Abstract
Our current understanding on how pathogens evolve relies on the hypothesis that pathogens' transmission is traded off against host exploitation. In this study, we surveyed the possibility that trade-offs determine the evolution of the bacterial insect pathogen, Xenorhabdus nematophila. This bacterium rapidly kills the hosts it infects and is transmitted from host cadavers to new insects by a nematode vector, Steinernema carpocapsae. In order to detect trade-offs in this biological system, we produced 20 bacterial lineages using an experimental evolution protocol. These lineages differ, among other things, in their virulence towards the insect host. We found that nematode parasitic success increases with bacteria virulence, but their survival during dispersal decreases with the number of bacteria they carry. Other bacterial traits, such as production of the haemolytic protein XaxAB, have a strong impact on nematode reproduction. We then combined the result of our measurements with an estimate of bacteria fitness, which was divided into a parasitic component and a dispersal component. Contrary to what was expected in the trade-off hypothesis, we found no significant negative correlation between the two components of bacteria fitness. Still, we found that bacteria fitness is maximized when nematodes carry an intermediate number of cells. Our results therefore demonstrate the existence of a trade-off in X. nematophila, which is caused, in part, by the reduction in survival this bacterium causes to its nematode vectors.
Keywords: evolution, virulence, Xenorhabdus, Steinernema
1. Introduction
Historically, our understanding of how virulence evolves in pathogens has been marked by two trends. Scientists have always recognized that pathogens have no direct interest in killing their host. The rationale for this prediction was that pathogen multiplication and transmission depend, in many circumstances, on the host. This view has led us to the first theory on how pathogens should evolve, the so-called ‘avirulence theory’ (see Alizon et al. [1] for a historical perspective on this point).
This theory was challenged in the 1980s by the seminal papers of Anderson & May [2,3] and Ewald [4]. These authors have proposed that the evolution of pathogens is driven, in part, by a positive link between their virulence and their capacity to contaminate new hosts. They have shown that, under some constraints on the shape of this link, pathogens can be poorly transmitted either because they are not contagious enough, or they are too virulent and therefore, kill their host before it has a chance to pass the infection to a susceptible conspecific. Selection should then favour intermediate virulence. This second theory on pathogen evolution has since then been called the ‘trade-off hypothesis’ (see again Alizon et al. [1] for a more detailed historical perspective).
Today, the empirical evidences that support the model of Anderson & May [3] exist but remain scarce. This is probably because it is difficult to measure pathogen transmission over the course of an infection (see earlier studies [1,5] for a review). The historical demonstration of the trade-off theory is provided by the case of Myxoma virus in European rabbits [6,7]. But one of the best-documented cases is probably that of the protozoan Ophryocystis elektroscirrha, an obligate-killing parasite that infects monarch butterflies [8]. The authors of this study have indeed shown that the number of spores produced by the pathogen is positively correlated to its virulence, whereas its transmission saturates with the quantity of spores emitted from an infected insect. As a result, the lifetime transmission of this pathogen is maximized at an intermediate level of virulence. Other recent empirical studies have detected a relationship between pathogens' virulence and transmission; a few of them have concluded that intermediate virulence was optimal for transmission [7–12].
One issue with the trade-off theory is that it assumes that pathogens' transmission linearly increases with the duration of the infection. This assumption does not hold in many biological systems. In the extreme case of obligate-killing parasites, free-living forms of the pathogen are released upon the host's death [13] and persist in the environment. The ‘sit and wait’ hypothesis then predicts that pathogens' virulence should increase with the persistence of their free-living form [14]. In other situations, the dependence between the pathogen and the host is relaxed because transmission is ensured by a vector. The prediction would then be that the vector-borne pathogens should be more virulent than pathogens that are directly transmitted from host to host. This prediction is in fact as old as the trade-off theory itself [4] and has some experimental support [4,15,16]. Still, the data collected by Ewald [4] reveal considerable variation in virulence among pathogens that have similar modes of transmission.
The conclusion that can be drawn from all these studies is probably that the trade-off theory provides a general frame-work to study pathogen evolution, but understanding why pathogens should hurt their hosts requires the accumulation of knowledge on many different pathogenic systems. In this study, we present results from experiments we performed on the vector-borne insect pathogen Xenorhabdus nematophila. The vector of this gammaproteobacteria is the nematode Steinernema carpocapsae. During the dispersal stage of their life cycle, nematodes are present in the soil as infective juveniles (IJs; a larval form of nematodes). Each IJ carries a small population of X. nematophila in its intestine. Once they have found an insect larvae to parasitize, they release these bacteria in the haemolymph of their host. Bacteria contribute to the rapid death of the insect, by a combination of septicaemia and toxemia [17]. This, in turn, increases the chances that nematodes successfully reproduce in the insect cadaver [18] and efficiently disperse bacteria. Figure 1 summarizes this life cycle. From the description mentioned above, it could seem that X. nematophila is an obligate-killer sensu [13]. This is in fact unclear, because nematodes reproduce only in dead insect hosts but can kill them without the help of any bacteria. Another way to consider this system is to depict X. nematophila as a mutualistic symbiont of its vector, as it provides clear benefits to nematodes during the parasitic stage of their life cycle. It is also not so clear, because bacteria have been demonstrated to decrease nematode survival during dispersal [19,20]. As the transmission of bacteria depends both on how well vectors reproduce in the insect host, and on how well they survive when dispersing, X. nematophila may face a trade-off.
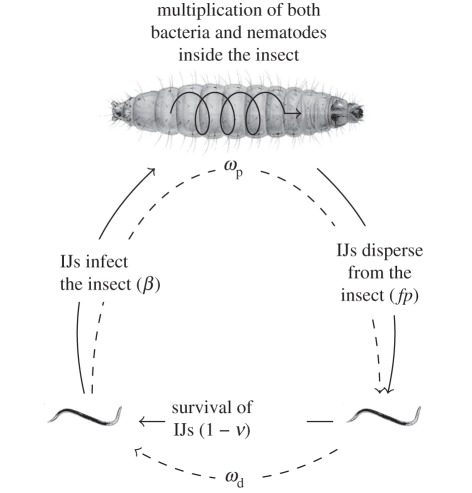
Figure 1.
The life cycle of Xenorhabdus nematophila and its vector Steinernema carpocapsae. Plain arrows indicate transitions between the different stages of the life cycle, with the corresponding life-history traits we have measured. Dashed arrows indicate the two fitness components we have estimated, 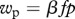 and
and 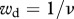 being, respectively, the parasitic and the dispersal component of bacteria fitness (see text for further details).
being, respectively, the parasitic and the dispersal component of bacteria fitness (see text for further details).
In this study, we tested the hypothesis that a trade-off exists between the parasitic and dispersal components of X. nematophila fitness. In a previous study [21], we produced 20 bacterial lineages that differ in their virulence towards insects. This was carried out by performing 20 independent serial passage experiments in an artificial culture medium. All these experiments were initiated from a unique single ancestral clone of X. nematophila strain Be06 (see Lenski and co-workers [22,23] for a similar selection method). We have demonstrated that the bacteria we have selected kill the insect host Galleria mellonella faster than their ancestral lineage, but that virulence varies among selected lineages [21]. Here, we re-associated evolved bacterial strains with their vectors and studied how differences in bacterial virulence towards insects impact nematode vectors. More precisely, we measured how bacteria modify nematode reproductive success, in the insect host, and survival during dispersal. We then used an epidemiological model to combine these measurements and estimate bacteria fitness. This last procedure allowed us to test for trade-offs in X. nematophila.
2. Material and methods
(a). The biological system
(i). The bacterial strains
We used X. nematophila (Enterobacteriaceae) strain Be06 as the ancestral lineage of our selection experiment. This particular strain was isolated in 2001 from a S. carpocapsae nematode from Belgium and stored since then [24]. We thus have little knowledge on the particular behaviour of this strain, compared with other more studied ones, but we have the guarantee that it has not experienced many generations of selection in laboratory conditions.
In a previous work, we performed 80 independent serial passage experiments, all starting from a unique clone of X. nematophila Be06. Each lineage was transferred every 24 h and 40 times, with a 1 : 10000 dilution factor for half of the lines (low-density inoculum (LDI) treatment, as in Lenski and co-workers [22,23]) and a 1 : 10 dilution factor for the remaining half (high-density inoculum (HDI) treatment). The whole procedure is described in detail by Chapuis et al. [21]. In this study, we have used a subset of 10 LDI and 10 HDI randomly chosen lineages.
We have shown in the same previous work [21] that evolved lineages were on average more virulent towards insects (lower lethal time 50 (LT50) which corresponds to the time at which 50% of the hosts injected with 2000 bacteria cells are dead) and had lower lag time in an artificial culture medium (lower medium lag time (ML)). We have also measured several phenotypic traits that probably contribute to the functioning of the symbiosis [25,17]. These traits are all under the control of a single master operon, flhDC, and are jointly expressed during late insect infection [26]. We found that evolved HDI strains have a drastically reduced mobility, and that both LDI and HDI lineages have an increased haemolytic activity, corresponding most probably to an increased expression of the xaxAB gene [21].
The 20 lineages, we have chosen to study, have probably accumulated mutations during the serial passage experiment. These caused part of the differences which we have found among lineages for all the traits we have measured. In this study, we will study how this variation impacts the way these bacterial lineages interact with their nematode vector.
(ii). The nematode strain
The vector of X. nematophila is the nematode species S. carpocapsae (Rhabditiae, Steinernematidae). Here, we used the SK27 strain of S. carpocapsae (Plougastel, Finistère, France), naturally associated with X. nematophila strain F1, as a vector of our selected bacteria. Aposymbiotic IJs (i.e. dispersing nematode larvae which are deprived of their bacterial symbionts) were available in the laboratory.
(iii). The experimental insect host
The symbiotic couple Xenorhabdus/Steinernema is known to attack a wide range of insects, but in laboratory conditions Lepidoptera are the most often used experimental hosts. Among the possible experimental hosts, G. mellonella (Lepidoptera, Pyralidae) is the most susceptible to infection. We chose this insect both to test the pathogenicity of our bacterial lineages and to study the impact of bacteria on nematodes parasitic success. Galleria mellonella was reared in the dark, on pollen and wax, at 28°C, in aerated glass jars, at the University of Montpellier, France. Pathogenicity tests were performed on the last instar, kept at 24°C.
(b). Re-associating nematodes and bacteria
In order to study how variations in bacterial traits affect nematodes' performances, we co-infected from 40 to 60 insect hosts with both aposymbiotic nematodes and each of the 20 bacterial lineages we obtained by the mean of experimental evolution. For this purpose, we followed the method developed by Sicard et al. [27].
Each G. mellonella larva was first infected by 20 nematodes IJ. This number was chosen because it guarantees a reasonably high probability of successful infection [19,27,28]. Infected insects were then incubated for 24 h at 24°C, after which they were injected with 2000 cells of Xenorhabdus. In order to control for potential bias in our measurements, insects were weighed prior to infection and the number of injected cells was controlled a posteriori by streaking appropriate volumes of the injected bacterial suspension onto nutrient bromothymol blue tetrazolium chloride agar (NBTA) plates, and by counting colony forming units (CFUs) after 48 h of incubation at 28°C.
(c). Symbiotic life-history traits measurements
After several days of incubation at 24°C, cadavers of the co-infested insects were placed in the classical ‘White traps’ device [29], so that newborn nematodes (IJs) can easily be collected when they migrate from the insect cadaver. We describe below the traits that we have measured on these nematode larvae. All analyses were performed using the R software [30]—using mostly simple non-parametric comparison and correlation tests. We describe below the analyses that required more sophisticated statistical tools.
(i). Parasitic success and nematode emergence
For each bacterial lineage, we estimated the proportion of successful infections (hereafter parasitic success) over the 40–60 co-infected insects. This was performed after 12, 14, 17, 21, 28, 35 and 42 days of incubation at 24°C. We considered an infection to be successful when at least 10 IJs emerged from the insect cadaver. We analysed these data using a Cox proportional hazard model with differences among replicate experiments modelled as a random block factor. This was performed using the function coxme in the library kinship [31] of the R software.
We then randomly sampled, for each bacterial lineage, six infections among the successful ones, on which we measured the following three traits.
(ii). Reproductive success
We evaluated reproductive success for each of the six infections, by collecting and counting all the IJs that emerged from the insect cadaver after 42 days of incubation. A single estimate of mean reproductive success was obtained by averaging these six values.
(iii). Number of bacteria carried per nematode infective juvenile
We sampled 100 IJs from each of the six infections to estimate how many bacteria one nematode larvae carries on average. These IJs were disinfected and rinsed following Sicard et al. [18]. They were then crushed together for 5 min with a microtube piston to liberate bacterial symbionts. Hundred-microlitre samples of appropriate dilutions were then streaked onto NBTA plates and incubated at 28°C for 48 h. Estimates of the number of bacteria carried for each selected lineage were obtained by averaging these six independent estimates.
(iv). Infective juveniles survival
For each of the six successful infection, we placed two batches of 500 newly collected IJs at 28°C and measured the proportion of dead IJs after 17 and 56 days. Dead and alive IJs were distinguished based on their morphology and their response to a tactile stimulus [19,20]. A first simple analysis of the proportion of dead nematodes at 56 days was performed on survival averaged for each bacterial lineage. A more complete analysis was also performed in which measurements obtained from each replicate co-infection were considered. The proportion of dead nematodes after 17 days was included as an offset, so that we can analyse the increase in the proportion of dead nematodes between day 17 and day 56. We modelled differences among replicate experiments as a random block effect. To account for pseudo-replication, we also included a random effect of bacterial lineage in this model, and a random interaction between bacterial lineage and the number of bacteria carried. This analysis was performed using the lmer routine in the library lme4 [32] of the R statistical software.
(d). A transmission model to estimate bacteria fitness from symbiotic life-history traits measurements
We combined nematodes' parasitic success, reproductive rate, survival and the number of bacteria carried per IJ to obtain estimates of bacteria fitness. For this purpose, we designed a model that reproduces bacteria life cycle (as illustrated by figure 1). The transmission of Xenorhabdus is ensured by a vector, but vectors are contaminated within the body of infected hosts. Therefore, the rate at which infected vectors are produced does not depend on the rate at which infected hosts and susceptible vectors encounter. Rather it is a function of how many infected vectors emerge from the cadaver of infected hosts. For this reason, our model is very similar to that of Caraco & Wang [33] which describes the epidemiology of pathogens with a free-living stage.
This model is described by:
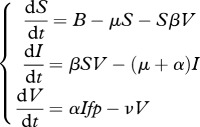 |
2.1 |
where S is the density in susceptible insects, B is their natural birth rate and μ is their natural death rate. I and V are the density of infected insects and of free-living bacteria-carrying nematodes, respectively. The rate at which susceptible hosts are infected is given by the product βV, β being thus a measure of the capacity of nematodes to infect insects. Infected cadavers die at a rate α, which is therefore a measure of bacteria virulence towards insects, and release, upon their death, a number f of nematode larvae. A fraction p of nematodes carry bacteria. Their death rate during dispersal is quantified by the parameter ν. From this equation, the transmission capacity of the bacteria can be estimated as:
| 2.2 |
where R0 S > 1 is the necessary condition for a population of bacteria to successfully establish, S being the density in susceptible insects. If μ is small compared to α, which is a reasonable assumption in the case of Xenorhabdus, then equation (2.2) reduces to:
| 2.3 |
R0 can then be written as the product of a parasitic component (wp = βfp) and a dispersal component (wd = 1/ν) which we estimated for the ancestral and selected lineages. From our experiments, we estimated the quantity β as the proportion of successful infections and f as the number of IJs emitted from an insect cadaver, which we estimated for six infections. For the same six infections, we estimated the average number N of bacteria carried per IJ. Assuming that this number follows a Poisson distribution, p can be estimated as 1 − e−N. For each strain, we first averaged f × p over the six infections we have studied and multiplied this by β which yielded as estimate of wp. Similarly, for each lineage, we obtained wd by averaging 1/ν over the six infections for which we have estimated IJs survival. Finally, R0 was obtained for each lineage by first averaging f × p/ν over the six studied infections and multiplying this quantity by β.
The trade-off between virulence and transmission, which is hypothesized in many models, should produce a negative correlation between wp and wd. We tested this hypothesis. Another prediction is that the fitness, estimated as R0, should be maximum for intermediate values of virulence. We therefore used linear models to test the influence of bacterial traits on fitness. When necessary, a Box–Cox transformation was applied to R0 to fulfil normality conditions.
3. Results
(a). Bacteria virulence and nematode parasitic success
Part of nematode fitness is determined by their ability to infect insects, and by their capacity to reproduce in the resulting cadaver. We found that, when deprived of any bacteria, nematodes successfully achieve these two steps in 14.6 per cent of the infections. When nematodes co-infect insects with the ancestral bacterial lineage, their parasitic success increases to 61.1 per cent. Selected bacteria also increased nematodes parasitic success, but to a lesser extent (parasitic success is 48.9% and 49.8% when nematodes are associated with HDI and LDI bacteria, respectively, see table 1 for a statistical analysis of these results). The traits that we have selected in HDI and LDI lineages have therefore reduced the average benefit bacteria provide to their vectors.
Table 1.
Analysis of the time at which nematode larvae emerge from infected insects. (This analysis is performed using a Cox proportional hazard model, with differences among replicate experiments modelled as a random block factor (see §2 for further details). In the first analysis, all lineages are considered and the difference between the ancestral and the selected lineages is tested. In the second analysis, only selected bacteria are considered and the effect of virulence (1/LT50), speed at which bacteria start multiplying (1/ML) and haemolytic activity (total haemolysis) are tested.)
| coefficient | s.e. | p-value | |
|---|---|---|---|
| all lineages (n = 874) | |||
| insect weight | 1.37 | 0.79 | 0.083 |
| injected CFU | 0.001 | 0.001 | 0.095 |
| HDI | −0.43 | 0.18 | 0.020 |
| LDI | −0.32 | 0.17 | 0.066 |
| selected lineages (n = 794) | |||
| insect weight | 1.69 | 0.85 | 0.047 |
| injected CFU | 0.002 | 0.001 | 0.037 |
| 1/LT50 | 35.09 | 12.52 | 0.005 |
| 1/ML5 | −51100 | 21960 | 0.020 |
| total haemolysis | 0.16 | 0.20 | 0.420 |
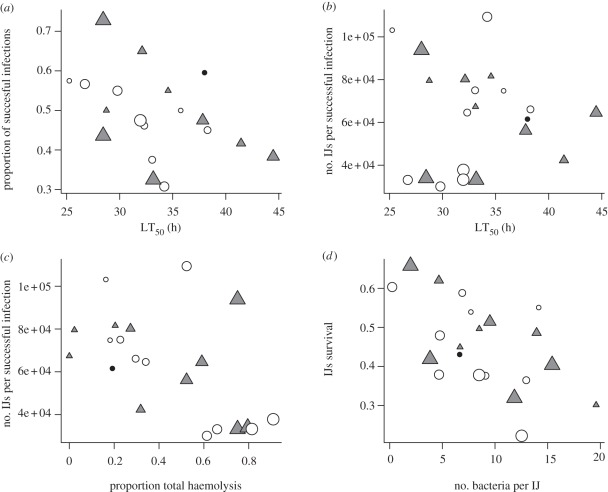
We found that the most virulent of the evolved bacterial lineages are those that provide the greatest increase in nematode parasitic success (Kendall correlation between LT50 and parasitic success, τ = −0.45, n = 20, p = 0.0057; figure 2a). We then conducted an analysis where the joint effects of several bacterial traits were tested (table 1, selected lineages). This analysis demonstrates that, all else being equal, the bacteria that increase the most nematodes' parasitic success are those that start multiplying the latest in an artificial culture medium (significant negative effect of 1/ML; table 1). However, even when taking this effect into account we still found that nematode parasitic success significantly increased with bacterial virulence (significant positive effect of 1/LT50; table 1). Our results therefore show that it is the capacity of bacteria to kill insects rapidly per se that determines differences in nematodes parasitic success among selected lineages, and not other factors that could correlate with bacterial virulence.
Figure 2.
(a) Proportion of successful infections as a function of bacteria virulence (indicated here by their lethal time 50 (LT50), highly virulent lineages having low LT50). (b) Reproductive success as a function of bacteria LT50. (c) Reproductive success as a function of XaxAB-dependent haemolytic activity. Haemolytic activity is measured as the proportion, for each lineage, of clones that display the full haemolysis phenotype. (d) Proportion of surviving IJs after 56 days spent at 28°C, as a function of the average number of bacteria they carry. In all figures, point size indicates haemolytic activity (filled circles, ancestral lineages; triangles, LDI; open circles, HDI).
(b). Bacteria influence nematode reproductive success in several different ways
A second component of nematodes' fitness is their reproductive success, i.e. the number of IJs produced per successful infection. When analysing all selected lineages, we found no correlation between nematode reproductive success and bacterial virulence (Kendall correlation between LT50 and reproductive success: τ = 0.02, n = 20, p = 0.7085; figure 2b). This is owing to a group of six highly virulent bacterial lineages, which increased nematode parasitic success but drastically reduced their reproductive success (see again figure 2b). Indeed, a highly significant negative correlation between LT50 and reproductive success (Kendall correlation: τ = −0.18, n = 14, p = 0.0057) appears when this group of bacteria is excluded from the analysis.
These six lineages differ from others by their low reproductive success, but also by a strong XaxAB-dependent haemolytic activity. More precisely, the frequency of clones that perform full haemolysis is higher in these six lineages than in the other selected bacteria (Fisher's exact test: p < 0.0001). Overall, XaxAB-dependent haemolytic activity is higher in selected bacteria than in the ancestral lineage (Fisher's exact test: p < 0.0001) and negatively impacts nematodes' reproductive success (Kendall correlation: τ = −0.23, n = 20, p < 0.0001; figure 2c). In §3a, we showed that nematodes' parasitic success was mostly related to bacterial virulence. Here, we found that nematodes' reproductive success depends on a complex interaction between bacterial virulence and XaxAB-dependent haemolytic activity.
(c). Carrying bacteria decreases nematode survival
Finally, a third component of nematode fitness is the survival of IJs during the dispersal stage. A first analysis demonstrated that the proportion of IJs that survived after 56 days spent at 28°C was higher in aposymbiotic nematodes than in those that are associated with the ancestral bacterial lineage (Fisher's exact test: p < 0.0001). We also found that, when nematodes were associated with selected bacteria, the proportion of dead IJs positively correlated to the average number of bacteria IJs carried (Kendall rank correlation: τ = 0.389, p=0.01641; figure 2d). The six lineages identified in §3b do not differ particularly from other lineages in this analysis.
Table 2 summarizes results of a more detailed analysis in which IJ mortality is analysed for each individual infection. Virulence can impact nematodes survival both directly, because virulent bacteria would be more costly to carry for IJs, and indirectly, because virulent bacteria kill insects faster which improves the quality of the environment in which nematodes were born [28]. In order to control for these indirect effects, we included as a covariate the number of IJs produced by each infection (see Emeliano et al. [19] and §2). Contrary to the previous analysis, where all measurements performed on a single lineage were pooled together, we did not detect any significant effect of the number of bacteria carried per IJ. This is probably because this effect was masked by the environmental variance between replicate measurements that are realized for the same lineage. Still, we detected a marginal interaction between bacteria virulence and the number of bacteria carried per nematode. This interaction suggests that the cost of carrying bacteria is lower with the least virulent of the selected bacterial lineages.
Table 2.
Analysis of infective juveniles (IJs) mortality after 52 days spent at 28°C when associated with selected bacterial lineages. (This analysis is performed using a logistic regression with the proportion of dead IJs after 17 days as an offset. The model therefore predicts the increase in the proportion of dead IJs between day 17 and day 52. Explanatory variables are the log-transformed reproductive success of IJs' parents (log(W)), the log-transformed number of carried bacteria per IJ (log(N)) and LT50. To insure model convergence, log(N) and log(W) have been centered and scaled and the quantitative variable LT50 has been transformed in a two levels factor. LT50sup, therefore, corresponds to bacteria in which LT50 is above median, that is to the least virulent of the bacteria. In addition to these fixed effects, we incorporated in our model a random block effect, as in the analysis presented in table 1.)
| coefficient | s.e. | p-value | |
|---|---|---|---|
| intercept | 3.63 | 0.47 | <10−4 |
| log(W) | −0.20 | 0.02 | <10−4 |
| LT50sup | 0.75 | 0.43 | 0.081 |
| log(N) | 0.35 | 0.33 | 0.296 |
| log(W) × log(N) | −0.06 | 0.02 | 0.008 |
| LT50sup × log(N) | −1.40 | 0.68 | 0.040 |
(d). Bacteria fitness is optimized when nematodes carry intermediate numbers of cells
From our life-history traits measurement and from our model (see §2), we estimated a parasitic (wp) and a dispersal (wd) component of bacterial fitness for each lineage. If a trade-off exists between transmission and host exploitation in Xenorhabdus, then we expect a negative correlation between these two components. We found no significant correlation between wp and wd (Kendall correlation: τ = −0.12, p = 0.5006). The correlation is still not significant if R0 is computed using nematodes parasitic success 14 days after infection instead of 42, which mimics more stringent transmission conditions (Kendall correlation: τ = − 0.14, p = 0.4223).
If bacteria have to face trade-offs, then it is also expected that their fitness is maximized for some intermediate value of life-history traits. We investigated this hypothesis by testing the influence of virulence and of the number of carried bacteria per IJ on bacteria fitness. We also included in our model quadratic terms for these two traits and a possible effect of haemolytic activity. When fitness is computed from the parasitic success measured 42 days after infection, the best model includes a positive linear effect of virulence (F1,18 = 111.45, p = 3.853e − 09) and a negative linear effect of haemolytic activity (F1,18 = 4.2986, p = 0.0528; figure 3a). We therefore did not detect any sign that bacteria fitness could be reduced when virulence is too high. When fitness incorporates parasitic success 14 days after infection, the best model incorporates only the linear (F1,18 = 60.546, p = 3.630e − 07) and quadratic (F1,18 = 11.983, p = 0.002784) effects of the number of bacteria carried per IJ. Bacteria fitness is therefore maximized at an intermediate number of bacteria carried per IJ and, quite remarkably, the ancestral lineage sits exactly at this optimal value (figure 3b).
Figure 3.
Bacteria fitness (R0) as a function of the average number of bacteria carried by a nematode IJ. Fitness includes either parasitic success (a) 14 days after infection or (b) after 42 days. The solid curve represents a quadratic regression, with a fixed intercept of zero, adjusted on all selected lineages. Dashed and dotted curves represents the same model adjusted on LDI and HDI lineages, respectively (plus, aposymbiotic; filled circles, ancestral lineages; triangles, LDI; open circles, HDI).
4. Discussion
In this study, we measured how several lineages of X. nematophila, which differ in their virulence towards the insect host G. mellonella, are transmitted by their nematode vector, S. carpocapsae. These bacterial lineages have been produced, from a single ancestral lineage, by a serial passage experiment in an artificial culture medium (as described in Chapuis et al. [21]).
We found that vectors have a higher parasitic success (i.e. succeed in reproducing in insects) and a higher reproductive success (i.e. produce a higher number of dispersing juveniles) when associated with the most virulent of the bacterial lineages. The interest of Xenorhabdus therefore seems to be aligned with that of its vector, at least for the parasitic phase of their life cycle. The mechanism of this alignment might come from the fact that the sooner the insect dies after infection, the less nematodes have to endure its immune defences. This confirms the previous demonstration that nematodes' parasitic success increases with the number of bacteria it inoculates an insect with [19].
Nematode parasitic success is probably determined by the success of very early stages of the parasitic phase, such as entering the insect and overcoming its defences. Conversely, reproductive success is determined by later stages where nematodes have to first colonize and exploit host tissues, and second, sexually reproduce [19]. This probably explains why parasitic success seems to be determined by virulence only, whereas nematode reproductive success is also impacted by other bacterial traits, such as haemolytic activity.
The haemolytic activity, we measured in our experiments, reflects the expression of the xaxAB gene [21,34,35]. This gene is co-regulated with other haemolysin genes, with protease and lipase genes and with the genes that permit the expression of flagellar motility [25]. These genes are all expressed relatively late during the infection process [26] and they probably allow bacteria to metabolize the insect's tissues after it has been killed [17]. A recent study showed that the inactivation of lipase genes reduces nematode reproductive success while having little impact on parasitic success [36]. Following these results, one would thus expect that any increase in bacteria haemolytic activity would benefit nematode reproduction. We found the exact opposite. One possible explanation of this unexpected result could be that, in selected lineages, xaxAB is strongly expressed but at the wrong time for bacteria. A dysregulation of this gene and subsequent early expression, could indeed render bacteria easier to detect by the insect host. This explanation is of course very speculative and deserves further investigation.
All together, we found that the number of nematode IJs produced by an infection increases with bacteria virulence. As these juvenile nematodes carry bacteria from insect to insect, our results suggest that X. nematophila infectivity (i.e. its capacity to contaminate susceptible insects) increases with its virulence. This connection is central to the trade-off theory of virulence [1,5].
The trade-off theory also assumes that being too virulent is not optimal from a pathogen's point of view, because killing its host too fast impedes transmission to susceptible hosts [2,3]. In the vector-borne X. nematophila, the cost of virulence could come from reduced vector survival [19,20]. We found only weak support for this hypothesis. One explanation for this is that nematode survival depends both on the environment they were born in, and on the conditions they experience during dispersal [28]. By killing the host rapidly, virulent bacteria provide a good quality parasitic environment to the nematodes that co-infect the insect. At the same time, virulent bacteria might be costly to carry for dispersing nematodes. These confounding factors could well explain, even if we tried to statistically account for them, why the link we detected between bacteria virulence and dispersing nematode survival is so weak. In spite of these difficulties, we found that the number of bacteria cells which dispersing nematodes carry varies from one bacterial lineage to another, and that nematode death rate increases with this number. This result is similar to that obtained in the case of Plasmodium chabaudii [16] and complete previous results obtained in Xenorhabdus [19], demonstrating the possibility of genetic variance in the cost bacteria impose on their vector.
Xenorhabdus increases both parasitic and reproductive success of its nematode vectors during the parasitic phase of their life cycle, and decreases their survival during the dispersal phase. This opens up the possibility of a trade-off for bacteria. We first tested this hypothesis by combining our life-history trait measurements into a parasitic component and a dispersal component of bacteria fitness. Contrary to what is expected under the hypothesis of a trade-off, we found no significant negative link between the two components of fitness. This might be because we underestimate the correlation between these fitness components. In a previous study, we indeed demonstrated that the more bacteria cells nematodes bring into an insect, the higher their parasitic success [19]. In natural conditions, there should, therefore, be a positive correlation between the number of bacteria nematodes carry and their parasitic success. This link does not exist in our experiment, as we did control the number of bacteria cells insects are infected with. Similarly, we fixed the number of IJs insects are infected by. A recent study of the protozoan O. elektroscirrha, a parasite that infects monarch butterflies, demonstrated the existence of a trade-off which is caused in part by the fact that transmission increases slower than linearly with the number of spores emitted from an infected insect [8]. In Xenorhabdus, the transmission could also increase slower than linearly with the number of nematode IJs emitted from an insect cadaver. Our experimental estimation of bacteria fitness does not incorporate this limitation of transmission.
In spite of these limitations, we found that Xenorhabdus fitness is maximized when nematodes carry intermediate numbers of bacteria cells. This is in part because bacteria transmission requires that nematodes carry some bacteria, whereas nematode survival decreases with the number of bacteria each nematode carries. However, if the trade-off was only between host exploitation and transmission, then we should have detected a negative correlation between the parasitic and the dispersal component of bacteria fitness. Part of the trade-off must therefore originate from constraints that play during the parasitic stage of Xenorhabdus life cycle. We found, in fact, a link between the parasitic success of nematodes and the number of bacteria they carry. The mechanism that could explain this link is so far unknown.
An optimum value of the number of carried bacteria exists if parasitic success is estimated 14 days after infection but not if it is estimated 42 days after infection. This is probably because 14 days represents a rather short period of time over which few infected insects have released nematode IJs. Said differently, estimating fitness this way probably mimics stringent environmental conditions for Xenorhabdus transmission [19], which facilitates the detection of trade-offs. When parasitic success is estimated 42 days after infection, bacteria fitness increases with virulence and decreases with haemolytic activity. Trade-offs, therefore, exist in Xenorhabdus, but they do not seem to constrain so much the evolution of its virulence. These conclusions should in fact be considered with caution. First, because the experimental conditions we used in our work might be very different from natural conditions and, second because we used in our experiments a single strain of nematode. Therefore, the generality of the trade-off we have demonstrated needs to be evaluated.
Most former demonstrations of trade-offs in pathogens are based on observations that spore production is maximized at intermediate values of virulence or replication [10–12]. Such trade-offs do not require that the contamination of susceptible hosts is impeded by high virulence. To our knowledge the only experimental demonstration of a trade-off where contamination is directly involved is that of de Roode et al. [8]. The trade-off we demonstrate here, as it is created in part by the mortality pathogens impose on their vectors, would be a second example of this kind.
Acknowledgements
The authors acknowledge Alain Givaudan and Franois Rousset for their comments on an early version of this manuscript, and Samuel Alizon for his numerous and insightful suggestions. The authors also thank two anonymous referees who helped improve the manuscript. This work has received financial support from French ANR (grant ANR-JC EvolNemBact).
References
- 1.Alizon S., Hurford A., Mideo N., van Baalen M. 2009. Virulence evolution and the trade-off hypothesis: history, current state of affairs and future. J. Evol. Biol. 22, 245–259 10.1111/j.1420-9101.2008.01658.x (doi:10.1111/j.1420-9101.2008.01658.x) [DOI] [PubMed] [Google Scholar]
- 2.Anderson R. M., May R. M. 1979. Population biology of infectious diseases. Nature 280, 361–367 10.1038/280361a0 (doi:10.1038/280361a0) [DOI] [PubMed] [Google Scholar]
- 3.Anderson R. M., May R. M. 1982. Coevolution of hosts and parasites. Parasitology 85, 411–426 10.1017/S0031182000055360 (doi:10.1017/S0031182000055360) [DOI] [PubMed] [Google Scholar]
- 4.Ewald P. W. 1983. Host–parasite relations, vectors, and the evolution of disease severity. Annu. Rev. Ecol. Evol. Syst. 14, 465–485 10.1146/annurev.es.14.110183.002341 (doi:10.1146/annurev.es.14.110183.002341) [DOI] [Google Scholar]
- 5.Froissart R., Doumayrou J., Vuillaume F., Alizon S., Michalakis Y. 2010. The virulence-transmission trade-off in vector-borne plant viruses: a review of (non-)existing studies. Phil. Tran. R. Soc. B 365, 1907–1918 10.1098/rstb.2010.0068 (doi:10.1098/rstb.2010.0068) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fenner F., Ratcliffe F. 1965. Myxomatosis. Cambridge, UK: Cambridge University Press [Google Scholar]
- 7.Bolker B. M., Nanda A., Shah D. 2010. Transient virulence of emerging pathogens. J. R. Soc. Interface 7, 811–822 10.1098/rsif.2009.0384 (doi:10.1098/rsif.2009.0384) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Roode J. C., Yates A. J., Altizer S. 2008. Virulence-transmission trade-offs and population divergence in virulence in a naturally occurring butterfly parasite. Proc. Natl Acad. Sci. USA 105, 7489–7494 10.1073/pnas.0710909105 (doi:10.1073/pnas.0710909105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fraser C., Hollingsworth T. D., Chapman R., de Wolf F., Hanage W. P. 2007. Variation in HIV-1 set-point viral load: epidemiological analysis and an evolutionary hypothesis. Proc. Natl Acad. Sci. USA 104, 17 441–17 446 10.1073/pnas.0708559104 (doi:10.1073/pnas.0708559104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jensen K. H., Little T., Skorping A., Ebert D. 2006. Empirical support for optimal virulence in a castrating parasite. PLoS Biol. 4, e197. 10.1371/journal.pbio.0040197 (doi:10.1371/journal.pbio.0040197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meyer S. E., Stewart T. E., Clement S. 2010. The quick and the deadly: growth vs virulence in a seed bank pathogen. New Phytol. 187, 209–216 10.1111/j.1469-8137.2010.03255.x (doi:10.1111/j.1469-8137.2010.03255.x) [DOI] [PubMed] [Google Scholar]
- 12.Berenos C., Schmid-Hempel P., Wegner K. M. 2009. Evolution of host resistance and trade-offs between virulence and transmission potential in an obligately killing parasite. J. Evol. Biol. 22, 2049–2056 10.1111/j.1420-9101.2009.01821.x (doi:10.1111/j.1420-9101.2009.01821.x) [DOI] [PubMed] [Google Scholar]
- 13.Ebert D., Weisser W. W. 1997. Optimal killing for obligate killers: the evolution of life histories and virulence of semelparous parasites. Proc. R. Soc. Lond. B 264, 985–991 10.1098/rspb.1997.0136 (doi:10.1098/rspb.1997.0136) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walther B. A., Ewald P. W. 2004. Pathogen survival in the external environment and the evolution of virulence. Biol. Rev. 79, 849–869 10.1017/S1464793104006475 (doi:10.1017/S1464793104006475) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ewald P., de Leo G. 2002. Alternative transmission modes and the evolution of virulence. In The adaptive dynamics of infectious diseases (Cambridge studies in adaptive dynamics) (eds Dieckmann U., Metz J. A. J., Sabelis M. W., Sigmund K.), pp. 10–25 Cambridge, UK: Cambridge University Press [Google Scholar]
- 16.Ferguson H. M., MacKinnon M. J., Chan B. H., Read A. F. 2003. Mosquito mortality and the evolution of malaria virulence. Evolution 57, 2792–2804 [DOI] [PubMed] [Google Scholar]
- 17.Richards G. R., Goodrich-Blair H. 2009. Masters of conquest and pillage: Xenorhabdus nematophila global regulators control transitions from virulence to nutrient acquisition. Cell. Microbiol. 11, 1025–1033 10.1111/j.1462-5822.2009.01322.x (doi:10.1111/j.1462-5822.2009.01322.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sicard M., Le Brun N., Pags S., Godelle B., Boemare N., Moulia C. 2003. Effect of native Xenorhabdus on the fitness of their Steinernema hosts: contrasting types of interaction. Parasitol. Res. 91, 520–524 10.1007/s00436-003-0998-z (doi:10.1007/s00436-003-0998-z) [DOI] [PubMed] [Google Scholar]
- 19.Emelianoff V., Chapuis E., Le Brun N., Chiral M., Moulia C., Ferdy J. B. 2008. A survival-reproduction trade-off in entomopathogenic nematodes mediated by their bacterial symbionts. Evolution 62, 932–942 10.1111/j.1558-5646.2008.00319.x (doi:10.1111/j.1558-5646.2008.00319.x) [DOI] [PubMed] [Google Scholar]
- 20.Emelianoff V., Sicard M., Le Brun N., Moulia C., Ferdy J. B. 2007. Effect of bacterial symbionts Xenorhabdus on mortality of infective juveniles of two Steinernema species. Parasitol. Res. 100, 657–659 10.1007/s00436-006-0284-y (doi:10.1007/s00436-006-0284-y) [DOI] [PubMed] [Google Scholar]
- 21.Chapuis E., Pags S., Emelianoff V., Givaudan A., Ferdy J. B. 2011. Virulence and pathogen multiplication: a serial passage experiment in the hypervirulent bacterial insect-pathogen Xenorhabdus nematophila. PLoS ONE 6, e15872. 10.1371/journal.pone.0015872 (doi:10.1371/journal.pone.0015872) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lenski R. E., Travisano M. 1994. Dynamics of adaptation and diversification. A 10000 generations experiment with bacterial populations. Proc. Natl Acad. Sci. USA 91, 6808–6814 10.1073/pnas.91.15.6808 (doi:10.1073/pnas.91.15.6808) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lenski R. E., Rose M. R., Simpson S. C., Tadler S. C. 1991. Long term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2000 generations. Am. Nat. 138, 1315–1341 10.1086/285289 (doi:10.1086/285289) [DOI] [Google Scholar]
- 24.Tailliez P., Pags S., Ginibre N., Boemare N. 2006. New insight into diversity in the genus Xenorhabdus, including the description of ten novel species. Int. J. Syst. Evol. Microbiol. 56, 2805–2818 10.1099/ijs.0.64287-0 (doi:10.1099/ijs.0.64287-0) [DOI] [PubMed] [Google Scholar]
- 25.Givaudan A., Lanois A. 2000. flhdc, the flagellar master operon of Xenorhabdus nematophilus: requirement for motility, lipolysis, extracellular hemolysis, and full virulence in insects. J. Bacteriol. 182, 107–115 10.1128/JB.182.1.107-115.2000 (doi:10.1128/JB.182.1.107-115.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jubelin G., Pages S., Lanois A., Boyer M. H., Gaudriault S., Ferdy J. B., Givaudan A. 2011. Studies of the dynamic expression of the Xenorhabdus FliAZ regulon reveal atypical iron-dependent regulation of the flagellin and haemolysin genes during insect infection. Environ. Microbiol. 13, 1271–1284 10.1111/j.1462-2920.2011.02427.x (doi:10.1111/j.1462-2920.2011.02427.x) [DOI] [PubMed] [Google Scholar]
- 27.Sicard M., Ferdy J. B., Pags S., Le Brun N., Godelle B., Boemare N., Moulia N. 2004. When mutualists are pathogens: an experimental study of the symbioses between Steinernema (entomopathogenic nematodes) and Xenorhabdus (bacteria). J. Evol. Biol. 17, 985–993 10.1111/j.1420-9101.2004.00748.x (doi:10.1111/j.1420-9101.2004.00748.x) [DOI] [PubMed] [Google Scholar]
- 28.Chapuis E., Emelianoff V., Paulmier V., Le Brun N., Pags S., Sicard M., Ferdy J.-B. 2009. Manifold aspects of specificity in a nematode-bacterium mutualism. J. Evol. Biol. 22, 2104–2117 10.1111/j.1420-9101.2009.01829.x (doi:10.1111/j.1420-9101.2009.01829.x) [DOI] [PubMed] [Google Scholar]
- 29.White G. 1927. A method for obtaining infective nematode larvae from culture. Science 66, 302–303 10.1126/science.66.1709.302-a (doi:10.1126/science.66.1709.302-a) [DOI] [PubMed] [Google Scholar]
- 30.R Development Core Team 2009. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing [Google Scholar]
- 31.Atkinson B., Therneau T. 2008 Kinship: mixed-effects Cox models, sparse matrices, and modeling data from large pedigrees: R package version 1.1.0-22. [Google Scholar]
- 32.Bates D., Maechler M. 2010 lme4: linear mixed-effects models using S4 classes: R package version 0.999375-33. See http://CRAN.R-project.org/package=lme4 . [Google Scholar]
- 33.Caraco T., Wang I. N. 2008. Free-living pathogens: life-history constraints and strain competition. J. Theor. Biol. 250, 569–579 10.1016/j.jtbi.2007.10.029 (doi:10.1016/j.jtbi.2007.10.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vigneux F., Zumbihl R., Jubelin G., Ribeiro C., Poncet J., Baghdiguian S., Givaudan A., Brehélin M. 2007. The xaxAB genes encoding a new apoptotic toxin from the insect pathogen Xenorhabdus nematophila are present in plant and human pathogens. J. Biol. Chem. 282, 9571–9580 10.1074/jbc.M604301200 (doi:10.1074/jbc.M604301200) [DOI] [PubMed] [Google Scholar]
- 35.Lanois A., Jubelin G., Givaudan A. 2008. FliZ, a flagellar regulator, is at the crossroads between motility, haemolysin expression and virulence in the insect pathogenic bacterium Xenorhabdus. Mol. Microbiol. 68, 516–533 10.1111/j.1365-2958.2008.06168.x (doi:10.1111/j.1365-2958.2008.06168.x) [DOI] [PubMed] [Google Scholar]
- 36.Richards G. R., Goodrich-Blair H. 2010. Examination of Xenorhabdus nematophila lipases in pathogenic and mutualistic host interactions reveals a role for xlpa in nematode progeny production. Appl. Environ. Microbiol. 76, 221–229 10.1128/AEM.01715-09 (doi:10.1128/AEM.01715-09) [DOI] [PMC free article] [PubMed] [Google Scholar]