 | G to C or A | Val139 | Leu or Met | 5 |
| 416 | T to C or G | Val139 | Ala or Gly | 6 |
| 421 | C to T | Gln141 | termination | 1 |
| 422 | A to C | Gln141 | Pro | 3 |
| 424 | A to C or G | Thr142 | Pro or Ala | 2 |
| 425 | C to A or T | Thr142 | Lys or Met | 8 |
| 402∼403 | CC insertion | Frameshift | 1 | |
| 407∼408 | C insertion | Frameshift | 1 | |
| 416∼417 | TG deletion | Frameshift | 1 | |
| 395∼411 | 17-bp deletion | Frameshift | 1 | |
Conclusion
These data provide a better understanding of the molecular basis of PZA resistance and expand the data on pncA mutations worldwide. Furthermore, it was demonstrated that adenine to guanine point mutations in the -11 upstream region are the most common type of pncA mutations. Because of the different RFLP patterns in the strains having the same mutations, the results of pncA mutations should be carefully interpreted for epidemiologic purposes.
Competing interests
Have you in the past five years received reimbursements, fees, funding, or salary from an organisation that may in any way gain or lose financially from the publication of this paper? No
Do you hold any stocks or shares in an organisation that may in any way gain or lose financially from the publication of this paper? No
Do you have any other financial competing interests? No
Are there any non-financial competing interests you would like to declare in relation to this paper? No
Pre-publication history
The pre-publication history for this paper can be accessed here:
http://www.biomedcentral.com/content/backmatter/1471-2334-1-4-b1.pdf
Contributor Information
Soon Kew Park, Email: snkpark@hyowon.cc.pusan.ac.kr.
Jung Yoo Lee, Email: ljy9039@hitel.net.
Chulhun Ludgerus Chang, Email: cchl@hyowon.cc.pusan.ac.kr.
Min Ki Lee, Email: mklee@hyowon.cc.pusan.ac.kr.
Han Chul Son, Email: hacson@hyowon.cc.pusan.ac.kr.
Cheol Min Kim, Email: kimcm@hyowon.cc.pusan.ac.kr.
Hyun Jung Jang, Email: biochiphyun@hotmail.com.
Hee Kyung Park, Email: dnachiphk@hotmail.com.
Seok Hoon Jeong, Email: kscpjsh@ns.kosinmed.or.kr.
References
- Heifets L, Lindholm-Levy P. Pyrazinamide sterilizing activity in vitro against semidormant Mycobacterium tuberculosis bacterial populations. Am Rev Respir Dis. 1992;145:1223–5. doi: 10.1164/ajrccm/145.5.1223. [DOI] [PubMed] [Google Scholar]
- Isenberg HD. Mycobacteriology: Identification tests for mycobacteria. In Clinical Microbiology Procedure Handbook Edited by Isenberg HD,vol 1 Washington DC: ASM Press, 1995;3:12–17. [Google Scholar]
- Scorpio A, Zhang Y. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat Med. 1996;2:662–7. doi: 10.1038/nm0696-662. [DOI] [PubMed] [Google Scholar]
- Cheng SJ, Thibert L, Sanchez T, Heifets L, Zhang Y. pncA mutations as a major mechanism of pyrazinamide resistance in Mycobacterium tuberculosis: spread of a monoresistant strain in Quebec, Canada. Antimicrob Agents Chemother. 2000;44:528–32. doi: 10.1128/AAC.44.3.528-532.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorpio A, Lindholm Levy P, Heifets L, Gilman R, Siddiqi S, Cynamon M, Zhang Y. Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1997;41:540–3. doi: 10.1128/aac.41.3.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Embden JD, Cave MD, Crawford JT, Dale JW, Eisenach KD, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick TM, et al. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–9. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno K, Feldmann FM, McDermott W. Pyrazinamide susceptibility and amidase activity of tubercle bacilli. Am Rev Respir Dis. 1967;95:461–9. doi: 10.1164/arrd.1967.95.3.461. [DOI] [PubMed] [Google Scholar]
- Hirano K, Takahashi M, Kazumi Y, Fukasawa Y, Abe C. Mutation in pncA is a major mechanism of pyrazinamide resistance in Mycobacterium tuberculosis. Tuber Lung Dis. 1997;78:117–22. doi: 10.1016/s0962-8479(98)80004-x. [DOI] [PubMed] [Google Scholar]
- Sreevatsan S, Pan X, Zhang Y, Kreiswirth BN, Musser JM. Mutations associated with pyrazinamide resistance in pncA of Mycobacterium tuberculosis complex organisms. Antimicrob Agents Chemother. 1997;41:636–40. doi: 10.1128/aac.41.3.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewlett D, Jr, Horn DL, Alfalla C. Drug-resistant tuberculosis: inconsistent results of pyrazinamide susceptibility testing. JAMA. 1995;273:916–7. doi: 10.1001/jama.273.12.916. [DOI] [PubMed] [Google Scholar]
- Marttila HJ, Marjamaki M, Vyshnevskaya E, Vyshnevskiy BI, Otten TF, Vasilyef AV, Viljanen MK. pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis isolates from northwestern Russia. Antimicrob Agents Chemother. 1999;43:1764–6. doi: 10.1128/aac.43.7.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre N, Sougakoff W, Truffot-Pernot C, Jarlier V. Characterization of new mutations in pyrazinamide-resistant strains of Mycobacterium tuberculosis and identification of conserved regions important for the catalytic activity of the pyrazinamidase PncA. Antimicrob Agents Chemother. 1999;43:1761–3. doi: 10.1128/aac.43.7.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlock GP, Crawford JT, Butler WR, Brim SE, Sikes D, Mazurek GH, Woodley CL, Cooksey RC. Phenotypic characterization of pncA mutants of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2000;44:2291–5. doi: 10.1128/AAC.44.9.2291-2295.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestdagh M, Fonteyne PA, Realini L, Rossau R, Jannes G, Mijs W, De Smet KA, Portaels F, Van den Eeckhout E. Relationship between pyrazinamide resistance, loss of pyrazinamidase activity, and mutations in the pncA locus in multidrug-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1999;43:2317–9. doi: 10.1128/aac.43.9.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown TJ, Tansel O, French GL. Simultaneous identification and typing of multi-drug-resistant Mycobacterium tuberculosis isolates by analysis of pncA and rpoB. J Med Microbiol. 2000;49:651–6. doi: 10.1099/0022-1317-49-7-651. [DOI] [PubMed] [Google Scholar]
- Escalante P, Ramaswamy S, Sanabria H, Soini H, Pan X, Valiente-Castillo O, Musser JM. Genotypic characterization of drug-resistant Mycobacterium tuberculosis isolates from Peru. Tuber Lung Dis. 1998;79:111–8. doi: 10.1054/tuld.1998.0013. [DOI] [PubMed] [Google Scholar]