Abstract
It is now well established that mature mammalian spermatozoa carry a population of mRNA molecules, at least some of which are transferred to the oocyte at fertilization, however, their function remains largely unclear. To shed light on the evolutionary conservation of this feature of sperm biology, we analysed highly purified populations of mature sperm from the fruitfly, Drosophila melanogaster. As with mammalian sperm, we found a consistently enriched population of mRNA molecules that are unlikely to be derived from contaminating somatic cells or immature sperm. Using tagged transcripts for three of the spermatozoal mRNAs, we demonstrate that they are transferred to the oocyte at fertilization and can be detected before, and at least until, the onset of zygotic gene expression. We find a remarkable conservation in the functional annotations associated with fly and human spermatozoal mRNAs, in particular, a highly significant enrichment for transcripts encoding ribosomal proteins (RPs). The substantial functional coherence of spermatozoal transcripts in humans and the fly opens the possibility of using the power of Drosophila genetics to address the function of this enigmatic class of molecules in sperm and in the oocyte following fertilization.
Keywords: Drosophila, spermatogenesis, sperm, spermatozoa, transcriptome
1. Introduction
Spermiogenesis, the production of a differentiated male gamete, is a remarkable example of cellular differentiation, yet despite the critical role the male gamete plays in the life cycle we still know relatively little about its full range of functions. In the past, it was widely believed that the only function of the spermatozoa was to deliver the male genome to the egg. However, it has been known for over 50 years that fertilization results in a complete fusion of the entire sperm and egg cells, and more recent genetic and functional studies have identified essential sperm cell components and molecules delivered to the zygote at fertilization [1–3]. These include a functional centrosome [4], an activation stimulus [5,6] and other paternal products [7,8]. In Drosophila, detailed molecular genetic studies have identified paternal gene products absolutely essential for successful fertilization [9–12] further lending credence to the idea that sperm-derived factors provide important functionality during, and perhaps following, fertilization and zygote formation.
Furthermore, a number of studies suggested that mature mammalian spermatozoa contain a variety of RNA species [13–15]. Several studies strengthened these observations and the presence of poly-adenylated messenger RNAs in spermatozoa [16,17] is now widely accepted. Work with both human and mouse demonstrated the delivery of some of these sperm transcripts to the oocyte [18,19]. Furthermore, comparisons between sperm from fertile and infertile males indicate that sperm transcripts may have diagnostic value and suggests a relationship between sperm transcripts and proper sperm function [20–22].
During normal spermatogenesis a large number of transcripts are produced, which encode the myriad of proteins, many sperm-specific, that are needed for spermiogenesis. Transcripts are often stored in the spermatocyte or spermatid cytoplasm for long periods before translation [23]. A critical aspect of spermiogenesis in mammals and Drosophila is the change in chromatin structure resulting from the replacement of somatic histones by sperm-specific protamines, which leads to a greater level of DNA compaction. Nuclear transcription shuts down during this process and as the sperm further mature there is a major loss of cytoplasm [24–26]. One explanation for the mRNAs found in mature sperm is that they represent remnants of the spermatogenesis programme that are left behind during sperm maturation. It was widely believed that sperm were translationally silent, however, it has recently been shown that labelled amino acids are incorporated into polypeptides during mammalian sperm capacitation, a process that occurs in the female reproductive tract [27]. Sperm translation is mediated by mitochondrial-type ribosomes and it is thus possible that sperm transcripts are substrates for this sperm protein production. There is mounting evidence that spermatozoal RNA is delivered into the oocyte and remains intact after fertilization. At least five sperm-specific mRNAs that are not detected in unfertilized oocytes are found post-fertilization [19]. In a recent study introducing human sperm into hamster oocytes by intracytoplasmic sperm injection, two human-specific transcripts with known roles in implantation (PSGI and HLA-E) that are not present in the hamster genome are detected 24 h after fertilization, suggesting that these paternal transcripts survive in the oocyte [28]. Together these studies point to functional roles for spermatozoal mRNAs in the oocyte.
The composition and quantity of sperm RNA is now considered to be a valuable diagnostic tool for male fertility. In individual human ejaculates, 3000–7000 different transcripts were detected in one microarray study [29] and 4000–5000 mRNA types were observed by serial analysis of gene expression in pooled sperm fractions from different ejaculates [15]. However, owing to the much lower concentration of spermatogenetic RNA compared with the maternal mRNA in the oocyte it has been assumed that sperm transcripts do not play a major role in fertilization and early embryogenesis. In support of a functional role for transfer of sperm RNA in development, a heritable paramutation-associated white tail phenotype was induced in mice by microinjection of total RNA from Kittm1Alf/+ heterozygotes into fertilized eggs [30]. Taken together, the evidence is mounting for the presence of functional poly-adenylated mRNAs in the mature sperm of some mammalian species but it is unclear whether this is a more universal feature of animal sperm.
Less is known about sperm RNA profiles in non-mammalian species, but recently de novo RNA transcription in Drosophila has been demonstrated in post-meiotic phases of spermatogenesis [31,32] suggesting mature Drosophila sperm may also provide mRNA to the egg during fertilization. Drosophila has proved to be a valuable model system for exploring conserved aspects of animal biology and, in the case of sperm biology, it is becoming clear that there are molecular and cell biological features conserved between mammals and flies [33,34]. Here we demonstrate that, as in mammals, poly-adenylated mRNA transcripts are present in mature Drosophila sperm cells and that paternal sperm transcripts are detected in the fertilized egg. The conservation in the functional annotations associated with sperm mRNAs found in the two species suggests that Drosophila may be an attractive alternative model system to explore the function of human sperm transcripts during and following fertilization.
2. Results
(a). Spermatozoal mRNA is reproducibly detected in purified Drosophila melanogaster sperm.
To explore the possibility that mature Drosophila sperm contain mRNA transcripts, we used DNA microarray analysis to assess the RNA content of isolated sperm samples. Highly purified sperm samples were obtained from dissected seminal vesicles, which are almost entirely composed of mature sperm, using methods described previously [35,36]. We also demonstrated that following removal of sperm, seminal vesicles contain very low levels of detectable soluble proteins by protein quantitation and two-dimensional gel electrophoresis (K. Chaney & T.L.K., 2010, unpublished data).
The main experiment used RNA extracted from three biological replicates of purified sperm as a template for oligo-dT-primed reverse transcription, amplification, labelling of dye-swapped technical replicates and hybridization to long oligonucleotides microarrays (see electronic supplementary material, supplementary methods). As a control, RNA from two biological replicates of dissected adult testis plus accessory glands was amplified, labelled (dye-swap technical replicate) and hybridized to similar arrays. After normalization and processing to remove low intensity values, we identified 5579 transcripts present in all three sperm replicates, 5358 transcripts from the testis/accessory gland samples and 4295 transcripts common to both datasets. To assess the reliability and reproducibility of the data, we calculated a Pearson correlation between sample pairs (electronic supplementary material, figure S1). For the purified sperm samples, technical replicates were highly reproducible (r = 0.97–0.99) as were biological replicates (r = 0.72–0.90). Testis/accessory gland samples were similarly highly correlated (r = 0.94–0.97 for technical replicate and r = 0.90–0.93 for biological replicates).
To determine if spermatozoal transcripts result from incidental capture of abundant testis transcripts or contamination from the accessory gland, we ranked normalized intensity values by the highest median value for both purified sperm and the whole tissue transcriptomes (table 1). If the spermatozoal transcripts result from contamination with seminal vesicle tissues, we would expect the most abundant testis/accessory gland transcripts to be high in the list of sperm transcripts. Similarly, if packaging of spermatozoal transcripts were purely a passive process, we would expect a correlation between the abundance of testis/accessory gland transcripts and those in sperm. Contrary to this expectation, the rankings are entirely distinct with the exception of two transcripts (CG31226 and CG10407), both encoding genes of unknown function. As expected, the ranking of the testis/accessory gland sample reveals that male-specific transcripts and accessory gland protein transcripts are particularly abundant. Their absence from the sperm ranking indicates that the purified sperm sample was not contaminated with accessory gland nor is the sperm transcript pool the result of passive transcript packaging.
Table 1.
Top ranked spermatozoal and testis/accessory gland mRNAs. On the left, the top 40 sperm transcripts based on normalized median intensity and their corresponding rank from the testis/accessory gland array (ribosomal proteins are highlighted in bold). On the right, the top ranked transcripts from the testis/accessory gland sample and corresponding rank in the sperm transcript list (Acps are highlighted in bold).
| sperm transcripts |
testis/accessory gland transcripts |
||||
|---|---|---|---|---|---|
| gene | sperm rank | testis rank | gene | sperm rank | testis rank |
| CG9336 | 1 | 304 | Acp98AB | 59 | 1 |
| CG8072 | 2 | 2344 | CG31226 | 36 | 2 |
| RpL37a | 3 | 99 | Acp62F | 650 | 3 |
| CG18628 | 4 | 83 | Mst57Dc | 292 | 4 |
| RpS20 | 4 | 85 | Acp53C14b | 443 | 5 |
| RpS11 | 6 | 155 | Acp70A | 232 | 6 |
| RpL31 | 7 | 157 | msopa | 1308 | 7 |
| RpL39 | 8 | 109 | CG10252 | 85 | 8 |
| CG6770 | 9 | 196 | Anp | 444 | 9 |
| Gdi | 9 | 260 | Jupiter | 124 | 10 |
| RpL36A | 11 | 129 | CG15841 | 410 | 11 |
| RpLP1 | 12 | 175 | Dup99B | 349 | 12 |
| CG5612 | 13 | 1604 | CG12861 | 51 | 13 |
| RpL41 | 14 | 51 | Acp26Aa | 2746 | 14 |
| Tsp42Ed | 15 | 742 | CG9284 | 102 | 15 |
| vir-1 | 16 | 1475 | CG18662 | 187 | 16 |
| mRpL27 | 17 | 463 | Acp53C14a | 743 | 17 |
| AnnIX | 18 | 716 | Mst57Db | 389 | 18 |
| RpL9 | 19 | 280 | CG31988 | 256 | 19 |
| eIF-5A | 20 | 293 | CG31659 | 624 | 20 |
| RpS8 | 21 | 285 | CG32240 | 238 | 21 |
| Cam | 22 | 110 | Acp53Ea | 1714 | 22 |
| AnnIX | 23 | 761 | CG4836 | 257 | 23 |
| RpS15 | 24 | 505 | CG17242 | 1064 | 24 |
| RpL7A | 25 | 501 | CG31468 | 177 | 25 |
| Idgf4 | 26 | 1301 | CG15657 | 239 | 26 |
| Ef1beta | 27 | 131 | CG1394 | 374 | 27 |
| CG10407 | 28 | 31 | CG8701 | 258 | 28 |
| Rack1 | 29 | 94 | CG14391 | 321 | 29 |
| RpS7 | 30 | 449 | CG32064 | 115 | 30 |
| RpS9 | 31 | 327 | CG10407 | 28 | 31 |
| RpL10Ab | 32 | 1014 | CG2127 | 234 | 32 |
| RpS5a | 33 | 392 | CG9016 | 242 | 33 |
| CG17549 | 34 | 508 | CG17376 | 76 | 34 |
| Tom7 | 35 | 45 | CG14926 | 260 | 35 |
| CG31226 | 36 | 2 | CG11106 | 408 | 36 |
| sop | 37 | 518 | CG7768 | 273 | 37 |
| Arc1 | 38 | 3047 | CG31820 | 348 | 38 |
| RpS18 | 39 | 491 | Acp32CD | 1125 | 39 |
| RpS4 | 40 | 255 | CG15425 | 427 | 40 |
(b). Spermatozoal RNA and the sperm proteome
To further address the possible persistence of residual transcripts that encode integral sperm components we compared the list of abundant spermatozoal transcripts to genes encoding the sperm proteome [35,37]. A survey of the 20 most abundant spermatozoal RNA transcripts reveals only one gene that encodes an integral sperm component. A more comprehensive analysis of the 200 and 500 most abundant sperm transcripts revealed that only 21 per cent (42 out of 200) and 18.4 per cent (92 out of 500) encode components of the sperm proteome. This observation is statistically indistinguishable from the results of an identical analysis of the most abundant testis transcripts, where 22.5 per cent (45 out of 200) and 19.6 per cent (98 out of 500) were found to encode sperm proteins. While this does not rule out the possible persistence of sperm proteome transcripts in mature sperm, it confirms that it does not contribute substantially to the identified abundant spermatozoal transcripts, nor does it exceed the overall proportion of abundant testis transcripts that encode sperm proteins. It is also noteworthy that among the 42 most abundant spermatozoal RNA transcripts that also encode sperm proteins, a significant enrichment (17 of 42; p = 4.99E − 11) are functionally involved in translation based on gene ontology (GO) annotation, including six components of the ribosomal large subunit, eight components of the small subunit and three elongation factors (see later).
(c). Expression of spermatozoal RNA
We next analysed the 200 most abundant transcripts from each of the sperm and testis/accessory gland sample sets using the FlyMine data warehouse [38]. We found 34 genes (17%) in common between both sperm and testis in the top 200 selection, further emphasizing the difference in the two populations of mRNA. When we consider the global properties of the lists, we again observe differences. Comparing the expression patterns of both datasets to the tissue-specific expression catalogue in the FlyAtlas [39], we find that sperm transcript genes are expressed in most tissues and, strikingly, only 20 per cent of the genes have high levels of testis expression, whereas 75 per cent have very low levels of testis expression. In contrast, with testis/accessory gland RNA, we find that 70 per cent of genes are highly expressed and 20 per cent have low expression levels in the FlyAtlas testis sample (electronic supplementary material, figure S2). We also noticed that 65 per cent of the genes in the sperm transcript list that have BDGP expression annotations (113 genes) are expressed in the early (stage 1–3) embryo, whereas only 25 per cent of the testis/accessory gland enriched genes have expression this early in development [40,41] (electronic supplementary material, figure S2). We conclude that the spermatozoal transcriptome does not include a substantial proportion of testis enriched spermatogenesis genes.
(d). Genomic distribution of spermatozoal RNA
As is well established for testis-specific genes, an analysis of the 200 most abundant testis/accessory gland transcripts reveals a significant under-representation of X-linked genes (32% of the expected value; χ2 = 15.81, p < 0.0001). In contrast, the abundant spermatozoal RNA genes show a slight, but non-significant under-representation of X-linked genes (74% of the expectation; χ2 = 2.35, p = 0.13). Additional analyses revealed significant co-localization of abundant testis transcripts within adjacent gene clusters (total clusters = 10; p < 0.01) and that clustering was restricted to the autosomes. This observation is consistent with previous studies of Drosophila sperm [35,37] and testis-overexpressed genes in Drosophila and the mouse [42–44]. However, analysis of spermatozoal RNA revealed no significant clustering on either the autosomes or X chromosome (p = 0.22), a finding which further reinforces the inherent differences in the properties of these two sets of transcripts.
(e). Functional enrichment of spermatozoal RNA
To assess the possible functions of sperm transcripts and highlight fundamental distinctions between these and the testis/accessory gland transcripts, we analysed molecular function and biological process GO on the most abundant 200 transcripts in both ranked lists. Strikingly, 33 per cent of spermatozoal RNA transcripts with GO molecular function annotations (47/142) encode components of the ribosome (p = 5E10–45). Additionally, 12 genes were identified with transmembrane transporter activity (p = 2E10−8) and five with translation elongation factor activity (p = 0.019). In contrast, highly expressed genes in the testis/accessory gland transcriptome are enriched solely for hormone activity (n = 8; p = 1E−4). When biological process ontology was analysed, the enriched categories were again found to be distinct between the groups. The largest sets of genes within enriched categories for abundant sperm transcripts include mitotic spindle organization (n = 30; p = 3E-16) and translation (n = 54; p = 3E−16). In contrast, testis/accessory gland transcripts are exclusively found within the reproduction category and 14 related daughter ontology categories (n = 29; p = 4E−5). Taken together, the global expression, genomic distribution and functional properties emphasize that the set of genes encoding sperm transcripts have distinct characteristics from the abundantly expressed genes in the testis and accessory gland.
(f). Spermatozoal RNA is transferred to the oocyte during fertilization
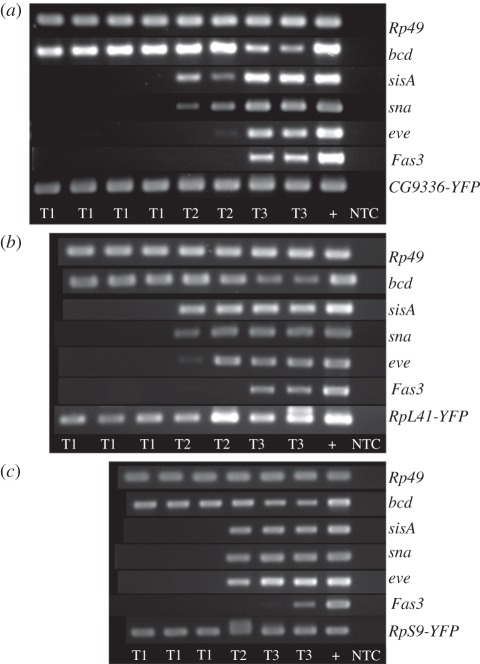
To determine if sperm transcripts are delivered into the oocyte, we used a collection of protein trap fly lines we have recently generated. The FlyProt project has created a set of fly lines in which endogenous genes are tagged with an in-frame artificial reporter exon encoding a yellow fluorescent protein (YFP) ([45,46]; www.flyprot.org). We identified three lines in our collection that carry protein traps in sperm transcripts present in our top 40 list: RpS9 (CPTI-000493), RpL41 (CPTI-002881) and CG9336 (CPTI-001654). Male flies heterozygous for each of these YFP-tagged genes were mated with wild-type females and embryos collected at three different time points after egg laying (T1: 0–15 min, T2: 60–90 min and T3: 180–210 min). We used reverse transcriptase- (RT-) PCR assays with RNA extracted from single embryos, employing paternal-specific primers that will only amplify the copy of the gene tagged with the YFP insertion. In order to control for detection of tagged transcripts that result from zygotic expression of the tagged gene from the paternal genome, the age of each embryo was determined using RT-PCR assays on the same RNA samples for genes known to be transcribed at different stages of early embryo development. Sisterless A (sisA) and snail (sna) are the earliest known zygotically transcribed genes in D. melanogaster, with expression first detected at nuclear cycle 8. This is followed by even skipped (eve) at nuclear cycle 9 [47–49]. Fasciclin-3 (Fas3) expression is initiated much later in development at stage 11–12 [50]. Bicoid (bcd) is a maternally deposited transcript that starts to degrade during the maternal-to-zygotic transition [51]. Rp49 was used as a ubiquitous control.
Detection of tagged sperm transcripts in an embryo along with bcd and the Rp49 controls, but in the absence of signals for the other transcripts, indicates that the sperm transcripts are either delivered paternally or represent zygotic expression considerably earlier than previous studies have found. We found 50–70% of T1 embryos were older than expected since they gave positive PCR results for one or more of the zygotically expressed genes (figure 1; electronic supplementary material, table S1). As expected, levels of paternal contributed mRNA in single embryos are extremely low, but we could detect the presence of the YFP-tagged mRNA for all three of the sperm transcripts assayed in approximately 30 per cent of the embryos confirmed not to have initiated zygotic expression. The detection of the YFP-tagged transcript increased with the age of the embryos but this is due to the onset of zygotic expression and transcription of the paternal allele. We can eliminate the possibility of genomic DNA contamination because the primers used to amplify the tagged paternal transcripts span an intron and the product we observe is of the size expected for a processed mRNA. We consider it extremely unlikely that the transcripts we detect in the early embryos are a result of precocious activation of the zygotic genome. In particular, each of the three independent paternal protein trap transcripts assayed contain an intron of at least 10 kb resulting from the insertion of the protein trap transposon: it is extremely unlikely that these genes could be transcribed and processed during the extremely short cell cycle times of the early embryo. We therefore conclude that in Drosophila, as in mammals, transcripts are specifically packaged into mature sperm, delivered to the oocyte and can be detected in the zygote.
Figure 1.
RT-PCR using staged embryos. Embryos from male-YFP x female Oregon-R were collected after 15 min laying (T1), after 30 min of laying plus 1 h ageing (T2), and after 30 min laying plus 3 h ageing (T3). The age of the embryos was determined using bcd, sisA, sna, eve and Fas3 to ensure embryos of T1 were collected before the onset of zygotic transcription. Rp49 was used to confirm presence of cDNA after RNA extraction and reverse transcription. NTC (no template control) and positive control for each gene product are shown. (a) CPTI001654 (CG9336-YFP), (b) CPTI002881 (RpL41-YFP) and (c) CPTI000493 (RpS9-YFP).
(g). Functional conservation of spermatozoal RNA between insects and humans
To assess parallels between insect and human spermatozoal RNA, we first characterized the relationship between abundant human spermatozoal RNA and both human testis mRNA expression and the human sperm proteome. Similar to our observations with Drosophila, human spermatozoal transcripts are, for the most part, not highly expressed in the testis, with only 36 of the most abundant 500 spermatozoal transcripts identified within the 10 per cent of probes displaying the highest levels of average testis expression [52] (electronic supplementary material, table S2). We next assessed the presence of proteins encoded by the same set of abundant spermatozoal transcripts and found that only 19.2 per cent (96 out of 500) have been identified in proteomic analysis of whole human sperm [53] or the human sperm nucleus [54]. This proportion is statistically indistinguishable from our observations in Drosophila (χ2 = 0.059, p = 0.81), although we note that a direct comparison does not account for potential differences in the extent of sperm proteome characterization between the taxa. We therefore conclude that spermatozoal transcripts in both humans and the fly are largely distinct from genes expressed at high levels during spermatogenesis and genes encoding the sperm proteome.
If spermatozoal RNAs function in the oocyte following fertilization, evolutionary conservation across taxa in RNA composition might be expected. Consistent with this predication, an analysis of the 500 most abundant transcripts in Drosophila to those previously identified in human spermatozoa [55] revealed that their functional composition is generally shared across 10 broadly defined GO functional categories (electronic supplementary material, figure S3). A more detailed analysis of molecular function GO enrichment in each species (relative to the composition of their respective genomes) reveals a highly significant and shared enrichment in structural constituents of the ribosome, including translation elongation and termination factors (table 2). Remarkably, these sets of spermatozoal RNA included 32.7 per cent (60 of 183) and 31 per cent (50 of 161) of the annotated structural constituents of the ribosome in the Drosophila and human genome, respectively. It is noteworthy that other than the molecular functional categories associated with translation, including structural molecule activity which largely comprises ribosomal components, there are no shared functional categories between the two datasets.
Table 2.
Molecular function enrichment for abundant Drosophila and human spermatozoal RNAs. Includes the 30 most significant molecular function categories in Drosophila and all significant categories in humans. Molecular functions specifically associated with translation are indicated in bold and are italics if they are identified in both Drosophila and humans.
|
Drosophila spermatozoa mRNA |
human spermatozoa mRNA |
||||
|---|---|---|---|---|---|
| GO ID | molecular function | p-value | GO ID | molecular function | p-value |
| 0003735 | structural constituent of ribosome | 6.84e−44 | 0003735 | structural constituent of ribosome | 5.07e−46 |
| 0005198 | structural molecule | 1.12e−22 | 0005198 | structural molecule | 2.62e−24 |
| 0015078 | hydrogen ion transmembrane transporter | 8.89e−16 | 0003723 | RNA binding | 2.36e−24 |
| 0015077 | monovalent inorganic cation transmembrane transporter | 9.94e−11 | 0019843 | rRNA binding | 5.38e−11 |
| 0008553 | hydrogen-exporting ATPase, phosphorylative mechanism | 3.24e−10 | 0008603 | cAMP-dependent protein kinase regulator | 1.36e−6 |
| 0022890 | inorganic cation transmembrane transporter | 5.41e−09 | 0003729 | mRNA binding | 3.33e−6 |
| 0015662 | ATPase, coupled to transmembrane movement of ions, phosphorylative mechanism | 1.50e−07 | 0003676 | nucleic acid binding | 5.47e−6 |
| 0016491 | oxidoreductase | 2.47e−07 | 0005515 | protein binding | 2.43e−6 |
| 0042625 | ATPase, coupled to transmembrane movement of ions | 5.69e−07 | 0008565 | protein transporter | 6.44e−5 |
| 0043022 | ribosome binding | 2.63e−06 | 0070063 | RNA polymerase binding | 0.00019 |
| 0004364 | glutathione transferase | 1.29e−05 | 0008135 | translation factor, nucleic acid binding | 0.00049 |
| 0004129 | cytochrome-c oxidase | 1.71e−05 | 0008139 | nuclear localization sequence binding | 0.00081 |
| 0015002 | haem-copper terminal oxidase | 1.71e−05 | 0008143 | poly(A) RNA binding | 0.0014 |
| 0016675 | oxidoreductase, acting on a haem group of donors | 1.71e−05 | 0070717 | poly-purine tract binding | 0.0030 |
| 0016676 | oxidoreductase, acting on a haem group, oxygen as acceptor | 1.71e−05 | 0045182 | translation regulator activity | 0.0041 |
| 0008135 | translation factor, nucleic acid binding | 2.24e−05 | 0005048 | signal sequence binding | 0.0041 |
| 0015399 | primary active transmembrane transporter | 8.62e−05 | 0005488 | binding | 0.0060 |
| 0015405 | P–P-bond-hydrolysis-driven transmembrane transporter | 8.62e−05 | 0003724 | RNA helicase | 0.0088 |
| 0046933 | hydrogen ion transporting ATP synthase, rotational mechanism | 8.70e−05 | 0008320 | protein transmembrane transporter | 0.0114 |
| 0016820 | hydrolase catalysing transmembrane movement of substances | 0.00016 | 0022884 | macromolecule transmembrane transporter | 0.0114 |
| 0042626 | ATPase, coupled to transmembrane movement of substances | 0.00016 | 0004726 | protein tyrosine phosphatase | 0.0175 |
| 0043492 | ATPase, coupled to movement of substances | 0.00016 | 0030371 | translation repressor | 0.0231 |
| 0009055 | electron carrier | 0.00034 | 0003743 | translation initiation factor | 0.0247 |
| 0046961 | proton-transporting ATPase, rotational mechanism | 0.00036 | 0019887 | protein kinase regulator | 0.0295 |
| 0043021 | ribonucleoprotein binding | 0.00050 | 0003746 | translation elongation factor | 0.0336 |
| 0003924 | GTPase | 0.00071 | 0015078 | hydrogen ion transmembrane transporter | 0.0394 |
| 0003743 | translation initiation factor | 0.00077 | 0031369 | translation initiation factor binding | 0.0417 |
| 0008121 | ubiquinol-cytochrome-c reductase | 0.00085 | 015450 | protein transmembrane transporter | 0.0417 |
| 0016681 | oxidoreductase, acting on diphenols as donors, cytochrome acceptor | 0.00085 | |||
| 0003746 | translation elongation factor | 0.0013 | |||
3. Discussion
In this report, we describe the characterization of an mRNA population carried by mature Drosophila melanogaster sperm cells, the first description of an invertebrate spermatozoal transcriptome. We further demonstrate that at least some of the spermatozoal transcripts are successfully delivered to the fertilized egg and can be detected prior to the onset of zygotic gene expression. Thus, in flies and vertebrates the mature sperm carries a defined set of mRNA transcripts into the egg at fertilization. A parallel analysis of testis and accessory gland mRNA indicates that sperm mRNAs are unlikely to result from residual waste remaining after sperm individualization, since the relative abundances of spermatozoal mRNAs are not reflective of transcript abundance in the reproductive tract. An analysis of published data from human sperm and testis indicate this property of spermatozoal RNA is conserved. We therefore suggest that particular mRNAs are either differentially located in developing sperm cells, such that they are retained during individualization, or that they are selectively sequestered to prevent loss. Our current study does not differentiate between these possibilities. High resolution mRNA in situ hybridization should allow this issue to be addressed using probes against the sperm transcripts identified in this study.
We found that the sperm mRNA population was particularly enriched for transcripts encoding ribosomal components and other proteins related to translation and that a similar enrichment is found for mRNAs carried by mature human sperm. This remarkable functional conservation hints at an underlying biological function for the spermatozoal transcriptome. Although sperm enter an environment rich in maternal mRNAs encoding the necessary components for constructing ribosomes, our observations raise the possibility of specific role(s) for ‘paternal’ ribosomes during and following fertilization. Another possibility is that RPs or their transcripts play another role in early development as has been previously shown for some RPs [56–58]. Intriguingly, RPS3, which is in the top 300 spermatozoal transcripts, has been shown to play a role in DNA repair and the possibility of such a function in pronuclear fusion is intriguing.
It has been suggested that spermatozoal transcripts may play a role in nuclear compaction, in some way marking regions of the paternal genome that do not undergo the histone to protamine replacement [20]. It is estimated that approximately 15 per cent of the DNA in human spermatozoa retain histones [59] and it is suggested that these regions of the genome may be regulatory or be important for epigenetic marking [60]. A histone to protamine-like protein transition has recently been described in Drosophila [26], however, the extent of the transition and whether regions of the genome remain associated with histones is not currently known in flies. Similarly, although epigenetic modification of parental chromosomes is not normally considered a feature of early Drosophila development, recent work indicates that the protein encoded by the paternal effect locus ms(3)k81 is required to protect paternal telomeres at fertilization [61]. Thus, there is a possibility that spermatozoal mRNA plays a role in marking the paternal genome that has hitherto been masked by the essential nature of the genes encoding the spermatozoal mRNAs. Whatever their role, the discovery of Drosophila spermatozoal mRNAs that encode a set of molecular functions conserved with those found in mammalian sperm opens the prospect of using the tools of fly genetics and developmental biology to explore the contribution these transcripts make to reproductive biology.
4. Methods
The experimental methods are summarized here with full details provided in the electronic supplementary material, supplementary methods.
(a). Tissue collection and RNA extraction for microarrays
Sperm were purified from adult D. melanogaster males essentially as described [35,36], and RNA extracted using the trizol method. For the purified sperm samples, RNA equivalent to the dissections from approximately 200 males were pooled to create three independent biological replicates. Testis/accessory gland samples were similarly processed.
(b). Microarray analysis
RNA samples were amplified using SMART method [62] and the resulting DNA labelled as technical dye-swap replicates for hybridization to long oligonucleotide microarrays printed in house (GEO platform accession GPL8244). Sperm and testis samples were cohybridized with labelled genomic DNA to aid spotfinding and the latter channel discarded for further analysis. After hybridization, data were normalized independently using a quantile method [63]. The median of the normalized intensity of each sample type was ranked using minimum ties method of the rank function in R, assigning the highest intensity value a rank of 1 (table 1).
(c). Data analysis
The chromosomal distribution of spermatozoal RNA genes was compared statistically with the distribution of annotated genes using a χ2-test with Yates correction. Analysis of gene clustering was conducted using a modified adjacent gene model [42,43]. Statistical analysis of GO molecular function enrichment of the 500 most abundant Drosophila and human sperm transcripts was conducted using a hypergeometric distribution and the Yekutieli (false discovery rate under dependency) multiple-test correction as implemented by the GOEAST toolkit [64].
(d). Drosophila stocks and embryo collections for RT-PCR confirmation
Drosophila stocks were: Oregon-R (OrR), CPTI000493 (RpS9-YFP), CPTI002881 (RpL41-YFP), CPTI001654 (CG9336-YFP), which contain a Venus (a YFP variant) exon insertion within an intron (http://www.flyprot.org; [45,46]).
(e). Reverse transcription and PCR reactions for RT-PCR confirmation
RNA was extracted from single embryos using the Trizol method (Invitrogen) and each sample analysed with the set of primer pairs described in the electronic supplementary material, supplementary methods.
Acknowledgements
This work was supported by BBSRC and Isaac Newton Trust Grants to S.R., a Ruth L. Kirschstein National Research Service Award (National Institutes of Health) and an Academic Research Fellowship from the Research Council of the United Kingdom to S.D., a Royal Society Wolfson Merit Award, the BBSRC and the Biodesign Institute at Arizona State University to T.L.K. We would also like to thank Elaine Wilkin for her assistance analysing gene clusters.
References
- 1.Fitch K. R., Yasuda G. K., Owens K. N., Wakimoto B. T. 1998. Paternal effects in Drosophila: implications for mechanisms of early development. Curr. Top. Dev. Biol. 38, 1–34 10.1016/S0070-2153(08)60243-4 (doi:10.1016/S0070-2153(08)60243-4) [DOI] [PubMed] [Google Scholar]
- 2.Karr T. L. 1996. Paternal investment and intracellular sperm-egg interactions during and following fertilization in Drosophila. Curr. Top. Dev. Biol. 34, 89–115 10.1016/S0070-2153(08)60709-7 (doi:10.1016/S0070-2153(08)60709-7) [DOI] [PubMed] [Google Scholar]
- 3.Sutovsky P., Schatten G. 2000. Paternal contributions to the mammalian zygote: fertilization after sperm-egg fusion. Int. Rev. Cytol. 195, 1–65 10.1016/S0074-7696(08)62703-5 (doi:10.1016/S0074-7696(08)62703-5) [DOI] [PubMed] [Google Scholar]
- 4.Simerly C., et al. 1995. The paternal inheritance of the centrosome, the cell's microtubule-organizing center, in humans, and the implications for infertility. Nat. Med. 1, 47–52 10.1038/nm0195-47 (doi:10.1038/nm0195-47) [DOI] [PubMed] [Google Scholar]
- 5.Kurokawa M., Yoon S. Y., Alfandari D., Fukami K., Sato K., Fissore R. A. 2007. Proteolytic processing of phospholipase Cζ and [Ca2+]i oscillations during mammalian fertilization. Dev. Biol. 312, 407–418 10.1016/j.ydbio.2007.09.040 (doi:10.1016/j.ydbio.2007.09.040) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saunders C. M., Larman M. G., Parrington J., Cox L. J., Royse J., Blayney L. M., Swann K., Lai F. A. 2002. PLC ζ: a sperm-specific trigger of Ca(2+) oscillations in eggs and embryo development. Development 129, 3533–3544 [DOI] [PubMed] [Google Scholar]
- 7.Loppin B., Lepetit D., Dorus S., Couble P., Karr T. 2005. Origin and neofunctionalization of a Drosophila paternal effect gene essential for zygote viability. Curr. Biol. 15, 87–93 10.1016/j.cub.2004.12.071 (doi:10.1016/j.cub.2004.12.071) [DOI] [PubMed] [Google Scholar]
- 8.Dubruille R., Orsi G., Delabaere L., Cortier E., Couble P., Marais G., Loppin B. 2010. Specialization of a Drosophila capping protein essential for the protection of sperm telomeres. Curr. Biol. 20, 2090–2099 10.1016/j.cub.2010.11.013 (doi:10.1016/j.cub.2010.11.013) [DOI] [PubMed] [Google Scholar]
- 9.Fitch K. R., Wakimoto B. T. 1998. The paternal effect gene ms(3)sneaky is required for sperm activation and the initiation of embryogenesis in Drosophila melanogaster. Dev. Biol. 197, 270–282 10.1006/dbio.1997.8852 (doi:10.1006/dbio.1997.8852) [DOI] [PubMed] [Google Scholar]
- 10.Ohsako T., Hirari K., Yamomoto M. T. 2003. The Drosophila misfire gene has an essential role in sperm activation during fertilization. Genes Genet. Syst. 78, 253–266 10.1266/ggs.78.253 (doi:10.1266/ggs.78.253) [DOI] [PubMed] [Google Scholar]
- 11.Perotti M. E., Cattaneo F., Pasini M. E., Vernì F., Hackstein J. H. 2001. Male sterile mutant casanova gives clues to mechanisms of sperm–egg interactions in Drosophila melanogaster. Mol. Reprod. Dev. 60, 248–259 10.1002/mrd.1085 (doi:10.1002/mrd.1085) [DOI] [PubMed] [Google Scholar]
- 12.Smith M. K., Wakimoto B. T. 2007. Complex regulation and multiple developmental functions of misfire, the Drosophila melanogaster ferlin gene. BMC Dev. Biol. 7, 21. 10.1186/1471-213X-7-21 (doi:10.1186/1471-213X-7-21) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feugang J. M., Rodriguez-Osorio N., Kaya A., Wang H., Page G., Ostermeier G. C., Topper E. K., Memili E. 2010. Transcriptome analysis of bull spermatozoa: implications for male fertility. Reprod. Biomed. Online 21, 312–324 10.1016/j.rbmo.2010.06.022 (doi:10.1016/j.rbmo.2010.06.022) [DOI] [PubMed] [Google Scholar]
- 14.Ostermeier G. C., Goodrich R. J., Diamond M. P., Dix D. J., Krawetz S. A. 2005. Toward using stable spermatozoal RNAs for prognostic assessment of male factor fertility. Fertil. Steril. 83, 1687–1694 10.1016/j.fertnstert.2004.12.046 (doi:10.1016/j.fertnstert.2004.12.046) [DOI] [PubMed] [Google Scholar]
- 15.Zhao Y., Li Q., Yao C., Wang Z., Zhou Y., Wang Y., Liu L., Wang L., Qiao Z. 2006. Characterization and quantification of mRNA transcripts in ejaculated spermatozoa of fertile men by serial analysis of gene expression. Hum. Reprod. 21, 1583–1590 10.1093/humrep/del027 (doi:10.1093/humrep/del027) [DOI] [PubMed] [Google Scholar]
- 16.Dadoune J. P. 2009. Spermatozoal RNAs: what about their functions? Microsc. Res. Tech. 72, 536–551 10.1002/jemt.20697 (doi:10.1002/jemt.20697) [DOI] [PubMed] [Google Scholar]
- 17.Miller D., Ostermeier G. C., Krawetz S. A. 2005. The controversy, potential and roles of spermatozoal, RNA. Trends Mol. Med. 11, 156–163 10.1016/j.molmed.2005.02.006 (doi:10.1016/j.molmed.2005.02.006) [DOI] [PubMed] [Google Scholar]
- 18.Hayashi S., Yang J., Christenson L., Yanagimachi R., Hecht N. B. 2003. Mouse preimplantation embryos developed from oocytes injected with round spermatids or spermatozoa have similar but distinct patterns of early messenger RNA expression. Biol. Reprod. 69, 1170–1176 10.1095/biolreprod.103.016832 (doi:10.1095/biolreprod.103.016832) [DOI] [PubMed] [Google Scholar]
- 19.Ostermeier G. C., Miller D., Huntriss J. D., Diamond M. P., Krawetz S. A. 2004. Reproductive biology: delivering spermatozoan RNA to the oocyte. Nature 429, 154. 10.1038/429154a (doi:10.1038/429154a) [DOI] [PubMed] [Google Scholar]
- 20.Lalancette C., Miller D., Li Y., Krawetz S. A. 2008. Paternal contributions: new functional insights for spermatozoal RNA. J. Cell. Biochem. 104, 1570–1579 10.1002/jcb.21756 (doi:10.1002/jcb.21756) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lalancette C., Platts A. E., Johnson G. D., Emery B. R., Carrell D. T., Krawetz S. A. 2009. Identification of human sperm transcripts as candidate markers of male fertility. J. Mol. Med. 87, 735–748 10.1007/s00109-009-0485-9 (doi:10.1007/s00109-009-0485-9) [DOI] [PubMed] [Google Scholar]
- 22.Wang H., Zhou Z., Xu M., Li J., Xiao J., Zu Z. Y., Sha J. 2004. A spermatogenesis-related gene expression profile in human spermatozoa and its potential clinical application. J. Mol. Med. 82, 317–324 10.1007/s00109-004-0526-3 (doi:10.1007/s00109-004-0526-3) [DOI] [PubMed] [Google Scholar]
- 23.Hecht N. B. 1998. Molecular mechanisms of male germ cell differentiation. BioEssays 20, 555–561 (doi:10.1002/(SICI)1521-1878(199807)20:7<555::AID-BIES6>3.0.CO;2-J) [DOI] [PubMed] [Google Scholar]
- 24.D'Occhio M. J., Hengstberger K. J., Johnston S. D. 2007. Biology of sperm chromatin structure and relationship to male fertility and embryonic survival. Anim. Reprod. Sci. 101, 1–17 10.1016/j.anireprosci.2007.01.005 (doi:10.1016/j.anireprosci.2007.01.005) [DOI] [PubMed] [Google Scholar]
- 25.Miller D., Brinkworth M., Iles D. 2010. Paternal DNA packaging in spermatozoa: more than the sum of its parts? DNA, histones, protamines and epigenetics. Reproduction 139, 287–301 10.1530/REP-09-0281 (doi:10.1530/REP-09-0281) [DOI] [PubMed] [Google Scholar]
- 26.Rathke C., Baarends W. M., Jayaramaiah-Raja S., Bartkuhn M., Renkawitz R., Renkawitz-Pohl R. 2007. Transition from a nucleosome-based to a protamine-based chromatin configuration during spermiogenesis in Drosophila. J. Cell Sci. 120, 1689–1700 10.1242/jcs.004663 (doi:10.1242/jcs.004663) [DOI] [PubMed] [Google Scholar]
- 27.Gur Y. 2006. Mammalian sperm translate nuclear-encoded proteins by mitochondrial-type ribosomes. Genes Dev. 20, 411–416 10.1101/gad.367606 (doi:10.1101/gad.367606) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Avendano C., Franchi A., Jones E., Oehninger S. 2009. Pregnancy-specific β-1-glycoprotein 1 and human leukocyte antigen-E mRNA in human sperm: differential expression in fertile and infertile men and evidence of a possible functional role during early development. Hum. Reprod. 24, 270–277 10.1093/humrep/den381 (doi:10.1093/humrep/den381) [DOI] [PubMed] [Google Scholar]
- 29.Ostermeier G. C., Dix D. J., Miller D., Khatri P., Krawetz S. A. 2002. Spermatozoal RNA profiles of normal fertile men. Lancet 360, 772–777 10.1016/S0140-6736(02)09899-9 (doi:10.1016/S0140-6736(02)09899-9) [DOI] [PubMed] [Google Scholar]
- 30.Rassoulzadegan M., Grandjean V., Gounon P., Vincent S., Gillot I., Cuzin F. 2006. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature 441, 469–474 10.1038/nature04674 (doi:10.1038/nature04674) [DOI] [PubMed] [Google Scholar]
- 31.Barreau C., Benson E., Gudmannsdottir E., Newton F., White-Cooper H. 2008. Post-meiotic transcription in Drosophila testes. Development 135, 1897–1902 10.1242/dev.021949 (doi:10.1242/dev.021949) [DOI] [PubMed] [Google Scholar]
- 32.Vibranovski M. D., Chalopin D. S., Lopes H. F., Long M., Karr T. L. 2010. Direct evidence for postmeiotic transcription during Drosophila melanogaster spermatogenesis. Genetics 186, 431–433 10.1534/genetics.110.118919 (doi:10.1534/genetics.110.118919) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dorus S., Wasbrough E. R., Busby J., Wilkin E. C., Karr T. L. 2010. Sperm proteomics reveals intensified selection on mouse sperm membrane and acrosome genes. Mol. Biol. Evol. 27, 1235–1246 10.1093/molbev/msq007 (doi:10.1093/molbev/msq007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White-Cooper H., Bausek N. 2010. Evolution and spermatogenesis. Phil. Trans. R. Soc. B 365, 1465–1480 10.1098/rstb.2009.0323 (doi:10.1098/rstb.2009.0323) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dorus S., Busby S. A., Gerike U., Shabanowitz J., Hunt D. F., Karr T. L. 2006. Genomic and functional evolution of the Drosophila melanogaster sperm proteome. Nat. Genet. 38, 1440–1445 10.1038/ng1915 (doi:10.1038/ng1915) [DOI] [PubMed] [Google Scholar]
- 36.Snook R. R., Cleland S. Y., Wolfner M. F., Karr T. L. 2000. Offsetting effects of Wolbachia infection and heat shock on sperm production in Drosophila simulans: analyses of fecundity, fertility and accessory gland proteins. Genetics 155, 167–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wasbrough E. R., Dorus S., Hester S., Howard-Murkin J., Lilley K., Wilkin E., Polpitiya A., Petritis K., Karr T. L. 2010. The Drosophila melanogaster sperm proteome-II (DmSP-II). J. Proteomics 73, 2171–2185 10.1016/j.jprot.2010.09.002 (doi:10.1016/j.jprot.2010.09.002) [DOI] [PubMed] [Google Scholar]
- 38.Lyne R., et al. 2007. FlyMine: an integrated database for Drosophila and Anopheles genomics. Genome Biol. 8, R129. 10.1186/gb-2007-8-7-r129 (doi:10.1186/gb-2007-8-7-r129) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chintapalli V. R., Wang J., Dow J. A. 2007. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 39, 715–720 10.1038/ng2049 (doi:10.1038/ng2049) [DOI] [PubMed] [Google Scholar]
- 40.Tomancak P., et al. 2002. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 3, RESEARCH0088-088.14. 10.1186/gb-2002-3-12-research0088 (doi:10.1186/gb-2002-3-12-research0088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomancak P., Berman B. P., Beaton A., Weiszmann R., Kwan E., Hartenstein V., Celniker S. E., Rubin G. M. 2007. Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 8, R145. 10.1186/gb-2007-8-7-r145 (doi:10.1186/gb-2007-8-7-r145) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boutanaev A. M., Kalmykova A. I., Shevelyov Y. Y., Nurminsky D. I. 2002. Large clusters of co-expressed genes in the Drosophila genome. Nature 420, 666–669 10.1038/nature01216 (doi:10.1038/nature01216) [DOI] [PubMed] [Google Scholar]
- 43.Divina P., Vlček Č., Strnad P., Pačes V., Forejt J. 2005. Global transcriptome analysis of the C57BL/6J mouse testis by SAGE: evidence for nonrandom gene order. BMC Genomics 6, 29. 10.1186/1471-2164-6-29 (doi:10.1186/1471-2164-6-29) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parisi M., et al. 2004. A survey of ovary-, testis-, and soma-biased gene expression in Drosophila melanogaster adults. Genome Biol. 5, R40. 10.1186/gb-2004-5-6-r40 (doi:10.1186/gb-2004-5-6-r40) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rees J. S., et al. 2011. In vivo analysis of proteomes and interactomes using parallel affinity capture (iPAC) coupled to mass spectrometry. Mol. Cell Proteomic 10 10.1074/mcp.M110.002386 (doi:10.1074/mcp.M110.002386) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ryder E., Spriggs H., Drummond E., Johnston St D., Russell S. 2009. The Flannotator: a gene and protein expression annotation tool for Drosophila melanogaster. Bioinformatics 25, 548–549 10.1093/bioinformatics/btp012 (doi:10.1093/bioinformatics/btp012) [DOI] [PubMed] [Google Scholar]
- 47.Erickson J. W., Cline T. W. 1993. A bZIP protein, sisterless-a, collaborates with bHLH transcription factors early in Drosophila development to determine sex. Genes Dev. 7, 1688–1702 10.1101/gad.7.9.1688 (doi:10.1101/gad.7.9.1688) [DOI] [PubMed] [Google Scholar]
- 48.Pritchard D. K., Schubiger G. 1996. Activation of transcription in Drosophila embryos is a gradual process mediated by the nucleocytoplasmic ratio. Genes Dev. 10, 1131–1142 10.1101/gad.10.9.1131 (doi:10.1101/gad.10.9.1131) [DOI] [PubMed] [Google Scholar]
- 49.ten Bosch J. R., Benavides J. A., Cline T. W. 2006. The TAGteam DNA motif controls the timing of Drosophila pre-blastoderm transcription. Development 133, 1967–1977 10.1242/dev.02373 (doi:10.1242/dev.02373) [DOI] [PubMed] [Google Scholar]
- 50.Patel N. H., Snow P. M., Goodman C. S. 1987. Characterization and cloning of fasciclin III: a glycoprotein expressed on a subset of neurons and axon pathway in Drosophila. Cell 48, 975–988 10.1016/0092-8674(87)90706-9 (doi:10.1016/0092-8674(87)90706-9) [DOI] [PubMed] [Google Scholar]
- 51.Tadros W., et al. 2007. SMAUG is a major regulator of maternal mRNA destabilization in Drosophila and its translation is activated by the PAN GU kinase. Dev. Cell 12, 143–155 10.1016/j.devcel.2006.10.005 (doi:10.1016/j.devcel.2006.10.005) [DOI] [PubMed] [Google Scholar]
- 52.Su A. I., et al. 2004. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl Acad. Sci. USA 101, 6062–6067 10.1073/pnas.0400782101 (doi:10.1073/pnas.0400782101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baker M. A., Reeves G., Hetherington L., Muller J., Baur I., Aitken R. J. 2007. Identification of gene products present in Triton X-100 soluble and insoluble fractions of human spermatozoa lysates using LC-MS/MS analysis. Proteomics Clin. Appl. 1, 524–532 10.1002/prca.200601013 (doi:10.1002/prca.200601013) [DOI] [PubMed] [Google Scholar]
- 54.de Mateo S., Castillo J., Estanyol J. M., Ballesca J. L., Oliva R. 2011. Proteomic characterization of the human sperm nucleus. Proteomics 11, 2714–2726 10.1002/pmic.201000799 (doi:10.1002/pmic.201000799) [DOI] [PubMed] [Google Scholar]
- 55.Platts A. E., et al. 2007. Success and failure in human spermatogenesis as revealed by teratozoospermic RNAs. Hum. Mol. Genet. 16, 763–773 10.1093/hmg/ddm012 (doi:10.1093/hmg/ddm012) [DOI] [PubMed] [Google Scholar]
- 56.Cramton S., Laski F. 1994. String of pearls encodes Drosophila ribosomal protein S2, has Minute-like characteristics, and is required during oogenesis. Genetics 137, 1039–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilson D., Deutsch W., Kelley M. 1994. Drosophila ribosomal protein S3 contains an activity that cleaves DNA at apurinic/apyrimidinic sites. J. Biol. Chem. 269, 25 359–25 364 [PubMed] [Google Scholar]
- 58.Fenton T., Gout I. 2011. Functions and regulation of the 70 kDa ribosomal S6 kinases. Int. J. Biochem. Cell Biol. 43, 47–59 10.1016/j.biocel.2010.09.018 (doi:10.1016/j.biocel.2010.09.018) [DOI] [PubMed] [Google Scholar]
- 59.Bench G., Friz A., Corzett M., Morse D., Balhorn R. 1996. DNA and total protamine masses in individual sperm from fertile mammalian subjects. Cytometry 23, 263–271 (doi:10.1002/(SICI)1097-0320(19960401)23:4<263::AID-CYTO1>3.0.CO;2-I) [DOI] [PubMed] [Google Scholar]
- 60.Bestor T., Bourc'his D. 2004. Transposon silencing and imprint establishment in mammalian germ cells. Cold Spring Harb. Symp. Quant. Biol. 69, 381–387 10.1101/sqb.2004.69.381 (doi:10.1101/sqb.2004.69.381) [DOI] [PubMed] [Google Scholar]
- 61.Gao G., Cheng Y., Wesolowska N., Rong Y. S. 2011. Paternal imprint essential for the inheritance of telomere identity in Drosophila. Proc. Natl Acad. Sci. USA 108, 4932–4937 10.1073/pnas.1016792108 (doi:10.1073/pnas.1016792108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seth D., Gorrell M. D., McGuinness P. H., Leo M. A., Lieber C. S., McCaughan G. W., Haber P. S. 2003. SMART amplification maintains representation of relative gene expression: quantitative validation by real time PCR and application to studies of alcoholic liver disease in primates. J. Biochem. Biophys. Methods 55, 53–66 10.1016/S0165-022X(02)00177-X (doi:10.1016/S0165-022X(02)00177-X) [DOI] [PubMed] [Google Scholar]
- 63.Bolstad B. M., Irizarry R. A., Astrand M., Speed T. P. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193 10.1093/bioinformatics/19.2.185 (doi:10.1093/bioinformatics/19.2.185) [DOI] [PubMed] [Google Scholar]
- 64.Zheng Q., Wang X. J. 2008. GOEAST: a web-based software toolkit for Gene Ontology enrichment analysis. Nucleic Acids Res. 36, W358–W363 10.1093/nar/gkn276 (doi:10.1093/nar/gkn276) [DOI] [PMC free article] [PubMed] [Google Scholar]