Abstract
Polyploidy is a major evolutionary process in eukaryotes—particularly in plants and, to a less extent, in animals, wherein several past and recent whole-genome duplication events have been described. Surprisingly, the incidence of polyploidy in other eukaryote kingdoms, particularly within fungi, remained largely disregarded by the scientific community working on the evolutionary consequences of polyploidy. Recent studies have significantly increased our knowledge of the occurrence and evolutionary significance of fungal polyploidy. The ecological, structural and functional consequences of polyploidy in fungi are reviewed here and compared with the knowledge acquired with conventional plant and animal models. In particular, the genus Saccharomyces emerges as a relevant model for polyploid studies, in addition to plant and animal models.
Keywords: polyploid, palaeopolyploid, whole-genome duplication, hybridization, reticulate evolution
1. Introduction
Polyploidy (definitions are given in box 1) has long been considered as a prominent process shaping eukaryotes evolution [1,2]. Several well-described natural polyploid organisms are known, such as oilseed rape (that combines both cabbage and turnip mustard [3]), cotton, wheat, goldfish or grey treefrog (for a review, see Otto & Whitton [2]). In addition to these recent polyploids, many ancient polyploidization events (also called palaeopolyploidization) were described in the evolutionary history of several taxa such as in angiosperms or vertebrates [4,5]. Although past and recent polyploidization events occurred repeatedly in the animal kingdom [2], it is particularly prominent in plants and especially in angiosperms. As pointed out by Soltis et al. [6], the actual question is no longer to know how many flowering plants are polyploids but how many polyploidization events occurred within each angiosperm lineage. Indeed, most of the knowledge regarding polyploid occurrence and evolution was obtained using plant models, and to a lesser extent using animals [2]. Surprisingly, the incidence of polyploidy in other large eukaryote kingdoms, such as the fungi, remains largely unknown despite numerous data collected for years. Polyploidy in fungi is usually evoked (and reduced to) the well-described whole-genome duplication (WGD) that occurred in yeast lineage about 100 Ma [7].
Box 1. Definitions.
Polyploidy is the state of having three or more sets of chromosomes in contrast to the two sets present in diploids (and one in haploids). The sets of chromosomes may originate from a single species (autopolyploidy) or from different ones, generally closely related (allopolyploidy). The polyploid status is heritable through the germ line: meiosis in polyploids leads to the formation of gametes having two or more chromosome sets.
Aneuploidy designs the occurrence of one or more extra or missing chromosomes by comparison with the normal haploid/diploid state of the species. When considering polyploids, the aneuploid level is intermediary between polyploid and diploid ones and may result from the diploidization process.
Endopolyploidy, or somatic polyploidy, arises through recurrent cycles of DNA replication without cellular division via either endoreduplication or endomitosis processes. Somatic polyploidy is generally associated with cellular differentiation or specific stages of life cycle (cyclic polyploidy) and results in genome content increase in the somatic line, not in the germ line. Thus, it is not heritable through sexual reproduction.
Diploidization is the process by which a polyploid organism returns to a diploid mode of chromosome pairing. Diploidization may involve various mechanisms, including partial or full chromosome losses, genome rearrangement, sequence divergence and deletion allowing the differentiation of the duplicated chromosomes and the apparition of diploid-like behaviour at meiosis. Diploidization leads to palaeopolyploid organisms that retain only traces of the past polyploidization event(s) on their genome.
Hybridization: merging of genomes from two different species (interspecific hybridization) or two different individuals of the same species (intraspecific hybridization).
Homoploid speciation is hybrid speciation without a change in chromosome number (without genome doubling).
Reticulate evolution is characterized by occasional hybridization, backcrosses and combination of two species. Reticulate evolution is frequently described in taxa prone to polyploidy.
Homologous: chromosomes or genes derived from a common ancestor.
Homeologous: paralogous chromosomes or genes merged within a single nucleus in allopolyploids. Homeologous genes are also referred to as homeoalleles.
Neopolyploid: newly generated polyploid individuals (also referred as synthetic polyploid), generally induced through artificial means (colchicine treatment, etc.).
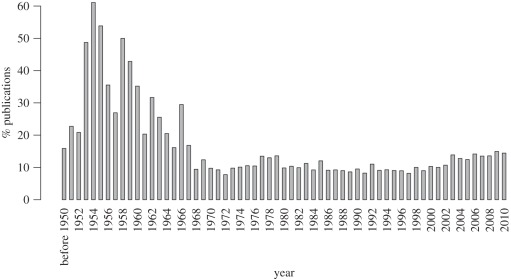
There are several reasons why the works dealing with fungal polyploids remain disregarded by non-specialists of mycology. Firstly, polyploidy in fungi has long been viewed as rare or absent [8], essentially because most reported haploid chromosome numbers were low, i.e. in the range of 4–8 [9]. Secondly, much of the available data were collected before the 1980s [10] and are poorly accessed now, whereas later works were published in very general fungal books so that the ‘polyploid section’ remains confidential for non-mycologists [11]. Thirdly, recent data were obtained, especially on polyploid yeasts of the Saccharomyces genus, but were hidden by the huge amount of publications dealing with non-polyploid yeasts. In fact, a few strains of Saccharomyces cerevisiae progressively came to dominate basic research the past four decades. These laboratory strains (S288C, W303, FL100, etc.) were initially selected for their simplified genetic manipulation, and were de facto chosen among diploid strains and/or their haploid derivatives. As a consequence, these laboratory haploid/diploid strains are now massively represented in yeast works. While the proportion of publications dedicated to polyploid Saccharomyces sp. represented about 30 per cent before the 1970s, it falls under 10 per cent until 2000 (figure 1). The past decade shows a little renewal in the interest in polyploid Saccharomyces with 13–15% of the total publications, yet falling within the scope of applied research rather than understanding evolutionary phenomena. For example, polyploid strains are used as models for cancer or cell cycle defects studies [12,13]. In addition, many industrial yeasts used in bakery, brewery, etc., are polyploids [14], so that several biotechnological-orientated works were described recently [15] but are not easily accessed by the scientific community working on the evolutionary consequences of polyploidy.
Figure 1.
Annual proportion of publications addressing polyploidy within the Saccharomyces genus. The annual number of publications addressing either ‘Saccharomyces tetraploid OR polyploid’ or ‘Saccharomyces haploid OR diploid -tetraploid -polyploid’ was estimated using Google Scholar (release of May 2011).
The aim of this work is thus to summarize the knowledge acquired on polyploid fungi, with a special emphasis on yeasts for which recent data are available. Issues that are traditionally studied with plant and animal models are focused on. In particular, the ecological, structural and functional consequences of polyploidy in fungi are addressed.
2. The occurrence of polyploidy in fungi
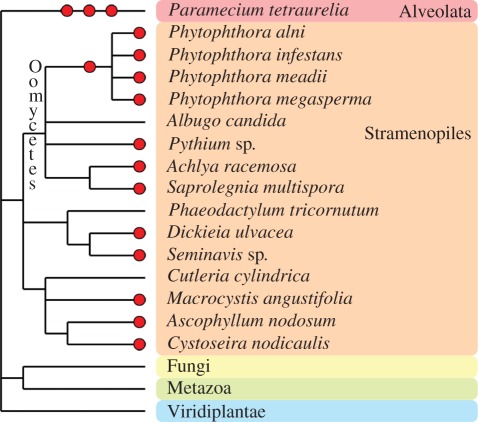
The fungal kingdom comprises more than 100 000 described species [16]. The precise taxonomy of fungi is constantly evolving alongside the acquisition of new genomic data, and some fungi-like taxa, such as the Oomycetes lineage, are no longer included among the fungi kingdom (box 2 and figure 2).
Box 2. Polyploidy in other eukaryotic taxa.
Besides plants, animals and fungi, other eukaryotic taxa have experienced one or more polyploidization events during their evolutionary history. The oomycetes, which are non-true fungi members, contain several examples of polyploid species, such as within the Phytophthora genus (figure 2 and electronic supplementary material, table S4). Some species of brown algae (Fucales, Laminariales and diatoms) contain apparent polyploid genomes. In the Alveolata group, the remarkable species Paramecium tetraurelia underwent three successive rounds of WGD [17] and established itself as a major model for palaeopolyploid studies. Thus, far from being restricted to plants and animals, polyploidy is more likely a general process of eukaryotic evolution.
Figure 2.
Inferred polyploidy and reticulate evolution in the Chromalveolata eukaryote supergroup. Schematic phylogeny and classification of the Chromalveolata eukaryote supergroup base on the taxonomy database maintained by the UniProt group newt [18] (release of May 2011). Branch lengths are not proportional to genetic distances. Red circles indicate suspected polyploidy. Full references are available in electronic supplementary material, table S4.
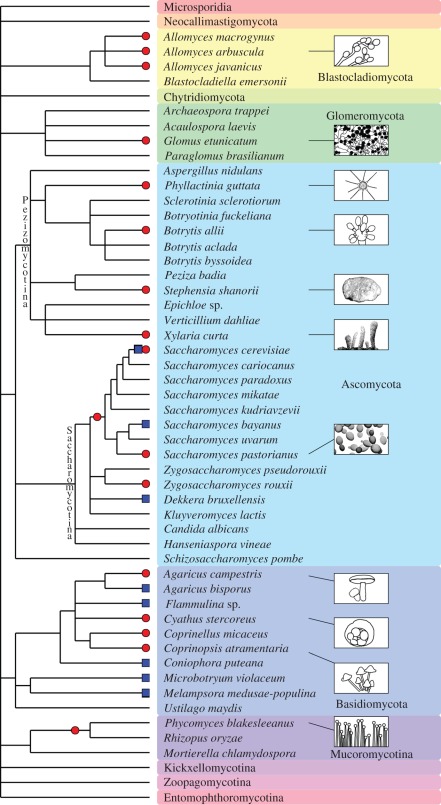
A recent phylogenetic classification of the true fungi described 11 phyla (figure 3), four of them having uncertain position (Mucoromycotina, Kickxellomycotina, Zoopagomycotina and Entomophthoromycotina are incertae sedis phyla) [19]. Several species were identified as natural polyploids (figure 3). For example Rhizopus oryzae, a human pathogen, was the first fungus from the early lineages of the fungi kingdom whose genome was fully sequenced [20]. Subsequent genomic analysis revealed that its evolutionary history was marked by a WGD followed by diploidization [20]. More recent fungal polyploidization events were identified within the Blastocladiomycota phylum, particularly among aquatic fungi (Allomyces sp.) that display polyploid series containing autotriploid, autotetraploid and allotetraploid representatives [21]. The Glomeromycota phylum comprises arbuscular mycorrhizal fungi that are thought to be the oldest group of asexual multicellular organisms. In a recent publication, Pawlowska & Tslor [22] demonstrated the existence of polyploid nucleus within Glomus etunicatum, and suggested that genome polyploidization might account for their long-term evolutionary persistence in the absence of sexual reproduction. Natural polyploidization events were also identified within the Basidiomycota phylum where several edible mushrooms and relatives may be polyploids (figure 3). For example, Cyathus stercoreus, commonly known as the dung-loving bird's nest, is a tetraploid species (possibly allotetraploid) displaying tetravalent formation at meiosis between its more closely related homeologous chromosomes [23]. Not surprisingly, the largest phylum of fungi, the Ascomycota, contains many polyploids: within both Pezizomycotina and Saccharomycotina subphyla, several studies suggested the existence of both inter and intraspecific polyploids within the Phyllactinia, Stephensia, Xylaria, Botrytis and Zygosaccharomyces genus (figure 3 and electronic supplementary material, table S1) [24,25]. However, to date, most of the evidenced polyploid fungi belong to the well-described Saccharomyces genus.
Figure 3.
Inferred polyploidy and reticulate evolution in fungi. Schematic phylogeny and classification of the fungi, based on Hibbet et al. [19]. Red circles indicate suspected polyploidy, blue squares indicate lineages with individuals having hybrid origin (reticulate evolution). Branch lengths are not proportional to genetic distances. Full references are available in electronic supplementary material, tables S1 and S2.
(a). Saccharomyces genus evolution: a polyploid story
The yeast S. cerevisiae has been exploited by humans for millennia to produce alcoholic beverages, including beer [26], wine [27] and spirits, or to leaven bread [28]. Besides its importance for several food industries, S. cerevisiae is one of the most intensively studied eukaryote models in molecular and cell biology, and was the first eukaryote whose genome was fully sequenced [29]. The analysis of the genome sequence revealed a WGD in the evolutionary history of the Saccharomyces genus [7], as first suggested by Smith [30]. The yeast WGD occurred after the divergence of Saccharomyces from Kluyveromyces around 100 Ma and was followed by subsequent diploidization, which is defined as the ‘process by which a polyploid genome turns into a diploid one’ [31]. In addition to this ancient WGD, several studies revealed that an important number of yeasts are recent polyploids [32]. For example, a genetic analysis of different S. cerevisiae food-processing strains revealed a noteworthy proportion of autotetraploids (10 of 26 strains) displaying tetrasomic inheritance at meiosis [33]. A polyploid population (possibly autotetraploid) was isolated from pearl millet beer in West Africa [34]. This population displays almost separate sexes, suggesting a shift from usual yeast hermaphroditism to a near-dioecious behaviour [34]. Many Saccharomyces strains used for wine-making were also proved to be polyploid such as for Tokaj wine-making in Slovakia and Hungary [35] or Spanish sherry-type wines [36]. A well-known example of allopolyploid speciation in yeast is the formation of Saccharomyces pastorianus, widely used in brewery to produce lager beer. Its allotetraploid origin was first suggested by Nilsson-Tillgren et al. [37], yet S. pastorianus progenitors were elucidated later as S. cerevisiae [38,39] and an unidentified species close to Saccharomyces bayanus [38,40,41]. Recently, Libkind et al. [42] established that the S. bayanus-like genome donor was actually a new species designed as Saccharomyces eubayanus. The identification of many polyploid Saccharomyces yeasts associated with different food-processing contexts led to their biotechnological exploitation in applied research [43,44] and hid the occurrence of polyploidy in non-industrial yeasts. However, recent works indicated that polyploidy is not restricted to food process, with the identification of polyploid series (ha-, di-, tri- and tetraploids) from soil isolates in Israel and opportunist Saccharomyces polyploids from clinical isolates [45,46].
(b). Polyploidy and hybridization
Polyploidization and hybridization are closely interrelated processes: allopolyploidy necessarily arises through interspecific hybridization associated with genome doubling. In addition, although autopolyploidy may arise without intraspecific hybridization (i.e. only through genome doubling), many autopolyploid species display higher heterozygosity levels than their diploid counterparts as in plants or yeasts [33,47], suggesting a hybrid origin. Evolution through hybridization, with or without genome doubling, is referred as reticulate evolution or reticulation [48] and may be the first step towards polyploidy. Until the 1990s, hybridization in fungi was considered to be rare [49] but several fungal hybrids have been described since then (electronic supplementary material, table S2). For example, interspecific fungal hybrids were described such as in the phytopathogen species Verticillium dahliae [50] and Melampsora × columbiana [51,52], the cultivated mushroom Agaricus bisporus [53] and other edible mushrooms from the Flammulina genus [54]. The Saccharomyces genus is particularly prone to interspecific hybridization: natural S. cerevisiae × Saccharomyces kudriavzevii and S. cerevisiae × S. bayanus hybrids have been repeatedly reported and may be much more frequent than initially thought [55–57]. A striking example of the Saccharomyces genus ability to mate is the strain CID1, used for cider production (a fermented beverage from apple juice) in France, which is a ‘triple’ hybrid having at least pieces of S. cerevisiae, S. kudriavzevii and S. bayanus genomes [58]. Saccharomyces bayanus is now established to be a complex hybrid species, with genome contributions from S. uvarum, S. eubayanus and to a less extent S. cerevisiae [42,59], explaining the difficulties and incongruities encountered for the identification of S. bayanus strains. The spoilage yeast Dekkera bruxellensis, which is responsible for the undesirable ‘Brett character’ in wine, has a complex and dynamic genome that originated through interspecific hybridization, aneuploidization and polyploidization [60]. An inter-family hybrid was also described between Hanseniaspora vineae and S. cerevisiae [61]. In addition to these natural hybrids, there are several reports of successful construction of interspecific and inter-genera fungal hybrids in the laboratory [62–64], illustrating the genome plasticity of fungi regarding genome merging.
Hybridization may be followed by backcrosses with one parental species, allowing the recovering of a parental-like species bearing a few introgressed genomic parts as in the wet rot fungus Coniophora puteana [65] or other Saccharomyces species [66,67], and sometimes uncovering the sterility associated with interspecific hybridization [68]. Finally, in the most extremes cases, hybridization may lead to hybrid speciation (also called homoploid speciation) as in many plant and animal taxa [69,70]. In fungi, some cases of homoploid hybrid speciation were described [71] as in the anther smut fungus Microbotryum violaceum (formerly Ustilago violacea) [72]. Indeed, the occurrence of hybridization in a given taxon gives another illustration of its tolerance to genome merging. It is not surprising that hybridization and reticulate evolution in fungi seem to occur in lineages also displaying polyploid members (figure 3) as in plants and animals [73,74].
(c). Factors affecting genome content
In addition to the species identified as actual polyploids (meaning that genome duplication persists in the germ line and is heritable through sexual reproduction), many other fungal species display large variation in their genome size (electronic supplementary material, table S3). In this regard, the fungal genome size database [75] provides freely accessible genome size data for more than 1000 fungal species (www.zbi.ee/fungal-genomesize). Variations in genome content may be associated with life cycle or cellular differentiation [76], such phenomena as somatic polyploidy (or endopolyploidy; box 1) rather than actual polyploidy. For example, the life cycle of C. albicans (the causal agent of candidiasis) is particularly atypical: C. albicans is a diploid yeast that has long been viewed as strictly asexual. However, a cryptic mating cycle (also referred as parasexual cycle; figure 4) has been described, through the fusion of diploid cells [77]. The resulting tetraploids then undergo random loss of multiple chromosomes, a process termed concerted chromosome loss [78]. As a consequence of such unconventional life cycle, haploid, diploid, triploid, tetraploid and aneuploid C. albicans populations coexist among clinical isolates [79]. Candida albicans may not be considered as a ‘true’ polyploid species (i.e. from an evolutionary viewpoint), but remains a remarkable example of variation of ploidy level associated with life cycle. A close relative of S. cerevevisiae, Candida glabrata, also displays a striking genome plasticity; although these yeasts have acquired an haploid lifestyle in comparison with other yeasts, frequent changes in the chromosomal complement have been evidenced, in relation with pathogenicity and adaptation to a fluctuating environment [80]. Thus, the yeasts exhibit a genome flexibility that may favour ploidy variations.
Figure 4.
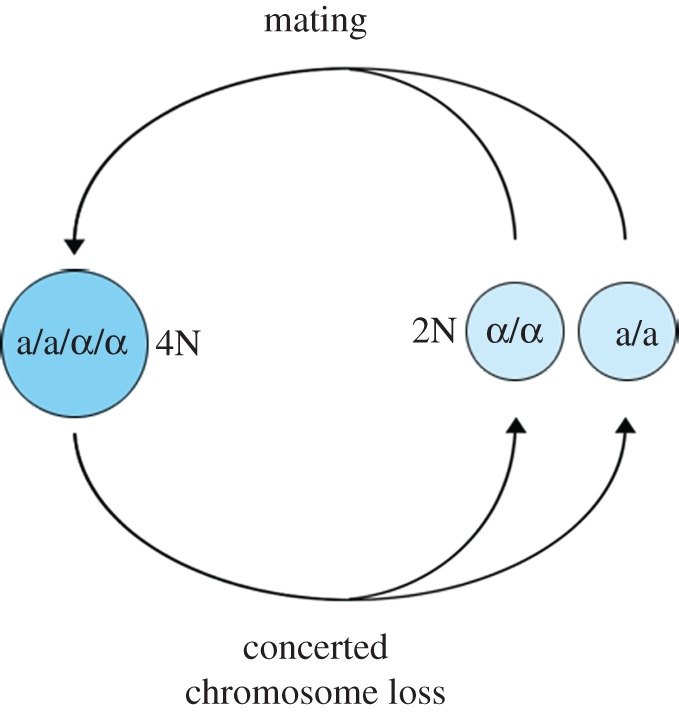
The parasexual cycle of Candida albicans.
Several environmental factors have been shown to induce variation of genome content and chromosomal complement in various fungal species, such as heat shock [81], saline stress [82], fungicides treatments [83], host–pathogen interactions [84], etc. (electronic supplementary material, table S3). Genetic factors may also be associated with variation of genome content: some genes are associated with ploidy variation when mutated, most of these being involved in spindle body structure or function [85,86] (electronic supplementary material, table S3). Although such variations in chromosomal complements and genome size may not be considered actual polyploidy (from an evolutionary viewpoint), they are the hallmark of the genomic plasticity that may support further polyploidization and subsequent fungal speciation.
3. Diversification in polyploid fungi
Following WGD, one would expect the newly formed polyploid to possess the sum of the parental genomes and display mid-parent patterns of relative expression [87]. This so-called additivity hypothesis has been verified in cases such as that in synthetic allopolyploid cotton where ‘structural genomic stasis’ has been described. However, deviation from the additivity hypothesis was evidenced for many polyploid species, and duplicated genes can undergo immediate structural and functional divergence [87]. Indeed, specific patterns of evolution were described in plant and animal polyploids and are supposed to facilitate evolution and adaptation. In fungi also, WGD is associated with long- or short-term structural, functional and phenotypical diversification.
(a). Genome evolution and diversification in polyploid fungi
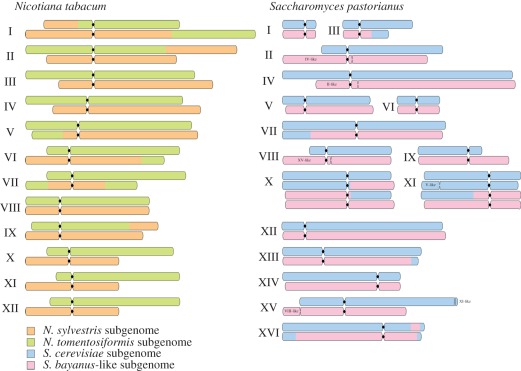
Any increase in chromosome number is expected to enhance meiotic and mitotic abnormalities, particularly in allopolyploid's meiosis where the chromosomal colinearity between closely related parental genomes merged within a single nucleus may result in improper meiotic pairing and homeologous recombination [88]. Indeed, the very first meiosis of synthetic Brassica napus allotetraploids acts as a genome blender and generates several chromosomal rearrangements [89], and many other examples of homeologous recombination were evidenced in both plant (figure 5) and animal polyploids [90,91]. In fungi, meiotic defects were observed in yeast polyploids with general instability [92], abnormal chromosomal disjunction [93] or atypical meiotic timing and topology [94]. Genome instability in yeast polyploids was also observed during mitosis, with the occurrence of chromosome loss 30-fold higher in triploids and approximately 1000-fold higher in tetraploids than in diploids [95]. Another autopolyploid yeast series, evolving experimentally over 1800 mitotic generations, converged towards diploidy [96] mainly through chromosomal loss and some additional chromosome mis-segregation events [97]. WGD in yeast was followed by a decrease in chromosome number mainly imputable to telomere-to-telomere fusion between chromosomes [98]. Exhaustive genomic studies of the lager yeast S. pastorianus revealed that, following allotetraploidization, several chromosomal translocations arose between the parental subgenomes [41] as well as large chromosomal rearrangements [40,99]. These chimaeric chromosomes appear currently stable within lager strains currently used in breweries (figure 5).
Figure 5.
Polyploid genomes of Nicotiana tabacum and Saccharomyces pastorianus. The schematic genomes illustrate the parental origin of the chromosomes. The different types of chromosomes are drawn. Black circles represent centromere position. The structure of N. tabacum genome was adapted from Chester et al. [48] and displayed eight inter-genomic rearrangements. For S. pastorianus, Nakao et al. [41] identified 10 inter-genomic rearrangements and six intra-genomic translocations between chromosomes II–IV and VIII–XV (S. bayanus-like subgenome, now identified as S. eubayanus) and V–XI and XI–XV (S. cerevisiae subgenome).
Transposable elements (TEs) and other repeated sequences are traditionally involved in structural and functional dynamics of plant and animal polyploids [100,101]. They seem also involved in fungi post-polyploid evolution. TEs represent a very little part of the total Saccharomyces sp. genome compared with plant and animal ones: only 3 per cent of yeast genome, i.e. around 300 TE per haploid genome [102]. However, some translocation breakpoints in lager yeast S. pastorianus are located near Ty retrotransposon elements [40], suggesting that TE mediated genomic rearrangements following allotetraploidization as in other eukaryote polyploids. From a long-term perspective, the WGD event in the Saccharomyces lineage 100 Ma was followed by reciprocal translocations resulting from ectopic recombination between Ty elements or other repeated sequences [103]. Other well-known repeated sequences associated with genome restructuring in polyploids are the cluster of ribosomal DNA (rDNA). In plants and animals, many synthetic and natural polyploids display partial or complete homogenization of their rDNA [104–106], suggesting concerted evolution. In polyploid fungi, rDNA are also associated with genetic modifications: in the lager yeast S. pastorianus, one parental rDNA cluster has undergone a significant reduction following polyploidization [41] and, in Botrytis allotetraploid, the lack of polymorphism in the rDNA may be explained by concerted evolution [24]. The subtelomeric regions were particularly prone to duplications and rearrangements in yeasts following WGD [107] or allotetraploidization in lager yeast [108]. Genetic diversification in polyploids may involve smaller sequences and encompass limited duplication and/or gene loss as in plants and animals [109–111]. Extensive gene loss following WGD in yeast lineage is described [112] and could be a driving force of speciation. In the lager yeast S. pastorianus, changes in copy number of specific repeated sequences (loss or duplication) are described, highlighting the dynamic nature of yeast polyploid genome [113]. Experimental evolution of synthetic yeast allotetraploid subjected to mutagenesis was associated with reciprocal gene loss [114]. In conclusion, polyploidization in fungi appears to be associated with various gross or restricted structural rearrangements as found in the plant and animal kingdoms. As a result, the genome of the allotetraploid S. pastorianus now displays highly chimaeric chromosomes that strikingly echo plant allopolyploid ones (figure 5).
(b). Evolution of gene expression in polyploid fungi
From a functional viewpoint, one plus one does not equal two in polyploids [87], and many plant- and animal-duplicated genomes transgress the additivity hypothesis (predicting mid-parent relative expression). In fungi, expression data in a polyploid context are available mainly within the Saccharomyces genus.
Microarray-based gene expression analysis of isogenic haploid, diploid, triploid and tetraploid S. cerevisiae strains allowed the identification of a few genes (17) displaying non-additive expression [115]. A recent analysis identified substantially more transgressive genes (65) but showed that cell size increase, rather than genome doubling itself, was the cause of gene expression alteration in yeast autotetraploids [116]. Altogether, these results suggest that genome doubling by itself may trigger few expression changes as in plant models [117,118]. By contrast, allopolyploidy in yeast is associated with several expression changes; an exhaustive expression analysis was recently conducted on S. pastorianus using microarray [119] and allowed to be distinguished most homeoallele pairs during the fermentation process. If 600 genes showed similar expression patterns between S. cerevisiae and S. bayanus-like (now known as S. eubayanus) parental genes, then 400 other homeologous genes show unequal contributions to the transcriptome of S. pastorianus [119]. Interestingly, the contributions of homeologous pairs vary along the fermentation process; for example, some homeoalleles display equal expression contribution at the very beginning of fermentation, and unequal contribution in the last fermentation steps [119]. Indeed, the transcriptomic profiling of S. pastorianus shares common features with other plant models; the unequal contributions of the homeoalleles were described, for the first time, in the allotetraploid cotton Gossypium hirsutum with organ-specificity [120]. Further analyses must be conducted to decipher the mechanisms underlying gene expression regulation in yeast polyploids. Functional changes may be related to the structural diversification associated with polyploidy as described in S. pastorianus, where a chromosomal rearrangement was coupled with a loss of function at breakpoints of the resulting hybrid GPH1 gene [99]. Dosage compensation, a process by which genes duplicated by polyploidy or aneuploidy show diploid-like expression as described in plants [121], also counted in the functional evolution of S. pastorianus [113]. Epigenetic regulation of gene expression in polyploids has received a great attention in plant polyploids essentially through DNA methylation studies [122,123]. In particular, the methylation state of TE may be related to a transposition burst following polyploidization [124–126] and may be associated with the deregulation of small RNA [127]. Cytosine methylation is absent in Saccharomyces genus that do not possess DNA methyltransferases [128], but other epigenetic mechanisms are known; for example, histone deacetylation is involved in the regulation of gene expression in yeast [129] and it could be interesting to test its putative occurrence in polyploid and hybrid context.
(c). Phenotypic diversification and ecological consequences
Polyploidization triggers several structural and/or functional changes that are assumed to favour phenotypic diversification and thus facilitate further evolution and adaptation in plants and animals [1,2] and also in fungi [22,130]. In Saccharomyces sp., genome doubling is associated with morphological variation such as cell size, shape, organization (colonies forming) and growth [131,132]. Metabolic changes are also observed. For example, metabolic fluxes increase with the ploidy level in autopolyploid series [131,133], and the allotetraploid S. pastorianus and its genome donor S. cerevisiae display highly different exometabolomes [134]. Because the productivity of yeast cultures seems to increase with the ploidy level in many cases [135], polyploidy in Saccharomyces has been much more studied from a biotechnological viewpoint than from an evolutionary perspective. However, recent data regarding the fitness of polyploid yeasts were described: in soil yeasts, high ploidy level may be a mechanism of adaptation to high solar radiation [136]. Baking is closely associated with autotetraploid S. cerevisiae, suggesting that autotetraploidy in yeast may promote adaptation to the harsh bakery environment [33]. In S. pastorianus allotetraploid, specific changes in sugar and sulphite metabolism were evidenced in comparison with its S. cerevisiae and S. eubayanus progenitors [42]. These modifications may have been crucial for domestication in the lager-brewing environment [42]. The WGD in the Saccharomyces lineage and subsequent preferential retention of duplicated glycolytic genes may have favoured glucose fermentation, adaptation to glucose-rich environments and occupation of new ecological niches [137]. Indeed, modelling the evolution of metabolic networks in post-WGD yeasts indicates that polyploidization is generally detrimental in the original (parental) environment, but has immediate fitness benefits in new environmental conditions [138]. This may explain why autopolyploid S. cerevisiae strains show reduced fitness under laboratory conditions [96]. In addition, the diploidization process by itself may be adaptative: aneuploidy and major chromosomal changes in yeast may be associated with increased fitness [139]. The partitioning of yeast co-expression networks after WGD [140] led to partial redundancy and functional overlapping, and is responsible, in part, for genetic network robustness [141] that may promote adaptative changes. Further analyses of polyploid and palaeopolyploid yeasts from an evolutionary viewpoint will increase our knowledge of the consequences and the fate of duplicated genomes in fungi.
4. Conclusions and future directions
Although fungal polyploidization has been long illustrated solely through yeast WGD, there is other evidence indicating that polyploidy has played a preeminent role in the evolutionary history of the fungi kingdom, as it has in plants and animals. It is highly probable that the non-exhaustive list of past and recent polyploidization events presented here will increase greatly in the future because until now fungi are less studied than plants and animals. The cytological and phylogenetic data already available could be used to infer the evolution of chromosome number in fungi and to estimate the occurrence of polyploidy using probabilistic models as described recently [142]. Such work may help to draw a more precise image of polyploidy in fungi.
It is noteworthy that Saccharomyces genus emerges as the fungal alter ego of widely studied plant taxa such as the Brassicaceae, the Triticeae, etc., which exhibit a complex pattern of polyploidy. The evolutionary history of Saccharomyces species are shaped by past and recent WGD events, associated with hybridization and reticulate evolution. The structural and functional outcomes of polyploid Saccharomyces genomes strikingly reflect the evolutionary fate of plant polyploid ones, designing yeast as a relevant complementary model for polyploid studies. The yeast model may offer several technical and laboratory facilities: in addition to the high number of large-scale molecular tools available (micro-array, whole-genome sequencing, proteomics, etc.), neopolyploids can be synthesized through protoplasts fusion [143], and can be compared and/or competed with their diploid progenitor and natural auto- and allo-polyploid counterparts to analyse fitness and adaptation features. Neopolyploid yeasts are pertinent models to study reproductive isolation through the establishment of post-zygotic barriers [114] and genetic incompatibilities [144]. Yeast's short generation time (a few hours) allows experimental evolution over hundreds or thousands of mitotic generations [145] that may give new insights into polyploid evolution and subsequent diploidization. For example, experimental evolution of neopolyploids could help unravel the role of TEs in polyploid evolution and, in particular, their impact on genome modifications from both structural and functional viewpoints. Yeast polyploids could be useful to explore another interesting issue: the role of nucleocytoplasmic interactions in polyploid formation and propagation. In plants, studies of reciprocal hybrids and polyploids have evidenced differential genome evolution as in Brassica polyploids [146] or differences in fitness as in Epilobium hybrids [147]. Several authors hypothesized that the merging of two nuclear genome components with a unique cytoplasmic component in an interspecific hybrid may unbalance the interactions between the nuclei and cytoplasm, and may favour the parental genome initially associated with the cytoplasmic one, i.e. the maternal one in plants [148]. Indeed, a nuclear gene from S. bayanus was shown incompatible with S. cerevisiae mitochondria, suggesting possible nucleo-cytoplasmic incompatibilities within the corresponding hybrids [149]. Such hypothesis could be tested using yeast as a model: in most cases, hybrids resulting from crosses involving the same parents may inherit either mitochondria [63], allowing the comparison of nuclei-identical hybrids, but having different mitochondrial DNA. Moreover, it could be interesting to test the tolerance of the yeast model to high ploidy level: although natural and synthetic triploid and tetraploid yeasts have been repeatedly described, it is not known whether higher ploidy levels could be generated as in plants and animals where several dodecaploids harbouring hundreds of chromosomes are known [150–152]. Finally, there is still a lack of knowledge on the relationships between structural and functional diversification in polyploids and further adaptation ability. Saccharomyces yeasts are suitable biological models for systems biology approaches that will help unravel the adaptative value of WGD and understand why polyploidy is such an evolutionary success among eukaryotes.
Acknowledgements
We are very grateful to Michel Aigle, Delphine Sicard, Hervé Thiellement and Dominique de Vienne for the strong scientific support and the careful reading of the manuscript. We thank the anonymous reviewers for their valuable comments and suggestions to improve the manuscript.
References
- 1.Wendel J. F. 2000. Genome evolution in polyploids. Plant Mol. Biol. 42, 225–249 10.1023/A:1006392424384 (doi:10.1023/A:1006392424384) [DOI] [PubMed] [Google Scholar]
- 2.Otto S. P., Whitton J. 2000. Polyploid incidence and evolution. Annu. Rev. Genet. 34, 401–437 10.1146/annurev.genet.34.1.401 (doi:10.1146/annurev.genet.34.1.401) [DOI] [PubMed] [Google Scholar]
- 3.U, N 1935. Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn. J. Bot. 7, 389–452 [Google Scholar]
- 4.Blanc G., Wolfe K. H. 2004. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell. 16, 1667–1678 10.1105/tpc.021345 (doi:10.1105/tpc.021345) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohno S. 1970. Evolution by gene duplication. Berlin, Germany: Springer [Google Scholar]
- 6.Soltis D. E., et al. 2009. Polyploidy and angiosperm diversification. Am. J. Bot. 96, 336–348 10.3732/ajb.0800079 (doi:10.3732/ajb.0800079) [DOI] [PubMed] [Google Scholar]
- 7.Wolfe K. H., Shields D. C. 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387, 708–713 10.1038/42711 (doi:10.1038/42711) [DOI] [PubMed] [Google Scholar]
- 8.Stebbins G. L. 1950. Variation and evolution in plants, pp. 251–379 New York, NY: Columbia University Press [Google Scholar]
- 9.Olive L. S. 1953. The structure and behavior of fungus nuclei. Bot. Rev. 19, 439–586 10.1007/BF02861808 (doi:10.1007/BF02861808) [DOI] [Google Scholar]
- 10.Rogers J. D. 1973. Polyploidy in fungi. Evolution 27, 153–160 10.2307/2407129 (doi:10.2307/2407129) [DOI] [PubMed] [Google Scholar]
- 11.Burnett J. H. 2003. Fungal populations and species, pp. 182–184 Oxford, UK: Oxford University Press [Google Scholar]
- 12.Storchova Z., Pellman D. 2004. From polyploidy to aneuploidy, genome instability and cancer. Nat. Rev. Mol. Cell Biol. 5, 45–54 10.1038/Nrm1276 (doi:10.1038/Nrm1276) [DOI] [PubMed] [Google Scholar]
- 13.Thorpe P. H., Gonzalez-Barrera S., Rothstein R. 2007. More is not always better: the genetic constraints of polyploidy. Trends Genet. 23, 263–266 10.1016/j.tig.2007.03.016 (doi:10.1016/j.tig.2007.03.016) [DOI] [PubMed] [Google Scholar]
- 14.Querol A., Bond U. 2009. The complex and dynamic genomes of industrial yeasts. FEMS Microbiol. Lett. 293, 1–10 10.1111/j.1574-6968.2008.01480.x (doi:10.1111/j.1574-6968.2008.01480.x) [DOI] [PubMed] [Google Scholar]
- 15.Hahn-Hagerdal B., Wahlbom C. F., Gardonyi M., van Zyl W. H., Cordero Otero R. R., Jonsson L. J. 2001. Metabolic engineering of Saccharomyces cerevisiae for xylose utilization. Adv. Biochem. Eng. Biotechnol. 73, 53–84 10.1186/1475-2859-7-36 (doi:10.1186/1475-2859-7-36) [DOI] [PubMed] [Google Scholar]
- 16.Aguileta G., Hood M. E., Refregier G., Giraud T. 2009. Genome evolution in plant pathogenic and symbiotic fungi. Adv. Bot. Res. 49, 151–193 10.1016/S0065-2296(08)00603-4 (doi:10.1016/S0065-2296(08)00603-4) [DOI] [Google Scholar]
- 17.Aury J. M., et al. 2006. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 444, 171–178 10.1038/nature05230 (doi:10.1038/nature05230) [DOI] [PubMed] [Google Scholar]
- 18.Phan I. Q., Pilbout S. F., Fleischmann W., Bairoch A. 2003. NEWT, a new taxonomy portal. Nucleic Acids Res. 31, 3822–3823 10.1093/nar/gkg516 (doi:10.1093/nar/gkg516) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hibbett D. S., et al. 2007. A higher-level phylogenetic classification of the fungi. Mycol. Res. 111(Pt 5), 509–547 10.1016/j.mycres.2007.03.004 (doi:10.1016/j.mycres.2007.03.004) [DOI] [PubMed] [Google Scholar]
- 20.Ma L.-J., et al. 2009. Genomic analysis of the basal lineage fungus Rhizopus oryzae reveals a whole-genome duplication. PLoS Genet. 5, e1000549. 10.1371/journal.pgen.1000549 (doi:10.1371/journal.pgen.1000549) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emerson R., Wilson C. M. 1954. Interspecific hybrids and the cytogenetics and cytotaxonomy of Euallomyces. Mycologia 46, 393–434 [Google Scholar]
- 22.Pawlowska T. E., Taylor J. W. 2004. Organization of genetic variation in individuals of arbuscular mycorrhizal fungi. Nature 427, 733–737 10.1038/nature02290 (doi:10.1038/nature02290) [DOI] [PubMed] [Google Scholar]
- 23.Lu B. 1964. Polyploidy in the Basidiomycete Cyathus stercoreus. Am. J. Bot. 51, 343–347 10.2307/2440307 (doi:10.2307/2440307) [DOI] [Google Scholar]
- 24.Nielsen K., Yohalem D. Y. 2001. Origin of a polyploid botrytis pathogen through interspecific hybridization between Botrytis aclada and B. byssoidea. Mycologia 93, 1064–1071 10.2307/3761668 (doi:10.2307/3761668) [DOI] [Google Scholar]
- 25.Gordon J. L., Wolfe K. H. 2008. Recent allopolyploid origin of Zygosaccharomyces rouxii strain ATCC 42981. Yeast 25, 449–456 10.1002/yea.1598 (doi:10.1002/yea.1598) [DOI] [PubMed] [Google Scholar]
- 26.Meussdoerffer F. G. 2009. A comprehensive history of beer brewing. In Handbook of brewing: processes, technology, markets (ed. Eßlinger H. M.), pp. 1–42 Weinheim, Germany: Wiley-VCH [Google Scholar]
- 27.Cavalieri D., McGovern P. E., Hartl D. L., Mortimer R., Polsinelli M. 2003. Evidence for S. cerevisiae fermentation in ancient wine. J. Mol. Evol. 57 (Suppl 1), S226–S232 10.1007/s00239-003-0031-2 (doi:10.1007/s00239-003-0031-2) [DOI] [PubMed] [Google Scholar]
- 28.Samuel D. 1996. Investigation of ancient Egyptian baking and brewing methods by correlative microscopy. Science 273, 488–490 10.1126/science.273.5274.488 (doi:10.1126/science.273.5274.488) [DOI] [PubMed] [Google Scholar]
- 29.Goffeau A., et al. 1996. Life with 6000 genes. Science 274, 546, 563–547 10.1126/science.274.5287.546 (doi:10.1126/science.274.5287.546) [DOI] [PubMed] [Google Scholar]
- 30.Smith M. M. 1987. Molecular evolution of the Saccharomyces cerevisiae histone gene loci. J. Mol. Evol. 24, 252–259 10.1007/BF02111238 (doi:10.1007/BF02111238) [DOI] [PubMed] [Google Scholar]
- 31.Wolfe K. H. 2001. Yesterday's polyploids and the mystery of diploidization. Nat. Rev. Genet. 2, 333–341 10.1038/35072009 (doi:10.1038/35072009) [DOI] [PubMed] [Google Scholar]
- 32.Naumov G. I., Naumova E. S., Masneuf I., Aigle M., Kondratieva V. I., Dubourdieu D. 2000. Natural polyploidization of some cultured yeast Saccharomyces sensu stricto: auto- and allotetraploidy. Syst. Appl. Microbiol. 23, 442–449 10.1016/S0723-2020(00)80076-4 (doi:10.1016/S0723-2020(00)80076-4) [DOI] [PubMed] [Google Scholar]
- 33.Albertin W., Marullo P., Aigle M., Bourgais A., Bely M., Dillmann C., De Vienne D., Sicard D. 2009. Evidence for autotetraploidy associated with reproductive isolation in Saccharomyces cerevisiae: towards a new domesticated species. J. Evol. Biol. 22, 2157–2170 10.1111/j.1420-9101.2009.01828.x (doi:10.1111/j.1420-9101.2009.01828.x) [DOI] [PubMed] [Google Scholar]
- 34.Al Safadi R., Weiss-Gayet M., Briolay J., Aigle M. 2010. A polyploid population of Saccharomyces cerevisiae with separate sexes (dioecy). FEMS Yeast Res. 10, 757–768 10.1111/j.1567-1364.2010.00660.x (doi:10.1111/j.1567-1364.2010.00660.x) [DOI] [PubMed] [Google Scholar]
- 35.Naumov G. I., Naumova E. S., Antunovics Z., Sipiczki M. 2002. Saccharomyces bayanus var. uvarum in Tokaj wine-making of Slovakia and Hungary. Appl. Microbiol. Biotechnol. 59, 727–730 10.1007/s00253-002-1077-6 (doi:10.1007/s00253-002-1077-6) [DOI] [PubMed] [Google Scholar]
- 36.Guijo S., Mauricio J. C., Salmon J. M., Ortega J. M. 1997. Determination of the relative ploidy in different Saccharomyces cerevisiae strains used for fermentation and ‘flor’ film ageing of dry sherry-type wines. Yeast 13, 101–117 10.1002/(SICI)1097-0061(199702)13:2%3C101::AID-YEA66%3E3.0.CO;2-H (doi:10.1002/(SICI)1097-0061(199702)13: 2<101::AID-YEA66>3.0.CO;2-H) [DOI] [PubMed] [Google Scholar]
- 37.Nilsson-Tillgren T., Gjermansen C., Kielland-Brandt M., Petersen J., Holmberg S. 1981. Genetic differences between Saccharomyces carlsbergensis and S. cerevisiae. Analysis of chromosome III by single chromosome transfer. Carlsberg Res. Commun. 46, 65–76 10.1007/bf02906199 (doi:10.1007/bf02906199) [DOI] [Google Scholar]
- 38.Pedersen M. 1986. DNA sequence polymorphisms in the genus Saccharomyces IV. Homoeologous chromosomes III of Saccharomyces bayanus, S. carlsbergensis, and S. uvarum. Carlsberg Res. Commun. 51, 185–202 10.1007/bf02907323 (doi:10.1007/bf02907323) [DOI] [PubMed] [Google Scholar]
- 39.Casaregola S., Nguyen H. V., Lapathitis G., Kotyk A., Gaillardin C. 2001. Analysis of the constitution of the beer yeast genome by PCR, sequencing and subtelomeric sequence hybridization. Int. J. Syst. Evol. Microbiol. 51, 1607–1618 [DOI] [PubMed] [Google Scholar]
- 40.Dunn B., Sherlock G. 2008. Reconstruction of the genome origins and evolution of the hybrid lager yeast Saccharomyces pastorianus. Genome Res. 18, 1610–1623 10.1101/gr.076075.108 (doi:10.1101/gr.076075.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakao Y., Kanamori T., Itoh T., Kodama Y., Rainieri S., Nakamura N., Shimonaga T., Hattori M., Ashikari T. 2009. Genome sequence of the lager brewing yeast, an interspecies hybrid. DNA Res. 16, 115–129 10.1093/dnares/dsp003 (doi:10.1093/dnares/dsp003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Libkind D., Hittinger C. T., Valerio E., Goncalves C., Dover J., Johnston M., Goncalves P., Sampaio J. P. 2011. Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. Proc. Natl Acad. Sci. USA 108, 14 539–14 544 10.1073/pnas.1105430108 (doi:10.1073/pnas.1105430108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamada R., Tanaka T., Ogino C., Kondo A. 2010. Gene copy number and polyploidy on products formation in yeast. Appl. Microbiol. Biotechnol. 88, 849–857 10.1007/s00253-010-2850-6 (doi:10.1007/s00253-010-2850-6) [DOI] [PubMed] [Google Scholar]
- 44.Kim H. R., Im Y. K., Ko H. M., Chin J. E., Kim I. C., Lee H. B., Bai S. 2011. Raw starch fermentation to ethanol by an industrial distiller's yeast strain of Saccharomyces cerevisiae expressing glucoamylase and α-amylase genes. Biotechnol. Lett. 33, 1643–1648 10.1007/s10529-011-0613-9 (doi:10.1007/s10529-011-0613-9) [DOI] [PubMed] [Google Scholar]
- 45.Muller L. A., McCusker J. H. 2009. Microsatellite analysis of genetic diversity among clinical and nonclinical Saccharomyces cerevisiae isolates suggests heterozygote advantage in clinical environments. Mol. Ecol. 18, 2779–2786 10.1111/j.1365-294X.2009.04234.x (doi:10.1111/j.1365-294X.2009.04234.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clemons K. V., Park P., McCusker J. H., McCullough M. J., Davis R. W., Stevens D. A. 1997. Application of DNA typing methods and genetic analysis to epidemiology and taxonomy of Saccharomyces isolates. J. Clin. Microbiol. 35, 1822–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soltis D. E., Soltis P. S. 1993. Molecular data and the dynamic nature of polyploidy. Crit. Rev. Plant Sci. 12, 243–273 [Google Scholar]
- 48.Chester M., Leitch A. R., Soltis P. S., Soltis D. E. 2010. Review of the application of modern cytogenetic methods (FISH/GISH) to the study of reticulation (polyploidy/hybridisation). Genes 1, 166–192 10.3390/genes1020166 (doi:10.3390/genes1020166) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brasier C. 2000. The rise of the hybrid fungi. Nature 405, 134–135 10.1038/35012193 (doi:10.1038/35012193) [DOI] [PubMed] [Google Scholar]
- 50.Collins A., Mercado-Blanco J., Jiménez-Diaz R. M., Olivares C., Clewes E., Barbara D. J. 2005. Correlation of molecular markers and biological properties in Verticillium dahliae and the possible origins of some isolates. Plant Pathol. 54, 549–557 10.1111/j.1365-3059.2005.01240.x (doi:10.1111/j.1365-3059.2005.01240.x) [DOI] [Google Scholar]
- 51.Spiers A. G., Hopcroft D. H. 1994. Comparative studies of the poplar rusts Melampsora medusae, M. larici-populina and their interspecific hybrid M.medusae-populin. Mycol. Res. 98, 889–903 10.1016/S0953-7562(09)80260-8 (doi:10.1016/S0953-7562(09)80260-8) [DOI] [Google Scholar]
- 52.Newcombe G., Stirling B., McDonald S., Bradshaw H. D., Jr 2000. Melampsora × columbiana, a natural hybrid of M. medusae and M. occidentalis. Mycol. Res. 104, 261–274 10.1017/S0953756299001665 (doi:10.1017/S0953756299001665) [DOI] [Google Scholar]
- 53.Callac P., Jacobe de Haut I., Imbernon M., Guinberteau J., Desmerger C., Theochari I. 2003. A novel homothallic variety of Agaricus bisporus comprises rare tetrasporic isolates from Europe. Mycologia 95, 222–231 10.2307/3762033 (doi:10.2307/3762033) [DOI] [PubMed] [Google Scholar]
- 54.Hughes K. W., Petersen R. H. 2001. Apparent recombination or gene conversion in the ribosomal ITS region of a Flammulina (fungi, Agaricales) hybrid. Mol. Biol. Evol. 18, 94–96 10.1093/oxfordjournals.molbev.a003724 (doi:10.1093/oxfordjournals.molbev.a003724) [DOI] [PubMed] [Google Scholar]
- 55.Belloch C., Orlic S., Barrio E., Querol A. 2008. Fermentative stress adaptation of hybrids within the Saccharomyces sensu stricto complex. Int. J. Food Microbiol. 122, 188–195 10.1016/j.ijfoodmicro.2007.11.083 (doi:10.1016/j.ijfoodmicro.2007.11.083) [DOI] [PubMed] [Google Scholar]
- 56.Lopandic K., et al. 2007. Genetically different wine yeasts isolated from Austrian vine-growing regions influence wine aroma differently and contain putative hybrids between Saccharomyces cerevisiae and Saccharomyces kudriavzevii. FEMS Yeast Res. 7, 953–965 10.1111/j.1567-1364.2007.00240.x (doi:10.1111/j.1567-1364.2007.00240.x) [DOI] [PubMed] [Google Scholar]
- 57.Liti G., Peruffo A., James S. A., Roberts I. N., Louis E. J. 2005. Inferences of evolutionary relationships from a population survey of LTR-retrotransposons and telomeric-associated sequences in the Saccharomyces sensu stricto complex. Yeast 22, 177–192 10.1002/yea.1200 (doi:10.1002/yea.1200) [DOI] [PubMed] [Google Scholar]
- 58.Masneuf I., Hansen J., Groth C., Piskur J., Dubourdieu D. 1998. New hybrids between Saccharomyces sensu stricto yeast species found among wine and cider production strains. Appl. Environ. Microbiol. 64, 3887–3892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nguyen H. V., Lepingle A., Gaillardin C. A. 2000. Molecular typing demonstrates homogeneity of Saccharomyces uvarum strains and reveals the existence of hybrids between S. uvarum and S. cerevisiae, including the S. bayanus type strain CBS 380. Syst. Appl. Microbiol. 23, 71–85 10.1016/S0723-2020(00)80048-X (doi:10.1016/S0723-2020(00)80048-X) [DOI] [PubMed] [Google Scholar]
- 60.Hellborg L., Piskur J. 2009. Complex nature of the genome in a wine spoilage yeast, Dekkera bruxellensis. Eukaryot. Cell. 8, 1739–1749 10.1128/EC.00115-09 (doi:10.1128/EC.00115-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cappello M. S., Poltronieri P., Blaiotta G., Zacheo G. 2010. Molecular and physiological characteristics of a grape yeast strain containing atypical genetic material. Int. J. Food. Microbiol. 144, 72–80 10.1016/j.ijfoodmicro.2010.08.013 (doi:10.1016/j.ijfoodmicro.2010.08.013) [DOI] [PubMed] [Google Scholar]
- 62.Foulongne-Oriol M., Dufourcq R., Spataro C., Devesse C., Broly A., Rodier A., Savoie J.-M. 2011. Comparative linkage mapping in the white button mushroom Agaricus bisporus provides foundation for breeding management. Curr. Genet. 57, 39–50 10.1007/s00294-010-0325-z (doi:10.1007/s00294-010-0325-z) [DOI] [PubMed] [Google Scholar]
- 63.Marinoni G., Manuel M., Petersen R. F., Hvidtfeldt J., Sulo P., Piskur J. 1999. Horizontal transfer of genetic material among Saccharomyces yeasts. J. Bacteriol. 181, 6488–6496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McCullough J., Herskowitz I. 1979. Mating pheromones of Saccharomyces kluyveri: pheromone interactions between Saccharomyces kluyveri and Saccharomyces cerevisiae. J. Bacteriol. 138, 146–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kauserud H., Svegarden I. B., Decock C., Hallenberg N. 2007. Hybridization among cryptic species of the cellar fungus Coniophora puteana (Basidiomycota). Mol. Ecol. 16, 389–399 10.1111/j.1365-294X.2006.03129.x (doi:10.1111/j.1365-294X.2006.03129.x) [DOI] [PubMed] [Google Scholar]
- 66.de Barros Lopes M., Bellon J. R., Shirley N. J., Ganter P. F. 2002. Evidence for multiple interspecific hybridization in Saccharomyces sensu stricto species. FEMS Yeast Res. 1, 323–331 10.1016/S1567-1356(01)00051-4 (doi:10.1016/S1567-1356(01)00051-4) [DOI] [PubMed] [Google Scholar]
- 67.Muller L. A., McCusker J. H. 2009. A multispecies-based taxonomic microarray reveals interspecies hybridization and introgression in Saccharomyces cerevisiae. FEMS Yeast Res. 9, 143–152 10.1111/j.1567-1364.2008.00464.x (doi:10.1111/j.1567-1364.2008.00464.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Antunovics Z., Nguyen H. V., Gaillardin C., Sipiczki M. 2005. Gradual genome stabilisation by progressive reduction of the Saccharomyces uvarum genome in an interspecific hybrid with Saccharomyces cerevisiae. FEMS Yeast Res. 5, 1141–1150 10.1016/j.femsyr.2005.04.008 (doi:10.1016/j.femsyr.2005.04.008) [DOI] [PubMed] [Google Scholar]
- 69.Baack E. J., Whitney K. D., Rieseberg L. H. 2005. Hybridization and genome size evolution: timing and magnitude of nuclear DNA content increases in Helianthus homoploid hybrid species. New Phytol. 167, 623–630 10.1111/j.1469-8137.2005.01433.x (doi:10.1111/j.1469-8137.2005.01433.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mallet J. 2005. Hybridization as an invasion of the genome. Trends Ecol. Evol. 20, 229–237 10.1016/j.tree.2005.02.010 (doi:10.1016/j.tree.2005.02.010) [DOI] [PubMed] [Google Scholar]
- 71.Greig D., Louis E. J., Borts R. H., Travisano M. 2002. Hybrid speciation in experimental populations of yeast. Science 298, 1773–1775 10.1126/science.1076374 (doi:10.1126/science.1076374) [DOI] [PubMed] [Google Scholar]
- 72.Devier B., Aguileta G., Hood M. E., Giraud T. 2010. Using phylogenies of pheromone receptor genes in the Microbotryum violaceum species complex to investigate possible speciation by hybridization. Mycologia 102, 689–696 10.3852/09-192 (doi:10.3852/09-192) [DOI] [PubMed] [Google Scholar]
- 73.Doyle J. J., Doyle J. L., Rauscher J. T., Brown A. H. D. 2003. Diploid and polyploid reticulate evolution throughout the history of the perennial soybeans (Glycine subgenus Glycine). New Phytol. 161, 121–132 10.1046/j.1469-8137.2003.00949.x (doi:10.1046/j.1469-8137.2003.00949.x) [DOI] [Google Scholar]
- 74.Dufresne F., Hebert P. D. N. 1994. Hybridization and origins of polyploidy. Proc. R. Soc. Lond. B 258, 141–146 10.1098/rspb.1994.0154 (doi:10.1098/rspb.1994.0154) [DOI] [Google Scholar]
- 75.Gregory T. R., Nicol J. A., Tamm H., Kullman B., Kullman K., Leitch I. J., Murray B. G., Kapraun D. F., Greilhuber J., Bennett M. D. 2007. Eukaryotic genome size databases. Nucleic Acids Research 35, D332–D338 10.1093/nar/gkl828 (doi:10.1093/nar/gkl828) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zaragoza O., Garcia-Rodas R., Nosanchuk J. D., Cuenca-Estrella M., Rodriguez-Tudela J. L., Casadevall A. 2010. Fungal cell gigantism during mammalian infection. PLoS Pathog. 6, e1000945. 10.1371/journal.ppat.1000945 (doi:10.1371/journal.ppat.1000945) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hull C. M., Raisner R. M., Johnson A. D. 2000. Evidence for mating of the ‘asexual’ yeast Candida albicans in a mammalian host. Science 289, 307–310 10.1126/science.289.5477.307 (doi:10.1126/science.289.5477.307) [DOI] [PubMed] [Google Scholar]
- 78.Bennett R. J., Johnson A. D. 2003. Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains. EMBO J. 22, 2505–2515 10.1093/emboj/cdg235 (doi:10.1093/emboj/cdg235) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ibrahim A. S., Magee B. B., Sheppard D. C., Yang M., Kauffman S., Becker J., Edwards J. E., Jr, Magee P. T. 2005. Effects of ploidy and mating type on virulence of Candida albicans. Infect. Immun. 73, 7366–7374 10.1128/IAI.73.11.7366-7374.2005 (doi:10.1128/IAI.73.11.7366-7374.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Polakova S., Blume C., Zarate J. A., Mentel M., Jorck-Ramberg D., Stenderup J., Piskur J. 2009. Formation of new chromosomes as a virulence mechanism in yeast Candida glabrata. Proc. Natl Acad. Sci. USA 106, 2688–2693 10.1073/pnas.0809793106 (doi:10.1073/pnas.0809793106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hilton C., Markie D., Corner B., Rikkerink E., Poulter R. 1985. Heat shock induces chromosome loss in the yeast Candida albicans. Mol. Gen. Genet. 200, 162–168 10.1007/BF00383330 (doi:10.1007/BF00383330) [DOI] [PubMed] [Google Scholar]
- 82.Dhar R., Sagesser R., Weikert C., Yuan J., Wagner A. 2011. Adaptation of Saccharomyces cerevisiae to saline stress through laboratory evolution. J. Evol. Biol. 24, 1135–1153 10.1111/j.1420-9101.2011.02249.x (doi:10.1111/j.1420-9101.2011.02249.x) [DOI] [PubMed] [Google Scholar]
- 83.Welker D. L., Williams K. L. 1980. Mitotic arrest and chromosome doubling using thiabendazole, cambendazole, nocodazole and Ben late in the slime mould Dictyostelium discoideum. J. Gen. Microbiol. 116, 397–407 10.1099/00221287-116-2-397 (doi:10.1099/00221287-116-2-397) [DOI] [Google Scholar]
- 84.Ou S. H. 1980. Pathogen variability and host resistance in rice blast disease. Annu. Rev. Phytopathol. 18, 167–187 10.1146/annurev.py.18.090180.001123 (doi:10.1146/annurev.py.18.090180.001123) [DOI] [Google Scholar]
- 85.Bernard P., Hardwick K., Javerzat J.-P. 1998. Fission yeast Bub1 is a mitotic centromere protein essential for the spindle checkpoint and the preservation of correct ploidy through mitosis. J. Cell Biol. 143, 1775–1787 10.1083/jcb.143.7.1775 (doi:10.1083/jcb.143.7.1775) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Baum P., Yip C., Goetsch L., Byers B. 1988. A yeast gene essential for regulation of spindle pole duplication. Mol. Cell. Biol. 8, 5386–5397 10.1128/mcb.8.12.5386 (doi:10.1128/mcb.8.12.5386) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Otto S. P. 2003. In polyploids, one plus one does not equal two. Trends Ecol. Evol. 18, 431–433 10.1016/S0169-5347(03)00213-1 (doi:10.1016/S0169-5347(03)00213-1) [DOI] [Google Scholar]
- 88.Comai L. 2005. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6, 836–846 10.1038/nrg1711 (doi:10.1038/nrg1711) [DOI] [PubMed] [Google Scholar]
- 89.Szadkowski E., et al. 2010. The first meiosis of resynthesized Brassica napus, a genome blender. New Phytol. 186, 102–112 10.1111/j.1469-8137.2010.03182.x (doi:10.1111/j.1469-8137.2010.03182.x) [DOI] [PubMed] [Google Scholar]
- 90.Osborn T. C., Butrulle D. V., Sharpe A. G., Pickering K. J., Parkin I. A., Parker J. S., Lydiate D. J. 2003. Detection and effects of a homeologous reciprocal transposition in Brassica napus. Genetics 165, 1569–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Allendorf F. W., Danzmann R. G. 1997. Secondary tetrasomic segregation of MDH-B and preferential pairing of homeologues in rainbow trout. Genetics 145, 1083–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Codon A. C., Benitez T., Korhola M. 1997. Chromosomal reorganization during meiosis of Saccharomyces cerevisiae baker's yeasts. Curr. Genet. 32, 247–259 10.1007/s002940050274 (doi:10.1007/s002940050274) [DOI] [PubMed] [Google Scholar]
- 93.Loidl J. 1995. Meiotic chromosome pairing in triploid and tetraploid Saccharomyces cerevisiae. Genetics 139, 1511–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Trelles-Sticken E., Loidl J., Scherthan H. 2003. Increased ploidy and KAR3 and SIR3 disruption alter the dynamics of meiotic chromosomes and telomeres. J. Cell Sci. 116, 2431–2442 10.1242/jcs.00453 (doi:10.1242/jcs.00453) [DOI] [PubMed] [Google Scholar]
- 95.Mayer V. W., Aguilera A. 1990. High levels of chromosome instability in polyploids of Saccharomyces cerevisiae. Mutat. Res. 231, 177–186 [DOI] [PubMed] [Google Scholar]
- 96.Gerstein A. C., Chun H. J., Grant A., Otto S. P. 2006. Genomic convergence toward diploidy in Saccharomyces cerevisiae. PLoS Genet. 2, e145. 10.1371/journal.pgen.0020145 (doi:10.1371/journal.pgen.0020145) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gerstein A. C., McBride R. M., Otto S. P. 2008. Ploidy reduction in Saccharomyces cerevisiae. Biol. Lett. 4, 91–94 10.1098/rsbl.2007.0476 (doi:10.1098/rsbl.2007.0476) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gordon J. L., Byrne K. P., Wolfe K. H. 2011. Mechanisms of chromosome number evolution in yeast. PLoS Genet. 7, e1002190. 10.1371/journal.pgen.1002190 (doi:10.1371/journal.pgen.1002190) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Usher J., Bond U. 2009. Recombination between homoeologous chromosomes of lager yeasts leads to loss of function of the hybrid GPH1 gene. Appl. Environ. Microbiol. 75, 4573–4579 10.1128/AEM.00351-09 (doi:10.1128/AEM.00351-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hanson R. E., Islam-Faridi M. N., Crane C. F., Zwick M. S., Czeschin D. G., Wendel J. F., McKnight T. D., Price H. J., Stelly D. M. 2000. Ty1-copia-retrotransposon behavior in a polyploid cotton. Chromosome Res. 8, 73–76 10.1023/A:1009239522541 (doi:10.1023/A:1009239522541) [DOI] [PubMed] [Google Scholar]
- 101.Liu D., You C., Liu S., Liu L., Duan W., Chen S., Yan J., Liu Y. 2009. Characterization of a novel Tc1-like transposon from bream (Cyprinidae, Megalobrama) and its genetic variation in the polyploidy progeny of bream–red crucian carp crosses. J. Mol. Evol. 69, 395–403 10.1007/s00239-009-9295-5 (doi:10.1007/s00239-009-9295-5) [DOI] [PubMed] [Google Scholar]
- 102.Kim J. M., Vanguri S., Boeke J. D., Gabriel A., Voytas D. F. 1998. Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 8, 464–478 [DOI] [PubMed] [Google Scholar]
- 103.Fischer G., James S. A., Roberts I. N., Oliver S. G., Louis E. J. 2000. Chromosomal evolution in Saccharomyces. Nature 405, 451–454 10.1038/35013058 (doi:10.1038/35013058) [DOI] [PubMed] [Google Scholar]
- 104.Wendel J. F., Schnabel A., Seelanan T. 1995. Bidirectional interlocus concerted evolution following allopolyploid speciation in cotton (Gossypium). Proc. Natl Acad. Sci. USA 92, 280–284 10.1073/pnas.92.1.280 (doi:10.1073/pnas.92.1.280) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Joly S., Rauscher J. T., Sherman-Broyles S. L., Brown A. H., Doyle J. J. 2004. Evolutionary dynamics and preferential expression of homeologous 18S-5.8S-26S nuclear ribosomal genes in natural and artificial glycine allopolyploids. Mol. Biol. Evol. 21, 1409–1421 10.1093/molbev/msh140 (doi:10.1093/molbev/msh140) [DOI] [PubMed] [Google Scholar]
- 106.Gromicho M., Coutanceau J.-P., Ozouf-Costaz C., Collares-Pereira M. 2006. Contrast between extensive variation of 28S rDNA and stability of 5S rDNA and telomeric repeats in the diploid-polyploid Squalius alburnoides complex and in its maternal ancestor Squalius pyrenaicus (Teleostei, Cyprinidae). Chromosome Res. 14, 297–306 10.1007/s10577-006-1047-4 (doi:10.1007/s10577-006-1047-4) [DOI] [PubMed] [Google Scholar]
- 107.Liti G., Louis E. J. 2005. Yeast evolution and comparative genomics. Annu. Rev. Microbiol. 59, 135–153 10.1146/annurev.micro.59.030804.121400 (doi:10.1146/annurev.micro.59.030804.121400) [DOI] [PubMed] [Google Scholar]
- 108.James T. C., Usher J., Campbell S., Bond U. 2008. Lager yeasts possess dynamic genomes that undergo rearrangements and gene amplification in response to stress. Curr. Genet. 53, 139–152 10.1007/s00294-007-0172-8 (doi:10.1007/s00294-007-0172-8) [DOI] [PubMed] [Google Scholar]
- 109.Kashkush K., Feldman M., Levy A. A. 2002. Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics 160, 1651–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Brunet F. d. R. G., Crollius H. R., Paris M., Aury J.-M., Gibert P., Jaillon O., Laudet V., Robinson-Rechavi M. 2006. Gene loss and evolutionary rates following whole-genome duplication in teleost fishes. Mol. Biol. Evol. 23, 1808–1816 10.1093/molbev/msl049 (doi:10.1093/molbev/msl049) [DOI] [PubMed] [Google Scholar]
- 111.LangkjAer R. B., Cliften P. F., Johnston M., Piskur J. 2003. Yeast genome duplication was followed by asynchronous differentiation of duplicated genes. Nature 421, 848–852 10.1038/nature01419 (doi:10.1038/nature01419) [DOI] [PubMed] [Google Scholar]
- 112.Scannell D. R., Byrne K. P., Gordon J. L., Wong S., Wolfe K. H. 2006. Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature 440, 341–345 10.1038/nature04562 (doi:10.1038/nature04562) [DOI] [PubMed] [Google Scholar]
- 113.Bond U., Neal C., Donnelly D., James T. C. 2004. Aneuploidy and copy number breakpoints in the genome of lager yeasts mapped by microarray hybridisation. Curr. Genet. 45, 360–370 10.1007/s00294-004-0504-x (doi:10.1007/s00294-004-0504-x) [DOI] [PubMed] [Google Scholar]
- 114.Maclean C. J., Greig D. 2011. Reciprocal gene loss following experimental whole-genome duplication causes reproductive isolation in yeast. Evolution 65, 932–945 10.1111/j.1558-5646.2010.01171.x (doi:10.1111/j.1558-5646.2010.01171.x) [DOI] [PubMed] [Google Scholar]
- 115.Galitski T., Saldanha A. J., Styles C. A., Lander E. S., Fink G. R. 1999. Ploidy regulation of gene expression. Science 285, 251–254 10.1126/science.285.5425.251 (doi:10.1126/science.285.5425.251) [DOI] [PubMed] [Google Scholar]
- 116.Wu C. Y., Rolfe P. A., Gifford D. K., Fink G. R. 2011. Control of transcription by cell size. PLoS Biol. 8, e1000523. 10.1371/journal.pbio.1000523 (doi:10.1371/journal.pbio.1000523) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Parisod C., Holderegger R., Brochmann C. 2010. Evolutionary consequences of autopolyploidy. New Phytol. 186, 5–17 10.1111/j.1469-8137.2009.03142.x (doi:10.1111/j.1469-8137.2009.03142.x) [DOI] [PubMed] [Google Scholar]
- 118.Albertin W., Brabant P., Catrice O., Eber F., Jenczewski E., Chevre A. M., Thiellement H. 2005. Autopolyploidy in cabbage (Brassica oleracea L.) does not alter significantly the proteomes of green tissues. Proteomics 5, 2131–2139 10.1002/pmic.200401092 (doi:10.1002/pmic.200401092) [DOI] [PubMed] [Google Scholar]
- 119.Minato T., Yoshida S., Ishiguro T., Shimada E., Mizutani S., Kobayashi O., Yoshimoto H. 2009. Expression profiling of the bottom fermenting yeast Saccharomyces pastorianus orthologous genes using oligonucleotide microarrays. Yeast 26, 147–165 10.1002/yea.1654 (doi:10.1002/yea.1654) [DOI] [PubMed] [Google Scholar]
- 120.Adams K. L., Cronn R., Percifield R., Wendel J. F. 2003. Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc. Natl Acad. Sci. USA 100, 4649–4654 10.1073/pnas.0630618100 (doi:10.1073/pnas.0630618100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Birchler J. A., Riddle N. C., Auger D. L., Veitia R. A. 2005. Dosage balance in gene regulation: biological implications. Trends Genet. 21, 219–226 10.1016/j.tig.2005.02.010 (doi:10.1016/j.tig.2005.02.010) [DOI] [PubMed] [Google Scholar]
- 122.Madlung A., Masuelli R. W., Watson B., Reynolds S. H., Davison J., Comai L. 2002. Remodeling of DNA methylation and phenotypic and transcriptional changes in synthetic Arabidopsis allotetraploids. Plant Physiol. 129, 733–746 10.1104/pp.003095 (doi:10.1104/pp.003095) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Salmon A., Ainouche M. L., Wendel J. F. 2005. Genetic and epigenetic consequences of recent hybridization and polyploidy in Spartina (Poaceae). Mol. Ecol. 14, 1163–1175 10.1111/j.1365-294X.2005.02488.x (doi:10.1111/j.1365-294X.2005.02488.x) [DOI] [PubMed] [Google Scholar]
- 124.Yaakov B., Kashkush K. 2011. Massive alterations of the methylation patterns around DNA transposons in the first four generations of a newly formed wheat allohexaploid. Genome 54, 42–49 10.1139/G10-091 (doi:10.1139/G10-091) [DOI] [PubMed] [Google Scholar]
- 125.Kraitshtein Z., Yaakov B., Khasdan V., Kashkush K. 2010. Genetic and epigenetic dynamics of a retrotransposon after allopolyploidization of wheat. Genetics 186, 801–812 10.1534/genetics.110.120790 (doi:10.1534/genetics.110.120790) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Parisod C., Salmon A., Zerjal T., Tenaillon M., Grandbastien M. A., Ainouche M. 2009. Rapid structural and epigenetic reorganization near transposable elements in hybrid and allopolyploid genomes in Spartina. New Phytol. 184, 1003–1015 10.1111/j.1469-8137.2009.03029.x (doi:10.1111/j.1469-8137.2009.03029.x) [DOI] [PubMed] [Google Scholar]
- 127.Kenan-Eichler M., Leshkowitz D., Tal L., Noor E., Melamed-Bessudo C., Feldman M., Levy A. A. 2011. Wheat hybridization and polyploidization results in deregulation of small RNas. Genetics 188, 263–272 10.1534/genetics.111.128348 (doi:10.1534/genetics.111.128348) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Colot V., Rossignol J. L. 1999. Eukaryotic DNA methylation as an evolutionary device. Bioessays 21, 402–411 (doi:10.1002/(SICI)1521-1878(199905)21:5<402::AID-BIES7>3.0.CO;2-B) [DOI] [PubMed] [Google Scholar]
- 129.Halme A., Bumgarner S., Styles C., Fink G. R. 2004. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell 116, 405–415 10.1016/S0092-8674(04)00118-7 (doi:10.1016/S0092-8674(04)00118-7) [DOI] [PubMed] [Google Scholar]
- 130.Querol A., Fernandez-Espinar M. T., del Olmo M., Barrio E. 2003. Adaptive evolution of wine yeast. Int. J. Food Microbiol. 86, 3–10 10.1016/S0168-1605(03)00244-7 (doi:10.1016/S0168-1605(03)00244-7) [DOI] [PubMed] [Google Scholar]
- 131.Salmon J.-M. 1997. Enological fermentation kinetics of an isogenic ploidy series derived from an industrial Saccharomyces cerevisiae strain. J. Biosci. Bioeng. 83, 253–260 [Google Scholar]
- 132.Townsend G. F., Lindegren C. C. 1954. Characteristic growth patterns of the different members of a polyploid series of Saccharomyces. J. Bacteriol. 67, 480–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lamprecht I., Schaarschmidt B., Welge G. 1976. Microcalorimetric investigation of the metabolism of yeasts. V. Influence of ploidy on growth and metabolism. Radiat. Environ. Biophys. 13, 57–61 10.1007/BF01323624 (doi:10.1007/BF01323624) [DOI] [PubMed] [Google Scholar]
- 134.Pope G. A., et al. 2007. Metabolic footprinting as a tool for discriminating between brewing yeasts. Yeast 24, 667–679 10.1002/yea.1499 (doi:10.1002/yea.1499) [DOI] [PubMed] [Google Scholar]
- 135.Kosikov K. V., Raevskaia O. G. 1976. Biological productivity of hybrids and strains of yeast cultures of different ploidy. Mikrobiologiia 45, 1040–1044 [PubMed] [Google Scholar]
- 136.Lidzbarsky G. A., Shkolnik T., Nevo E. 2009. Adaptive response to DNA-damaging agents in natural Saccharomyces cerevisiae populations from ‘Evolution Canyon’, Mt. Carmel, Israel. PLoS ONE 4, e5914. 10.1371/journal.pone.0005914 (doi:10.1371/journal.pone.0005914) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Conant G. C., Wolfe K .H. 2007. Increased glycolytic flux as an outcome of whole-genome duplication in yeast. Mol. Syst. Biol. 3, 129. 10.1038/msb4100170 (doi:10.1038/msb4100170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.van Hoek M. J., Hogeweg P. 2009. Metabolic adaptation after whole genome duplication. Mol. Biol. Evol. 26, 2441–2453 10.1093/molbev/msp160 (doi:10.1093/molbev/msp160) [DOI] [PubMed] [Google Scholar]
- 139.Pavelka N., Rancati G., Zhu J., Bradford W. D., Saraf A., Florens L., Sanderson B. W., Hattem G. L., Li R. 2010. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature 468, 321–325 10.1038/nature09529 (doi:10.1038/nature09529) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Conant G. C., Wolfe K. H. 2006. Functional partitioning of yeast co-expression networks after genome duplication. PLoS Biol. 4, e109. 10.1371/journal.pbio.0040109 (doi:10.1371/journal.pbio.0040109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Blank L., Kuepfer L., Sauer U. 2005. Large-scale 13C-flux analysis reveals mechanistic principles of metabolic network robustness to null mutations in yeast. Genome Biol. 6, R49. 10.1186/gb-2005-6-6-r49 (doi:10.1186/gb-2005-6-6-r49) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mayrose I., Barker M. S., Otto S. P. 2010. Probabilistic models of chromosome number evolution and the inference of polyploidy. Syst. Biol. 59, 132–144 10.1093/sysbio/syp083 (doi:10.1093/sysbio/syp083) [DOI] [PubMed] [Google Scholar]
- 143.Takagi A., Harashima S., Oshima Y. 1985. Hybridization and polyploidization of Saccharomyces cerevisiae strains by transformation-associated cell fusion. Appl. Environ. Microbiol. 49, 244–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Greig D., Borts R. H., Louis E. J., Travisano M. 2002. Epistasis and hybrid sterility in Saccharomyces. Proc. R. Soc. Lond. B 269, 1167–1171 10.1098/rspb.2002.1989 (doi:10.1098/rspb.2002.1989) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Andalis A. A., Storchova Z., Styles C., Galitski T., Pellman D., Fink G. R. 2004. Defects arising from whole-genome duplications in Saccharomyces cerevisiae. Genetics 167, 1109–1121 10.1534/genetics.104.029256 (doi:10.1534/genetics.104.029256) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Song K., Lu P., Tang K., Osborn T. C. 1995. Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc. Natl Acad. Sci. USA 92, 7719–7723 10.1073/pnas.92.17.7719 (doi:10.1073/pnas.92.17.7719) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Michaelis P. 1954. Cytoplasmic inheritance in Epilobium and its theoretical significance. Adv. Genet. 6, 287–401 10.1016/S0065-2660(08)60132-7 (doi:10.1016/S0065-2660(08)60132-7) [DOI] [PubMed] [Google Scholar]
- 148.Leitch A., Lim K. A. R., Skalicka K., Kovarik A., Cigna A., Durante M. 2006. Nuclear cytoplasmic interaction hypothesis and the role of translocations in Nicotiana allopolyploids. In Radiation risk estimates in normal and emergency situations (eds Cigna A. A., Durante M.), pp. 319–326 Dordrecht, The Netherlands: Springer [Google Scholar]
- 149.Lee H.-Y., Chou J.-Y., Cheong L., Chang N.-H., Yang S.-Y., Leu J.-Y. 2008. Incompatibility of nuclear and mitochondrial genomes causes hybrid sterility between two yeast species. Cell 135, 1065–1073 10.1016/j.cell.2008.10.047 (doi:10.1016/j.cell.2008.10.047) [DOI] [PubMed] [Google Scholar]
- 150.Ainouche M. L., Baumel A., Salmon A., Yannic G. 2003. Hybridization, polyploidy and speciation in Spartina (Poaceae). New Phytol. 161, 165–172 10.1046/j.1469-8137.2003.00926.x (doi:10.1046/j.1469-8137.2003.00926.x) [DOI] [Google Scholar]
- 151.Evans B. J., Kelley D. B., Tinsley R. C., Melnick D. J., Cannatella D. C. 2004. A mitochondrial DNA phylogeny of African clawed frogs: phylogeography and implications for polyploid evolution. Mol. Phylogenet. Evol. 33, 197–213 10.1016/j.ympev.2004.04.018 (doi:10.1016/j.ympev.2004.04.018) [DOI] [PubMed] [Google Scholar]
- 152.Grivet L., D'Hont A., Roques D., Feldmann P., Lanaud C., Glaszmann J. C. 1996. RFLP mapping in cultivated sugarcane (Saccharum spp.): genome organization in a highly polyploid and aneuploid interspecific hybrid. Genetics 142, 987–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]