Abstract
During neural development, patterning, neurogenesis and overall growth are highly regulated and coordinated between different brain regions. Here, we show that primary cilia and the regulation of Gli activity, are necessary for the normal expansion of the cerebral cortex. We show that loss of Kif3a, an important functional component of primary cilia, leads to the degeneration of primary cilia, marked overgrowth of the cortex, and altered cell cycle kinetics within cortical progenitors. The G1 phase of the cell cycle is shortened through a mechanism likely involving reduced Gli3 activity and a resulting increase in expression of cyclin D1 and Fgf15. The defects in Gli3 activity alone are sufficient to accelerate cell cycle kinetics and cause the molecular changes seen in brains that lack cilia. Finally, we show that levels of full-length and repressor Gli3 proteins are tightly regulated during normal development and correlate with changes in expression of two known Shh-target genes, CyclinD1 and Fgf15, and with the normal lengthening of the cell cycle during corticogenesis. These data suggest that Gli3 activity is regulated through the primary cilium to control cell cycle length in the cortex and thus determine cortical size.
Keywords: Kif3a, cilia, Gli3, proliferation, cell cycle
INTRODUCTION
Different brain regions arise from a simple neuroepithelial tube, which even at early stages of development exhibits distinct molecular expression patterns and variable rates of proliferation, cell cycle exit, and apoptosis (Haydar et al., 1999; Donovan and Dyer, 2005). Little is known about the mechanisms that direct the differential growth of progenitors in different regions of the neuraxis. Exploring these mechanisms may provide insight into how growth rates were adjusted during evolution to enable the selective expansion of the forebrain, which coincided with increased intellectual capacity.
During development, the ventricular zone (VZ) houses mitotically active progenitor cells, each of which extends a single primary cilium into the ventricle and contacts cerebral spinal fluid (CSF) (Dahl, 1963; Rohatgi et al., 2007). Primary cilia can act as chemical or mechanical sensors, enabling cells to detect cues such as Sonic hedgehog (Shh) or the motion of fluids, as in the developing kidney (Praetorius and Spring, 2001). Most cells possess a cilium, which localizes specific receptors and signaling components in different cell types (Whitfield, 2004; Schneider et al., 2005). In spinal cord progenitors, many components of the Shh signaling pathway localize to cilia, and this localization is necessary for Shh-dependent patterning (Huangfu and Anderson, 2005; May et al., 2005; Rohatgi et al., 2007).
Recent studies have elucidated the contribution of primary cilia to Shh signaling. Shh binds to Patched (Ptc), a multipass transmembrane receptor that in its unbound and inactive state localizes to the primary cilium (Rohatgi et al., 2007). Upon binding, Ptc moves out of the cilium allowing Smo translocation to the ciliary membrane, accumulation of full-length Gli proteins, activation of Gli transcription factors, and the upregulation of Shh target genes (Murone et al., 1999; Rohatgi et al., 2007; Goetz and Anderson, 2010). Of the three mammalian Gli family members, two (Gli2 and Gli3) are proteolytically processed to produce a transcriptional repressor (GliRep) from a longer activator form (Gli full-length or GliFL) (Pan et al., 2006), which requires phosphorylation to elicit transcriptional changes in the nucleus (Humke et al., 2010; Wang et al., 2010). Many Shh signaling components depend on cilia for their proper function (Haycraft et al., 2005; Huangfu and Anderson, 2005; Liu et al., 2005; May et al., 2005). In the absence of Shh, Gli2/3 proteins are phosphorylated and processed by the proteasome to produce the repressor forms.
ENU screens and studies of Gli3 mutant mice suggest that cilia and Gli3 play important roles in forebrain development. Gli3-deficient brains are often exencephalic and show attenuated growth and abnormal patterning, in which ventrolateral gene expression expands dorsally with only a small remnant of cortical tissue remaining (Theil et al., 1999; Tole et al., 2000). Mutations that disrupt primary cilia phenocopy many of these forebrain abnormalities and reduce Gli3 proteolytic processing (Willaredt et al., 2008; Ashique et al., 2009). Because Gli3 plays sequential roles during hindbrain development (Blaess et al., 2008), here we explore the roles of cilia and Gli3 during post-patterning stages of forebrain development.
METHODS
Animals
Mice were used according to the protocols approved by the Institutional Animal Care and Use Committee of Stanford University and Weill Medical School at Cornell University, and in accordance with NIH guidelines. Timed-pregnant females were maintained in a 12 hour light/dark cycle and obtained by overnight breeding with noon of the following day considered E0.5. Conditional Kif3a mice were made by Larry Goldstein and obtained from the MMRRC (Lin et al. 2003). Gli3Δ68 mice were generated by Baolin Wang (Wang et al., 2007). Nestin-cre transgenic mice were generated by Weimann Zhong (Petersen et al., 2002). Gli3 conditional mutants were obtained from Dr. Alexandra Joyner (Blaess et al., 2008). Wildtype Swiss Webster mice were obtained from Jackson Labs.
Histochemistry and In Situ Hybridization
Radioactive in situ hybridization was carried out using S35-labeled riboprobes as described previously (Hébert et al. 2003). Standard immunohistochemical methods were used (Okada et al., 2006). A Sodium Citrate antigen-retrieval protocol was used to expose Ki67 and BrdU antigens for detection. Protein localization and cell cycle analysis were analyzed on 14 μm cryosections of 4% paraformaldehyde fixed, sucrose protected embryos. Nuclei were visualized using propidium iodide or syto11 (both Molecular Probes). All antibodies were used at 1:500 unless otherwise noted. Rabbit anti-Polaris (gift of B. Yoder), Rat anti-BrdU (Accurate Chemical & Scientific), mouse anti-Ki67 (BD Pharmingen), mouse anti-γ-tubulin (Sigma), mouse anti-β-catenin (BD Pharmingen), anti-calretinin (1:1K, Chemicon), mouse anti-TuJ1 (Covance), rabbit anti-Tbr1 (1:2K, Hevner), rabbit anti-phospho H3 (Upstate Biotechnology), chicken anti-Myc (Molecular Probes), rabbit anti-GFP (Molecular Probes), mouse anti-Nestin (BD Pharmingen).
Stereology
E13.5 embryos were decapitated in cold PBS, heads were fixed in 4% PFA overnight, cryopreserved in 30% sucrose overnight, and embedded in OCT (Tissuetek). Heads were cryosectioned and 14 μm sections were collected on Fisherbrand Superfrost Plus slides. Sections were dried 2–3 days at RT and stained with cresyl violet. Every 12th section was imaged, photomontages were created using Adobe Photoshop CS, and 4 cortical areas were outlined based on cell density and cellular organization: ventricular zone (VZ), intermediate zone/cortical plate (IZ/CP), and ectopic ventricular zone (ectopic VZ) (found only in Kif3acko mice). ImageJ software was used to calculate areas of traced regions and volumes were calculated.
Scanning Electron Microscopy
Embryos were dissected in warm PBS, cortical hemispheres were removed, placed in standard EM fixative (2% Gluteraldehyde, 4% Paraformaldehyde in 100 mM Na Cacodylate, pH 7.2) and any tissue impeding a clear view to the cortical apical surface was removed. Standard processing for scanning electron microscopy was used. Samples were imaged on a Hitachi S-3400N VP-SEM.
Western blotting
All Western blots were conducted on E12.5 or E14.5 dorsal cortical tissue. Embryos were dissected in cold PBS. Cortical caps from an individual embryo (region of cortical hemispheres just dorsal to the lateral ganglionic eminence to the medial curve of the cortex) were collected, placed in an eppendorf tube and snap frozen in liquid nitrogen. After genotyping, 2–4 sets of caps (depending on age) were pooled, homogenized on dry ice and ground to fine powder. Modified RIPA buffer (150 mM NaCl, 10 mM Tris-Hcl, 0.1% SDS, 1% TritonX, 1% Sodium deoxycholate, 5 mM EDTA; pH 7.5) and protease and phosphatase inhibitor cocktails were added to 8x (both Sigma) and sample was pulled through a 30-gauge syringe 5 times. A BCA assay (Pierce) was performed as per manufacturer’s instructions on all samples to normalize protein concentrations, 4x NuPAGE LDS buffer (Invitrogen) was added to 1x, samples were aliquoted and stored at −80°C until use.
Standard methods for Western blotting were used. Primary antibodies were diluted in 5% casein, 1% BSA in PBST containing the antimicrobial agent thimerisol at 0.05%. Secondary antibodies conjugated to HRP (1:8K–1:10K) from Jackson ImmunoResearch were used and detected using ECL (Amersham). The following primary antibodies were used: Rabbit anti-Ptch (1:500, gift of R. Rohatgi), rabbit anti-Gli3 (1:200, B. Wang), rabbit anti-Cyclin D1 (1:1K, Santa Cruz Biotechnology), mouse anti-PCNA (1:1K, Sigma), mouse anti-TuJ1 (1:1K, Covance), mouse anti-tubulin (1:1K, Sigma), rabbit monoclonal anti-phospho p42/44 (ERK) (1:1.5K, Cell Signaling), activated-Notch (1:1K Cell Signaling), and activated β-catenin (1:400, Upstate).
Flow cytometry
The dorsal cerebral cortex was collected from individual animals, transferred to serum-free medium on ice, resuspended in Trypsin-EDTA (Invitrogen) for 10 min at 37°C, mechanically dissociated into a single-cell suspension and fixed in ice-cold 70% ethanol. Fixed cells were stored at −20°C for up to 1 week and animals of the same genotype were pooled and stained with 40 μg/ml Propidium Iodide in phosphate-buffered saline containing 10 μg/ml Rnase for 30–45 min. Samples were filtered through mesh caps of flow cytometry tubes and run immediately on the cell sorter (Flasher II, Stanford Shared FACS Facility).
In utero electroporation
Standard mouse in utero electroporation techniques were used (Ohtsuka et al., 2001).
RESULTS
Primary cilia are lost in the brains of Kif3a mutants
Prior studies have shown that loss or attenuation of primary cilia signaling leads to abnormal forebrain patterning and disruption of cortical integrity (Besse et al., 2011; Willaredt et al., 2008; Ashique et al., 2009). To examine the role of primary cilia in the developing forebrain after the completion of early patterning, we utilized the Cre-loxP system and a conditional (floxed) allele of Kif3a (Kif3af) (Marszalek et al., 1999), which encodes an essential component of Kinesin II, the main anterograde motor of the primary cilium. In Nestin-Cre transgenic mice, Cre is expressed in CNS progenitors by E10.5, thus enabling us to eliminate Kif3a gene function in cortical progenitors at the start of neurogenesis. Analysis of Kif3a expression by Western blot and in situ hybridization showed a complete loss of the Kif3a gene product by E13.5 in Nestin-Cre/+; Kif3af/f (referred to as Kif3acko) brains compared to Nestin-Cre/+; Kif3af/+ (referred to as Kif3ahet) and Kif3af/f or Kif3af/+ (referred to as Kif3awt) cortices (data not shown).
To assess the presence of primary cilia in cortical progenitors, we first performed immunohistochemistry on E13.5 coronal sections and visualized the subcellular localization of γ-tubulin, which marks the basal body, and the intraflagellar transport (IFT) protein Polaris, which marks the cilium (Taulman et al., 2001). Consistent with previous analyses, γ-tubulin immunoreactivity appeared as puncta along the apical surfaces of VZ cells (Chenn et al., 1998) and at the spindle poles of mitotic figures, where centrosomes are located (Hinds and Ruffett, 1971). In contrast, polaris immunoreactivity labeled the cilia, which appeared as small fingerlike structures that extended from the γ-tubulin staining into the ventricle (Suppl. Fig. 1A–C). Punctate polaris immunoreactivity was also seen throughout the VZ, likely reflecting the labeling of transport vesicles. The localization of γ-tubulin in Kif3ahet and Kif3acko mice was identical to that in wildtype mice, indicating the basal body was localized normally in mutant embryos (Suppl. Fig. 1G,K). Although polaris immunoreactivity in Kif3acko brains was still localized apically, in a domain of intense β-catenin staining, the polaris staining did not extend into the ventricle (Suppl. Fig. 1D–F, H–J).
These data strongly suggested that centrosomes are correctly positioned in Kif3acko cortical progenitors, but that primary cilia were no longer present. Scanning electron microscopy (SEM) of the apical (lumenal) surface of the cortical VZ at E12.5 and E14.5 in Kif3awt and Kif3ahet embryos confirmed the lack of cilia in Kif3a mutants (Suppl. Fig. 1L,M,O,P). At E12.5, most Kif3awt cells had a single small projection (the primary cilium) that extended about 0.75 μm into the ventricle while the primary cilia of Kif3acko progenitors were largely gone; nearly all cells possessed a shorter than normal cilium, with a length around 0.1 μm, only rarely was a longer cilium seen. By E14.5, a primary cilium of wildtype length was almost never seen in Kif3acko embryos, and nearly all cilia were 0.1 μm in length or shorter (Suppl. Fig. 1Q). Together, these data demonstrate that the conditional deletion of Kif3a in the developing brain leads to a loss of primary cilia in cortical progenitor cells.
Loss of primary cilia leads to larger brains
Kif3acko mice are born in Mendelian ratios, but they die within the first few hours after birth and are phenotypically distinct from littermates, possessing a bulging forehead (not shown). The overall dorsal-ventral patterning of Kif3a mutant brains was assessed by in situ hybridization at E13.5 and revealed normal dorsal expression of Pax6, Emx1 and Emx2 in both Kif3acko and control brains (Suppl. Fig. 2A–C), while Mash1, Nkx2.1 and Dlx2 were localized normally to ventral structures (Suppl. Fig. 2D–F). Thus cells in the Kif3a mutant cortex have retained their dorsal cortical identity.
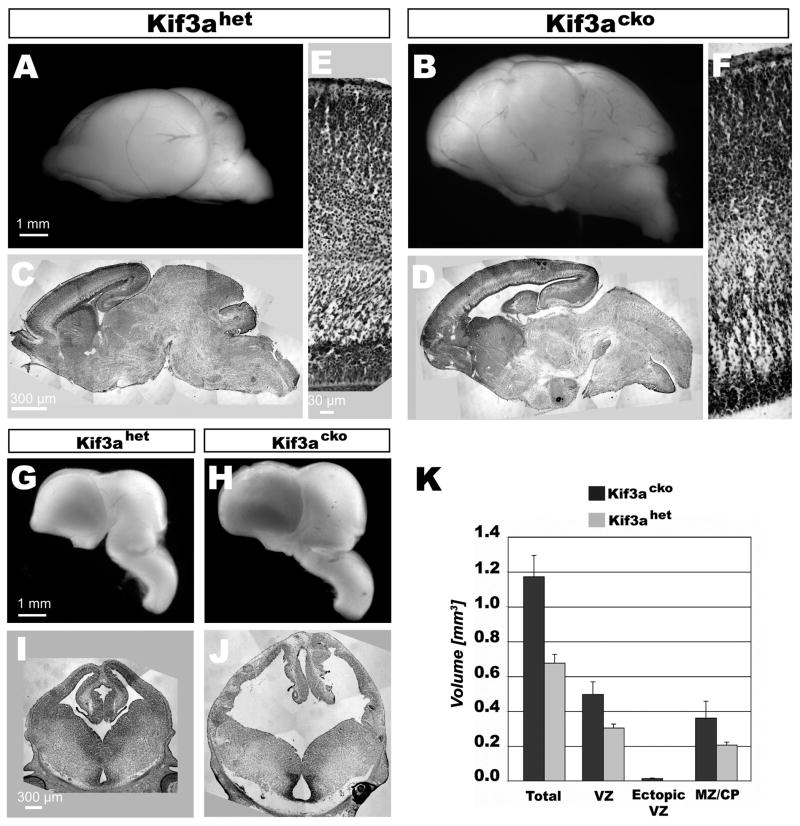
To investigate whether primary cilia play a functional role in cortical development, we analyzed the size and morphology of Kif3acko brains. The brains of Kif3a mutants at birth (P0) were larger than those of control mice, and both the size and morphology of many brain regions, including the olfactory bulbs, cerebral cortex, hippocampus, superior colliculus and cerebellum, were affected (Fig. 1A–F). We focused our subsequent analyses on the cerebral cortex, which was not only strikingly larger than in wildtype mice, but also displayed a bumpy surface that contrasted with the smooth pial surface of the normal cortex.
Figure 1. Kif3acko cortices are 1.75x larger than in Kif3ahet mice.
(A and B) Kif3acko cortices at P0 are large and bumpy. (C and D) Nissl staining of sagittal sections of Kif3acko brains reveal ectopic cell clusters in the cortical plate. (E and F) 20x magnification of region in (C and D) shows that Kif3acko cortices contain relatively normal layers in regions without ectopic cells disruptions. (G–J) Mature phenotype is present by E13.5. (K) Stereological calculation of volume increase in Kif3acko compared to Kif3ahet. Error bars are standard deviations of three brains.
Analysis of the organization of Kif3acko cerebral hemispheres revealed multiple defects: first, Kif3acko cortices appeared much larger than those of control animals (Fig. 1A,B); second, the thickness of the cortical plate varied widely but with no consistent pattern (Fig. 1C,D); third, ectopic groups of cells periodically disrupted the layered organization of the cortical plate (Fig. 1D), which was otherwise relatively normal (Fig. 1F); and fourth, the lateral ventricles appeared enlarged (Fig. 1C,D).
To assess brain size quantitatively and in a manner that excluded the effects of the ventricular enlargement, we conducted a stereological analysis to measure the volume of cortical tissue within the cerebral hemispheres. Brains were analyzed at E13.5, a time at which the increase in the size of the cortex was clearly visible (Fig. 1G–J). Coronal sections of mouse brains spanning the anterior-posterior extent of the cerebral cortex were Nissl stained to visualize its cellular organization. For consistency, cortical volume was determined by measuring from the dorsalmost point of the cortex to the junction between the cortex and the lateral ganglionic eminence. To glean more information regarding the affects of the mutation on different cell populations, the cortex was divided into three subdomains, identifiable through their characteristic cellular organization: the ventricular zone (VZ), which possesses mitotically-active progenitors; the ectopic ventricular zone (ectopic VZ), found only in the mutants and containing proliferative cells outside of the VZ; and the cortical plate and intermediate zone (IZ/CP), which contain postmitotic neurons. The volume of each subdomain was calculated from these measurements, revealing that each of the domains in Kif3acko animals showed a relative increase of ~1.75-fold increase in volume (Fig. 1K). Despite the increase in volume, the densities and sizes of cells in the VZ and the IZ/CP were similar between Kif3ahet and Kif3acko mice (data not shown), suggesting that the increased volume of the Kif3a mutant brain is likely the result of a proportional increase in the numbers of both progenitor cells and neurons.
Changes in polarity, cell death, or neurogenesis cannot explain the increased brain size in Kif3a mutants
The disruption of genes that regulate cell polarity can alter many processes in neural progenitor cells, including proliferation and adhesion (Klezovitch et al., 2004; Cappello et al., 2006; Imai et al., 2006; Lien et al., 2006). However, the overall polarity, junctional contacts, and morphology of progenitor cells were intact in Kif3acko brains (Suppl. Fig. 3). Despite this, the ventricular epithelium of Kif3a mutants sometimes invaginated toward the pia (Suppl. Fig. 4A–C), and in adjacent sections rosette-like structures were observed in the cortical plate (Suppl. Fig. 4D–F). Similar structures have been observed in Gli3 mutants (Fotaki et al., 2006) and other Shh-pathway mutants. Apart from their abnormal positions, the rosettes maintained the normal cellular organization of the developing cerebral wall, with actively dividing cells adjacent to a lumen and TuJ1-positive cells located toward the periphery (Suppl. Fig. 4A–F), suggestive of ongoing neurogenesis. Analysis of apical markers (Suppl. Fig. 4D–F) and SEM of the ventricular surface (Suppl. Fig. 4G,H) suggest that an expansion of the neuroepithelium in Kif3a mutants leads to invaginations of the VZ and the formation of loops or rosettes, from which young postmitotic neurons migrate outward. Because we were not able to identify a specific cause of rosette formation, we focus the remainder of our analyses on the overall expansion of cortical size that occurs in Kif3a mutants.
The enlarged brains and proportional increases in both the proliferative and postmitotic zones in Kif3acko mice suggest that primary cilia are required for the normal regulation of proliferation in cortical progenitors. However, such a phenotype might also arise if cortical progenitors that would normally undergo programmed cell death were retained in the mutant animals, or if primary cilia are required for cells to sense cues necessary for cell cycle exit. To ascertain whether the increased cortical size in Kif3acko mice is due to a reduction in cell death, we performed TUNEL staining on E13.5 tissue. In contrast to the prediction that larger brain size would be caused by a decrease in programmed cell death, Kif3acko embryos (Suppl. Fig. 5B,D,F,G) had about twice the number of TUNEL-positive cells than in Kif3ahet controls (Suppl. Fig. 5A,C,E,G). These results indicate that reduced cell death cannot explain the increased cortical volume observed in mutants.
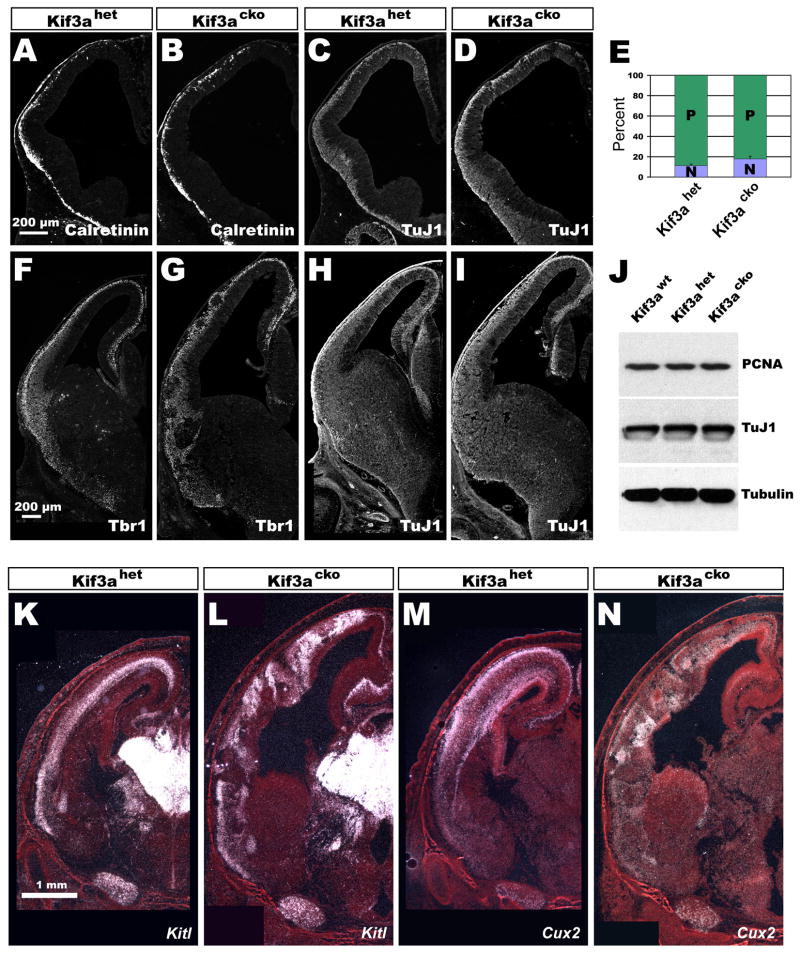
To determine whether the increase in brain size was due to a delay in neurogenesis, which might allow an expansion of the early progenitor pool, we first analyzed the expression of neuronal markers and the rate of neuronal production during early stages of corticogenesis. Immunostaining at E11.5 for calretinin and reelin, which mark early-generated Cajal-Retzius cells, and for the pan-neuronal protein TuJ1 showed that Cajal-Retzius cells and the developing preplate formed normally in Kif3acko animals (Fig. 2A–D). At E13.5, immunostaining for TuJ1 and Tbr1, a transcription factor expressed strongly by preplate and deep-layer projection neurons (Hevner et al. 2001), also revealed no differences in thickness of the cortical plate or the density of the nascent neuronal population in mutants compared to controls (Fig. 2F–I).
Figure 2. Neurogenesis occurs normally in Kif3acko cortices.
(A–D) Immunostaining of E11.5 coronal sections of Kif3ahet (A and C) and Kif3acko (B and D) brains. Calretinin labels Cajal Retzius cells and TuJ1 is a pan-neuronal marker. (E) Quit fraction analysis of E12.5 cortices. About 7% more neurons are produced in Kif3acko than Kif3ahet cortices in a 24-hour period. (F–I) Immunostaining of E13.5 coronal sections of Kif3ahet (F and H) and Kif3acko (G and I) brains. Tbr1 labels preplate and layer 6 neurons. (K–N) In situ hybridizations of E18.5 coronal sections showing that fate specification is normal in Kif3acko brains. Kitl marks deep-layer neurons and Cux2 labels superficial neurons.
To further test the hypothesis that delayed neurogenesis might contribute to the larger brain size in Kif3a mutants, we assessed the rate of neurogenesis quantitatively by determining the quit fraction, or fraction of cells that exit the cell cycle, at E12.5. A single injection of BrdU was administered to pregnant dams during their 11th day of pregnancy, 24 hrs prior to sacrifice. Brains were then stained with antibodies against Ki67, an antigen present in all proliferating cells, and BrdU, to identify the cohort of labeled cells. The quit fraction was determined by dividing the number of BrdU+;Ki67− cells by the total number of BrdU+ cells (Chenn and Walsh 2002). This analysis showed a slight but significant (p<0.02) increase in neuron generation in Kif3acko embryos (17.9% ± 2.6%) compared to Kif3ahet embryos (11.3% ± 1.5%) (Fig. 2E). Interestingly, Western blot analysis suggested that the overall ratio between progenitors and neurons was unchanged (Fig. 2J), consistent with the possibility that the overall balance between proliferation and differentiation was not altered in mutants. Although we did not directly examine whether the mutation in Kif3a altered the fraction of asymmetric divisions that produce neurons, our data are suggest the possibility that the increased quit fraction may have been caused by progenitor cells that cycled more rapidly than normal and thus underwent an additional cell cycle in Kif3acko embryos. We test this possibility below.
Since different classes of neurons are normally generated in a predictable temporal manner, we also conducted in situ hybridization to ascertain whether neurons adopted normal laminar fates in the mutants. Riboprobes specific for deep layer (e.g., Kitl: Fig. 2K,L), and superficial layer (Cux2: Fig. 3M,N) neurons revealed that both early-and late-born neurons appear to have been specified normally. However, not all cells migrated into their normal destinations, presumably because of the cellular rosettes that formed within the cerebral wall. Collectively these data indicate that a delay in neurogenesis, and an expansion of the progenitor pool at the expense of the earliest-generated neurons, was not the cause of increased cortical size in Kif3acko mice.
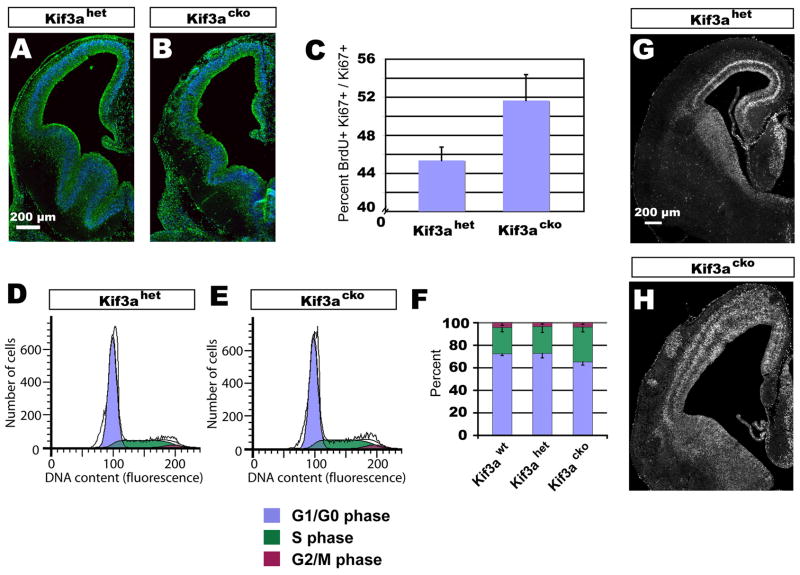
Figure 3. Kif3acko cortical progenitors cycle faster with a reduction in G1.
(A and B) Representative immunostaining of E12.5 Labeling Index analysis. Ki67 in green and BrdU in blue. (C) Kif3acko progenitors cycle faster than Kif3ahet progenitors shown by quantification of E12.5 labeling index. (D and E) Representative flow cytometry traces of progenitors sorted based on DNA content using propidium iodide and fit by ModfitLT. (F) Graph of ModfitLT values showing G1 is shorter in Kif3acko progenitors with a corresponding increase in S phase. (G and H) E14.5 Kif3acko embryos administered a BrdU pulse 24-hours earlier show three bands or BrdU + cells whereas Kif3ahet embryos only show two.
Loss of primary cilia speeds up the cell cycle
In light of the increased cell death and enhanced neurogenesis in Kif3a mutants – both of which should decrease rather than increase the overall size of the cortex – we hypothesized that mutant progenitor cells might progress more rapidly through the cell cycle, leading to increased cell numbers over time. To assess the length of the cell cycle, E12.5 embryos were pulsed for 30 min with BrdU, labeling S-phase precursor cells. A modified labeling index (LI) was calculated by staining brain sections for the cycling cell marker Ki67, counting the number of BrdU+, Ki67+ cells, then dividing this by the total number of Ki67+ progenitors. Calculating the fraction of BrdU+ cells within the Ki67+ population enables an estimate of cell cycle length, since Ki67− cells that have exited the cell cycle are excluded from the analysis (Chenn and Walsh, 2002). In addition, previous studies using cortical progenitors showed that the lengths of S, G2 and M-phase remain relatively constant while the length of G1 is variable and determines proliferation time (Caviness and Takahashi, 1995; Caviness et al., 1999). If the cell cycle is shortened, the relative fraction of cells labeled by BrdU will increase, corresponding to a decrease in the length of G1. The LI for Kif3acko embryos was 51.7% ± 2.7%, a significant increase from that observed in Kif3ahet embryos (45.4% ± 1.4%; p< 0.03), indicating an increase of almost 7% in the fraction of mutant cells in S phase (Fig. 3A–C). Although this difference might seem minor, a 7% change in the LI is predicted to decrease the total length of the cell cycle by ~1.5 hours, from roughly 10.2 hours at E12 (Takahashi et al., 1995) to 8.7 hours, assuming that the length of G1 is affected.
To independently assay changes in the cell cycle in Kif3acko mice, the dorsal cerebral cortex was dissected from E12 embryos (a stage at which the vast majority of cells are cycling progenitors), dissociated into a cell suspension, labeled with propidium iodide, and analyzed by flow cytometry based on DNA content. These studies enabled us to identify the percentage of cells in the G1/G0, S, and G2/M phases of the cell cycle. The proportion of cells in S phase was increased in Kif3acko cortices, as predicted by the LI studies above. In addition, the percentage of cells with G1/G0 DNA content was decreased proportionately (Fig. 3D–F). These data are consistent with the interpretation that Kif3a mutant progenitors were cycling more rapidly due to a shortened G1 phase of the cell cycle.
The alterations in cell cycle time predicted from the data above should result in an overall increase in the output of the VZ over time. Indeed, when progenitors were labeled with BrdU at E14.5, a stage at which the cell cycle normally spans ~15.1 hours (Takahashi et al., 1995), and embryos survived for 24 hours, we observed two bands of BrdU-labeled cells in Kif3ahet brains, one in the VZ and one in the intermediate zone (IZ) (Fig. 3G). This pattern is consistent with the interpretation that VZ cells completed one full cycle and produced a single cohort of postmitotic, migratory cells during a 24 hour period. In contrast, similar labeling experiments in Kif3acko embryos resulted in the production of three bands of BrdU+ cells (Fig. 3H), with one in the VZ and two in the IZ, suggesting that mutant progenitors went through two rounds of cell division during the course of 24 hours.
Shh target genes are upregulated in Kif3a mutants
To explore the mechanisms that link primary cilia and cell cycle length, we examined Gli3 processing due to a large degree of overlap between cilia mutants and Shh-pathway mutants and the established role of Shh influencing proliferation in the CNS (Donovan and Dyer, 2005; Fuccillo et al., 2006). However, many other signaling pathways (such as those mediated by Wnt and Notch) are also known to influence proliferation in the developing brain, some of which have been suggested to be regulated by the primary cilium. For example, overexpression of an activated form of β-catenin, a downstream component in the Wnt signaling pathway, causes a dramatic expansion of the cerebral cortex (Chenn and Walsh, 2002). Ectopic expression of the intracellular domain Notch (NotchAct) biases cell divisions toward the production of two progenitors and thus expands the progenitor pool at the expense of neurons (Gaiano et al., 2000), and both loss-and gain-of-function analyses of IGF and FGF signaling support a role in cortical size control (Vaccarino et al., 1999; Hodge et al., 2004). Recently, a cortex-specific deletion of Smo shows a reduction in cortical progenitor proliferation suggesting an active role for Shh signaling in cortical expansion (Komada et al., 2008). Additionally, mice that lack α-E-catenin show an upregulation in components of the Shh pathway (Smo, Gli1, Fgf15) in the cortex, which is enlarged due to more rapidly cycling progenitors and decreases in cell death (Lien et al., 2006). However, the latter study did not show a direct link between Shh signaling and cortical size in normal development.
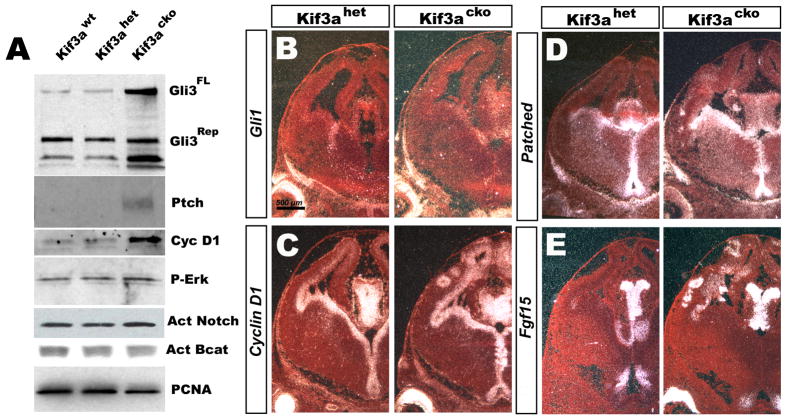
To investigate whether activation of any of the pathways above may have contributed to a shortening of the cell cycle in Kif3a mutants, we examined key downstream components for each that would provide evidence for pathway activation. The cerebral hemispheres of E12.5 in mutant, heterozygous and wildtype mice were analyzed by Western blots for the expression of NotchAct, phospho-ERK (p42/44, activated in growth factor signaling), β-catenin, and Gli3 (both the full length and repressor forms, Gli3FL and Gli3Rep, which have distinct molecular weights). Similar levels of β-catenin, phospho-ERK, NotchAct were detected in control and Kif3acko tissue; however, Gli3 processing was already reduced, as represented by an increased ratio of Gli3FL toh Gli3Rep (Fig. 4). These data suggested that components of the Shh signaling pathway may be activated in Kif3a mutant brains. To test this, we performed in situ hybridization using probes against critical members and targets of the Shh pathway, including cyclin D1 (Kenney and Rowitch, 2000), Ptc (Marigo and Tabin, 1996), Fgf15 (Saitsu et al., 2005), Gli1 (Dai et al., 1999), and Smo (Kalderon, 2005) on E13.5 tissue. No changes were observed in the expression of Gli1 or Smo (Fig. 4B and data not shown), but we saw a dramatic increase in the cortical expression of cyclin D1 (Fig. 4C), and an expansion of Ptc (Fig. 4D) and Fgf15 (Fig. 4E) expression into the cortex, where they are normally not seen at this age. The increase in cyclin D1 and Ptc mRNA levels were supported by the observation of increased protein levels in Western blots of cortical tissue (Fig. 4A). Together, these data suggest that an increased ratio of Gli3FL to Gli3Rep result in increased expression of cyclin D1 and FGF15, each of which might contribute to the reduction in cell cycle length seen in Kif3acko mice.
Figure 4. Decreased processing of Gli3 and upregulation of Shh target genes in Kif3acko mice.
(A) Western blot analysis of Gli3FL/Gli3Rep and CyclinD1 normalized to PCNA levels show an increase in Gli3FL/Gli3Rep ratio and Shh target genes (Ptc and Cyclin D1) in Kif3acko cortices. There is no change in phospho-ERK, β-catenin, or NotchAct expression. The PCNA loading control was used to correct for the number of progenitors. (B–E) In situ hybridization shows many Shh-target genes are upregulated in Kif3acko cortices including CyclinD1 (C), Ptc (D), and Fgf15 (E) while Gli1 (B) is not.
Attenuated Gli3 processing or loss of Gli3 phenocopies changes in cell cycle length
We reasoned that if the Kif3a phenotype arose in whole or in part from defects in Gli3 processing, mice that are unable to process Gli3 efficiently should show similar molecular and proliferative changes to those observed in Kif3a mutants. To address this, we utilized mice engineered to express a hypermorphic allele of Gli3 (Gli3Δ68), in which a 68-residue deletion encompassing the proteolytic processing site leads to reduced processing and increased transcriptional activity of the full-length (activator) mutant protein (Wang et al., 2007). This allele was used in combination with a null allele of Gli3 (Gli3Xt) because Gli3Δ68/Xt mice have a limb phenotype consistent with an increased Gli3 activator:Gli3 repressor ratio (Wang et al., 2007).
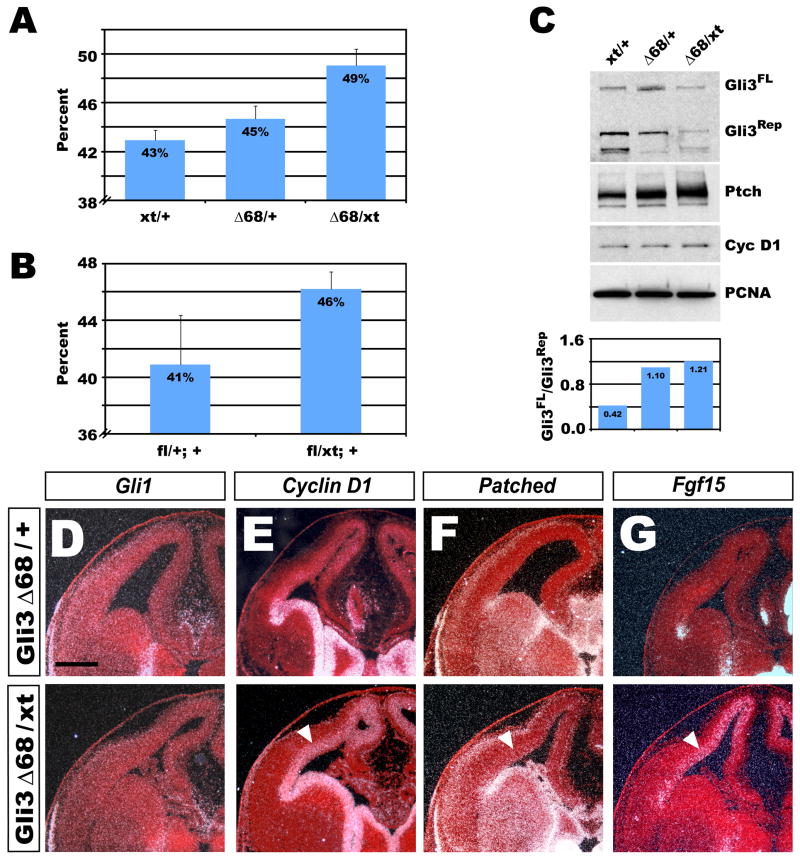
To ascertain whether Gli3Δ68/Xt mutants displayed a proliferation defect similar to that observed in Kif3a mutants, BrdU was injected at E12.5 and the LI was calculated. The LI for Gli3Δ68/Xt mutants (49.1% ± 1.4%) was significantly higher than that in Gli3Xt/+ controls (42.9% ± 0.8%; p< 0.003) (Fig. 5A). This increase of ~6% was strikingly similar to that observed in Kif3acko mice. Interestingly, Gli3Δ68/Xt mutants did not show as extreme an expansion of cortical tissue as did the Kif3a mutants (not shown), suggesting that Kif3a may be required for other processes besides the proteolytic processing of Gli3.
Figure 5. Increased Gli3FL/Gli3Rep ratio is sufficient to phenocopy cilia mutant.
(A and B) Graphs of labeling indices calculated for (A) Gli3+/xt, Gli3+/Δ68 and Gli3Δ68/xt and (B) Nestin Cre +; Gli3fl/+ and Nestin Cre +; Gli3fl/xt of E12.5 embryos showing an increased cycling rate for Gli3Δ68/xt and Nestin Cre +; Gli3fl/xt cortical progenitors. (C) Western blot analysis of E14.5 cortical caps showing an increased Gli3FL/Gli3Rep in Gli3Δ68/xt mice with a corresponding increase in Ptc and CyclinD1. Quantitation of Gli3FL/Gli3Rep normalized to PCNA levels above. (D–G) In situ hybridization for Shh target genes showing an increase in Cyclin D1 (E), Ptc (F), and Fgf15 (G) with no increase in Gli1 (D) expression.
Western blots confirmed that Gli3 processing is disrupted in cortices of Gli3Δ68/Xt mice, which displayed an increased Gli3FL/Gli3Rep ratio (Fig. 5B,C), as predicted. Western analyses also revealed that the levels of the Shh downstream effectors Ptc and cyclin D1 (Fig. 5B,C) were increased, similar to the changes observed in Kif3a mutant brains. Gli3Δ68/Xt mutants were further examined for changes in the expression of other Shh pathway members and targets that were altered in Kif3acko mice. In situ hybridization revealed no change in the expression of Gli1 or Smo (Fig. 5D and data not shown), an increase in cyclin D1 expression (Fig. 5E), and an expansion of Ptc (Fig. 5F) and Fgf15 (Fig. 5G) into the cortex, as seen in Kif3a mutants. Collectively these data suggest that the phenotype observed in Kif3acko mice is due primarily to an increase in the ratio of Gli3FL/Gli3Rep. However, we could not rule out that the loss of Gli3Rep at the expense of Gli3FL was the primary cause of increased cortical size in Kif3a mutants. To address this we used a conditional allele of Gli3 (Gli3f) to generate animals lacking any functional Gli3 protein in the developing cortical neuroepithelium (Gli3cko) and conducted a similar analysis of LI. The LI for Gli3cko mutants (46.2% ± 1.2%) was significantly higher than that in Gli3f/+; Nestin-cre+ controls (40.9% ± 3.5%; p< 0.027) (Fig. 5A). An increase of ~5% was similar to that observed in both the Kif3acko and Gli3Δ68/Xt mice, suggesting that the increased cortical size observed in Kif3acko mice may be due primarily to a loss in Gli3Rep. Again, although we did perform a detailed analysis of cortical size in Gli3cko mutants, we did not observe as extreme an expansion as in Kif3a mutants, suggesting that Kif3a has additional roles besides the regulation of Gli3 processing.
Fgf15 overexpression leads to faster cycling kinetics of cortical progenitors
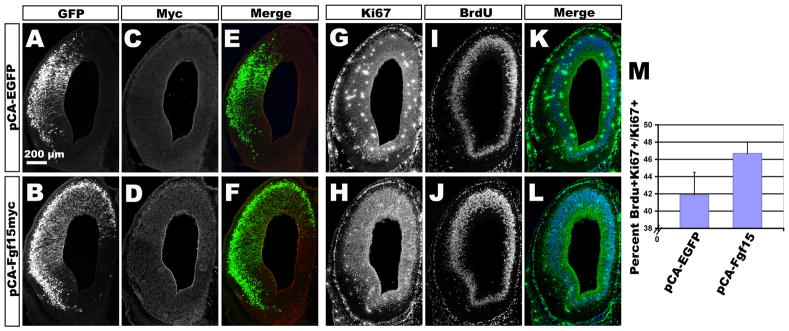
The role of Fgf15 in mouse brain development has more controversial. Recently it was reported that FGF15 suppresses proliferation in the developing forebrain (Vincentz et al., 2005; Borello et al., 2008). However, knockdown studies of Fgf19, a homolog of Fgf15 in zebrafish, have revealed reduced levels of proliferation of the forebrain neuroepithelium, suggesting the opposite role (Miyake et al., 2005). To determine whether the ectopic expression of Fgf15 observed in both Kif3acko and Gli3Δ68/Xt mice is sufficient to shorten the cell cycle length of cortical progenitors, we used in utero electroporation to overexpress Fgf15 in the cortices of wildtype embryos. E12 mice were co-electroporated with plasmids encoding FGF15 tagged with a C-terminal myc epitope and GFP, or GFP alone, and brains were analyzed 2 days later at E14. Immunostaining for myc and GFP in co-electroporated brains revealed the expression of FGF15 within the domain of GFP-positive cells (Fig. 6B,D,F), whereas no myc staining was observed in control brains (Fig. 6A,C,E).
Figure 6. Overexpression of Fgf15 causes faster cell cycle kinetics.
(A–E) Electroporation of pCA-EGFP (A,C,E) or pCA-Fgf15myc (B,D,F) into E12 cortices and analyzed 2 days later at E14 by immunostaining with GFP and myc antibodies. (G–L) Immunostaining of serial section in (A–F) with antibodies against Ki67 and BrdU to calculate a labeling index. (M) Quantification of labeling index showing that progenitors in pCA-Fgf15myc electroporated brains are cycling faster than pCA-EGFP progenitors with a 5% increase in labeling index.
To ascertain whether the ectopic expression of Fgf15 is sufficient to alter cell cycle kinetics, electroporated embryos were injected with BrdU 30 min prior to sacrifice. Reasoning that FGF15 is a secreted protein that should affect the behavior of many cells within the electroporated field (whether GFP+ or not), we assessed the LI for all Ki67+ cells located within the GFP and/or myc expression domain (Fig. 6G–L). The LI in domains that overexpressed Fgf15 was 46.7% ± 1.3%, a significant increase of ~5% compared to that in control embryos (41.9% ± 2.6%; p< 0.05). These data indicate that increased expression of Fgf15 is sufficient to alter cell cycle dynamics in a manner similar to that seen in both Kif3acko and Gli3Δ68/Xt mice (Fig. 6M).
Gli3FL:Gli3Rep ratios correlate with cell cycle lengths during normal development
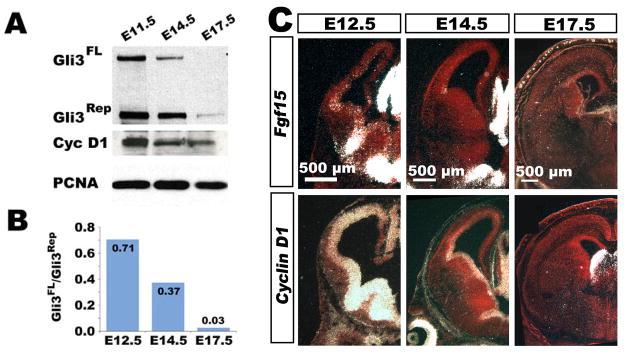
The previous experiments show that primary cilia are required for the normal regulation of Gli3 processing in the cortex, and in turn, that this processing event is important for controlling the length of the cell cycle. Interestingly, cell cycle length gradually increases over cortical development: at E11.5, the cell cycle is only about 8 hours long, while at the end of neurogenesis, at E17.5, it is nearly 20 hours long (Caviness and Takahashi, 1995; Caviness et al., 1995; Caviness et al., 1996). To investigate the possibility that Gli3 normally plays a role in regulating these temporal changes in cell cycle dynamics, we surveyed the expression of the different forms of Gli3 and their effectors over the time course of neurogenesis in the cortex. These studies revealed a marked correlation between the ratio of Gli3FL to Gli3Rep and cell cycle length during normal development. Early in corticogenesis, when the cell cycle is short, the ratio of Gli3FL to Gli3Rep is high, but this ratio decreases as the cell cycle lengthens (Fig. 7A,B). Additionally, two of the genes (Fgf15 and cyclin D1) that are regulated by Gli3 follow the pattern of high expression levels early and low expression late (Fig. 7A,C).
Figure 7. Changes in Gli3FL/Gli3Rep ratio correspond with changes in cell cycle length over cortical development.
(A) Gli3FL/Gli3Rep ratio and Cyclin D1 protein levels decrease over cortical development analyzed by western blot. (B) Expression of Shh target genes in E12.5, E14.5 and E17.5 coronal sections. Fgf15 and CyclinD1 decrease over cortical development evident by radioactive in situ hybridization. (C) Model of cell cycle length control in the cerebral cortex. A high Gli3FL/Gli3Rep ratio present during early corticogenesis leads to high levels of CyclinD1 and Fgf15 expression and a short G1 phase. A decreased Gli3FL/Gli3Rep ratio present later in development leads to a decrease in Cyclin D1 and no Fgf15 expression and a much longer G1 phase. Kif3acko or Gli3 mutants no longer undergo normal developmental changes and mimic the early state.
Collectively our observations and many others suggest a model whereby the primary cilium regulates the processing of Gli proteins during normal development (Fig. 7D). Gli3 transcription factors translocate to the nucleus where they regulate expression of the cyclin D1 and/or Fgf15 loci, which in turn control cell cycle length. It is tempting to speculate that cilia are responding to cue(s) in the CSF that regulate the balance of proliferation vs. differentiation during development. Interestingly, a recent study ascertained that Shh is present in embryonic CSF, which provides support for this model (Huang et al., 2010b). Early in cortical development, when signals promoting proliferation are high and/or those favoring differentiation are low, there is little processing of Gli3 from its full length form to the repressor form; high Gli3FL and low Gli3Rep promotes the expression of cyclin D1 and Fgf15, which lead to more rapid progress through the cell cycle. At later stages of development, a reduction in proliferative signals and/or increased signals promoting differentiation trigger an increase in Gli3 processing (higher Gli3Rep), which reduces cyclin D1 and Fgf15 levels and slows the cell cycle. In Kif3a mutants, this natural progression is no longer effectual because the loss of the primary cilium favors the production of Gli3FL (possibly by unlinking the cell cycle from external cues sensed by cilia or as a result of no functional Gli3 protein), thus promoting the expression of factors that cause cells to cycle at their earliest, most rapid rate throughout corticogenesis.
DISCUSSION
Here we report that the primary cilia of cortical progenitors are necessary for regulation of cortical growth during development. The absence of functional cilia in Kif3a conditional mutants disrupts Gli3 processing in the cerebral cortex and is accompanied by alterations in the ratio of Gli3FL to Gli3Rep, an upregulation of Gli3 target genes, and a shortening of the cell cycle. These phenotypes can be recapitulated in part by altering Gli3FL to Gli3Rep ratios (observed in Gli3Δ68/Xt mice) or by upregulating Gli3 target genes alone. These data suggest that the disruption of Gli3 processing in Kif3a mutant mice mimics the early, rapidly proliferative state of cortical progenitors and suggests that developmental alterations in the ratio of GliFL to GliRep proteins provide a potential mechanism by which progenitors control the length of the cell cycle during development.
Cilia and cortical development
A strong link for the necessity of cilia for proper Shh signaling has been shown in many developing organs, including the limbs (Liu et al., 2005), spinal cord (Huangfu et al., 2003b; Huangfu et al., 2003a; Huangfu and Anderson, 2005; Liu et al., 2005) and brain (Herron et al., 2002; May et al., 2005; Willaredt et al., 2008). Much of the data has come from analysis of mouse mutants generated from ENU screens and therefore create defects in genetic code present at the onset of development (Herron et al., 2002). This approach has biased discovery towards the initial phenotype, or the first role for primary cilia in a particular developing tissue or organ. Only a few studies have investigated later roles for cilia in brain development (Besse et al., 2011; Chizhikov et al., 2007; Alvarez-Buylla et al., 2008; Han et al., 2008; Spassky et al., 2008). However, Shh signaling is often reused for many aspects of a maturing organ, for example in the spinal cord, Shh signaling patterns the dorso-ventral axis, expands specific progenitor pools (Cayuso et al., 2006; Ulloa and Briscoe, 2007) and then guides commissural axons in crossing the midline (Charron et al., 2003). Similarly, analysis of forebrains in ENU mutants has uncovered a role for cilia through Gli3 function in patterning the cortex, and consistently these brains strongly phenocopy Gli3Xt brains (Willaredt et al., 2008). Interestingly, recent work on another ciliopathy gene, Ftm, and its early role in telencephalic patterning has also suggested that a primary function of cilia is to regulate Gli3 processing (Besse et al., 2011). Here we identified a previously unidentified role for Gli3 and primary cilia in controlling the size of the cerebral cortex. It appears Gli3 is required first in patterning the forebrain and, second after progenitor allocation, in the expansion of those progenitor domains through the regulation of cell cycle length.
Gli3 in cortical development
A role for Gli3 in the expansion of the cortical progenitor pool may have been masked because of its pleiotrophic role in cortical development: earlier patterning defects obscure the later proliferative role. Gli3 mutants display morphological defects by E9.5 with a failure to establish the telencephalic-diencephalic boundary correctly (Theil et al., 1999). Interestingly, this results in expansion of the Pax6 and Otx1 diencephalic expression domains and a reduction in cortical size—a phenotype opposite to that which we observed, and likely due to the early patterning defect. Additionally, a comprehensive study analyzing the role of Gli3 in development of the mid-hindbrain revealed temporally separable roles for Gli3 (Blaess et al., 2008). By inactivating Gli3 at successive timepoints, the authors determined that Gli3 is required first to specify boundaries and cytoarchitecture in the posterior tectum, isthmus and cerebellum, second for defining the cerebellar foliation pattern and lastly in regulating growth and cell viability. Together with our data, this suggests a common mechanism whereby Gli3 acts first to pattern the brain region and second to influence the final size.
Gli3 processing and cell cycle progression
Signaling pathways can affect the size of the developing cerebral cortex through two distinct mechanisms: biasing cells toward a progenitor fate or controlling the length of the cell cycle. Increased Notch signaling (Gaiano et al., 2000) and the activation of β-catenin via Wnt signaling (Chenn and Walsh, 2002) bias progenitors towards symmetric, progenitor-producing divisions that delay the onset of neurogenesis and dramatically expand the progenitor pool. Shh, in contrast, can act as a mitogen to promote proliferation by altering cell cycle kinetics. Shh regulates the growth of many organs and has a strong proliferative role in several regions of the CNS, including the retina, spinal cord and cerebellum (Donovan and Dyer, 2005). In spinal cord progenitor cells, the ectopic expression of positive regulators and application of pathway inhibitors have revealed that Shh signaling regulates the length of the G1 phase of the cell cycle. In addition, overexpression of the Shh effector Gli3Act decreases cell cycle length and increases the expression of cyclin D1 and N-myc, which regulate cell cycle progression (Cayuso et al., 2006). The role of Shh in retinal development is a bit more complex; activation of Shh signaling in Xenopus retinal progenitors shortens both the G1 and G2 phases, but these more rapidly cycling cells exit the cell cycle earlier (Locker et al., 2006).
In contrast to the tissues above, a mitogenic role for Shh in the cerebral cortex is more controversial. Blocking Shh signaling by removing Smo at E11.5 or disrupting Gli1 and Gli2 activity (in Gli1−/−;Gli2−/− mutant mice) had no impact on embryonic forebrain development (Machold et al., 2003), and Gli3 has not previously been reported to influence cortical cell proliferation. Here, we show that Gli3Rep functions to lengthen cell cycle kinetics to slow neuronal production. Interestingly, it was recently reported that loss of Smo function at E10.5 using Emx1cre shows a reduction in cortex size by E18.5 caused by a slowing of cell cycle kinetics (Komada et al., 2008). Although it appears Shh itself regulates Gli protein processing in cortical progenitors, the extent of Shh signaling is not clear. Our data provide support for greater Shh activation early in corticogenesis during the progenitor expansion phase at E12.5 when we see the highest levels of CyclinD1 and the lowest Gli3FL: Gli3Rep ratio, and a vast reduction at the end of neurogenesis at E17.5 where we see a very low level of CyclinD1 and no Gli3FL. Kif3a mutants appear to exhibit a shortened G1 phase of the cell cycle, similar to the changes seen in retina and spinal cord when Shh signaling is disrupted; however, we observed no alterations in progression through G2. In addition, the expression of both cyclin D1 and Fgf15 were upregulated in Kif3a mutants, similar to studies cited above, whereas the expression of N-myc was not obviously affected. The differences between the changes resulting from the disruption of Kif3a in the developing cortex and those resulting from altered Shh signaling in other neural tissues suggest that some downstream targets of Gli signaling are conserved in different brain regions, but variations in the specific targets may enable different growth rates in distinct areas of the brain. Alternatively, the differences may reflect the convergence of other signaling pathways on these loci in different brain regions.
Our data support both a direct effect on cell cycle kinetics through the upregulation of CyclinD1 and an indirect effect through the upregulation of Fgf15. In our overexpression studies, we show that Fgf15 is capable of increasing proliferation on its own, a result that has previously been reported in zebrafish. Surprisingly, loss of Fgf15 in the developing mouse cortex has been reported to promote proliferation (Borello et al., 2008), a result opposite to that obtained here. However, patterning defects are also observed in these mutants and could be responsible for the contradictory results, as appears to be the case for a reduction in cortex size seen in Gli3Xt brains.
Significance
Most progenitor cells in the developing central nervous system line the CSF-filled ventricles. Interestingly, each progenitor possesses a single primary cilium that contacts the CSF where it likely receives important growth information. Recently, Shh protein was identified in embryonic CSF (Huang et al., 2010a). Here we show that loss of the primary cilia lead to a shortened cell cycle as a result of defective Gli3 processing. Gli3 processing is tightly regulated over cortical development such that the changes in processing follow the known slowing of the cell cycle observed in cortical development. This in conjunction with the observed overgrowth of the midbrain in Gli3 conditional mutants suggests a tempting hypothesis whereby cilia and Shh-signaling globally regulate differential growth of brain regions. A common primary cilium thus allows for the coordinated growth across morphologically diverse brain regions to produce the correct proportion of neurons to create a fully functional brain.
Supplementary Material
Acknowledgments
We thank C. Kaznowski, J. Perrino and the Stanford Shared FACS Facility for technical support; A. Okada and J. Weimann for experimental advice; R. Rohatgi, B. Yoder, and R. Hevner for antibodies; W. Zhong for Nestin-cre mice; J. Hébert, F. Charron, J. Rubenstein, E. Alcamo, P. Gruss, and A. Joyner for in situ probes, A. Joyner for Gli3 conditional mutant embryos, and L. Schaevitz for critical reading the manuscript. Supported by NIH grants MH51864 (S.K.M.) and CA111673 (B.W.). S.L.W. was supported by NIH training grant GM007276.
References
- Alvarez-Buylla A, Mirzadeh Z, Han YG. Cilia in postnatal germinal niches and its function in neural stem cells. Febs Journal. 2008;275:53–53. [Google Scholar]
- Ashique AM, Choe Y, Karlen M, May SR, Phamluong K, Solloway MJ, Ericson J, Peterson AS. The Rfx4 transcription factor modulates Shh signaling by regional control of ciliogenesis. Sci Signal. 2009;2:ra70. doi: 10.1126/scisignal.2000602. [DOI] [PubMed] [Google Scholar]
- Besse L, Neti M, Anselme I, Gerhardt C, Ruther U, Laclef C, Schneider-Maunoury S. Primary cilia control telencephalic patterning and morphogenesis via Gli3 proteolytic processing. Development. 2011;138:2079–2088. doi: 10.1242/dev.059808. [DOI] [PubMed] [Google Scholar]
- Blaess S, Stephen D, Joyner AL. Gli3 coordinates three-dimensional patterning and growth of the tectum and cerebellum by integrating Shh and Fgf8 signaling. Development. 2008;135:2093–2103. doi: 10.1242/dev.015990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borello U, Cobos I, Long JE, McWhirter JR, Murre C, Rubenstein JLR. FGF15 promotes neurnogenesis and opposes FGF8 function during neocortical development. Neural Development 3. 2008;3:17. doi: 10.1186/1749-8104-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappello S, Attardo A, Wu X, Iwasato T, Itohara S, Wilsch-Brauninger M, Eilken HM, Rieger MA, Schroeder TT, Huttner WB, Brakebusch C, Gotz M. The Rho-GTPase cdc42 regulates neural progenitor fate at the apical surface. Nat Neurosci. 2006;9:1099–1107. doi: 10.1038/nn1744. [DOI] [PubMed] [Google Scholar]
- Caviness VS, Jr, Takahashi T. Proliferative events in the cerebral ventricular zone. Brain Dev. 1995;17:159–163. doi: 10.1016/0387-7604(95)00029-b. [DOI] [PubMed] [Google Scholar]
- Caviness VS, Jr, Takahashi T, Nowakowski RS. Numbers, time and neocortical neuronogenesis: a general developmental and evolutionary model. Trends Neurosci. 1995;18:379–383. doi: 10.1016/0166-2236(95)93933-o. [DOI] [PubMed] [Google Scholar]
- Caviness VS, Jr, Takahashi T, Nowakowski RS. The G1 restriction point as critical regulator of neocortical neuronogenesis. Neurochem Res. 1999;24:497–506. doi: 10.1023/a:1022579712262. [DOI] [PubMed] [Google Scholar]
- Caviness VS, Jr, Takahashi T, Miyama S, Nowakowski RS, Delalle I. Regulation of normal proliferation in the developing cerebrum potential actions of trophic factors. Exp Neurol. 1996;137:357–366. doi: 10.1006/exnr.1996.0037. [DOI] [PubMed] [Google Scholar]
- Cayuso J, Ulloa F, Cox B, Briscoe J, Marti E. The Sonic hedgehog pathway independently controls the patterning, proliferation and survival of neuroepithelial cells by regulating Gli activity. Development. 2006;133:517–528. doi: 10.1242/dev.02228. [DOI] [PubMed] [Google Scholar]
- Charron F, Stein E, Jeong J, McMahon AP, Tessier-Lavigne M. The morphogen Sonic hedgehog is an axonal chemoattractant that collaborates with Netrin-1 in midline axon guidance. Cell. 2003;113:11–23. doi: 10.1016/s0092-8674(03)00199-5. [DOI] [PubMed] [Google Scholar]
- Chenn A, Walsh CA. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002;297:365–369. doi: 10.1126/science.1074192. [DOI] [PubMed] [Google Scholar]
- Chenn A, Zhang YA, Chang BT, McConnell SK. Intrinsic polarity of mammalian neuroepithelial cells. Mol Cell Neurosci. 1998;11:183–193. doi: 10.1006/mcne.1998.0680. [DOI] [PubMed] [Google Scholar]
- Chizhikov VV, Davenport J, Zhang Q, Shih EK, Cabello OA, Fuchs JL, Yoder BK, Millen KJ. Cilia proteins control cerebellar morphogenesis by promoting expansion of the granule progenitor pool. Journal of Neuroscience. 2007;27:9780–9789. doi: 10.1523/JNEUROSCI.5586-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl HA. Fine structure of cilia in rat cerebral cortex. Z Zellforsch Mikrosk Anat. 1963;60:369–386. doi: 10.1007/BF00336612. [DOI] [PubMed] [Google Scholar]
- Dai P, Akimaru H, Tanaka Y, Maekawa T, Nakafuku M, Ishii S. Sonic Hedgehog-induced activation of the Gli1 promoter is mediated by GLI3. J Biol Chem. 1999;274:8143–8152. doi: 10.1074/jbc.274.12.8143. [DOI] [PubMed] [Google Scholar]
- Donovan SL, Dyer MA. Regulation of proliferation during central nervous system development. Semin Cell Dev Biol. 2005;16:407–421. doi: 10.1016/j.semcdb.2005.02.012. [DOI] [PubMed] [Google Scholar]
- Fotaki V, Yu T, Zaki PA, Mason JO, Price DJ. Abnormal positioning of diencephalic cell types in neocortical tissue in the dorsal telencephalon of mice lacking functional Gli3. Journal of Neuroscience. 2006;26:9282–9292. doi: 10.1523/JNEUROSCI.2673-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuccillo M, Joyner AL, Fishell G. Morphogen to mitogen: the multiple roles of hedgehog signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:772–783. doi: 10.1038/nrn1990. [DOI] [PubMed] [Google Scholar]
- Gaiano N, Nye JS, Fishell G. Radial glial identity is promoted by Notch1 signaling in the murine forebrain. Neuron. 2000;26:395–404. doi: 10.1016/s0896-6273(00)81172-1. [DOI] [PubMed] [Google Scholar]
- Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han YG, Spassky N, Romaguera-Ros M, Garcia-Verdugo JM, Aguilar A, Schneider-Maunoury S, Alvarez-Buylla A. Hedgehog signaling and primary cilia are required for the formation of adult neural stem cells. Nature Neuroscience. 2008;11:277–284. doi: 10.1038/nn2059. [DOI] [PubMed] [Google Scholar]
- Haycraft CJ, Banizs B, Aydin-Son Y, Zhang Q, Michaud EJ, Yoder BK. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 2005;1:e53. doi: 10.1371/journal.pgen.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydar TF, Kuan CY, Flavell RA, Rakic P. The role of cell death in regulating the size and shape of the mammalian forebrain. Cereb Cortex. 1999;9:621–626. doi: 10.1093/cercor/9.6.621. [DOI] [PubMed] [Google Scholar]
- Herron BJ, Lu WN, Rao C, Liu SM, Peters H, Bronson RT, Justice MJ, McDonald JD, Beier DR. Efficient generation and mapping of recessive developmental mutations using ENU mutagenesis. Nature Genetics. 2002;30:185–189. doi: 10.1038/ng812. [DOI] [PubMed] [Google Scholar]
- Hinds JW, Ruffett TL. Cell proliferation in the neural tube: an electron microscopic and golgi analysis in the mouse cerebral vesicle. Z Zellforsch Mikrosk Anat. 1971;115:226–264. doi: 10.1007/BF00391127. [DOI] [PubMed] [Google Scholar]
- Hodge RD, D’Ercole AJ, O’Kusky JR. Insulin-like growth factor-I accelerates the cell cycle by decreasing G1 phase length and increases cell cycle reentry in the embryonic cerebral cortex. J Neurosci. 2004;24:10201–10210. doi: 10.1523/JNEUROSCI.3246-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Liu J, Ketova T, Fleming JT, Grover VK, Cooper MK, Litingtung Y, Chiang C. Transventricular delivery of Sonic hedgehog is essential to cerebellar ventricular zone development. Proceedings of the National Academy of Sciences of the United States of America. 2010a;107:8422–8427. doi: 10.1073/pnas.0911838107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Liu J, Ketova T, Fleming JT, Grover VK, Cooper MK, Litingtung Y, Chiang C. Transventricular delivery of Sonic hedgehog is essential to cerebellar ventricular zone development. Proc Natl Acad Sci U S A. 2010b;107:8422–8427. doi: 10.1073/pnas.0911838107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D, Anderson KV. Cilia and Hedgehog responsiveness in the mouse. Proc Natl Acad Sci U S A. 2005;102:11325–11330. doi: 10.1073/pnas.0505328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu DW, Rakeman A, Anderson K. Mouse intraflagellar transport proteins regulate hedgehog signaling. Developmental Biology. 2003a;259:513–514. [Google Scholar]
- Huangfu DW, Liu AM, Rakeman AS, Murcia NS, Niswander L, Anderson KV. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003b;426:83–87. doi: 10.1038/nature02061. [DOI] [PubMed] [Google Scholar]
- Humke EW, Dorn KV, Milenkovic L, Scott MP, Rohatgi R. The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes Dev. 2010;24:670–682. doi: 10.1101/gad.1902910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai F, Hirai S, Akimoto K, Koyama H, Miyata T, Ogawa M, Noguchi S, Sasaoka T, Noda T, Ohno S. Inactivation of aPKClambda results in the loss of adherens junctions in neuroepithelial cells without affecting neurogenesis in mouse neocortex. Development. 2006;133:1735–1744. doi: 10.1242/dev.02330. [DOI] [PubMed] [Google Scholar]
- Kalderon D. The mechanism of hedgehog signal transduction. Biochem Soc Trans. 2005;33:1509–1512. doi: 10.1042/BST0331509. [DOI] [PubMed] [Google Scholar]
- Kenney AM, Rowitch DH. Sonic hedgehog promotes G(1) cyclin expression and sustained cell cycle progression in mammalian neuronal precursors. Mol Cell Biol. 2000;20:9055–9067. doi: 10.1128/mcb.20.23.9055-9067.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klezovitch O, Fernandez TE, Tapscott SJ, Vasioukhin V. Loss of cell polarity causes severe brain dysplasia in Lgl1 knockout mice. Genes Dev. 2004;18:559–571. doi: 10.1101/gad.1178004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komada M, Saitsu H, Kinboshi M, Miura T, Shiota K, Ishibashi M. Hedgehog signaling is involved in development of the neocortex. Development. 2008;135:2717–2727. doi: 10.1242/dev.015891. [DOI] [PubMed] [Google Scholar]
- Lien WH, Klezovitch O, Fernandez TE, Delrow J, Vasioukhin V. alphaE-catenin controls cerebral cortical size by regulating the hedgehog signaling pathway. Science. 2006;311:1609–1612. doi: 10.1126/science.1121449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Wang B, Niswander LA. Mouse intraflagellar transport proteins regulate both the activator and repressor functions of Gli transcription factors. Development. 2005;132:3103–3111. doi: 10.1242/dev.01894. [DOI] [PubMed] [Google Scholar]
- Locker M, Agathocleous M, Amato MA, Parain K, Harris WA, Perron M. Hedgehog signaling and the retina: insights into the mechanisms controlling the proliferative properties of neural precursors. Genes Dev. 2006;20:3036–3048. doi: 10.1101/gad.391106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machold R, Hayashi S, Rutlin M, Muzumdar MD, Nery S, Corbin JG, Gritli-Linde A, Dellovade T, Porter JA, Rubin LL, Dudek H, McMahon AP, Fishell G. Sonic hedgehog is required for progenitor cell maintenance in telencephalic stem cell niches. Neuron. 2003;39:937–950. doi: 10.1016/s0896-6273(03)00561-0. [DOI] [PubMed] [Google Scholar]
- Marigo V, Tabin CJ. Regulation of patched by sonic hedgehog in the developing neural tube. Proc Natl Acad Sci U S A. 1996;93:9346–9351. doi: 10.1073/pnas.93.18.9346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marszalek JR, Ruiz-Lozano P, Roberts E, Chien KR, Goldstein LS. Situs inversus and embryonic ciliary morphogenesis defects in mouse mutants lacking the KIF3A subunit of kinesin-II. Proc Natl Acad Sci U S A. 1999;96:5043–5048. doi: 10.1073/pnas.96.9.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May SR, Ashique AM, Karlen M, Wang B, Shen Y, Zarbalis K, Reiter J, Ericson J, Peterson AS. Loss of the retrograde motor for IFT disrupts localization of Smo to cilia and prevents the expression of both activator and repressor functions of Gli. Dev Biol. 2005;287:378–389. doi: 10.1016/j.ydbio.2005.08.050. [DOI] [PubMed] [Google Scholar]
- Miyake A, Nakayama Y, Konishi M, Itoh N. Fgf19 regulated by Hh signaling is required for zebrafish forebrain development. Dev Biol. 2005;288:259–275. doi: 10.1016/j.ydbio.2005.09.042. [DOI] [PubMed] [Google Scholar]
- Murone M, Rosenthal A, de Sauvage FJ. Sonic hedgehog signaling by the patched-smoothened receptor complex. Curr Biol. 1999;9:76–84. doi: 10.1016/s0960-9822(99)80018-9. [DOI] [PubMed] [Google Scholar]
- Ohtsuka T, Sakamoto M, Guillemot F, Kageyama R. Roles of the basic helix-loop-helix genes Hes1 and Hes5 in expansion of neural stem cells of the developing brain. J Biol Chem. 2001;276:30467–30474. doi: 10.1074/jbc.M102420200. [DOI] [PubMed] [Google Scholar]
- Okada A, Charron F, Morin S, Shin DS, Wong K, Fabre PJ, Tessier-Lavigne M, McConnell SK. Boc is a receptor for sonic hedgehog in the guidance of commissural axons. Nature. 2006;444:369–373. doi: 10.1038/nature05246. [DOI] [PubMed] [Google Scholar]
- Pan Y, Bai CB, Joyner AL, Wang B. Sonic hedgehog signaling regulates Gli2 transcriptional activity by suppressing its processing and degradation. Mol Cell Biol. 2006;26:3365–3377. doi: 10.1128/MCB.26.9.3365-3377.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen PH, Zou K, Hwang JK, Jan YN, Zhong W. Progenitor cell maintenance requires numb and numblike during mouse neurogenesis. Nature. 2002;419:929–934. doi: 10.1038/nature01124. [DOI] [PubMed] [Google Scholar]
- Praetorius HA, Spring KR. Bending the MDCK cell primary cilium increases intracellular calcium. J Membr Biol. 2001;184:71–79. doi: 10.1007/s00232-001-0075-4. [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- Saitsu H, Komada M, Suzuki M, Nakayama R, Motoyama J, Shiota K, Ishibashi M. Expression of the mouse Fgf15 gene is directly initiated by Sonic hedgehog signaling in the diencephalon and midbrain. Dev Dyn. 2005;232:282–292. doi: 10.1002/dvdy.20236. [DOI] [PubMed] [Google Scholar]
- Schneider L, Clement CA, Teilmann SC, Pazour GJ, Hoffmann EK, Satir P, Christensen ST. PDGFRalphaalpha signaling is regulated through the primary cilium in fibroblasts. Curr Biol. 2005;15:1861–1866. doi: 10.1016/j.cub.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Spassky N, Han YG, Aguilar A, Strehl L, Besse L, Laclef C, Ros MR, Garcia-Verdugo JM, Alvarez-Buylla A. Primary cilia are required for cerebellar development and Shh-dependent expansion of progenitor pool. Developmental Biology. 2008;317:246–259. doi: 10.1016/j.ydbio.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Nowakowski RS, Caviness VS., Jr The cell cycle of the pseudostratified ventricular epithelium of the embryonic murine cerebral wall. J Neurosci. 1995;15:6046–6057. doi: 10.1523/JNEUROSCI.15-09-06046.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taulman PD, Haycraft CJ, Balkovetz DF, Yoder BK. Polaris, a protein involved in left-right axis patterning, localizes to basal bodies and cilia. Mol Biol Cell. 2001;12:589–599. doi: 10.1091/mbc.12.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theil T, Alvarez-Bolado G, Walter A, Ruther U. Gli3 is required for Emx gene expression during dorsal telencephalon development. Development. 1999;126:3561–3571. doi: 10.1242/dev.126.16.3561. [DOI] [PubMed] [Google Scholar]
- Tole S, Ragsdale CW, Grove EA. Dorsoventral patterning of the telencephalon is disrupted in the mouse mutant extra-toes(J) Dev Biol. 2000;217:254–265. doi: 10.1006/dbio.1999.9509. [DOI] [PubMed] [Google Scholar]
- Ulloa F, Briscoe J. Morphogens and the control of cell proliferation and patterning in the spinal cord. Cell Cycle. 2007;6:2640–2649. doi: 10.4161/cc.6.21.4822. [DOI] [PubMed] [Google Scholar]
- Vaccarino FM, Schwartz ML, Raballo R, Nilsen J, Rhee J, Zhou M, Doetschman T, Coffin JD, Wyland JJ, Hung YT. Changes in cerebral cortex size are governed by fibroblast growth factor during embryogenesis. Nat Neurosci. 1999;2:246–253. doi: 10.1038/6350. [DOI] [PubMed] [Google Scholar]
- Vincentz JW, McWhirter JR, Murre C, Baldini A, Furuta Y. Fgf15 is required for proper morphogenesis of the mouse cardiac outflow tract. Genesis. 2005;41:192–201. doi: 10.1002/gene.20114. [DOI] [PubMed] [Google Scholar]
- Wang C, Pan Y, Wang B. A hypermorphic mouse Gli3 allele results in a polydactylous limb phenotype. Dev Dyn. 2007;236:769–776. doi: 10.1002/dvdy.21082. [DOI] [PubMed] [Google Scholar]
- Wang C, Pan Y, Wang B. Suppressor of fused and Spop regulate the stability, processing and function of Gli2 and Gli3 full-length activators but not their repressors. Development. 2010;137:2001–2009. doi: 10.1242/dev.052126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield JF. The neuronal primary cilium--an extrasynaptic signaling device. Cell Signal. 2004;16:763–767. doi: 10.1016/j.cellsig.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Willaredt MA, Hasenpusch-Theil K, Gardner HA, Kitanovic I, Hirschfeld-Warneken VC, Gojak CP, Gorgas K, Bradford CL, Spatz J, Wolfl S, Theil T, Tucker KL. A crucial role for primary cilia in cortical morphogenesis. J Neurosci. 2008;28:12887–12900. doi: 10.1523/JNEUROSCI.2084-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.