SUMMARY
microRNAs (miRNAs), small interfering RNAs (siRNAs), and piwi-interacting RNAs (piRNAs) impact numerous biological processes in eukaryotes. In addition to biogenesis, turnover contributes to the steady-state levels of small RNAs. One major factor that stabilizes miRNAs and siRNAs in plants as well as siRNAs and piRNAs in animals is 2′;-O-methylation on the 3′; terminal ribose by the methyltransferase HUA ENHANCER1 (HEN1) [1–6]. Genetic studies with Arabidopsis, Drosophila and zebrafish hen1 mutants show that 2′-O-methylation protects small RNAs from 3′-to-5′ truncation and 3′ uridylation, the addition of non-templated nucleotides, predominantly uridine [2, 7, 8]. Uridylation is a widespread phenomenon that is not restricted to small RNAs in hen1 mutants, and is often associated with their reduced accumulation ([7, 9, 10]; reviewed in [11]). The enzymes responsible for 3′ uridylation of small RNAs when they lack methylation in plants or animals have remained elusive. Here, we identify the Arabidopsis HEN1 SUPPRESSOR1 (HESO1) gene as responsible for small RNA uridylation in hen1 mutants. HESO1 exhibits terminal nucleotidyl transferase activity, prefers uridine as the substrate nucleotide, and is completely inhibited by 2′-O-methylation. We show that uridylation leads to miRNA degradation, and the degradation is most likely through an enzyme that is distinct from that causing the 3′ truncation in hen1 mutants.
Keywords: miRNA, siRNA, HESO1, nucleotidyl transferase, methylation, uridylation
RESULTS AND DISCUSSION
Loss of function in HESO1 suppresses the morphological defects of a partial loss-of-function hen1 mutant
We hypothesized that a terminal nucleotidyl transferase uridylates miRNAs in Arabidopsis hen1 mutants and that uridylation serves as a signal to trigger miRNA degradation. Hence, we predicted that loss of function in this nucleotidyl transferase gene would lead to increased miRNA accumulation in hen1 mutants and may rescue the morphological phenotypes associated with reduced miRNA accumulation in hen1. To identify the nucleotidyl transferase, we first searched for Arabidopsis proteins with sequence similarities to known nucleotidyl transferases with uridylation activity such as the human TUT4 [12], fission yeast cid-1 [13], and Chlamydomonas MUT-68 [9]. Ten putative Arabidopsis nucleotidyl transferases of unknown biological function were identified. Next, we obtained T-DNA mutants in each of the ten putative nucleotidyl transferase genes and crossed each mutant with hen1-8, a partial loss-of-function hen1 mutant in the Col accession [14], the same accession in which the nucleotidyl transferase mutants were generated. We found that only the mutation in At2g39740 (Figures S1A and S1B) partially rescued the morphological phenotypes of hen1-8 (Figures 1A and S1C). Therefore, we named this gene HEN1 SUPPRESSOR1 (HESO1). The hen1-8 heso1-1 double mutant was larger and had better fertility than the hen1-8 single mutant (Figures 1A and S1C). To ensure that the heso1-1 mutation was solely responsible for the phenotypic rescue of hen1-8, we introduced a HESO1 genomic fragment fused in-frame to GFP (pHESO1:HESO1-GFP) into the hen1-8 heso1-1 double mutant. Many independent transgenic lines showed phenotypes that resembled hen1-8 mutants (Figure S1C), indicating that it was the heso1-1 mutation that suppressed the defects of hen1-8. Some transgenic lines showed phenotypes even more severe than hen1-8 mutants. It is possible that these lines had higher levels of HESO1 expression, which enhanced hen1-8 defects.
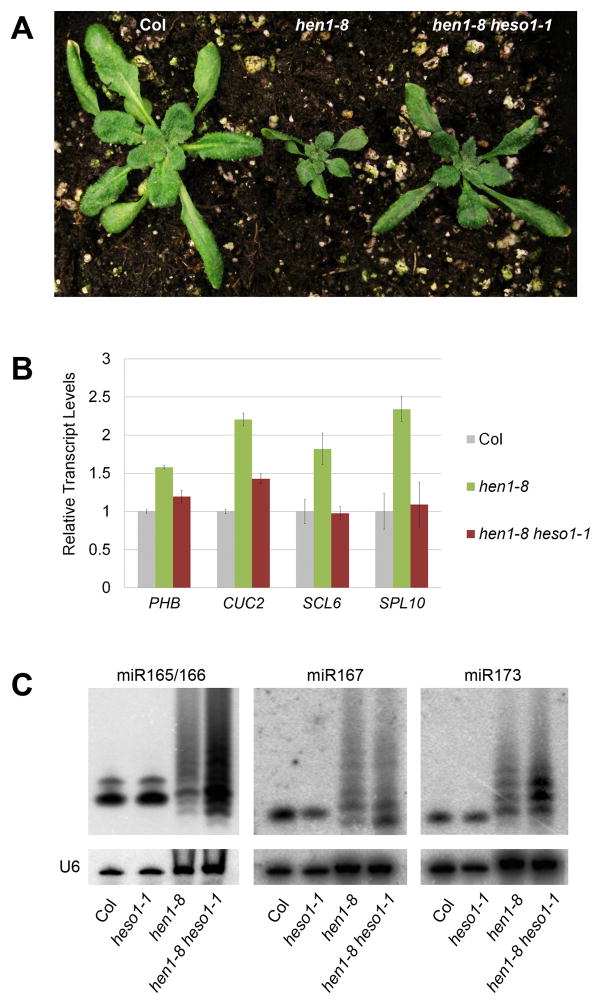
Figure 1.
The heso1-1 mutation partially rescues the developmental and molecular defects of hen1-8 plants. (A) 20-day-old plants of wild-type (Col), hen1-8, and hen1-8 heso1-1 genotypes, as indicated. (B) Relative transcript levels from four miRNA target genes in the three genotypes as determined by real-time RT-PCR. The de-repression of the genes in hen1-8, due to reduced miRNA accumulation, was fully or partially rescued by heso1-1. PHB, CUC2, SCL6, and SPL10 are targets of miR165/166, miR164, miR171, and miR156/157, respectively. (C) The accumulation of three miRNAs in the four genotypes as determined by northern blotting. Note that 50 μg of total RNA was used for hen1-8 and hen1-8 heso1-1 whereas 5 μg of total RNA was used for Col and heso1-1.
Loss of function in HESO1 results in an increase in miRNA abundance
Suppression of the morphological defects of hen1-8 by heso1-1 suggested that miRNA activities were partially rescued by heso1-1. One activity of plant miRNAs is the cleavage of their target transcripts to lead to their degradation. We examined the levels of four miRNA target transcripts in wild type, hen1-8, and hen1-8 heso1-1 by real-time RT-PCR. As expected, in hen1-8, these transcripts accumulated to higher levels as compared to wild type (Figure 1B), due to the reduced levels of miRNAs [14]. Indeed, the increase in the transcript levels in hen1-8 was partially or fully rescued by heso1-1 (Figure 1B).
Next, we tested whether the heso1-1 mutation resulted in higher abundance of miRNAs. Northern blotting was performed to detect miR166, miR167, and miR173 in wild type, heso1-1, hen1-8, and hen1-8 heso1-1. Like other hen1 alleles [8], the hen1-8 mutation resulted in both a reduction in miRNA levels and the accumulation of heterogeneous species - bands corresponding to 3′ truncated as well as 3′ tailed species were present (Figure 1C). The heso1-1 mutation did not affect miRNA accumulation in the wild-type background, but it resulted in an obvious increase in the abundance of the heterogeneous miRNA species in the hen1-8 background (Figure 1C). The increase in miRNA levels in the double mutant was rescued by the pHESO1::HESO1-GFP transgene (Figure S1D). For miR167 and miR173, it was also apparent that the increase in abundance was restricted to normal-sized species, 3′ truncated species, or species with short tails (Figure 1C). This suggested that 3′ tailing was reduced in the hen1-8 heso1-1 double mutant relative to hen1-8.
Our previous studies show that mutations in RNA polymerase IV (Pol IV) subunit genes, NRPD1 and NRPD2, rescue the morphological, as well as miRNA accumulation, defects of a weak hen1 allele, hen1-2, which harbors the same molecular lesion as hen1-8 but is in the Landsberg accession [14]. Pol IV is a key factor in the biogenesis of a predominant class of endogenous siRNAs, the 24 nucleotide (nt) siRNAs [15–18]. As natural substrates of HEN1 [8], the 24 nt siRNAs compete with miRNAs for methylation by the mutant HEN1-2 protein such that elimination of these siRNAs in nrpd1 or nrpd2 mutants allows miRNAs to gain methylation and thus increase in abundance in the hen1-2 background. With β-elimination followed by northern blotting [6], miR165/166 and miR173 were found to be unmethylated in both hen1-8 and hen1-8 heso1-1 (Figure S1E). Therefore, unlike nrpd1 or nrpd2 mutations, the heso1-1 mutation led to an increase in miRNA accumulation without affecting their methylation status.
Loss of function in HESO1 results in reduced 3′ uridylation without affecting 3′ truncation
Although northern blotting suggested that the heso1-1 mutation caused a reduction in 3′ tailing, the concomitant increase in miRNA accumulation made it difficult to determine the degree of reduction in 3′ tailing. To obtain a global and quantitative view of 3′ modifications of miRNAs, we constructed small RNA libraries from wild type, heso1-1, hen1-8, and hen1-8 heso1-1 and subjected them to deep sequencing. Two biological replicates were performed for each genotype. An algorithm was developed to classify reads corresponding to a known miRNA into four categories: full-length, 3′ truncated only, 3′ tailed only, and 3′ truncated and tailed (see Experimental Procedures).
We first focused on the status of 3′ tailing of miRNAs. For each biological replicate, 44 miRNAs that were represented by at least 50 transcripts per million (TPM) in all four libraries were included in the analysis. 3′ tailing was quantified for each miRNA as a percentage of (3′ tailed only reads + 3′ truncated and tailed reads)/total reads. In this analysis, the lengths of the tails were not taken into account as long as the read had at least one non-templated nucleotide, it was considered a tailed species. For each of the miRNAs, representatives of which are shown in Figure 2A (replicate 2) and Figure S2A (replicate 1), the percentage of tailed species was reduced in hen1-8 heso1-1 relative to hen1-8, indicating that HESO1 was partially responsible for 3′ tailing in hen1-8. The heso1-1 mutation did not affect the tailing status of most miRNAs in the wild-type background, with the exception of miR319a, which showed higher levels of tailing relative to other miRNAs in wild type (Figures 2A and S2A).
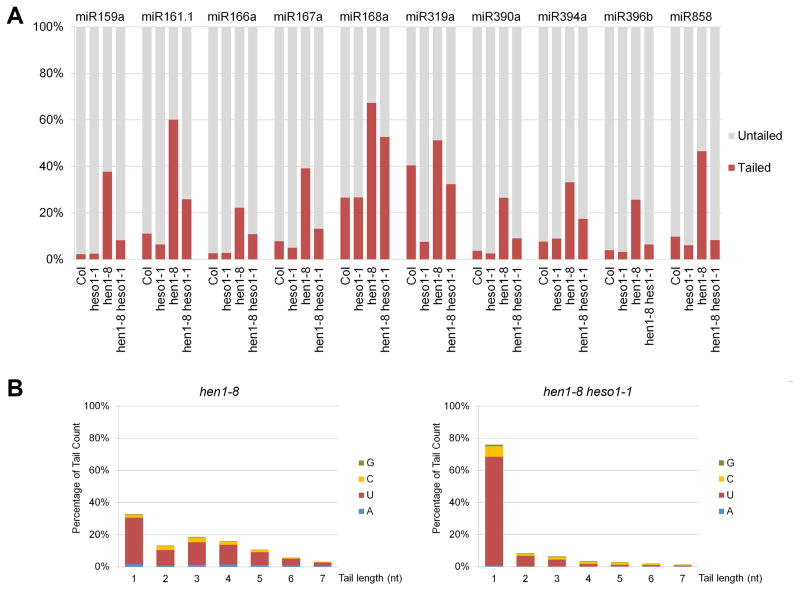
Figure 2.
The heso1-1 mutation reduces 3′ uridylation of miRNAs in hen1-8 as revealed by small RNA high throughput sequencing. Small RNA reads corresponding to known miRNAs were categorized into four classes: full-length (class 1), tailed only (full-length reads plus tails) (class 2), truncated only (class 3), and truncated and tailed (class 4). (A) The proportion of tailed and untailed reads for ten miRNAs in hen1-8 and hen1-8 heso1-1. The proportions of tailed and untailed species were calculated as %(sum of classes 2 and 4 read numbers/total read number) and %(sum of classes 1 and 3 read numbers/total read number), respectively. Data from the second biological replicate were used for the calculations here. Similar results were obtained from biological replicate one (Figure S2A). (B) Tail length distribution and nucleotide frequencies in the tails of miR166a. All tailed miR166a reads (classes 2 and 4) were analyzed for frequencies of tail length. The proportions of miR166a species with tails of 1-7 nt are shown. Nucleotide frequencies in the tails were calculated as % (number of a nucleotide in the tail/tail length).
We next examined the nature of the non-templated nucleotides in the tails and the lengths of the tails. For each miRNA, all tailed reads (both full-length species with tails and 3′ truncated species with tails) were categorized by tail length, and the frequencies of the four nucleotides in the tails were calculated. Although the tailing patterns differed to some extent among the miRNAs, two observations were obvious (Figures 2B and S2B). First, uridine was the predominant nucleotide of the tails in hen1-8 as previously reported for other hen1 alleles [8]. Second, there was a shift towards shorter tails in hen1-8 heso1-1 as compared to hen1-8. For miR166a and many other miRNAs, tails of 1 to 7 nt were found in hen1-8, but 1 nt tails were the most predominant in hen1-8 heso1-1 (Figures 2B and S2B). These results demonstrated that HESO1 was responsible for 3′ uridylation of miRNAs in hen1.
We next examined the status of 3′ truncation in relationship to 3′ tailing by HESO1 for all 44 miRNAs abundantly represented in the libraries. We found similar results for nearly all miRNAs; ten representative miRNAs from the two biological replicates are shown in Figures 3 and S3. In wild type and the heso1-1 single mutant, in which miRNA methylation was unaffected, most miRNA reads belonged to the full-length category. In hen1-8, the lack of methylation led to an increase in truncated only, tailed only, and truncated and tailed species. In hen1-8 heso1-1, 3′ tailing was greatly reduced, such that species with 0 or 1 nt tails were most abundant. Intriguingly, reads representing 3′ truncation were largely unaffected by the heso1-1 mutation, which was inconsistent with the hypothesis that 3′ uridylation triggers 3′ truncation.
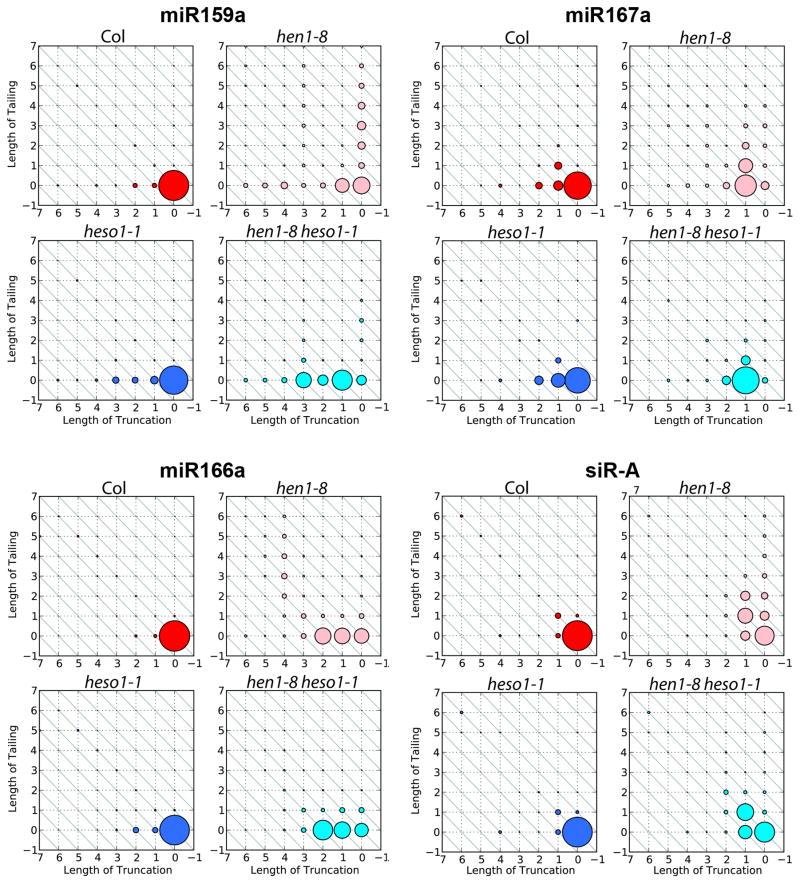
Figure 3.
The heso1-1 mutation reduces 3′ tailing of three miRNAs and an siRNA without much effects on 3′ truncation in hen1-8. The status of 3′ truncation and/or 3′ tailing for each small RNA is represented by a two dimensional matrix in which the X-axis represents the extents of 3′ truncation, and the Y-axis represents the extents of 3′ tailing. The sizes of the circles indicate the relative abundance of the small RNA species. In hen1-8, both 3′ truncation and 3′ tailing occurred at a much higher frequency than in wild type. In hen1-8 heso1-1, 3′ tailing was drastically reduced but 3′ truncation was largely unaffected. Data from biological replicate two were used for the analysis here. Results from biological replicate one as well as results for more small RNAs from biological replicate two are shown in Figure S3.
Endogenous siRNAs are also 3′ uridylated in hen1 mutants [8]. To determine whether HESO1 was responsible for the uridylation of 24 nt siRNAs, we focused on three abundantly represented 24 nt siRNAs (Table S1). All three siRNAs were truncated and tailed in hen1-8; In hen1-8 heso1-1, an obvious reduction in 3′ tailing was observed but 3′ truncated species were still present (Figures 3 and S3). Therefore, siRNAs and miRNAs were similarly affected by loss of function in HESO1.
HESO1 exhibits terminal nucleotidyl transferase activity
The above genetic evidence suggested that HESO1 is responsible for 3′ uridylation of unmethylated miRNAs and siRNAs in hen1 mutants. We sought biochemical evidence that HESO1 uridylated small RNAs with a recombinant His-HESO1 protein produced in E. coli. His-HESO1 was incubated with an RNA oligonucleotide corresponding to unmethylated miR173 that was radiolabeled at its 5′ end. HESO1 was able to add a U tail to miR173 that lengthened with time and reached up to approximately 60 nt after 40 min of incubation (Figure 4, lanes 1–6). To rule out that the terminal nucleotidyl transferase activities were due to a contaminating protein from E. coli, we mutated two aspartate residues in His-HESO1 that are part of the metal binding pocket in the nucleotidyl transferase domain to alanine (Figure S4). The mutant His-HESO1m protein was similarly purified and assayed for activity. The His-HESO1m protein was unable to add a U tail to unmethylated miR173 (Figure 4, lanes 13–14).
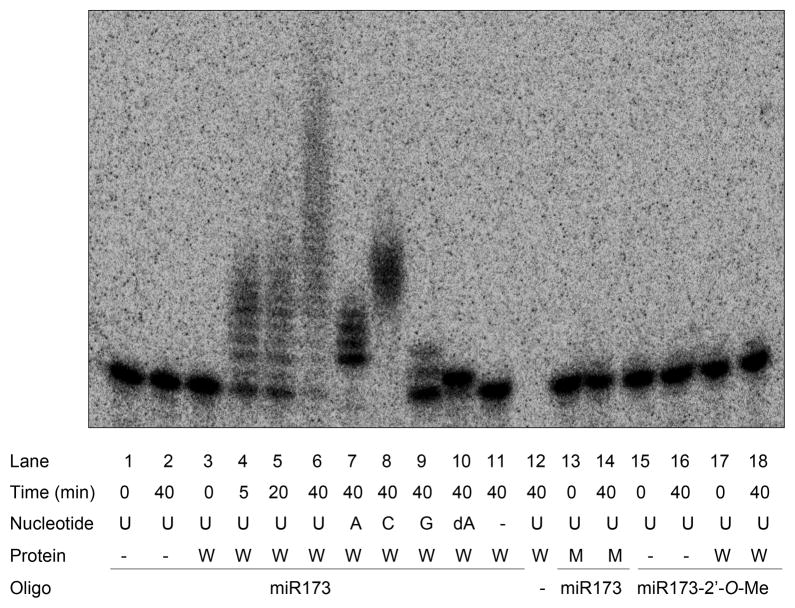
Figure 4.
HESO1 exhibited terminal nucleotidyl transferase activity. The nucleotidyl transferase assay was conducted with 5′ radiolabeled miR173 without or with 2′-O-methylation. Recombinant wild-type (W) or mutant His-HESO1, in which two catalytic residues were mutated (M; Figure S4), was incubated with the miRNA in the presence of various nucleotide triphosphates. Nucleotidyl transferase activity is represented by the presence of higher molecular weight bands relative to the input miRNA. The “-” signs indicate the absence of the nucleotide triphosphate, protein, or RNA oligonucleotide.
Next, we examined the enzymatic properties of HESO1. HESO1 was able to use ATP, CTP, or GTP, but the tail lengths with these nucleotides were much shorter than U tails, indicating that HESO1 preferred uridine (Figure 4, lanes 7–9). When dATP was used in the reaction, only one nucleotide was added (Figure 4, lane 10), suggesting that the 3′ OH of the substrate RNA was the site of phosphodiester bond formation. More importantly, when 2′-O-methylated miR173 was incubated with HESO1 in the presence of UTP, no tailing was observed (Figure 4, lanes 15–18). This indicated that HESO1 activity was completely inhibited by 2′-O-methylation.
CONCLUSIONS
Genetic and biochemical evidence in this study supports HESO1 as the nucleotidyl transferase that uridylates miRNAs and siRNAs when they lack 2′-O-methylation. The large increase in miRNA abundance in hen1-8 heso1-1 vs. hen1-8 demonstrates that uridylation leads to degradation. The fact that the heso1-1 mutation caused a large reduction in 3′ uridylation without strong effects on 3′ truncation suggests that 3′ truncation observed in hen1 mutants is largely independent of 3′ uridylation. Two implications from these observations are: 1) uridylation triggers the degradation of small RNAs by a nuclease distinct from the one that causes 3′ truncation; and 2) if the nuclease that degrades uridylated small RNAs is a 3′-to-5′ exonuclease, then it should be highly processive such that few or no degradation intermediates (i.e. 3′ truncated species) would be present in vivo. The lack of an effect on miRNA accumulation by heso1-1 in wild type suggests that HESO1 may not be part of the normal small RNA degradation machinery. However, given the presence of other nucleotidyl transferase genes with potentially overlapping functions, a role of HESO1 in miRNA degradation in wild-type plants cannot be excluded. Although HESO1 activity is completely inhibited by 2′-O-methylation, it is possible that HESO1 acts cooperatively with SDN1, which truncates small RNAs [19], or another nucleotidyl transferase that is not inhibited by 2′-O-methylation, to degrade miRNAs. The activities of these enzymes would generate unmethylated miRNAs on which HESO1 can uridylate.
EXPERIMENTAL PROCEDURES
Supplementary Material
Acknowledgments
We thank Elizabeth Luscher for comments on this manuscript and Dr. Detlef Weigel for sharing the pJL-blue plasmid. We thank Lei Gao, Tammy Luu and Renee Zamora for technical assistance. This work was supported by grants from National Science Foundation (NSF MCB-1021465) to X.C., Chinese National Science Foundation (30970265) to B.M., and NSF (0701745) to B.C.M. Thanh D. was supported by an NSF ChemGen IGERT fellowship (DGE0504249), and Jixian Z. was supported by a University of Delaware graduate fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 2.Kamminga LM, Luteijn MJ, den Broeder MJ, Redl S, Kaaij LJ, Roovers EF, Ladurner P, Berezikov E, Ketting RF. Hen1 is required for oocyte development and piRNA stability in zebrafish. The EMBO journal. 2010;29:3688–3700. doi: 10.1038/emboj.2010.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kirino Y, Mourelatos Z. The mouse homolog of HEN1 is a potential methylase for Piwi-interacting RNAs. RNA. 2007;13:1397–1401. doi: 10.1261/rna.659307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurth HM, Mochizuki K. 2′-O-methylation stabilizes Piwi-associated small RNAs and ensures DNA elimination in Tetrahymena. RNA. 2009;15:675–685. doi: 10.1261/rna.1455509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saito K, Sakaguchi Y, Suzuki T, Suzuki T, Siomi H, Siomi MC. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi- interacting RNAs at their 3′ ends. Genes & development. 2007;21:1603–1608. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–1539. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J, Yang Z, Yu B, Liu J, Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ibrahim F, Rymarquis LA, Kim EJ, Becker J, Balassa E, Green PJ, Cerutti H. Uridylation of mature miRNAs and siRNAs by the MUT68 nucleotidyltransferase promotes their degradation in Chlamydomonas. Proc Natl Acad Sci U S A. 2010;107:3906–3911. doi: 10.1073/pnas.0912632107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Wolfswinkel JC, Claycomb JM, Batista PJ, Mello CC, Berezikov E, Ketting RF. CDE-1 affects chromosome segregation through uridylation of CSR-1-bound siRNAs. Cell. 2009;139:135–148. doi: 10.1016/j.cell.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 11.Kim YK, Heo I, Kim VN. Modifications of small RNAs and their associated proteins. Cell. 2010;143:703–709. doi: 10.1016/j.cell.2010.11.018. [DOI] [PubMed] [Google Scholar]
- 12.Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 13.Rissland OS, Mikulasova A, Norbury CJ. Efficient RNA polyuridylation by noncanonical poly(A) polymerases. Mol Cell Biol. 2007;27:3612–3624. doi: 10.1128/MCB.02209-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu B, Bi L, Zhai J, Agarwal M, Li S, Wu Q, Ding SW, Meyers BC, Vaucheret H, Chen X. siRNAs compete with miRNAs for methylation by HEN1 in Arabidopsis. Nucleic Acids Res. 2010;38:5844–5850. doi: 10.1093/nar/gkq348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- 16.Kanno T, Huettel B, Mette MF, Aufsatz W, Jaligot E, Daxinger L, Kreil DP, Matzke M, Matzke AJ. Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat Genet. 2005;37:761–765. doi: 10.1038/ng1580. [DOI] [PubMed] [Google Scholar]
- 17.Onodera Y, Haag JR, Ream T, Costa Nunes P, Pontes O, Pikaard CS. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 18.Pontier D, Yahubyan G, Vega D, Bulski A, Saez-Vasquez J, Hakimi MA, Lerbs-Mache S, Colot V, Lagrange T. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes & development. 2005;19:2030–2040. doi: 10.1101/gad.348405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramachandran V, Chen X. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science. 2008;321:1490–1492. doi: 10.1126/science.1163728. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.