Abstract
Soils are complex ecosystems and the pore-scale physical structure regulates key processes that support terrestrial life. These include maintaining an appropriate mixture of air and water in soil, nutrient cycling and carbon sequestration. There is evidence that this structure is not random, although the organizing mechanism is not known. Using X-ray microtomography and controlled microcosms, we provide evidence that organization of pore-scale structure arises spontaneously out of the interaction between microbial activity, particle aggregation and resource flows in soil. A simple computational model shows that these interactions give rise to self-organization involving both physical particles and microbes that gives soil unique material properties. The consequence of self-organization for the functioning of soil is determined using lattice Boltzmann simulation of fluid flow through the observed structures, and predicts that the resultant micro-structural changes can significantly increase hydraulic conductivity. Manipulation of the diversity of the microbial community reveals a link between the measured change in micro-porosity and the ratio of fungal to bacterial biomass. We suggest that this behaviour may play an important role in the way that soil responds to management and climatic change, but that this capacity for self-organization has limits.
Keywords: soil structure, self-organization, microbial diversity, modelling, biophysics
1. Introduction
The three-dimensional structure of the porous matrix of soil affects aeration, the flow of other gases and liquids and the storage of water, while at the same time provides the range of microscale habitats for the most diverse and functionally important component of the terrestrial biosphere [1,2]. The physical architecture of this matrix is known to be highly geometrically complex, yet organized. While many technological advances have made it possible to characterize the microbial diversity in soil, we know relatively little about the physical and biological interactions that are key to many soils processes [3]. We do know that soil structure is not static in space or time, and that spatio-temporal heterogeneity is exhibited in most processes [4], driven by changes in porosity and particularly by how much porosity there is and how connected it is to the atmosphere and/or to groundwater flow [5,6]. The interactions between this porosity and the microbes in soil are of fundamental importance because the biophysical properties of soil are the product of both genotypic and micro-environmental diversity [6–8]. It is important to understand if these properties emerge from random combinations of genotype and environment, or if there is some underlying organizing principle.
Particle aggregation is a widely studied phenomenon and in many cases particle–scale interactions, if left on their own, maintain the system in a state of higher organization that is highly unlikely to be reached by random chance [9]. In these situations, the system is said to be self-organizing. The aggregation of complexes of physical particles and biological cells has been explored only once before in a comparatively well-mixed system [10]. The presence of a biological component is significant because it introduces a potential for structural organization that is conducive to supporting life (e.g. through the provision of resources). It also means the system will be subject to natural selection of the extended phenotype [11].
We have previously proposed that the soil–microbe complex is self-organizing [1]. There is experimental evidence for soil restructuring in the vicinity of plants roots where microbial activity is likely to be enhanced [12,13]. However, to date, there have been no studies that isolate the feedback between microbes and structure. Furthermore, both the consequences of a self-organized state for the dynamics of the soil–microbe system, and the potential to manage it remain unclear.
It is important to distinguish between the well-known conceptual model for the impact of microbes on structure [14], and self-organization. In the former case, we have a causal effect of microbial activity on structure without the feedback of structure on microbial activity. Therefore, microbes cause structure change, but there is no account of the effect this change has on microbial activity. Furthermore, if there are no changes in resources, then a steady state is reached. In the latter case, we include the feedback, and then not only is a dynamical state likely even in the absence of resource changes as we shall see, but also because the structure now affects the activity, the aggregated state of the physical particles causes the biological state and vice versa.
The potential for self-organization in soil is significant for a number of additional reasons. Firstly, because self-organization arises from internal processes that regulate the pore-scale arrangement of microbes and structure over time, it could play a central role in sustaining the properties of soil over time. Secondly, since interactions are the causal agents of the self-organized state rather than the individual components, an understanding of how soil changes over time will only be achieved by studying the integrated behaviour of the biology and physics. Finally, to identify targets for sustainable management of soil and for understanding the consequences of environmental change, a systems approach to soil science will be required that accounts for how external factors impact on the interactions to direct the self-organized state.
To search for evidence of self-organization, we studied the dynamics of soil using a combination of theoretical and experimental approaches including the use of X-ray microtomography to image soil structure. This necessitated the development of a new microcosm design to experimentally isolate the effects of the interaction between microbes and soil structure generation. Finally, a model for the interaction between microbial activity and particle aggregation in soil was developed to help in the interpretation of the data.
2. Methodology
We used two experiments that contrasted in their degree of complexity. The first experiment used reconstituted field soil, previously sterilized using gamma-ray irradiation and re-inoculated with prescribed single strains of micro-organism. The samples were held at a constant moisture content. This experiment allowed us to isolate the effects of a typical soil bacterium and a typical soil fungus on structure. The second experiment used unsterilized field soil that was diluted with sterile field soil at a range of dilution rates. The samples underwent a single wet–dry cycle. These manipulations were intended to create conditions closer to the field, and to study the effect of manipulating the community structure.
2.1. Experiments using simple microcosms
Soil was collected from the top 30 cm of an arable field at Shuttleworth Agricultural College, Bedfordshire, UK (Ordinance Survey Grid Ref 513 242) in February 2005. The soil is a Ludford series sandy clay loam comprising 590 g kg−1 sand, 230 g kg−1 silt, 180 g kg−1 clay, with a pH of 6.9, and an organic matter content of 10 g kg−1 [15]. After collection, the soil was stored at 4°C in the dark until use. It was then passed through a 4 mm sieve and was packed into 50 ml polypropylene syringe bodies (i.e. microcosms) to 16 cm3 by uni-axial compression to give a gravimetrically adjusted dry bulk density of 1.4 g cm−3. The microcosms were gamma-irradiated to sterilize them (Isotron PLC, Swindon, UK) at a minimum of 25 kGy and allowed to settle for eight weeks.
The microcosms were then inoculated with 1 ml of 1/4 strength Ringer's solution containing a suspension of fungus (Rhizoctonia solani Kühn) or bacterium (Pseudomonas fluorescens Migula) separately, and in combination, onto the surface of the soil in the microcosm, and allowed to infiltrate into the soil. These organisms are common soil microbes that are known to release metabolic by-products including mucilage into the soil environment. The soil was held at a 14 per cent gravimetric moisture content, once the inoculum was added, which was the value centred round the inflection point of the moisture retention curve. The microcosms were then incubated in natural light conditions at 20 ± 2°C and moisture loss minimized by capping the core with a HEPA-filter mounted in a Suba-seal (typical moisture loss 0.06 ml d−1). A control treatment comprises microcosms packed with sterile soil and incubated under otherwise identical conditions. After 25 days incubation, microcosms were plunged into liquid N and freeze-dried in order to preserve the structural properties prior to structural determination using a 160 keV X-ray microtomography scanner as described in the appendix. The soils were then scanned in the microtomograph and reconstructed at two voxel resolutions (53 and 9 µm) and analysed to quantify the impact of microbial-induced changes on the pore structure at the two scales.
2.2. Experiments using field soil and manipulating diversity
Soil as above was passed through a 4 mm sieve and sub-samples of 400 g at 10.8 per cent gravimetric moisture content were placed in sterilized mesocosms. These comprised an autoclavable 1 l plastic pot with a screw-lid. HEPA filters were placed into a 7 mm diameter hole in the lid and silicone-sealant was used to seal around the HEPA filter above and below the hole. Mesocosms were then autoclaved at 121°C for 15 min at 15 psi and allowed to cool. Mesocosms with soil were then gamma-irradiated to sterilize them (Isotron PLC, Swindon, UK) at a minimum of 25 kGy and allowed to settle for eight weeks.
Subsamples of soil collected at the same time as above was used to re-inoculate the sterilized mesocosms, but with a serial dilution of the soil biomass. Seven replicate innocula were set up at each dilution by using a different soil subsample to create the dilution in order to avoid pseudo-replication. For example, the five replicates for first dilution were formed by taking a distinct 25 g sample of field soil at 10.8% m/c and placing this in to 100 ml 1/4 strength Ringer's solution in a sterilized medical flat bottle—making the 100 dilution. The slurry was mixed flat on an orbital shaker for 15 min at 140 r.p.m. This was then diluted by taking 10 ml of the 100 dilution and placing it into 90 ml of 1/4 Ringer's solution and shaking it for 15 min as previously to produce a 10−1 dilution inoculum. This process was repeated for each dilution, using a different 25 g starting subsample, up to a dilution factor of 10−10.
Aliquots (10 ml) from each independent dilution were inoculated into sterilized mesocosms, and this was repeated for each of the 10−2, 10−4, 10−6, 10−8, 10−10 dilutions. Both sterile and unsterilized (‘field’) mesocosms were set up (1/4 strength Ringer's added). The final moisture content of the mesocosms was at 13.3 per cent after addition of the inoculum. The five independent dilution series and seven replicates gave rise to 35 mesocosms. These were incubated at 20 ± 2°C under natural light conditions until the biomass as determined by substrate-induced respiration (SIR) [16] of each mesocosms had equilibrated to statistically similar concentrations. This occurred after 14 months. During this period, the mesocosms were regularly agitated to disturb the structure and hasten the equilibration point, and were kept at constant moisture content by the addition of sterile water. They were subsequently harvested and soil was passed through a 2 mm sieve under aseptic conditions (keeping each replicate and treatment uncontaminated). Subsamples of soil were preserved by freeze-drying [17] and phenotypic and genotypic profiles determined as described in the appendix using terminal restriction fragment length polymorphism (T-RFLP) to measure relative (as opposed to absolute) diversity and phospholipid fatty-acid (PLFA) to measure biomass.
A study of the impact of the different dilutions on physical structure was then determined by packing the sieved soil into 50 ml polypropylene syringe bodies (i.e. microcosms as above) to 16 cm3 by uni-axial compression to give a gravimetrically adjusted dry bulk density of 1.4 g cm−3 at 13.3 per cent gravimetric moisture content. The soils were adjusted to prescribed matric potential using a tension table system that maintained the respective microbiological statuses of each microcosm independently. Microcosms were placed on sterile tension tables and 1 ml glucose solution was added at the start of a drying cycle to reach 1 mg C g−1 soil. Microcosms were placed at −2 kPa and drawn down to −5 kPa after 10 days. After 3 days at this tension, microcosms were returned to −2 kPa for a further 12 days. Microcosms were incubated in natural light conditions at 20 ± 2°C and moisture loss minimized by capping the core with a HEPA-filter mounted in a Suba-seal (typical moisture loss 0.06 ml d−1). They were then plunged into liquid N and freeze-dried in order to preserve the structural properties prior to structural determination by X-ray computed tomography as described in the appendix.
2.3. Modelling feedback between structure and microbial activity
The model comprises a regular cubic lattice of cells that can contain soil particles or pore space and is developed from our earlier work [18]. The basis of the model is that stabilizing agents are produced as a consequence of local biological activity such as hyphae, and/or extracellular polysaccharides [19] act to bind the soil particles together and reduce the probability of local disruption. In particular, for two particles, i and j, in the lattice, we define an affinity, αij, between them by a linear function of the form,
where μi is the concentration of stabilizing agents in the voxel i, and Kij is a constant depending on the state of the couple ij (e.g. pore–solid and solid–solid). In the absence of data, we make the simplest assumption, and take the production of these stabilizing agents to be proportional to the local respiration rate (i.e. oxygen consumption rate). This results in more favourable microenvironments being preferentially stabilized relative to less favourable ones. The same agents are assumed to decay at a constant relative rate. For any solid voxel, summing this affinity measure over its immediate neighbours gives a measure of stability for that voxel. Comparing this stability measure across solid voxels in the simulation is the key to structure evolution. Since microsites are connected in the larger matrix, instabilities in structure at one location can lead to changes in the oxygen concentration at all connected sites, potentially affecting the stability of these sites. The resulting competitive or cooperative interaction between microsites gives rise to novel dynamical behaviour.
Oxygen movement through the pore space is assumed to be by Fickian diffusion from the upper surface into the pore space. The uptake of oxygen is a constant in all pore cells that are adjacent to a soil particle, provided that these cells have non-zero oxygen concentration. When the oxygen concentration is zero in these cells (technically less that 10−6 of the surface concentration), the uptake rate is assumed to be zero. We consider the stability of particle clusters defined as those 33 nearest-neighbour cells in the first rank Moore neighbourhood of each solid particle in the lattice. The strength of each such cluster is derived by summing over the affinities of the individual particle–particle bonds in the cluster and adding a contribution to the strength resulting from the microbial activity in the cluster pore cells. At the beginning of each iteration, the model calculates the rate of oxygen consumption in each pore cell and updates the oxygen concentration accordingly. For each particle cell in turn, the total strength of the associated cluster is calculated taking account of both the increase associated with respiration and the decay. Once this has been completed, the 1 per cent of particle cells with associated clusters having the lowest strength is selected. Each of these particle cells are relocated in their respective cluster neighbourhood such that the stability of the new cluster is maximized. This is intended to mimic random local reorganization of soil particles resulting from preferential fragmentation of weaker cluster structures relative to structures of higher stability [18]. At the end of the restructuring, the next iteration of the model begins. The qualitative behaviour of the model is not sensitive to the values for the parameters, which are currently unknown. The values used in the model are calibrated to determine if the model can match the changes in porosity observed in the experiments.
3. Results
3.1. A simple model suggests soil has the capacity for self-organization
The model was run with an initial condition corresponding to a disordered arrangement of particles, and with a range of porosities (defined as the pore volume divided by the total volume). We define the surface-connected porosity as the ratio of the volume of porosity that is connected to the top surface of the simulated soil volume, to the total volume. At the start of the simulation, the disordered soil structure has a zero surface-connected porosity with probability close to 1 for all initial porosities used. We ran the simulation to compare the case where there was a feedback between microbial activity and microenvironment stabilization, with the case where there was no feedback. This was repeated for each of the initial porosity values.
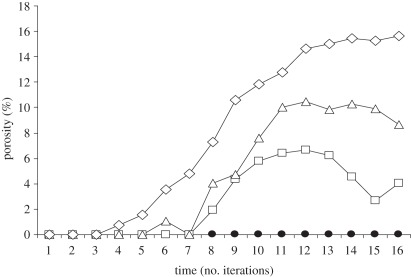
Figure 1 shows the change in the connected porosity predicted by the model for each of the selected starting porosities. For the case where there is a feedback between microbial activity and microenvironment stabilization, the structure begins to show evidence of organization after a short number of iterations, and the connected porosity increases significantly above the value for the random structure. This increase levels off, and the connected porosity reaches a maximum value and then remains in a dynamical state close to this value for the remainder of the simulation.
Figure 1.
Model predictions for the change in surface-connected porosity for initially random structure of porosity 23, 24 and 25%, along with the corresponding curve for the case with no microbial activity. In all cases, the initial surface-connected porosity was 0%. The curve for 22% was indistinguishable from the curve corresponding to no microbial activity. Filled circles with continuous line, no microbes; open squares with continuous line, microbes p = 23%; open triangles with continuous line, microbes p = 24%; open diamonds with continuous line, microbes p = 25%.
If the model is run with no feedback, no such organization of the structure occurs. This supports the proposition that the interaction between microbial activity, particle aggregation and resources flows leads to self-organization that is manifest in an increase in surface-connected porosity over time, compared with a random (disorganized) structure. The maximum value of the surface-connected porosity attained in the model is sensitive to the starting porosity. For higher porosities, the resulting surface-connected porosity is higher. Interestingly, there reaches a value of starting porosity below which the value for the connected porosity cannot increase above that for the random structure. Self-organization is no longer possible, and the system behaves as though the feedback between microbial activity and structure is removed.
3.2. Experimental evidence for self-organization in simple microcosms
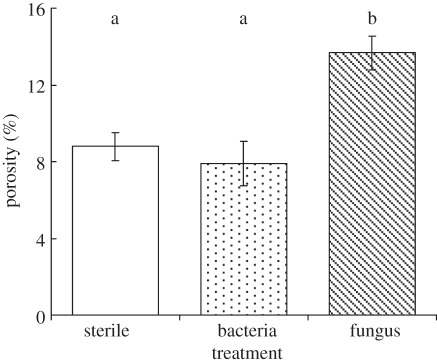
Following the 25 day incubation of the simple microcosms described in §2.1, we found that the porosity of the soil in the samples containing the fungus was significantly greater than that in the control sample (figure 2). The surface-connected porosity (i.e. the percentage of the sample volume that comprised pore space that was connected to the boundary of the volume) showed similar trends and was 12 ± 1% in the samples containing fungi, compared with 8 ± 1% in the sterile samples. Thus not only the porosity, but its connectivity was increased in the samples with microbial activity. Interestingly, when the same samples were scanned at a lower resolution of 53 µm, there was no significant difference in porosity or surface-connected porosity. Thus, over the timescales used for the incubations in this study, fungal activity impacted only on pore features that were smaller than 53 µm. It is the pore features at these scales that hold water under normal field conditions, determine flow rates and regulate microbial activity and function [1].
Figure 2.
Porosity of soil inoculated with fungi and bacteria compared with sterilized field soil. All treatments were incubated for 25 days under conditions of constant matric potential. Bars show means (n = 3), whiskers denote s.e. Bars labelled by the same letters are not significantly different using the Fisher multiple-ranges test in ANOVA.
In contrast with the fungal microcosm, we found no significant effect of bacterial activity on soil structure over the timescales and conditions associated with the incubations (figure 2). Neither did we find any significant difference between the fungal microcosm and microcosms containing both bacteria and fungi (data not shown).
3.3. Self-organization is important for soil water
The direct measurement of hydraulic conductivity of soil samples generally induces structural changes in itself. Therefore, to study the impact of the observed structure changes on physical processes, we adopted a modelling approach. We discretized the three-dimensional pore architecture images to provide a numerical mesh for the computational modelling of transport coefficients. Using the lattice Boltzmann approach, which has been successfully applied to the prediction of the saturated hydraulic conductivity of soil directly from images of structure [20], we calculated the conductivity of the images corresponding to both the sterile and the fungal-inoculated soil. We assume in the modelling that the presence of microbial activity had no effect on the properties of the water, and so any calculated differences are solely due to structure changes. The soil inoculated with fungi had a mean-simulated-saturated conductivity of 1.52 ± 0.52 cm d−1 compared with 0.27 ± 0.09 cm d−1 for the sterile soil samples. These results show that, owing to the nonlinear dependence of conductivity on the porosity and the fact that the connectivity of the pore space is also changed, fungal-induced re-organization of the soil structure can lead to a disproportionate increase in the associated flow rates. This is despite the fact that the changes in structure occur at scales less than 53 µm. We expect that the impact of self-organization on the unsaturated hydraulic conductivity will be even more dramatic.
3.4. Manipulation of microbial community structure directs self-organization
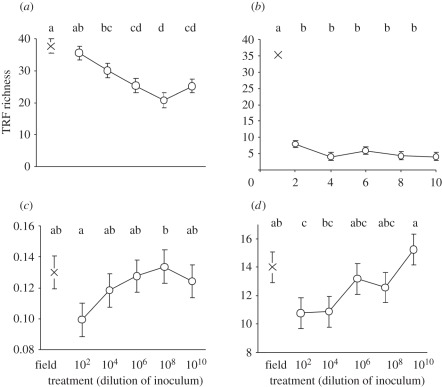
As hypothesized, the T-RFLP analysis confirmed that there was a steady and significant decline in bacterial diversity with increased dilution (figure 3a). In contrast, however, fungal diversity declined significantly at the first dilution, and remained low and constant across all subsequent dilution rates (figure 3b). The PLFA profiles indicate that although the diversity remained constant, there was a concomitantly steady increase in fungal biomass relative to microbial biomass in the community phenotype with increasing dilution (figure 3c). Thus, as well as impacting on the bacterial diversity, the dilution treatment altered the ratio of fungal to bacterial biomass, with the latter being highest at the lowest dilutions.
Figure 3.
Genotypic, phenotypic and functional profiles of soils experimentally manipulated to produce a graded series of biodiversity by inoculating sterile soil with a dilution series of non-sterile (source) soil across 10 orders-of-magnitude and incubating for 14 months [Symbol O]. The symbol X denotes equivalent data for the unsterilized soil (‘field’) from which the dilution series was derived. (a) Number of distinct eubacterial T-RFLP fragments; (b) number of distinct fungal T-RFLP fragments; (c) fungal : bacterial biomass ratio derived from phospholipid fatty acid (PLFA) profile; (d) porosity at 9 µm scale derived from X-ray tomography imaging of undisturbed soils. Points show means (n = 5); bars show standard errors (in part (b), these fall within the confines of the symbols). Within each panel, points labelled by the same letters are not significantly different using the Fisher multiple-ranges test in ANOVA.
The data at the end of the 10 day incubation show that porosity at the lowest dilution is significantly below the value for soil-containing field-levels of diversity. Subsequent values then show an increase in the porosity with dilution such that the porosity of the most dilute soils was significantly greater than that for the least dilute soils (figure 3d). Indeed, the porosity of the most dilute soil was not significantly different from the soil containing the undiluted field community. This trend in porosity with dilution is different from the trend in bacterial diversity, which is consistent with a monotonic decline with dilution from its level in the field. Therefore, it is unlikely that the observed porosity trend arises as a consequence of changes to the bacterial diversity alone. Instead, the trend in porosity is similar to that of the fungal : bacterial ratio shown in figure 3c and is consistent with the observation from the first experiment that fungi are the dominant agent in self-organization in soil. The magnitude of the porosity response is dependent on the relative biomass proportions of fungi to bacteria over the timescales studied here.
4. Discussion and conclusions
This study aims to elucidate the link between microbial activity and the physical structure of soil. We present experimental results from both biologically simple microcosms and from microcosms containing microbial communities that are representative of field conditions. A simple model for particle aggregation is introduced to support the interpretation of these results. Both the experimental and theoretical work presented here provide evidence that the soil–microbe system is self-organizing as a consequence of the feedback between microbial activity and particle aggregation. This feedback results in an increase in porosity at scales relevant to water storage and flow and to gas diffusion (i.e. smaller than 53 µm). Our experiments indicate that fungi are the most important agents of self-organization, at least over the range of conditions studied here. Importantly, we show the resulting self-organized state is altered, with consequences for aeration, respiration and hydrology, when the ratio of fungal to bacterial biomass in the microbial community is changed.
The evidence supporting the self-organized state comes from both the modelling and the experimental work. Firstly, the model demonstrates that self-organization requires both an effect of microbes on structure, and a feedback between changes in structure and microbial activity. Our experiments clearly show that microbes affect structure. The evidence of feedback between structure and microbial activity comes from the observed increase in connected porosity. If there was no feedback of structure on microbial activity, then no particular organized state would be selected. The fact that the state that does emerge is one that increases resource flow into the structure and towards the microbes shows that there is feedback between structure and microbial activity. We conclude, therefore, that the observations support the idea that the soil–microbe system is self-organizing.
It is important to point out the distinction between the behaviour that is reported here, and the existing knowledge of the effects of microbes on soil structure. For example, Tisdall & Oades [14] proposed, nearly three decades ago, that by-products of microbial activity, along with plant roots, are involved in soil aggregation. A considerable volume of research has confirmed the effects of microbes on aggregate size and stability. However, none of this work considered the feedback on microbial activity that might arise from such changes in structure. Without feedback, there can be no dynamics or self-organization of structure. The modelling results presented here show that the feedback gives rise to a dynamical behaviour that allows recovery of connected porosity towards a quasi-stable dynamical equilibrium that maintains the connected porosity close to a fixed value. Since our results show that changes are not dependent on external forcing (e.g. wet–dry cycles), self-organization is a dynamical state of soil that can arise from purely endogenous processes. The state that emerges is dependent on internal factors including microbial community structure, but the modelling also suggests that external factors such as compaction can also play an important role.
An unanticipated consequence of our manipulation of the microbial community through successive dilutions of the field community was that the ratio of fungal to bacterial biomass increased with increase in dilution, and that this gave rise to an increase in porosity with dilution of the field community. Our interpretation is that fungal relative dominance was compromised with respect to bacteria when re-inoculated into the sterile soil. However, at the greater dilutions, fungal diversity persisted while the bacterial diversity continued to fall, allowing the fungi to re-assert dominance and therefore successfully restructure the soil. A mechanism for this could be that the lower bacterial diversity resulted in a competitive release of the fungi, allowing their dominance in the high-dilution treatments. Mille-Lindblom et al. [21] report such success of established fungi in out-competing bacteria for substrate.
It is perhaps not surprising that microbes have evolved the capacity to influence their abiotic environment in a manner that preserves features that are essential for supporting life. The complexity of the soil physical structure at scales of 50 µm and less ensures the availability of air and water over a broad range of environmental conditions. Others [9,11] have suggested that the interactions between biotic and abiotic components in soil give rise through unidentified evolutionary processes to properties that are more conducive to supporting life. We also find evidence of self-organization in the oldest form of life discovered in the fossil record, but which is still found growing today in a number of locations including Perth, Australia. Stromatolites are formed by blue-green algae that cement sedimenting particles at a rate dependent on local microbial activity. The resulting self-organized structures ensure the most favourable microbial environments (i.e. those that promote highest microbial activity) remain in optimal light conditions [10,22]. Therefore, even 3.5 billion years ago, progenitor ‘soil’–microbe systems were capable of self-organization.
We can only speculate at this stage what the significance of self-organization is for the soil–microbe system. Perhaps, the most important consequence is that it is a dynamical state, and so is relevant to understanding and managing the resilience of soil to perturbation. It results in an opening up of the pore space at scales that increase both convective and diffusive transport rates. The assumption of the model that microbes are distributed throughout the soil volume means that an increase in surface-connected porosity results in an increase in microbial respiration. Under this assumption, self-organization of soil will result in an increase in microbial respiration rate, provided that oxygen is the only constraint on microbial activity. This, in turn will increase nutrient cycling rates. The scales at which we observed structural changes in the experiments are relevant to the water-holding capacity of soil. Given the connected porosity increased, we can anticipate that self-organization will also result in improved drainage of soil. All of these consequences act to increase the potential productivity of the soil, however considerable further experimentation will be required to verify this.
Given the importance of soil for carbon storage, it will be important to determine the impact of self-organizing behaviour for carbon turnover. The model predicts that structure is continuously reorganizing. This means that the environment of carbon in soil will change over time. There will be a range of carbon turnover timescales associated with the range in structure turnover times that expose carbon to microbial degradation in an oxic environment. Further work with a more sophisticated model is currently underway to gain a quantitative understanding of the consequences for the stabilization of carbon in soil.
The results presented here identify the feedback between structure, microbial activity and particle consolidation as an essential feature of soil that could be subject to evolution. Williams & Lenton [19] have suggested a link between microbial networks that engineer their environments to achieve equitable conditions and evolutionary success because these communities would have stability over time and would be more likely to disperse into new environments. Given the important role in soil of the ratio of fungal to bacterial biomass reported here, this view may also explain why late successional ecosystem soils tend to be dominated by fungi [23]. Under this interpretation, soil should be viewed as an extended phenotype of the resident microbial community [11] and selection should be regarded as acting on the soil metagenome. Given the importance of soil for primary production and climate regulation, this has implications, which extend beyond soil ecology, for our understanding of the whole of the Earth system.
Acknowledgements
We thank three anonymous reviewers for their comments and suggestions that substantially improved the manuscript. This research was supported by the BBSRC.
Appendix A. Material and methods
A.1. Molecular methods
A.1.1. Phenotypic community structure
PLFA analysis was used to determine the phenotypic properties of the soils. PLFAs were extracted from 10 g freeze-dried soil, using a method based on Frostegård et al. [24], after Bligh & Dyer [25]. Resulting fatty-acid methyl esters (FAMEs) were identified using a gas chromatogram (Agilent Technologies 6890N fitted with an HP-5 capillary column; 30 m length, 0.32 mm internal diameter and 0.25 µm film) and retention time compared with standard qualitative bacterial acid methyl ester mix (26 standard; Supelco, UK). A standard flame ionization detector was used to detect FAMEs as they were released from the column. Results were expressed as a percentage of identified peak area on chromatograms, normalized to unity within each sample. The fungal : bacterial ratio was calculated by dividing the proportion of the profile ascribed to 18 : 2w6 by the sum of those proportions ascribed to the bacterial PLFAs i15 : 0, a15 : 0, 15 : 0, i16 : 0, i17 : 0, cy17 : 0, 17 : 0, 18 : 1ω7.
A.1.2. Genotypic community structure
T-RFLP is a well-established method in characterizing microbial community structure but is not reliable for measuring absolute bacterial diversity [26]. However, as a method for examining comparative diversity as required here, it is robust [27]. DNA was extracted from 0.25 g of each soil sample using a PowerSoil DNA kit according to manufacturer's instruction (Mo Bio Laboratories, Carlsbad, CA, USA). 16S rRNA genes from bacterial communities and ITS region from fungal communities were amplified separately by PCR using the following primers: (i) 16S-63F (Bacteria) CAG GCC TAA CAC ATG CAA GTC; (ii) 16S-1087R (Bacteria) CTC GTT GCG GGA CTT AAC CC; (iii) ITS1F FAM CTT GGT CAT TTA GAG GAA GTA A; (iv) ITS4R (fungi) TCC TCC GCT TAT TGA TAT GC. The fluorescent dyes 6-carboxyfluorescein (FAM; blue), and VIC (green), were used for fungal and bacteria, respectively. Concentrations of reagents in the PCR reaction mix and PCR conditions were as described in Singh et al. [28]. PCR products were purified using charge Switch PCR clean up kit (Invitrogen Ltd, UK) according to manufacturer instructions. About 200 ng of each PCR product was digested at 37°C for 3 h with the restriction enzyme Hha I (Promega, UK) and this reaction was stopped by further incubating of samples at 95°C for 15 min. Digested DNA products from the two different communities (bacteria and fungi) from the same sample were combined together and an aliquot (2 µl) was used to mix with 12 µl of formamide and 0.3 µl of Liz labelled GS5000 (−250) internal size standard. Samples were then run on a DNA sequencer (ABI3130xl; Applied Biosystem Instruments) for fragment size analysis. T-RFLP data profiles were obtained using GeneMapper software (ABI) and relative abundance and binary data were obtained as described by Singh et al. [28] and Singh & Thomas [29].
A.2. X-ray computed tomography
All soils were scanned using a high-resolution Metris X-TEK Benchtop CT system (http://www.xtekxray.com/products/systems.html#bt: 160 kV X-ray source, 5 µm focal spot reflection target), using a tungsten filament and a molybdenum target at 110 kV and 115 µA. Aluminium filter of 0.1 mm thickness was used for reducing beam-hardening artefacts. Initially, the packed cores were non-destructively scanned and reconstructed (isotropic voxel size of 53 µm). In order to scan and reconstruct at higher resolutions (voxel size 9 µm), cores were carefully disassembled and small aggregates (10 reps) were randomly selected and then scanned using Metris X-TEK Inspect-X v. 1.1 acquisition software. Ring artefacts were minimized and centre of rotation corrections were applied to each radiograph. Acquired datasets consisted of 1169 angular projections with each radiograph averaged over 32 frames. Reconstruction and initial visualization of the three-dimensional datasets were achieved using Metris X-TEK software CT-Pro v. 1.1 and VGStudioMAX v.1.2 software, respectively. Filtered back-projection algorithm was used for three-dimensional reconstruction in CT-Pro. VGStuidoMAX was used for adjusting contrast in reconstructed volumes, which were then exported into voxel-thick TIFF (tagged image file format) image stacks. Image stacks were then imported into ImageJ v.1.38 and 140 × 140 pixel regions of interest were selected in each of 200 slices. Three-dimensional analysis was achieved using SCAMP software [30,31] a plug-in for ImageJ (http://rsbweb.nih.gov/ij/). The three-dimensional volume was initially read in as a series of slices to ImageJ (typically 200 slices) and segmented into binary structure using SCAMP thresholding tool. Threshold parameters were calculated for each image stack after thorough investigation of greyscale values corresponding to small (10–50 voxels in size) connected and ‘thin valley’ pores. Measurements of total porosity, connected porosity and fractal dimension were obtained.
A.3. Statistical analysis
Peaks within PLFA and T-RFLP profiles were normalized to percentage of total areas within each profile. Mean values for all properties were compared using one-way ANOVA using Statistica v. 8.0 (Statsoft Inc., Tulsa, OK, USA).
References
- 1.Young I. M., Crawford J. W. 2004. Interactions and self-organization in the soil-microbe complex. Science 304, 1634–1637 10.1126/science.1097394 (doi:10.1126/science.1097394) [DOI] [PubMed] [Google Scholar]
- 2.Ritz K. 2008. Soil as a paradigm of a complex system. In Complexity and security (eds Ramsden J. J., Kervalishvili P. J.), pp. 103–119 Amsterdam, The Netherlands: IOS Press [Google Scholar]
- 3.Young I. M., Crawford J. W., Nunan N., Otten W., Spiers A. 2008. Microbial distribution in soils: physics and scaling. In Advances in agronomy, vol. 100 (ed. Sparks D. L.), pp. 81–121 San Diego, CA: Elsevier Academic Press Inc [Google Scholar]
- 4.Roger-Estrade J., Richard G., Dexter A. R., Boizard H., De Tourdonnet S., Bertrand M., Caneill J. 2009. Integration of soil structure variations with time and space into models for crop management. Agronomy Sust. Dev. 29, 135–142 10.1051/agro:2008052 (doi:10.1051/agro:2008052) [DOI] [Google Scholar]
- 5.Or D., Smets B. F., Wraith J. M., Dechesne A., Friedman S. P. 2007. Physical constraints affecting bacterial habitats and activity in unsaturated porous media—a review. Adv. Water Resour. 30, 1505–1527 10.1016/j.advwatres.2006.05.025 (doi:10.1016/j.advwatres.2006.05.025) [DOI] [Google Scholar]
- 6.Ruamps L. S., Nunan N., Chenu C. 2011. Microbial biogeography at the soil pore scale. Soil Biol. Biochem. 43, 280–286 10.1016/j.soilbio.2010.10.010 (doi:10.1016/j.soilbio.2010.10.010) [DOI] [Google Scholar]
- 7.Coleman D. C., Crossley D. A., Hendrix P. F. 2004. Fundamentals of soil ecology, 2 edn. London, UK: Academic Press [Google Scholar]
- 8.O'Donnell A. G., Young I. M., Rushton S. P., Shirley M. D., Crawford J. W. 2007. Visualization, modelling and prediction in soil microbiology. Nat. Rev. Microbiol. 5, 689–699 10.1038/nrmicro1714 (doi:10.1038/nrmicro1714) [DOI] [PubMed] [Google Scholar]
- 9.van Breeman N. 1993. Soils as biotic constructs favouring net primary productivity. Geoderma 57, 183–211 10.1016/0016-7061(93)90002-3 (doi:10.1016/0016-7061(93)90002-3) [DOI] [Google Scholar]
- 10.Dupraz C., Pattisina R., Verrecchia E. P. 2006. Translation of energy into morphology: simulation of stromatolite morphospace using a stochastic model. Sedimentary Geol. 185, 185–203 10.1016/j.sedgeo.2005.12.012 (doi:10.1016/j.sedgeo.2005.12.012) [DOI] [Google Scholar]
- 11.Phillips J. D. 2009. Soils as extended composite phenotypes. Geoderma 149, 143–151 10.1016/j.geoderma.2008.11.028 (doi:10.1016/j.geoderma.2008.11.028) [DOI] [Google Scholar]
- 12.Feeney D. S., Crawford J. W., Daniell T., Hallett P. D., Nunan N., Ritz K., Rivers M., Young I. M. 2006. Three-dimensional microorganization of the soil-root-microbe system. Microb. Ecol. 52, 151–158 10.1007/s00248-006-9062-8 (doi:10.1007/s00248-006-9062-8) [DOI] [PubMed] [Google Scholar]
- 13.Whalley W. R., Riseley B., Leeds-Harrison P. B., Bird N. R. A., Leech P. K., Adderley W. P. 2005. Structural differences between bulk and rhizosphere soil. Eur. J. Soil Sci. 56, 353–360 10.1111/j.1365-2389.2004.00670.x (doi:10.1111/j.1365-2389.2004.00670.x) [DOI] [Google Scholar]
- 14.Tisdall J. M., Oades J. M. 1982. Organic-matter and water-stable aggregates in soils. J. Soil Sci. 33, 141–163 10.1111/j.1365-2389.1982.tb01755.x (doi:10.1111/j.1365-2389.1982.tb01755.x) [DOI] [Google Scholar]
- 15.Brown C. D., Hollis J. M., Bettinson R. J., Beulke S., Fryer C. J. 1997. Pesticide mobility: lysimeter study to validate the relative leaching potential of UK soils. Research Report for MAFF Project PL0510 to the UK Ministry of Agriculture, Food and Fisheries. Soil Survey and Land Research Centre, Cranfield University, Bedfordshire, UK. [Google Scholar]
- 16.Anderson T.-H., Domsch K. H. 1978. Mineralization of bacteria and fungi in chloroform-fumigated soils Soil. Biol. Biochem. 10, 215–221 10.1016/0038-0717(78)90099-8 (doi:10.1016/0038-0717(78)90099-8) [DOI] [Google Scholar]
- 17.Deacon L. J., Grinev D., Crawford J. W., Harris J. A., Ritz K., Young I. M. 2008. Simultaneous preservation of soil structural properties and phospholipid profiles: a comparison of three drying techniques. Pedosphere 18, 284–287 10.1016/S1002-0160(08)60018-1 (doi:10.1016/S1002-0160(08)60018-1) [DOI] [Google Scholar]
- 18.Crawford J. W., Verrall S., Young I. M. 1997. The origin and loss of fractal scaling in simulated soil aggregates. Eur. J. Soil Sci. 48, 643–650 10.1046/j.1365-2389.1997.00118.x (doi:10.1046/j.1365-2389.1997.00118.x) [DOI] [Google Scholar]
- 19.Williams H. T. P., Lenton T. M. 2008. Environmental regulation in a network of simulated microbial ecosystems. Proc. Natl Acad. Sci. USA 105, 10 432–10 437 10.1073/pnas.0800244105 (doi:10.1073/pnas.0800244105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X. X., Deeks L. K., Bengough A. G., Crawford J. W., Young I. M. 2005. A mass balance based numerical method for the fractional advection–dispersion equation: theory and application. J. Hydrol. 306, 59–70 10.1016/j.jhydrol.2004.08.039 (doi:10.1016/j.jhydrol.2004.08.039) [DOI] [Google Scholar]
- 21.Mille-Lindblom C., Fischer H., Tranvik L. J. 2006. Antagonism between bacteria and fungi: substrate competition and a possible tradeoff between fungal growth and tolerance towards bacteria. Oikos 113, 233–242 10.1111/j.2006.0030-1299.14337.x (doi:10.1111/j.2006.0030-1299.14337.x) [DOI] [Google Scholar]
- 22.Batchelor M. T., Burne R. V., Henry B. I., Jackson M. J. 2004. A case for biotic morphogenesis of coniform stromatolites. Phys. A Stat. Mech. Appl. 337, 319–326 10.1016/j.physa.2004.01.065 (doi:10.1016/j.physa.2004.01.065) [DOI] [Google Scholar]
- 23.Harris J. 2009. Soil microbial communities and restoration ecology: facilitators or followers? Science 325, 573–574 10.1126/science.1172975 (doi:10.1126/science.1172975) [DOI] [PubMed] [Google Scholar]
- 24.Frostegard A., Tunlid A., Baath E. 1991. Microbial biomass measured as total lipid phosphate in soils of different organic content. J. Microb. Methods 14, 151–163 10.1016/0167-7012(91)90018-L (doi:10.1016/0167-7012(91)90018-L) [DOI] [Google Scholar]
- 25.Bligh E. G., Dyer W. J. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 10.1139/o59-099 (doi:10.1139/o59-099) [DOI] [PubMed] [Google Scholar]
- 26.Liu W. T., Marsh T. L., Cheng H., Forney L. J. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63, 4516–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schutte U. M. E., Abdo Z., Bent S. J., Shyu C., Williams C. J., Pierson J. D., Forney L. J. 2008. Advances in the use of terminal restriction fragment length polymorphism (T-RFLP) analysis of 16S rRNA genes to characterize microbial communities. Appl. Microbiol. Biotechnol. 80, 365–380 10.1007/s00253-008-1565-4 (doi:10.1007/s00253-008-1565-4) [DOI] [PubMed] [Google Scholar]
- 28.Singh B. K., Nazaries L., Munro S., Anderson I., Campbell C. 2006. Use of multiplex terminal restriction fragment length polymorphism for rapid and simultaneous analysis of different components of the soil microbial community. Appl. Environ. Microbiol. 72, 7278–7585 10.1128/AEM.00510-06 (doi:10.1128/AEM.00510-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh B. K., Thomas N. 2006. Multiplex-terminal restriction fragment length polymorphism. Nat. Protoc. 1, 2428–2433 10.1038/nprot.2006.392 (doi:10.1038/nprot.2006.392) [DOI] [PubMed] [Google Scholar]
- 30.Bird M. I., Ascough P. L., Young I. M., Wood C. V., Scott A. C. 2008. X-ray microtomographic imaging of charcoal. J. Arch. Sci. 35, 2698–2706 10.1016/j.jas.2008.04.018 (doi:10.1016/j.jas.2008.04.018) [DOI] [Google Scholar]
- 31.Hallet P. D., Feeney D. S., Bengough A. G., Rillig M. C., Scrimgeour C. M., Young I. M. 2009. Disentangling the impact of AM fungi versus roots on soil structure and water transport. Plant Soil 314, 183–196 10.1007/s11104-008-9717-y (doi:10.1007/s11104-008-9717-y) [DOI] [Google Scholar]