Abstract
Nonhydrolyzable aminoacyl-adenylates that inhibit protein synthesis provide a promising route towards the development of novel antibiotics whose mechanism of action limits the appearance of bacterial drug resistance. The ‘Trojan horse’ antibiotic microcin C (McC) consists of a nonhydrolyzable aspartyl–adenylate that is efficiently imported into bacterial cells owing to a covalently attached peptide carrier. Once inside the cell, the carrier is removed by proteolytic processing to release a potent aspartyl tRNA synthetase inhibitor. The focus of this article is on the mechanism of biosynthesis of McC. We also examine the strategies utilized by McC-producing strains to overcome toxicity due to unwanted, premature processing of the drug. This article will discuss how McC biosynthesis can be systematically manipulated for the development of derivatives that will target the entire battery of aminoacyl tRNA synthetases in various bacteria.
Keywords: acetyltransferase, aminoacyl-tRNA synthetase, microcin C, Trojan-horse inhibitors
Background
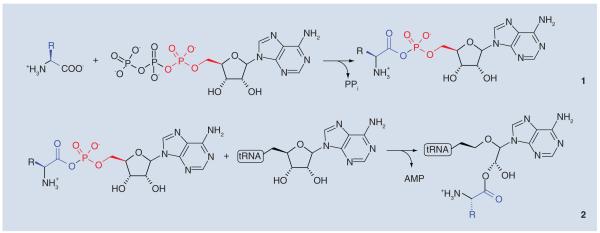
Bacterial drug resistance is a very significant medical problem and identification of novel antibacterials and new drug targets is an important goal. Among promising targets are the 20 aminoacyl-tRNA synthetases (aaRSs) that are essential for protein synthesis [1]. aaRSs catalyze the attachment of correct amino acids to cognate tRNAs and thus play a fundamental role in translating the genetic code [2]. The acylation reaction consists of two steps, the formation of an activated aminoacyl adenylate and subsequent nucleophilic attack of the terminal 2′ or 3′-hydoxyl of a tRNA molecule to generate the aminoacyl-tRNA (Figure 1).
Figure 1. Two-step mechanism for the production of charged tRNA molecules by aminoacyl-tRNA synthetases.
The acylation reaction first involves the formation of an enzyme-bound aminoacyl-adenylate complex. Nucleophilic attack of the 2′ or 3′-hydroxyl of the terminal ribose of the tRNA on the adenylate forms the aminoacyl-tRNA (the substrate for polypeptide synthesis) and adenosine monophosphate.
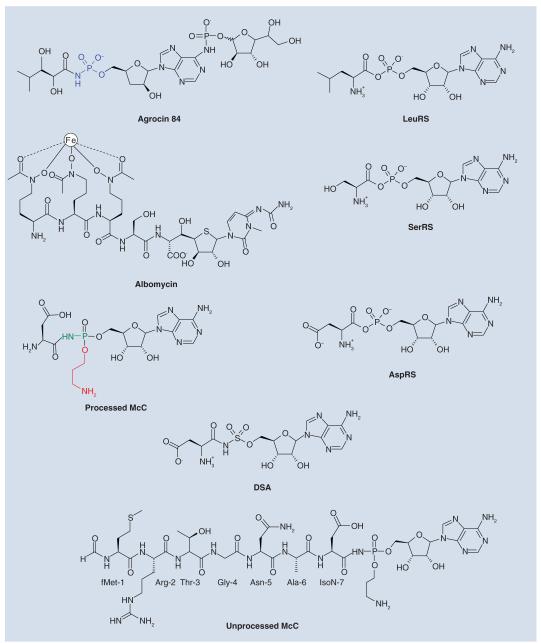
Several classes of naturally occurring anti-microbials target aaRSs [3,4]. One class of aaRS inhibitors consists of nonhydrolyzable analogs of the aminoacyl-adenylate intermediate generated during the aaRS reaction [5]. Since aminoacyl-adenylates generally poorly penetrate into bacterial cells, natural compounds that mimic aminoacyl-adenylate intermediates often employ a ‘Trojan-horse’ strategy, wherein the active component is conjugated with a specialized module that facilitates its active transport into bacterial cell. The transport module is subsequently removed, releasing the inhibitor inside the cell. Examples of naturally occurring aminoacyl-adenylate-based antibiotics that employ this strategy include the leucyl-tRNA synthetase inhibitor agrocin 84 [6], the seryl-tRNA synthetase inhibitor albomycin [7], and the subject of this review, aspartyl-tRNA synthetase inhibitor microcin C (McC) (Figure 2) [8].
Figure 2. Natural and synthetic nonhydrolyzable aminoacyl-adenylate analogs and the corresponding aminoacyl-adenylates that are mimicked by these molecules.
Also shown are the synthetic aspartyl-tRNA synthetase inhibitor DSA and the chemical structure of the unprocessed McC.
AspRS: Aspartyl-adenylate synthesized by aspartyl-tRNA synthetase; DSA: Aspartyl sulfamoyl adenylate; LeuRS: Leucyl-adenylate synthesized by leucil-tRNA synthetase; McC: Microcin C; SerRS: Seryl-adenylate synthesized by seryl-tRNA synthetase.
McC structure & mechanism of function
Microcins are small (<10 kDa) ribosomally synthesized antibacterial peptides produced by some enterobacteria [9,10]. Some microcins, like McC, are subject to complex post-translational modifications (Box 1) [11]. Full-length McC consists of a MRTGNAD heptapeptide with a modified AMP attached to the α carboxyl group of the aspartate through an N-acyl phosphoramidate linkage (Figure 2) [12]. The N-terminal methionine is formylated, and the phosphate group is decorated with a propylamine group [12].
Box 1. Microcin C: a note on nomenclature.
▪ McC is an antibiotic
▪ MccA is a precursor of McC peptide part encoded by the mccA gene
▪ MccC (a product of mccC gene) is an McC efflux pump
McC is produced by some Escherichia coli strains carrying plasmids with the mccABCDEF gene cluster. The peptide moiety of McC is encoded by mccA, a 21 bp-long gene that is probably among the shortest protein-coding bacterial genes known [13]. The mccB, mccD and mccE genes are responsible for the synthesis of antibiotic. The products of mccC, mccE and mccF make cells immune to both endogenously produced and externally added McC [14]. The mccABCDE genes constitute an operon and are transcribed from a single promoter activated by nutrient deprivation [14,15]. The divergent mccF gene is transcribed separately [14].
McC synthesis
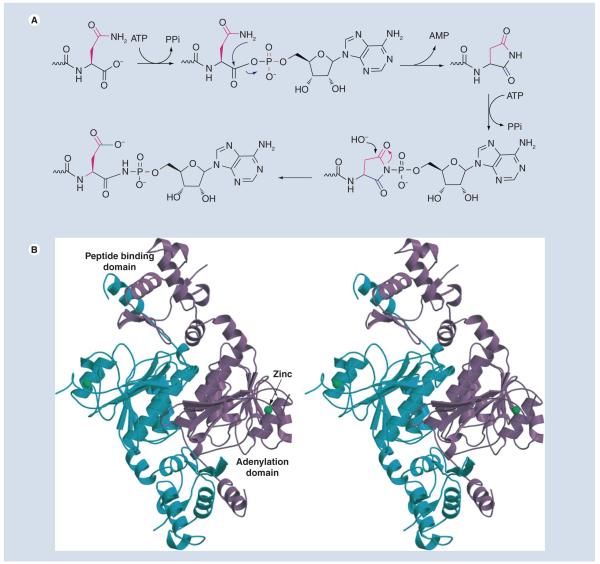
The biosynthesis of McC begins with the attachment of AMP to the MccA (fMRT-GNAN) peptide by the MccB protein [16]. The reaction results in the conversion of MccA carboxy-terminal Asn to an Asp amide, with the nitrogen linked to AMP through a phosphoramidate bond. Conjugation consumes two molecules of ATP: the first stage of the reaction generates an acyl-adenylate that undergoes an intermolecular rearrangement to expel AMP and generate a peptide–succinimide intermediate (Figure 3A). The succinimidyl nitrogen of this intermediate then attacks the α-phosphate of the second ATP to generate a hydrolytically stable phosphoramidate-linked adenylate that is biologically active [16].
Figure 3. Adenylation of MccA peptide by MccB.
(A) Mechanism for phosphoramidate (N–P) bond formation by MccB. For clarity, only the carboxy terminal Asn-7 of MccA substrate is shown. (B) The crystal structure of the MccB dimer (in blue and purple) The location of the adenylation domain and the peptide-binding domain are indicated.
The crystal structure of MccB reveals a dimeric protein with a bidomain architecture [17]. The central 260 residues (approximation) of MccB form a domain similar to the adenylation domain of UBL protein-activating enzymes [18]. The N-terminal 90 amino acid region of each MccB monomer packs against 20 C-terminal amino acids of the other monomer forming a ‘peptide clamp’ that holds the MccA peptide in place (Figure 3B) [17].
Adenylation reactions catalyzed by UBL protein-activating enzymes typically proceed through a covalent enzyme-linked intermediate such as a thioester of acyl-disulfide formed via an active site thiol. By contrast, the reaction catalyzed by MccB apparently does not require such an intermediate. MccB is further distinguished from UBL protein-activating enzymes in that it modifies a C-terminal Asn, rather than a Gly residue [17,18].
Crystal structures of MccB in complex with different ligands reveal considerable conformational differences in the orientation of the substrate owing to local flexibility at the peptide-binding domain [17]. This flexibility is consistent with systematic mutational analysis that showed that the MccB active site is flexible enough to tolerate substitutions at every position of the MccA peptide with the exceptions of Thr-3 and Asn-7 [19]. Surprisingly, analysis of MccB–ligand complex structures reveals that Asn-7 of bound MccA or the equivalent succinimide moiety of synthetic substrate analog are too far away from the α-phosphate of ATP for efficient catalysis [17]. Significant rearrangement of the bound ligand and/or the peptide-binding domain must therefore occur in order to bring the two substrates (ATP and activated MccA) at a distance that would favor nucleophilic attack.
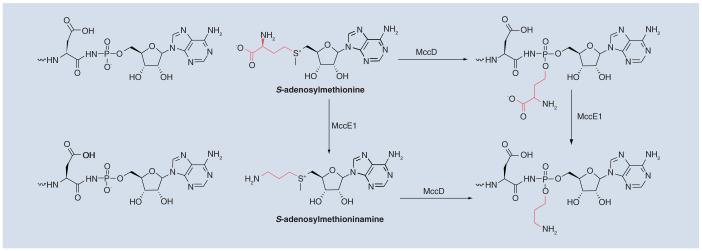
The product of a MccB-catalyzed reaction, adenylated heptapeptide, is further modified in a reaction that requires MccD and the N-terminal domain of MccE (MccENTD). This modification results in the attachment of an aminopropyl moiety to the phosphate [20]. MccENTD is similar to pyridoxal 5′-phosphate-dependent decarboxylases, and MccD shows signature motifs found in S-adenosylmethionine (SAM)-dependent methyltransferases. Therefore, the following hypothetical route of aminopropyl attachment can be envisioned: MccD transfers the 3-amino-3-carboxypropyl group (rather than the methyl group) from SAM onto the product of the MccB-catalyzed reaction, followed by MccENTD-catalyzed decarboxylation to yield mature McC containing the 3-aminopropyl modification (Figure 4). The modification increases the biological activity approximately tenfold.
Figure 4.
Possible biosynthetic routes for the attachment of the aminopropyl moiety onto unprocessed McC by MccD and MccE.
McC is exported from producing cells through a major facilitator superfamily efflux pump encoded by the mccC gene. Most McC produced by E. coli cells contains a formyl group; a deformylated derivative, as well as maturation intermediates lacking the propylamine group (both formylated and deformylated) are also produced to a varying degree.
McC uptake, processing & target inhibition
McC is taken up by sensitive bacterial strains via the inner membrane transporter YejABEF, which recognizes the peptide part of McC [21,22]. The biological function of YejABEF is not known. It may be involved in transport of oligopeptides, particularly oligopeptides containing an N-terminal formyl-methionine, inside the cell. Consistent with this idea, the YejAEF transporter contributes to Salmonella virulence [23] and yej mutants demonstrate superior vaccine properties [24]. McC variants, which contain shortened peptide parts (six amino acids or less) are poorly recognized by YejABEF [22].
Once inside the target cell, McC is processed: first, the formyl group is removed from the N-terminal methionine by peptide deformylase. Deformylated McC is then progressively digested in the N–C direction by any one of three cellular aminopeptidases, PepA, PepB or PepN [25]. Upon hydrolysis of the peptide bond connecting the sixth and seventh residues of the peptide, toxic processed McC, a nonhydrolyzable analogue of aspartyl-adenylate, is released [8]. The presence of the aminopropyl group increases the efficiency of processed McC interaction with its target approximately tenfold [20].
While unprocessed McC demonstrates good antibacterial activity against a range of enteric bacteria at low micromolar concentrations, addition of millimolar concentrations of processed McC to growth medium is needed to observe even traces of antibacterial activity [8]. Thus, McC is a bona fide Trojan-horse inhibitor: it is actively transported into sensitive cells, where the part responsible for facilitated transport is removed, releasing the toxic part that by itself is incapable of cell entry.
Processed McC analogs such as nonhydrolyzable aminoacyl sulfamoyl adenylates (aaSAs, where ‘aa’ denotes an amino acid in a single-letter code), such as aspartyl sulfamoyl adenylate (DSA), shown in Figure 2 exhibit antibacterial activity only at millimolar concentrations owing to poor uptake. aaSAs were successfully synthetically coupled to the C-terminus of a peptide corresponding to the first six amino acids of McC. The resulting synthetic McC analogs were biologically active at micromolar concentrations, acted through a Trojan-horse mechanism, but targeted aaRSs specified by amino acid attached to the sulfamoyl moiety [22,26,27].
Putative McC-like compounds encoded by mcc operons from bacteria other than E. coli
The processing machinery, as well as the processed McC target are universally conserved and processed McC readily inhibits aaRS from every bacterium tested, as well as eukaryotic aaRSs [8]. Yet, the spectrum of McC action is rather narrow. Evidently, the specificity is controlled at the level of McC uptake by the YejABEF transporter [8,22]. Bioinformatics analysis identifies clusters of genes homologous to E. coli mccB, mccE, mccD and mccE (the latter sometimes present as two separate genes coding for proteins corresponding to N-terminal and C-terminal domains of E. coli MccE) in the genomes of various bacteria [11]. Inspection of DNA sequences upstream of the mccB homologs led to identification of short open reading frames that might correspond to E. coli mccA [11]. In cases where mccBCDE gene homologs are present as complete open reading frames, the putative MccA peptides contain a C-terminal asparagine residue, indicating that the mechanism of MccB-catalyzed adenylation reaction is conserved. While the terminal residue of putative MccA analogs is conserved, their length varies. Analysis of the structure of E. coli MccB complexed with the MccA peptide, as well as in vitro adenylation data indicate that E. coli MccB is capable of interaction with MccA-based peptides containing N-terminal extensions [Severinov K & Nair SK, Unpublished Data] [17]. Thus, one can hypothesize that there exist McC-like molecules containing extensive peptidyl parts (predicted to be as long as 65 amino acids for some strains of Synechococcus containing putative mcc operons, [11]). If such McC analogs are indeed produced, they must enter sensitive cells through routes that are different from YejABEF. Yet, once inside the cell, they should function through the same Trojan-horse mechanism.
Mechanisms of McC resistance
Processed McC must inevitably accumulate inside a producing cell, leading to cessation of cell growth. This, however, is not observed as McC-producing cells continue to grow. Internally generated, processed McC is detoxified by at least two different mechanisms. The C-terminal domain of the MccE protein (MccECTD) is an acetyl-CoA-dependent acetyltransferase that acetylates the primary amino group of the aminoacyl moiety of processed McC; acetylated processed McC no longer inhibits aspartyl-tRNA synthetase [28]. MccECTD also acetylates and provides resistance to a wide spectrum of processed McC analogs such as aminoacyl sulfamoyl adenylates.
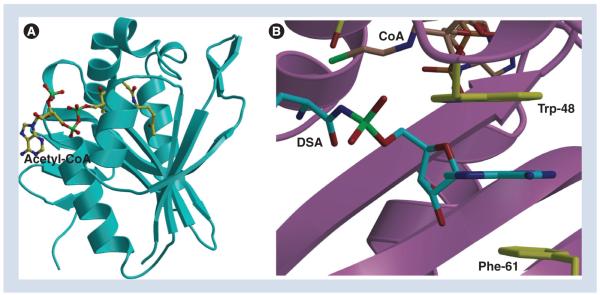
The crystal structures of MccECTD, in complex with acetyl-CoA and various substrate ligands, have been determined to resolutions greater than 1.5 Å [29]. The overall structure of MccECTD consists of a core β-strand structure that is surrounded by α-helices, which is typical of a canonical acetyltransferase but with additional decorations near the active site for substrate specificity (Figure 5A). The structure reveals a hydrophobic pocket located adjacent to the active site that serves as the major determinant for substrate recognition. Only minimal contacts are made with the aminoacyl moiety of the substrate. By contrast, the adenine ring of the substrate is sandwiched between two aromatic residues, Trp-453 and Phe-466, resulting in tight binding through strong π-stacking interactions (Figure 5B). This mode of purine base engagement is reminiscent of recognition mechanisms used by eukaryotic and viral proteins that recognize the 7-methylguanosine cap at the 5′ end of host mRNA [30], and by bacterial and viral nucleotidyltransferases, such as DNA/ RNA ligases and RNA capping enzymes [31]. Acetylation of the α-amino group of the amino acid adenylate would both disrupt favorable hydrogen bond interactions and result in steric clashes with the backbone of aspartyl-tRNA synthetase, which provides structural rationale for MccECTD protective function.
Figure 5. The structure of the MccE acetyltransferase.
(A) The C-terminal domain of the MccE protein-acetyl-CoA structure. The acetyl CoA is shown as a stick figure. (B) Close-up view of the active site in the MccE–CoA–DSA structure showing the stacking of the substrate adenine.
CoA: Coenzyme A; DSA: Aspartyl sulfamoyl adenylate.
The MccE acetyltransferase is homologous to bacterial N-terminal acetyltransferases of the Rim family [28]. The E. coli genome encodes three Rim proteins, RimI, RimJ and RimL, which acetylate ribosomal proteins S18, S5 and L12, respectively. The functions of these N-terminal acetyltransferases, and the significance of ribosomal protein acetylation for cell physiology are not entirely clear. One of the E. coli Rim proteins, RimL, contributes to the basal level of McC resistance through the same mechanism as the MccE acetyltransferase [Severinov K & Kazakov T, Unpublished Data].
The second mechanism of producing cell resistance to internally accumulating processed McC is provided by the MccF protein [32]. MccF is a serine protease that hydrolyses the isopeptide bond connecting the aspartate residue of intact or processed McC with the AMP moiety. Among the closest MccF homologs are E. coli LdcA, a d-l carboxypeptidase involved in the turnover of murein peptides [32,33] and AgrD, a protein encoded by agrocin 84 biosynthetic cluster [32,34]. The function of AgrD is not known. However, it is noteworthy that agrocin 84 is a Trojan horse inhibitor of leucyl-tRNA synthetase, whose active part is a nonhydrolyzable leucyl adenylate with a phosporamide bond in the same position as in McC [6]. Neither LdcA nor AgrD hydrolyze McC [32]. Genes encoding proteins closely related to E. coli MccF can be found in genomes of various bacteria. These genes are not associated with homologs of other mcc genes. The product of such a standalone gene from Bacillus anthracis cleaves McC in vitro and leads to McC-resistance when overexpressed in E. coli [Severinov K & Tikhonov A, Unpublished Data]. Since bacilli are resistant to McC, the physiological function of standalone mccF genes remains to be determined.
Future perspective
McC is an attractive versatile platform for design of terminally adenylated biologically active peptides. McC variants with altered peptide parts can be prepared by multiple strategies: mutagenesis of cloned mccA gene, total chemical synthesis, enzymatic synthesis by MccB, or by heterologous coexpression of mcc operons from bacteria other than E. coli. Peptide-adenylates that will become available as a result of these efforts will target various aaRSs and will share a common Trojan-horse mechanism of action but will penetrate cells through transporters other than the YejABEF transporter utilized by McC. Thus, expansion of the narrow specificity of McC and even custom design of McC-like molecules targeting specific classes of bacteria should become possible.
Executive summary.
Background
▪ Aminoacyl-tRNA synthetases are attractive drug targets.
▪ Many antibacterials targeting aminoacyl-tRNA synthetases employ a Trojan-horse strategy.
McC structure & mechanism of function
▪ Microcin C (McC) consists of a ribosomally synthesized heptapeptide C-terminally attached to modified adenosine monophosphate.
▪ McC biosynthesis and immunity requires six genes.
McC synthesis
▪ The peptide precursor of McC is adenylated by an ancestor of ubiquitin-E1 activating enzyme.
▪ A propylamine modification increases antibacterial activity of and target inhibition by McC.
McC uptake, processing & target inhibition
▪ The YejABEF peptide transporter provides the only route of McC entry into enteric bacteria.
▪ McC is processed inside sensitive cells by aminopeptidases with release of a processed McC, toxic nonhydrolyzable aspartyl-adenylate.
▪ McC analogs with an altered terminal amino acid retain the Trojan-horse mechanism of action but target aminoacyl-tRNA synthetases specified by an amino acid coupled to AMP.
Putative McC-like compounds encoded by mcc operons from bacteria other than E. coli
▪ Clustered homologs of E. coli mcc genes can be identified in diverse bacteria.
▪ Genes coding for putative McC-like peptides of different length can be identified.
Mechanisms of McC resistance
▪ Inside producing cells, processed McC that accumulates inside the producing cell is inactivated by acetylation catalyzed by the C-terminal domain of the MccE protein acetyltransferase.
▪ Both unprocessed and processed McC is inactivated by the MccF peptidase that cleaves an amide bond connecting the peptidyl (aminoacyl) and nucleotidyl part of McC.
Acknowledgments
Work in K Severinov’s laboratories is supported by NIH/NIAID North Eastern Biodefense Center grant U54 AI057158, Rutgers University Technology Commercialization Fund, and Russian Academy of Sciences Molecular and Cell Biology Program grant.
Footnotes
Financial & competing interests disclosure The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Ibba M, Soll D. Aminoacyl-tRNA synthesis. Ann. Rev. Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 2.Ling J, Reynolds N, Ibba M. Aminoacyl-tRNA synthesis and translational quality control. Ann. Rev. Microbiol. 2009;63:61–78. doi: 10.1146/annurev.micro.091208.073210. [DOI] [PubMed] [Google Scholar]
- 3.Tao J, Schimmel P. Inhibitors of aminoacyl-tRNA synthetases as novel anti-infectives. Exp. Opin. Invest. Drugs. 2000;9:1767–1775. doi: 10.1517/13543784.9.8.1767. [DOI] [PubMed] [Google Scholar]
- 4.Tao J, Wendler P, Connelly G, et al. Drug target validation: lethal infection blocked by inducible peptide. Proc. Natl Acad. Sci. USA. 2000;97:783–786. doi: 10.1073/pnas.97.2.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schimmel P, Tao J, Hill J. Aminoacyl tRNA synthetases as targets for new anti-infectives. FASEB J. 1998;12:1599–1609. [PubMed] [Google Scholar]
- 6.Reader JS, Ordoukhanian PT, Kim JG, et al. Major biocontrol of plant tumors targets tRNA synthetase. Science. 2005;309:1533. doi: 10.1126/science.1116841. [DOI] [PubMed] [Google Scholar]
- 7.Stefanska AL, Fulston M, Houge-Frydrych CS, Jones JJ, Warr SR. A potent seryl tRNA synthetase inhibitor SB-217452 isolated from a Streptomyces species. J. Antibiot. (Tokyo) 2000;53:1346–1353. doi: 10.7164/antibiotics.53.1346. [DOI] [PubMed] [Google Scholar]
- 8.Metlitskaya A, Kazakov T, Kommer A, et al. Aspartyl-tRNA synthetase is the target of peptide nucleotide antibiotic microcin C. J. Biol. Chem. 2006;281:18033–18042. doi: 10.1074/jbc.M513174200. [DOI] [PubMed] [Google Scholar]
- 9.Asensio C, Perez-Diaz JC, Martinez MC, Baquero F. A new family of low molecular weight antibiotics from enterobacteria. Biochem. Biophys. Res. Comm. 1976;69:7–14. doi: 10.1016/s0006-291x(76)80264-1. [DOI] [PubMed] [Google Scholar]
- 10.Baquero F, Bouanchaud D, Martinez-Perez MC, Fernandez C. Microcin plasmids: a group of extrachromosomal elements coding for low-molecular-weight antibiotics in Escherichia coli. J. Bacteriol. 1978;135:342–347. doi: 10.1128/jb.135.2.342-347.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Severinov K, Semenova E, Kazakov A, Kazakov T, Gelfand MS. Low-molecular-weight post-translationally modified microcins. Mol. Microbiol. 2007;65:1380–1394. doi: 10.1111/j.1365-2958.2007.05874.x. [DOI] [PubMed] [Google Scholar]
- 12.Guijarro JI, Gonzalez-Pastor JE, Baleux F, et al. Chemical structure and translation inhibition studies of the antibiotic microcin C7. J. Biol. Chem. 1995;270:23520–23532. doi: 10.1074/jbc.270.40.23520. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez-Pastor JE, San Millan JL, Moreno F. The smallest known gene. Nature. 1994;369:281. doi: 10.1038/369281a0. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez-Pastor JE, San Millan JL, Castilla MA, Moreno F. Structure and organization of plasmid genes required to produce the translation inhibitor microcin C7. J. Bacteriol. 1995;177:7131–7140. doi: 10.1128/jb.177.24.7131-7140.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fomenko DE, Metlitskaya AZ, Peduzzi J, et al. Microcin C51 plasmid genes: possible source of horizontal gene transfer. Antimicrob. Agents Chemother. 2003;47:2868–2874. doi: 10.1128/AAC.47.9.2868-2874.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roush RF, Nolan EM, Lohr F, Walsh CT. Maturation of an Escherichia coli ribosomal peptide antibiotic by ATP-consuming N–P bond formation in microcin C7. J. Am. Chem. Soc. 2008;130:3603–3609. doi: 10.1021/ja7101949. ▪▪ Describes the unusual mechanism of MccA peptide adenylation by the MccB synthase.
- 17.Regni CA, Roush RF, Miller DJ, Nourse A, Walsh CT, Schulman BA. How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic. EMBO J. 2009;28:1953–1964. doi: 10.1038/emboj.2009.146. ▪▪ Describes the structure of microcin C (McC) synthase, alone and in complex with McC precursor prepetide.
- 18.Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat. Rev. Mol. Cell. Biol. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kazakov T, Metlitskaya A, Severinov K. Amino acid residues required for maturation, cell uptake, and processing of translation inhibitor microcin C. J. Bacteriol. 2007;189:2114–2118. doi: 10.1128/JB.01609-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Metlitskaya A, Kazakov T, Vondenhoff GH, et al. Maturation of the translation inhibitor microcin C. J. Bacteriol. 2009;191:2380–2387. doi: 10.1128/JB.00999-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Novikova M, Metlitskaya A, Datsenko KA, et al. The Escherichia coli Yej transporter is required for the uptake of translation inhibitor microcin C. J. Bacteriol. 2007;189:8361–8365. doi: 10.1128/JB.01028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vondenhoff GH, Dubiley S, Severinov K, Lescrinier E, Rozenski J, Van Aerschot A. Extended targeting potential and improved synthesis of Microcin C analogs as antibacterials. Bioorg. Med. Chem. 2011;19:5462–5467. doi: 10.1016/j.bmc.2011.07.052. [DOI] [PubMed] [Google Scholar]
- 23.Eswarappa SM, Panguluri KK, Hensel M, Chakravortty D. The yejABEF operon of Salmonella confers resistance to antimicrobial peptides and contributes to its virulence. Microbiology. 2008;154:666–678. doi: 10.1099/mic.0.2007/011114-0. [DOI] [PubMed] [Google Scholar]
- 24.Hummel S, Apte RN, Qimron U, Vitacolonna M, Porgador A, Zöller M. Tumor vaccination by Salmonella typhimurium after transformation with a eukaryotic expression vector in mice: impact of a Salmonella typhimurium gene interfering with MHC Class 1 presentation. J. Immunother. 2005;28:467–479. doi: 10.1097/01.cji.0000170359.92090.8b. [DOI] [PubMed] [Google Scholar]
- 25.Kazakov T, Vondenhoff GH, Datsenko KA, et al. Escherichia coli peptidase A, B, or N can process translation inhibitor microcin C. J. Bacteriol. 2008;190:2607–2610. doi: 10.1128/JB.01956-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novikova M, Kazakov T, Vondenhoff GH, et al. MccE provides resistance to protein synthesis inhibitor microcin C by acetylating the processed form of the antibiotic. J. Biol. Chem. 2010;285:12662–12669. doi: 10.1074/jbc.M109.080192. ▪ A novel mechanism of McC resistance due to the action of MccE acetyltransferase is described.
- 27.Van de Vijver P, Vondenhoff GH, Kazakov TS, et al. Synthetic microcin C analogs targeting different aminoacyl-tRNA synthetases. J. Bacteriol. 2009;191:6273–6280. doi: 10.1128/JB.00829-09. ▪▪ A synthetic strategy allowing for the generation of McC variants that target an aminoacyl-tRNA synthetase of choice is described.
- 28.Vondenhoff GH, Blanchaert B, Geboers S, et al. Characterization of peptide chain length and constituency requirements for YejABEF-mediated uptake of microcin C analogues. J. Bacteriol. 2011;193:3618–3623. doi: 10.1128/JB.00172-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agarwal V, Metlitskaya A, Severinov K, Nair SK. Structural basis for microcin C7 inactivation by the MccE acetyltransferase. J. Biol. Chem. 2011;286:21295–21303. doi: 10.1074/jbc.M111.226282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fechter P, Brownlee GG. Recognition of mRNA cap structures by viral and cellular proteins. J. Gen. Virol. 2005;86:1239–1249. doi: 10.1099/vir.0.80755-0. [DOI] [PubMed] [Google Scholar]
- 31.Shuman S, Lima CD. The polynucleotide ligase and RNA capping enzyme superfamily of covalent nucleotidyltransferases. Curr. Opin. Struct. Biol. 2004;14:757–764. doi: 10.1016/j.sbi.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 32.Tikhonov A, Kazakov T, Semenova E, et al. The mechanism of Microcin C resistance provided by the MccF peptidase. J. Biol. Chem. 2010;285:12662–12669. doi: 10.1074/jbc.M110.179135. ▪ A novel mechanism of McC resistance due to the action MccF peptidase is described.
- 33.Metz R, Henning S, Hammes WP. LD-carboxypeptidase activity in Escherichia coli. II. Isolation, purification and characterization of the enzyme from E. coli K 12. Arch. Microbiol. 1986;144(2):181–186. doi: 10.1007/BF00414732. [DOI] [PubMed] [Google Scholar]
- 34.Kim JG, Park BK, Kim SU, et al. Bases of biocontrol: sequence predicts synthesis and mode of action of agrocin 84, the Trojan horse antibiotic that controls crown gall. Proc. Natl Acad. Sci. USA. 2006;103:8846–8851. doi: 10.1073/pnas.0602965103. [DOI] [PMC free article] [PubMed] [Google Scholar]