Abstract
Meroterpenoids are a class of fungal natural products that are produced from polyketide and terpenoid precursors. An understanding of meroterpenoid biosynthesis at the genetic level should facilitate engineering of second-generation molecules and increasing production of first-generation compounds. The filamentous fungus Aspergillus nidulans has previously been found to produce two meroterpenoids, austinol and dehydroaustinol. Using targeted deletions that we created, we have determined that, surprisingly, two separate gene clusters are required for meroterpenoid biosynthesis. One is a cluster of four genes including a polyketide synthase gene, ausA. The second is a cluster of ten additional genes including a prenyltransferase gene, ausN, located on a separate chromosome. Chemical analysis of mutant extracts enabled us to isolate 3,5-dimethylorsellinic acid and ten additional meroterpenoids that are either intermediates or shunt products from the biosynthetic pathway. Six of them were identified as novel meroterpenoids in this study. Our data, in aggregate, allow us to propose a complete biosynthetic pathway for the A. nidulans meroterpenoids.
INTRODUCTION
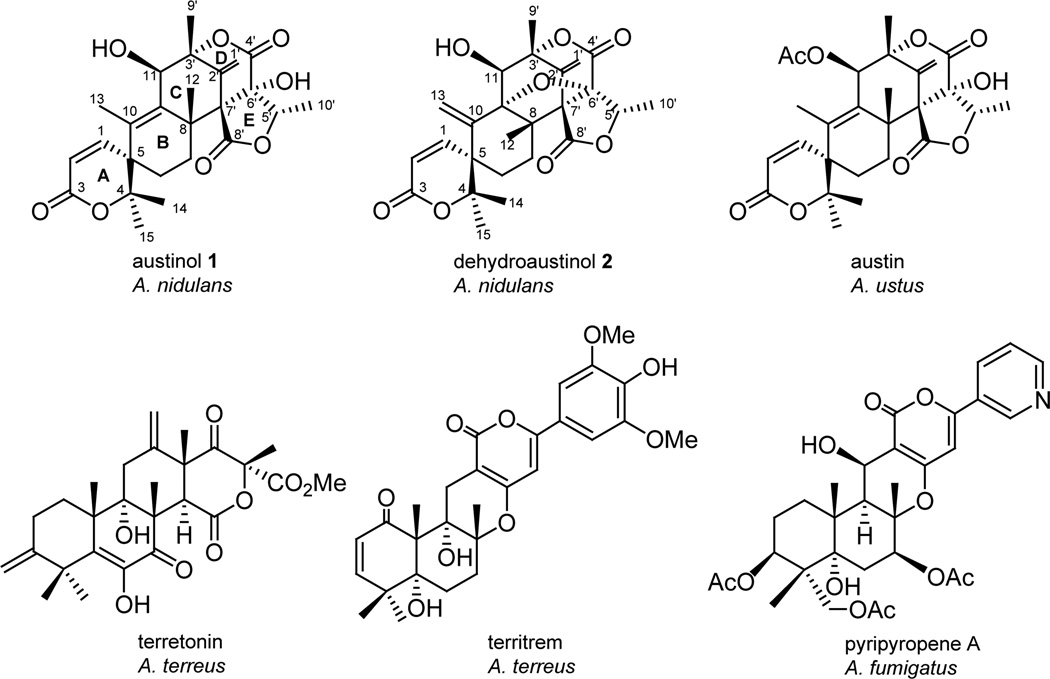
Fungal species are known to produce a variety of structurally complex secondary metabolites many of which have important relevance to human health.1 Members of the genus Aspergillus can combine polyketide and terpenoid precursors to produce structurally complex secondary metabolites called meroterpenoids.2 Examples of known meroterpenoids include terretonin and territrem produced by A. terreus,3,4 pyripyropene A produced by certain strains of A. fumigatus,5,6 austin produced by A. ustus7 and austinol (1) and dehydroaustinol (2) produced by A. nidulans (Fig. 1).8,9 Territrem is a potent irreversible inhibitor of acetylcholinesterase (AChE) and is a candidate for drug development for treating Alzheimer’s disease.4 Pyripyropene is a potent selective inhibitor of acyl-CoA cholesterol acyltransferase (ACAT), an enzyme that catalyzes intracellular esterification of cholesterol and is being developed for treating and preventing atherosclerosis.5,6
Figure 1.
Meroterpenoids from Aspergillus species.
A series of labeling studies in the eighties provided a framework for the biosynthetic mechanism of meroterpenoids.10,11,12 Recently the biosynthetic gene cluster of pyripyropene was identified in A. fumigatus and verified using a heterologous fungal expression approach.13 The pyripyropene gene cluster encodes one nonreducing polyketide synthase (NR-PKS), one prenyltransferase, one CoA ligase, two cytochrome P450s, one FAD-dependent monooxygenase, one terpene cyclase and two acetyltransferases.
In this study we attempted to locate the biosynthesis genes for austinol (1) and dehydroaustinol (2) in A. nidulans by searching the A. nidulans genome for regions where a NR-PKS gene and a prenyltransferase gene are in proximity. Interestingly no prenyltransferase genes were located near NR-PKS genes. This suggested to us that the genes responsible for austinol (1) and dehydroaustinol (2) biosynthesis in A. nidulans might be separated in the genome. Although secondary metabolite biosynthesis genes are generally clustered in fungal genomes,1 we recently showed that the genes that encode the prenyl xanthone biosynthesis pathway in A. nidulans are located in three separate regions of the genome.14
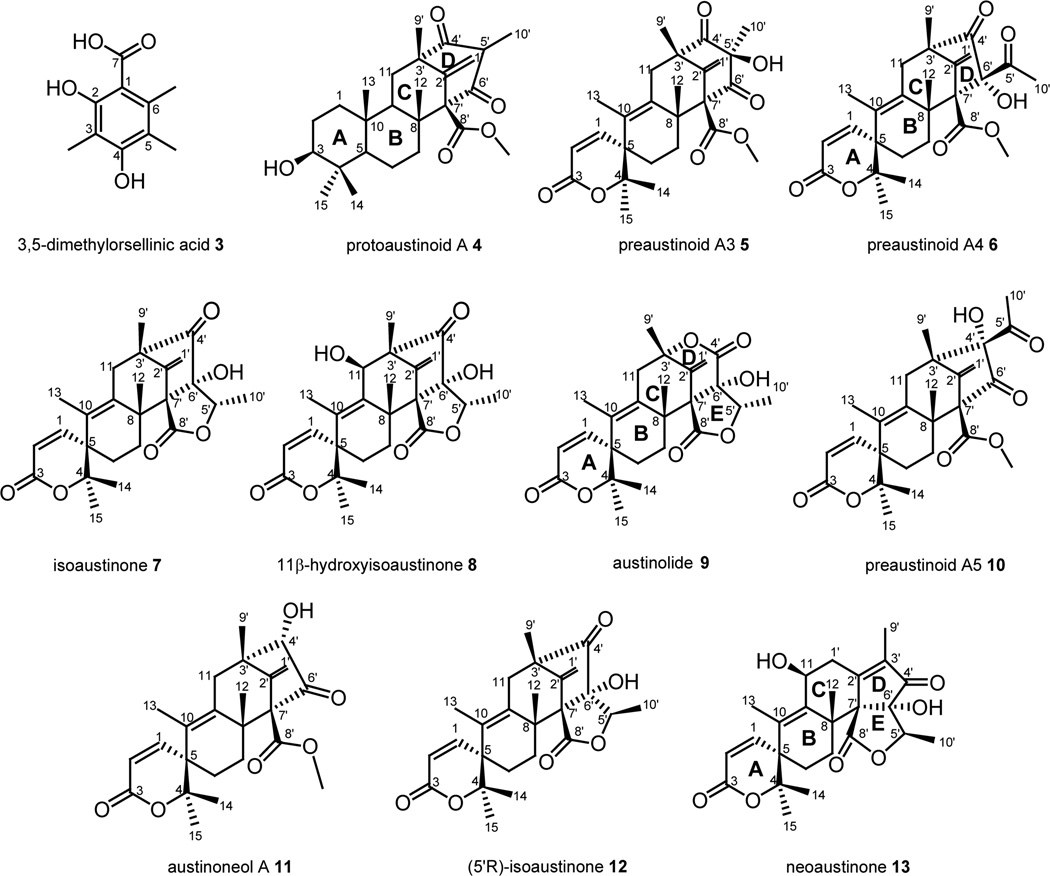
We identified, using a set of deletions of all non-reducing PKS genes, the NR-PKS responsible for the biosynthesis of the meroterpenoids in A. nidulans. This NR-PKS has been independently identified by Nielsen et al. and designated ausA.15 Next we selected two candidate prenyltransferase genes located elsewhere in the genome for deletion and from metabolome analysis of mutant extracts, we found that one was involved in meroterpenoid synthesis. To identify additional genes involved in the pathway we targeted the genes surrounding the NR-PKS gene and the prenyltransferase gene for deletion. The gene deletion experiments allowed us to identify twelve additional genes involved in meroterpenoid biosynthesis, that are located in two gene clusters, one containing the NR-PKS and the other containing the prenyltransferase. Large-scale cultures of the deletion strains enabled us to isolate several stable intermediates (Fig. 2) including six novel compounds allowing us to propose a biosynthesis pathway for austinol (1) and dehydroaustinol (2) biosynthesis.
Figure 2.
Intermediates isolated from gene deletion mutants.
RESULTS
Identification of the polyketide synthase responsible for austinol (1) and dehydroaustinol (2) biosynthesis
When cultivated on solid yeast agar glucose (YAG) medium A. nidulans produces two meroterpenoids, austinol (1) and dehydroaustinol (2) (Fig. 1).8,9 The structural similarity of these two compounds suggests that they may have the same biosynthetic origin. For this study we used an A. nidulans strain containing the nkuAΔ and stcJΔ mutations. The nkuAΔ deletion allows high homologous recombination rates and facilitates gene targeting. stcJΔ prevents production of the major A. nidulans polyketide sterigmatocystin. The rationale for the use of an stcJΔ strain is that the elimination of sterigmatocystin frees up the common polyketide precursor malonyl-CoA and also facilitates the detection and isolation of other metabolites. Early studies using labeled precursors showed that austin, which is very similar to austinol (1), is formed by the C-alkylation of 3,5-dimethylorsellinic acid, an aromatic polyketide, with farnesyl pyrophosphate.10 Because fungal secondary metabolite biosynthesis genes are usually clustered, we first looked for a PKS near a prenyltransferase in the A. nidulans genome in an effort to identify the meroterpenoid gene cluster. Surprisingly, we found no PKS near a prenyltransferase on any chromosome, leading us to conclude that the PKS and prenyltransferase genes required for austinol (1) and dehydroaustinol (2) biosynthesis must be separated in the genome. We consequently used a genome-wide gene targeting approach to locate the austinol (1) and dehydroaustinol (2) biosynthesis genes.
To locate the PKS responsible for austinol (1) and dehydroaustinol (2) we first determined, based on a comparison to other previously characterized fungal aromatic polyketides, that the aromatic structure of the predicted precursor 3,5-dimethylorsellinic acid (3) (Fig. 2) would likely require a non-reducing PKS (NR-PKS). Previously, we created NR-PKS knock out libraries in two different genetic backgrounds in A. nidulans to elucidate products of these NR-PKSs. Using these libraries we identified the NR-PKS involved in the biosynthesis of asperthecin,9 emodin,16 the prenylated xanthones,14 orsellinic acid and F-9775A/B.16,17 Separately we demonstrated that the NR-PKS gene AN1034.4 is involved in asperfuranone biosynthesis inducing a nearby transcription factor located within the gene cluster.18 The products of the remaining six NR-PKS (encoded by AN0523.4, AN2032.4, AN3230.4, AN3386.4, AN6448.4, AN7071.4) were unknown. Strains carrying these deletions were cultivated under austinol (1) and dehydroaustinol (2) producing conditions and metabolites were analyzed by liquid chromatography-diode array-mass spectrometry (LC-DAD-MS). We found that all these strains continued to produce the two meroterpenoids (data not shown) and, thus, that none of these NR-PKS is involved in meroterpenoid production.
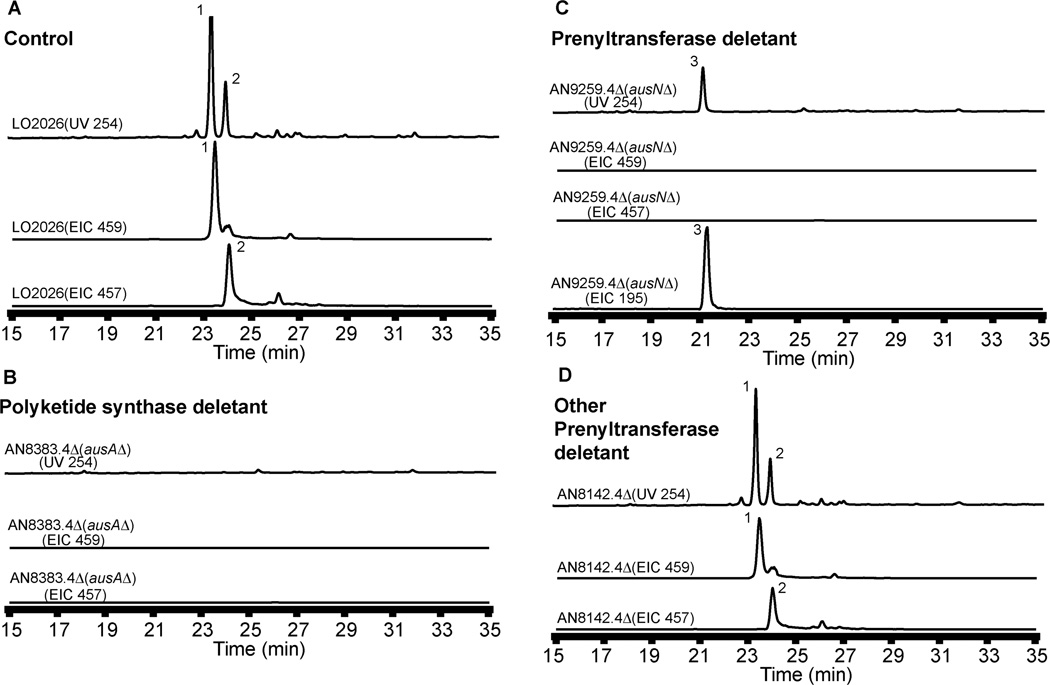
We reexamined the A. nidulans genome using the gene annotations of the Central Aspergillus Data Repository (CADRE) (http:www.cadre-genomes.org.uk) and the Aspergillus genome database (http://www.aspgd.org) and located AN8383.4, encoding a putative NR-PKS gene for which we had not created a deletant mutant in our previous studies due to an error in the initial annotation. Expression of AN8383.4 in another project revealed that it is responsible for the synthesis of 3,5-dimethylorsellinic acid, a likely precursor of the meroterpenoids (M. Ahuja et al., submitted). We consequently created deletion strains for AN8383.4 (AN8383.4Δ) using the gene-targeting procedures we have recently developed for A. nidulans involving a nkuAΔ strain and fusion PCR.19,20 Diagnostic PCR using three different primer sets confirmed that AN8383.4 was deleted correctly and we have used the same technique to verify the deletions in all strains used in this study (Fig.S3 and S4). The AN8383.4Δ strain was cultivated under austinol/dehydroaustinol producing conditions and LC-DAD-MS analysis showed that it failed to produce the two meroterpenoids (Fig. 3). These data allow us to identify AN8383.4 as the gene responsible for the biosynthesis of austinol (1) and dehydroaustinol (2).
Figure 3.
HPLC profile of extracts from (A) LO2026 (background control strain), (B) AN8383.4Δ PKS deletant (C) AN9259.4Δ prenyltransferase deletant and (D) AN8142.4Δ as detected by UV at 254 nm and mass spectrometry. Austinol 1 [M+H]+ m/z = 459 and dehydroaustinol 2 [M+H]+ m/z = 457 are detected using positive ion mode and 3,5-dimethylorsellinic acid 3 [M-H]− m/z = 195 is detected using negative ion mode. EIC = Extracted Ion Chromatogram.
AN8383.4 is located on chromosome V at bp 307999-315732. AN8383.4 is predicted to encode a 2476 amino acid NR-PKS which contains starter unit ACP transacylase (SAT), β-ketoacyl synthase (KS), acyltransferase (AT), product template (PT), acyl carrier protein (ACP), C-methyltransferase (C-MeT) and thiolesterase (TE) domains. During the course of this work, Nielson et al. reported the independent discovery that AN8383.4 is involved in austinol/dehydroaustinol synthesis and designated it ausA.15 For consistency, we will also use this designation.
Identification of the prenyltransferase involved in austinol and dehydroaustinol biosynthesis
The prenylation of 3,5-dimethylorsellinic acid produced by the NR-PKS AusA (the protein product of the ausA gene) must be catalyzed by a prenyltransferase. Since none of the genes located near ausA have homology to any known prenyltransferase, we looked to other parts of the genome to locate the necessary prenyltransferase. We performed a BLAST search of the A. nidulans genome using the amino acid sequence of UbiA as a query to search for its homologs. The UbiA enzyme is involved in the biosynthesis of prenylated quinones such as ubiquinone in E. coli.21 This enzyme catalyzes the transfer of all-trans prenyl moieties to the acceptor molecule 4-hydroxybenzoate (4HB). This enzyme was selected because it catalyzes C-C bond formation between an aromatic substrate and an isoprenoid diphosphate.
The top two candidate genes in the BLAST search were AN9259.4 and AN8142.4. Deletions were created and verified for each. Cultivation and analysis of the deletant strain by LC-DAD-MS showed that the AN8142.4 deletant (AN8142.4Δ) continued to synthesize austinol (1) and dehydroaustinol (2) while AN9259.4Δ failed to produce the two meroterpenoids (Fig. 3). We deduced therefore that AN9259.4 is required for austinol (1) and dehydroaustinol (2) biosynthesis and we designate it ausN.
From an ausNΔ strain we isolated, and characterized by NMR and mass spectrometry, 3,5-dimethylorsellinic acid (3) the polyketide precursor of austin biosynthesis predicted from earlier labeling studies.10 We also isolated this precursor in our AN8383.4 overexpression studies (M. Ahuja et al., submitted) indicating that ausA makes 3 which is subsequently modified by the product of ausN. Both the genetic data and the isolation of 3,5-dimethylorsellinic acid (3), thus, demonstrate that ausN is the prenyltransferase involved in biosynthesis of the two meroterpenoids. It is located on chromosome VIII while ausA is on chromosome V.
Targeted gene deletions reveal additional genes required for meroterpenoid biosynthesis
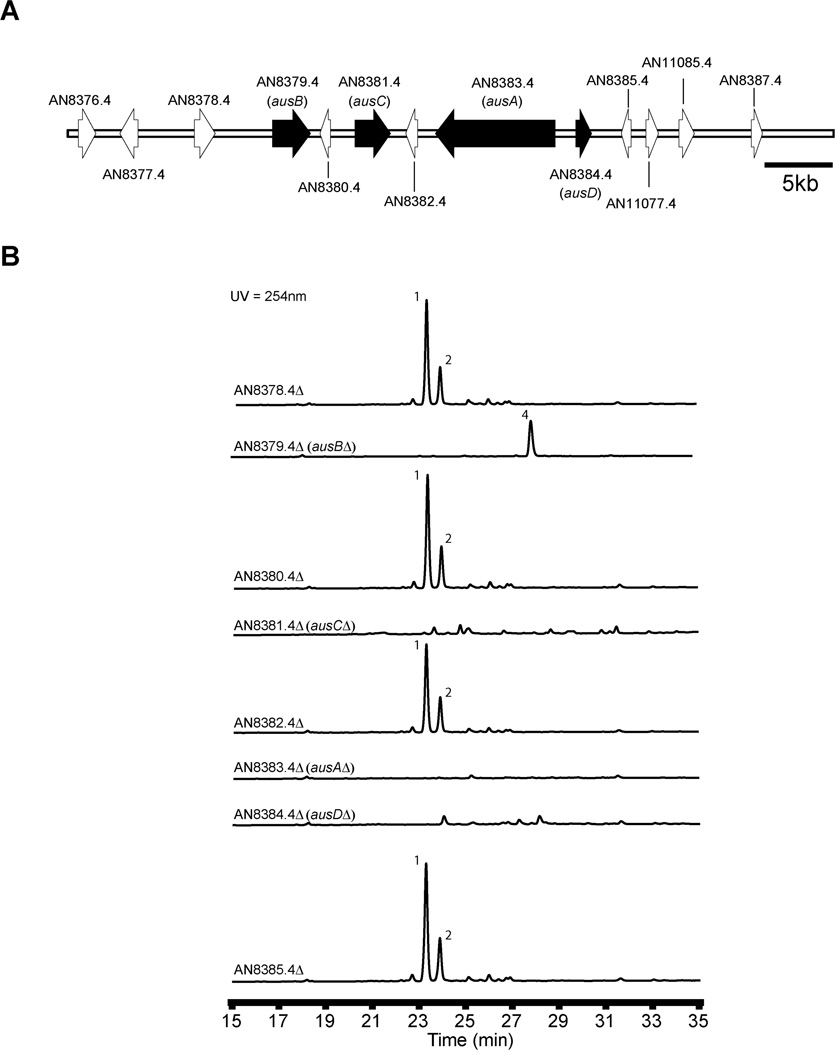
Early labeling studies showed that a series of oxidation steps are necessary for the biosynthesis of the austinol related molecule austin.12 Since the genes involved in the biosynthesis of particular secondary metabolites are usually clustered in fungi,22 we wished to delete genes surrounding ausA and ausN to identify other genes required for austinol and dehydroaustinol biosynthesis. A problem with respect to the ausA region was that there was a gap near ausA in the genome (i.e. the contiguous sequence containing ausA could not be connected to the adjacent contiguous sequence apparently because sequences were missing). To fill in this gap we amplified A. nidulans genomic DNA using primers flanking the gap. We sequenced the resulting fragment and found that the apparent gap was actually due to the insertion of an incorrect sequence. Once this sequence was eliminated the sequences on the two sides of the “gap” overlapped and matched perfectly and the gap was, thus, eliminated. We found that AN8381.4 extended across the previously incorrectly annotated gap. We communicated this information to CADRE and the gap has now been corrected in the CADRE and ASPGD databases.
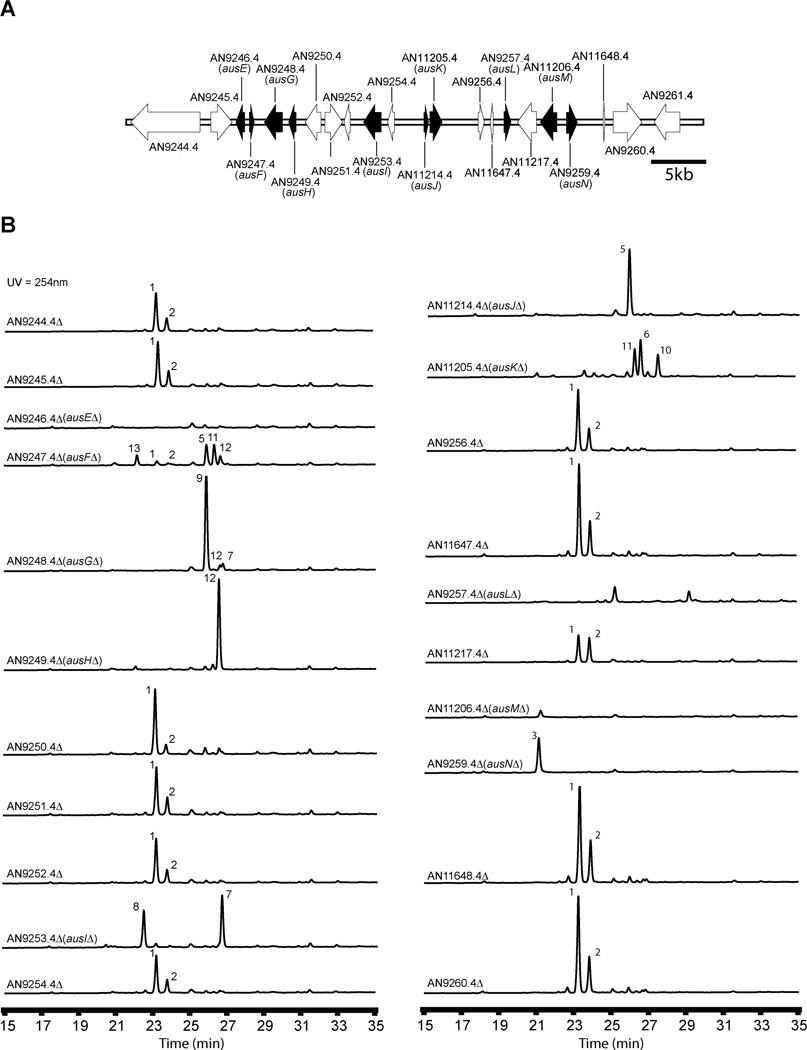
We deleted twelve genes surrounding ausA (AN8376.4, AN8377.4, AN8378.4, AN8379.4, AN8380.4, AN8381.4, AN8382.4, AN8384.4, AN8385.4, AN11077.4, AN11085.4, and AN8387.4) (Fig. 4A). Three putative deletants for each gene were verified by diagnostic PCR using three different primer sets. For each gene at least two of the three deletants proved correct (Fig. S4). At least two strains per gene were cultivated under austinol/dehydroaustinol producing conditions and the culture extracts screened by LC/MS. Deletions of AN8379.4, AN8381.4, AN8384.4 fully eliminated the production of both meroterpenoids (Fig. 4B) and we designate these genes ausB, ausC, and ausD respectively. We were able to detect by LC/MS a new UV active compound 4 from the ausBΔ strains. An ausBΔ strain was grown at a large scale and compound 4 was isolated by flash chromatography and preparative scale HPLC. Using both 1D and 2D NMR spectroscopy we determined the structure of compound 4 (NMR data shown in supporting information), which we named protoaustinoid A (4). Protoaustinoid A (4) has a tetracyclic structure with a 1,3-diketone functional group. A similar compound was proposed earlier as a key early intermediate in austin biosynthesis.12 Protoaustinoid A contains a methyl ester connected to carbon 8' instead of the carboxylic function group in the previously proposed structure.12
Figure 4.
(A) Organization of the polyketide synthase gene cluster (cluster A) for austinol/dehydroaustinol biosynthesis in A. nidulans. Each arrow indicates relative size and the direction of transcription of the ORFs deduced from analysis of nucleotide sequences. On the basis of our gene deletion results, genes in black are involved in austinol/dehydroaustinol biosynthesis while those in white are not. (B) HPLC profile of extracts of strains in the cluster as detected by UV absorption at 254 nm. Numbering on peaks corresponds to the intermediates shown in Figure 2.
From both AN8381.4Δ and AN8384.4Δ strains we were unable to obtain sufficient amounts of stable intermediates to elucidate the roles of the two genes in the biosynthetic pathway. We believe we have established one border of the gene cluster surrounding the PKS using the AN8376.4Δ, AN8377.4Δ, and AN8378.4Δ strains since these strains continue to produce the two meroterpenoids. The other border is established by the AN8385.4Δ, AN11077.4Δ, AN11085.4Δ, and AN8387.4Δ strains since these strains also continue to produce the two meroterpenoids. Our deletion data have, thus, allowed us to identify three genes surrounding the PKS gene ausA that are involved in the pathway.
The structural complexity of austinol/dehydroaustinol led us to believe that additional genes are necessary for their biosynthesis. We hypothesized that they are located adjacent to the prenyltransferase gene ausN and that deletion of genes in this region would reveal the remaining genes required for meroterpenoid biosynthesis. Initially we targeted nine genes surrounding the prenyltransferase gene ausN. The nine genes were AN11205.4, AN9256.4, AN11647.4, AN9257.4, AN11217.4 AN11206.4, AN11648.4, AN9260.4, and AN9261.4 (Fig. 5A). Three putative deletants for each gene were tested by diagnostic PCR using three different primer sets. At least two correct deletants per gene were cultivated under meroterpenoid producing conditions and screened by reverse-phase LC/MS. Metabolite profiles of AN11648.4, AN9260.4, and AN9261.4 deletants showed that they are not involved in austinol/dehydroaustinol biosynthesis and they define one of the boundaries for this cluster (Fig. 5 and Fig. S2). Because LC/MS analysis showed that AN11205.4 (ausK) is involved in the biosynthesis of the two meroterpenoids, we created additional gene deletants beyond AN11205.4 to look for the left boundary. We deleted twelve additional genes AN9244.4, AN9245.4, AN9246.4, AN9247.4, AN9248.4, AN9249.4, AN9250.4, AN9251.4, AN9252.4, AN9253.4, AN9254.4, AN11214.4, a total of 22 genes for this cluster, including ausN. We verified at least two deletants for each gene and used them for LC/MS analysis. The results show that AN9246.4 (ausE), AN9247.4 (ausF), AN9248.4 (ausG), AN9249.4 (ausH), AN9253.4 (ausI), AN11214.4 (ausJ), AN11025.4 (ausK), AN9257.4 (ausL) and AN11206.4 (ausM), are involved in the biosynthesis pathway since they fully eliminated the production of both meroterpenoids, except that deletion of AN9247.4 (ausF) permitted minor production of austinol/dehydroaustinol (Fig. 5B). The low production of austinol/dehydroasutinol by AN9247.4 deletion strains was confirmed by MS/MS (data not shown). This suggests that AN9247.4 might be involved in a late step of the biosynthesis pathway or, more likely, may have a regulatory function. Our gene deletion data in total allow us to deduce that AN9246.4 and AN9259.4 define the borders of the gene cluster surrounding ausN.
Figure 5.
(A) Organization of the prenyltransferase gene cluster (cluster B) for austinol/dehydroaustinol biosynthesis in A. nidulans. Each arrow indicates the relative size and the direction of transcription of the ORFs deduced from analysis of nucleotide sequences. On the basis of our gene deletion results, genes in black are involved in austinol/dehydroaustinol biosynthesis while those in white are not. (B) HPLC profile of extracts of strains in the cluster as detected by UV absorption at 254 nm. Numbering on peaks corresponds to the intermediates shown in Figure 2.
Several of our deletion strains produced sufficient amounts of chemically stable metabolites to allow us to isolate them and determine their structures using 1D and 2D NMR spectroscopy (NMR data for all the intermediates are shown in supporting information.). Compounds 5 – 13 isolated from large-scale cultures of the deletant strains are all meroterpenoids that are either proposed intermediates or shunt products in the austinol (1) and dehydroaustinol (2) biosynthesis pathways. Preaustinoid A3 (5), isoaustinone (7), austionolide (9) and austinoneol A (11) have been previously isolated and identified from Penicillium sp and have been proposed to be intermediates in austin biosynthesis.23 Our NMR analysis of these four compounds are in agreement with published NMR data of these compounds. The remaining chemicals that accumulated in the deletion strains were structurally characterized and found to be novel materials. We designate them preaustinoid A4 (6), 11β-hydroxyisoaustinone (8), preaustinoid A5 (10), (5'R)-isoaustinone (12), and neoaustinone (13). X-ray crystallographic analysis confirmed the absolute configuration of (5'R) – isoaustinone (12) (Fig. S5).
DISCUSSION
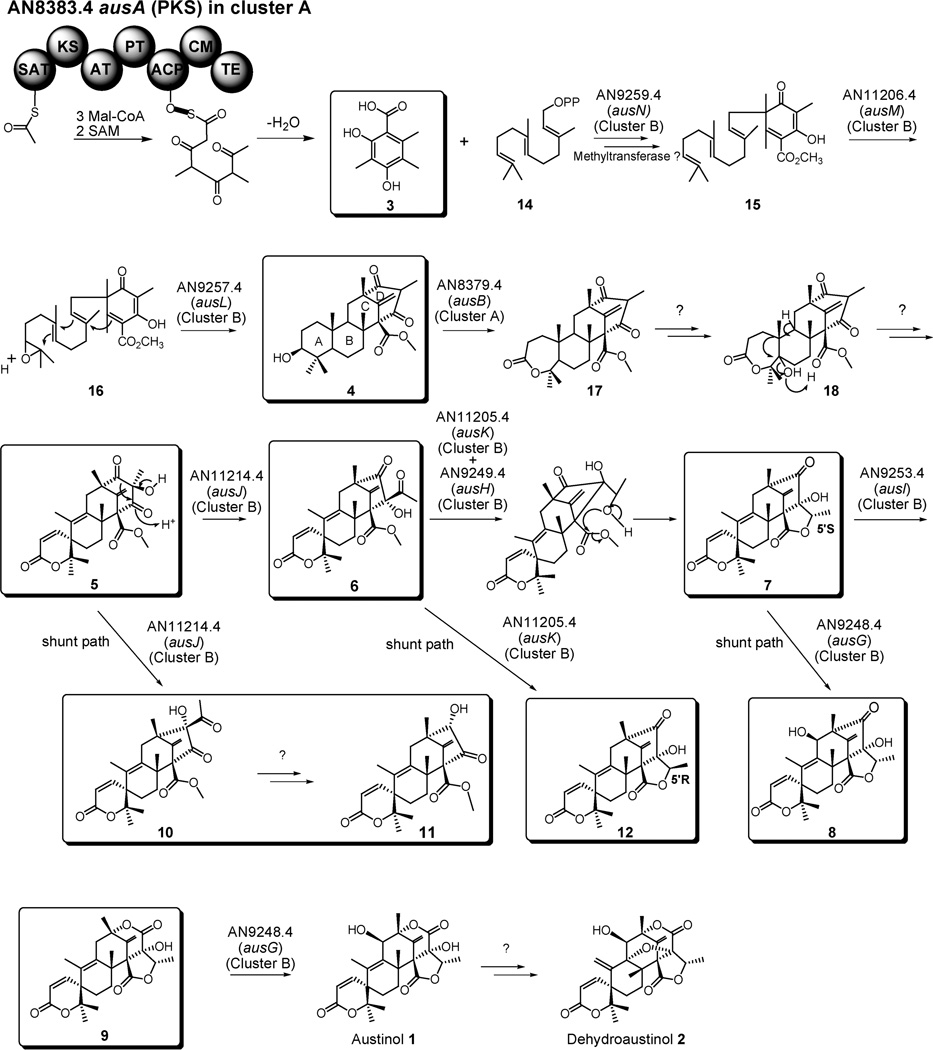
Using a series of targeted gene deletions, we have identified the genes encoding the biosynthesis pathway for the meroterpenoids austinol (1) and dehydroaustinol (2) in A. nidulans (Fig. 6). The pathway involves at least 14 genes in two separate gene clusters. Three genes are clustered around the polyketide synthase gene AN8383.4 (ausA) located on chromosome V and nine genes are clustered around the prenyltransferase gene AN9259.4 (ausN) located on chromosome VIII. For ease of reference we will call these two clusters, cluster A and cluster B respectively. Our findings illustrate the power of combining genomics, efficient gene targeting and natural products chemistry. Without combining these approaches, it would not have been possible to identify the genes involved in austinol and dehydroaustinol biosynthesis nor to obtain adequate data to propose a biosynthetic pathway for these compounds.
Figure 6.
Proposed biosynthesis pathway of austinol/dehydroaustinol. The clusters in which the genes are found (Cluster A or Cluster B) are indicated.
In our proposed pathway, the first gene, ausA, encodes a polyketide synthase for 3,5-dimethylorsellinic acid (3) synthesis. AusA contains the SAT, KS, AT, PT, ACP, CMet, and TE domains, and the lack of ER, DH, and KR domains indicates that it is an NR-PKS.24 This NR-PKS may utilize one acetyl-CoA, three malonyl-CoA, and two SAM (S-adenosyl methionine) to produce compound 3.
In the next step, C-alkylation of compound 3, an aromatic polyketide, by farnesyl pyrophosphate is catalyzed by an aromatic prenyltransferase AusN. We were able to isolate 3,5-dimethylorsellinic acid (but not austinol, dehydroaustinol nor any other intermediate) from the ausN deletion strain, which provides solid evidence that ausN encodes the prenyltransferase in the pathway and that the prenylation step occurs very early in the pathway, before other intermediates are synthesized. BLAST analysis revealed that AusN has 49% protein sequence similarity to LePGT-1 an aromatic prenyltransferase (Table 1). The enzymatic activity of LePGT-1 was examined by Yazaki et al.25 They showed that LePGT-1, which is a homologous to UbiA, makes 4-hydroxybenzoate an intermediate in shikonin biosynthesis in Lithospermum erythrorhizon. Analysis of the predicted protein sequence of AusN led to the identification of an aspartate-rich motif (88-NDLVDVDID-96) similar to LePGT-1. This NDXXDXXXD motif, which binds the prenyl diphosphate via a Mg2+ ion, is highly conserved among all prenyltransferases involved in lipoquinone biosynthesis.26 Unlike prenyltransferase enzymes such as protein prenyltransferases or fungal indole prenyltransferases, which are water soluble, lipoquinone prenyltransferases are membrane bound.26 In addition, AusN was predicted to have nine transmembrane helices by MEMPACK [on the PsiPred server (http://bioinf.cs.ucl.ac.uk/psipred/)]. Therefore, AusN could be a membrane bound aromatic prenyltransferase that catalyzes the transfer of a prenyl moiety to an aromatic acceptor molecule.
Table 1.
The genes required for austinol/dehydroaustinol biosynthesis in A. nidulans
| ORFs | Homologs | ||
|---|---|---|---|
| Gene | Predicted size (aa) | Match from BLAST search at NCBI (accession no.) | Identity/s imilarity (%) |
| AN8379.4 (ausB) | 745 | cyclopentadecanone 1,2-monooxygenase [Pseudomonas sp. HI-70] (BAE93346) | 37/54 |
| AN8381.4 (ausC) | 638 | cyclododecanone monooxygenase [Rhodococcus ruber] (AAL14233) | 43/56 |
| AN8383.4 (ausA) | 2476 | methylorcinaldehyde synthase [Acremonium strictum] (CAN87161) | 32/51 |
| AN8384.4 (ausD) | 282 | Hypothetical protein | |
| AN9246.4 (ausE) | 298 | Fumonisin C-5 hydroxylase Fum3p [Gibberella moniliformis] (AAG27131) | 34/52 |
| AN9247.4 (ausF) | 177 | Hypothetical protein | |
| AN9248.4 (ausG) | 529 | P450 monooxygenase [Sphaceloma manihoticola] (CAP07651) | 40/59 |
| AN9249.4 (ausH) | 174 | Hypothetical protein | |
| AN9253.4 (ausI) | 501 | P450 monooxygenase [Sphaceloma manihoticola] (CAP07653) | 32/55 |
| AN11214.4 (ausJ) | 162 | Hypothetical protein | |
| AN11205.4 (ausK) | 398 | Norsolorinic acid reductase [Aspergillus parasiticus] (Q00258) | 48/66 |
| AN9257.4 (ausL) | 204 | Terpene cyclases pyr4 [Aspergillus fumigatus F37]13 | 32/55 |
| AN11206.4 (ausM) | 479 | a. PaxM [Penicillium paxilli] (AAK11530) b. FAD-dependent monooxygenase pyr5 [Aspergillus fumigatus F37]13 |
a. 46/65 b. 42/58 |
| AN9259.4 (ausN) | 330 | a. 4-hydroxybenzoate geranyltransferase [Lithospermum erythrorhizon] (BAB84122) b. Prenyltransferase pyr6 [Aspergillus fumigatus F37]13 |
a. 30/49 b. 30/46 |
The next steps include the epoxidation of the prenylated polyketide intermediate possibly by the epoxidase encoded by the gene ausM followed by cyclization, catalyzed by a terpene cyclase to form the tetracyclic intermediate. We were unable to isolate analyzable amounts of the proposed prenylated polyketide 15 and the proposed epoxidated intermediate 16 presumably because these two intermediates are not sufficiently stable in our purification scheme (Fig. 6).
Although no intermediate could be isolated from ausM and ausL deletant strains, we can propose functions for them based on homology. BLAST searches with AusM showed that it has 65% similarity to an epoxidase, PaxM, involved in paxilline biosynthesis.27 It also has 58% similarity to another epoxidase encoded by pyr5 which catalyzes the epoxidation of the terpenoid moiety of farnesyl 4-hydroxy-6-(3-pyridinyl)-2H-pyran-2-one (HPPO) in pyripyropene A biosynthesis.13 Based on the epoxidation functions of its homologs, we propose that AusM catalyzes the epoxidation of the terminal alkene of terpenoid moiety of 15 to form 16.
We propose that the terpene cyclization step is catalyzed by AusL. This enzyme has 55% similarity to the enzyme encoded by pyr4, which is a terpene cyclase involved in pyripyropene A biosynthesis in Aspergillus fumigatus.13 Both AusL and Pyr4 are annotated as “integral membrane protein” by the Broad Institute website (http://www.broadinstitute.org/) and their sizes are smaller than any other terpene cyclases identified so far. This smaller size of pyr4 led Itoh et al. to describe it as a novel terpene cyclase.13 When we examined AusL, we identified a conserved residue (E62) in its sequence that, in other terpene cyclases, is proposed to be involved in the protonation of the epoxide ring that may trigger the following polyene cyclization step.28
Next, the lactone system in the ring A of 4 is formed via a Baeyer-Villiger oxidation by AusB. We were able to isolate large quantities of compound 4 from the ausBΔ strain providing evidence that the lactone formation is catalyzed by AusB. BLAST analysis showed that AusB has 54% amino acid sequence similarity to CpdB of Pseudomonas sp. strain HI-70, which was demonstrated to be a Baeyer-Villiger monooxygenase (BVMO).29 Some conserved motifs are shared between all the members of the BVMO family.30 Analysis of the predicted amino acid sequence of AusB, revealed that it, indeed, has conserved motifs found in typical BVMO sequences. They are two Rossmann folds (178-GAGFGG-183 and 374-GTGSTA-379) for binding FAD and NADPH, a BMVO fingerprint (338-FQGHIFHTARWD-349) and two other conserved motifs (210-GG-211 and 561-DLVVLATG-568). The underlined sequences are conserved among members of this class of enzyme. Based on homology, conserved motifs and the accumulation of compound 4 in the ausB deletant, we deduce that ausB encodes a BVMO that catalyzes the formation of Ɛ-lactone A ring of austinol/dehydroaustinol. The methyl ester connected to carbon 8' of protoaustinoid A (4) suggests that a methyltransferase should be involved in the early stage of the pathway. Since we were unable to isolate the proposed compound 15 and 16 from our mutant strains, we cannot say whether the esterification step occurs before or after cyclization by AusL.
The next steps involve the cleavage and rearrangement of the tetraketide portion of the intermediates from compound 5 to compound 9. Simpson’s group proposed that this rearrangement involves hydroxylation of the C-5', followed by α-ketol rearrangement and reduction of the ketone with lactone formation by the attack of the hydroxyl group on the carboxy group.12 Our results show that these steps of the pathway involve at least four genes ausJ, ausK, ausH, ausI. The ausJ deletant accumulated compound 5 (Fig. 5B), suggesting that this enzyme is responsible for the acid-catalyzed keto-rearrangement and ring contraction of the tetraketide portion to generate compound 6. The ausK deletant produced three compounds 6, 10, and 11. We propose that compounds 10 and 11 are shunt products of the pathway. BLAST analysis showed that AusK has 66% amino acid similarity to Nor1 which converts the 1′ keto group of norsolorinic acid to the 1’ hydroxyl group of averantin in A. parasiticus.31 We speculate that this enzyme is responsible for reducing the C-5′ keto of compound 6 to hydroxyl, and this hydroxyl oxygen then attacks the carboxyl to form the γ-lactone ring in the triketide portion of the molecule. A similar mechanism has been proposed in Simpson’s labeling study.12
1D and 2D NMR revealed that compounds 6 and 10 are constitutional isomers. Acetyl and hydroxyl groups are at C-6′ in compound 6 and at C-4′ in compound 10. (Please see supplemental information for details.) The presence of two isomers indicates to us that the keto rearrangement doesn’t occur in a stereoselective manner. The alkyl shift can be from C-5′ to either C-4′ or C-6′ to form compound 10 and 6 respectively. 11 was also found in this deletant and might be a shunt product made by oxidizing C-5′ of compound 10 to carboxylic acid followed by beta-keto-acid decarboxylation oxidation by other endogenous enzymes. Since we didn’t detect conversion of compound 6 to any compound similar to 11, the enzymes may prefer only compound 10 as substrate rather than compound 6. Among all the deletants, compound 10 was only found in the ausK deletion strain. Thus the consumption of compound 6 by AusK and the enzymes downstream in the pathway may lead the reaction to go through the major route of the pathway instead of the shunt path.
When we analyzed the metabolites of ausH deletants, we found an accumulation of compound 12 (5′ R configuration), which proved to be a stereoisomer of 7 (5′ S configuration) by 1D and 2D NMR and X-ray crystallographic analysis. AusH is annotated as a hypothetical enzyme on both Broad Institute and CADRE databases. BLAST analysis showed that none of its homologs have been studied so far. Our study suggests that AusH may play a role in altering AusK’s stereospecificity for its product. When there is no AusH present, AusK produces the 5′ R γ-hydroxyl ester precursor of compound 12. When AusH is present, AusK produces the 5′ S γ-hydroxyl ester precursor of compound 7. Therefore, AusH could function as an accessory enzyme working in tandem with AusK, to make compound 7.
BLAST analysis revealed that AusI has 52% similarity to SmP450-2, a P450 monooxygenase, which mediates the lactone formation of GA9 in gibberellin biosynthesis in Sphaceloma manihoticola.32 The ausI deletant produces isoaustinone (7). The accumulation of this compound suggests that ausI encodes a Baeyer-Villiger monooxygenase (BVMO) which inserts an oxygen atom between the C-4’ and vicinal carbon at C-3’ to create a lactone ring. Another intermediate, compound 8, was isolated along with compound 7 from this gene deletant strain. Compound 8 has an additional hydroxyl on C-11 to compound 7. It is possible that this modification is performed by a downstream enzyme AusG encoded by AN9248.4 which we propose to be a hydroxylase. AusG has 59% similarity to SmP450-1, a P450 monooxygenase, that mediates four oxidation steps in gibberellin biosynthesis in Sphaceloma manihoticola.32 Austinolide (9) was isolated from our ausG deletant strain. This suggests ausG encodes a hydroxylase that catalyzes C-11 hydroxylation to form austinol. Finally, we have been unable to locate a gene that we can show is responsible for converting austinol to dehydroaustinol. It is possible that additional genes involved in the pathway have yet to be identified or the conversion is spontaneous and not enzyme catalyzed.
Interestingly, although the LC/MS profile of AN11217.4 mutant suggests that this gene is not required for meroterpenoid biosynthesis, we found that the metabolite profile of this mutant is different from other meroterpenoid producing mutants and LO2026 (control strain). In LO2026, the amount of austinol is greater than dehydroaustinol, but the amount of austinol and dehydroaustinol are about the same in the AN11217.4 mutant (Fig. 3A and 5B). BLAST analysis showed that this gene has 45% identity to Bcmfs1 from Botryotinia fuckeliana (Table S11). Bcmfs1 is an efflux pump, which belongs to major facilitator superfamily (MFS). This enzyme can protect B. fuckeliana against natural toxins and fungicides by secreting them out.33
Several MFS coding genes have been found in natural product gene clusters and are responsible for transporting specifically these metabolites out of producer organisms34 such as TOXA in the HC toxin pathway of C. carbonum,35 TRI12 in trichothecene biosynthesis of F. sporotrichioides36 and CFP in the cercosporin pathway of C. kikuchii.37 Loss of TRI12 and CFP leads to the reduction in toxin production, suggesting that feedback inhibition may turn off the pathway genes when the toxic level is too high to be tolerated by these organisms.34,36,37
The MFS encoded by AN11217.4 could, therefore, be involved in the transportation of austinol/dehydroaustionl pathway metabolites. One, admittedly speculative, possibility is that the lower level of austinol in the mutant is due to the toxicity of austinol to its producer organism, A. nidulans. In addition, accumulation of austinol in the cell might allow more austinol to be converted to dehydroaustinol. Both effects may lead to the observed difference in metabolite profile of AN11217.4 deletion strains relative to the parental strain. However, whether the austinol is toxic to A. nidulans is still unknown and although it has been seen in many cases that pathway specific efflux is responsible for secreting pathway metabolites out of the host, there are exceptions. For example, AflT, a putative MFS transporter in the aflatoxin gene cluster in A. parasiticus does not play a significant role in aflatoxin secretion.34 In addition, it has been shown that some non-pathway specific transporters in some cases can compensate for the function of an MFS.34 The role of the product of AN11217.4 in austinol/dehydroaustinol secretion is, thus, by no means certain and awaits further investigation.
Regarding the organization of the austinol/dehydroaustinol biosynthetic pathway genes in A. nidulans, one of the most interesting aspects of these genes is that they are located in two separate gene clusters. In fungi, the genes that encode the proteins of a particular secondary metabolite biosynthetic pathway are generally clustered together as is the case in prokaryotes.1,38 Although secondary metabolite genes in plants are not known to be clustered, there is growing evidence that in certain systems they are indeed clustered.38 There is strong evidence for horizontal transfer of a secondary metabolite gene cluster from Aspergillus to Penicillium39 and it has been suggested that clusters of secondary metabolite biosynthetic genes might confer selective advantages to themselves with evolution favoring their propagation and survival as a unit.40 There is some precedence, however, for split gene clusters in fungi. The genes for T-toxin biosynthesis in Cochliobolus heterostrophus are in two unlinked loci41 and the genes for prenylated xanthone biosynthesis in A. nidulans are in three separate regions of the genome.14 It is worth noting, moreover, that without the sequenced genome and efficient gene targeting techniques, it would not have been possible to elucidate the austinol/dehydroaustionol clusters. It is possible that split or fragmented clusters are more common in fungi than has been thought and that they are being discovered now because tools have been developed that facilitate their discovery.
We wondered if the two austinol/dehydroaustinol clusters might be functionally specialized (e.g. the early steps of the pathway might require products of genes in cluster A and late steps require products in cluster B). As Figure 6 demonstrates, however, this is not the case.
In cluster B (the prenyltransferase cluster) there is a sequence between AN11205.4 and AN9256.4 (nucleotides 76655 to 77031 on linkage group VIII) that does not appear to be part of a functional gene but has strong nucleotide identity with a portion of AN8383.4 (in SAT domain of AN8383.4) (P = 6.7e-44). We speculate that the two clusters were once part of a single contiguous cluster that has been split by chromosomal rearrangement involving a partial duplication of AN8383.4 and translocation of AN8383.4, AN8379.4, AN8380.4, AN8381.4, AN8382.4 and AN8384.4, leaving only a dysfunctional remnant of AN8383.4 behind in the prenyltransferase cluster. If this speculation is correct, it follows that the translocation did not disrupt the co-regulation of the genes of the separated gene clusters.
We searched for synteny with secondary metabolite clusters in other species of Aspergillus to determine if any of these species might have an ancestral cluster syntenic with clusters A and B. We found a gene cluster in A. terreus extending from ATEG10077.1 to ATEG10085.1 (http://www.broadinstitute.org/) that exhibits substantial synteny with clusters A and B. (Fig. S21). It is highly speculative but possible that the putative meroterpenoid cluster in A. terreus shares a common ancestor with the meroterpenoid clusters A and B in A. nidulans. The product of the A. terreus cluster is currently unreported although A. terreus is known to produce the meroterpenoids, terretonin and territrem.3,4
Another interesting aspect of the two A. nidulans clusters is that there are a total of eight annotated genes internal to the clusters that are not required for austinol or dehydroaustionol biosynthesis. While it is possible that some of them are not actual genes and are simply missanotated sequences, others are clearly real and some of them are, on the basis of homology, predicted to have a role in secondary metabolism (Table S11). While we can only speculate as to the functions of these genes, it is worth noting that in A. nidulans the genes required for monodictyphenone synthesis are a subset of the prenyl xanthone biosynthetic pathway and that one gene, mdpE, is required for monodictyphenone biosynthesis but not prenyl xanthone biosynthesis and a second gene, mdpD, is required for prenyl xanthone biosynthesis but not monodictyphenone biosynthesis.14 This raises the highly speculative but intriguing possibility that fungi may use differential regulation of genes within secondary metabolite gene clusters to multiply the number of secondary metabolites produced by the clusters.
CONCLUSION
Pioneering work primarily using isotopic precursors in the 80's enabled the Simpson group to propose biosynthetic mechanisms for meroterpenoids involving the C-alkylation of 3,5-dimethylorsellinic acid.12 The alkylated intermediate is epoxylated and cyclized to form a tetracyclic intermediate, which then undergoes drastic modifications to form austinol and dehydroaustinol. Targeted gene deletion studies of genes flanking the genes encoding the NR-PKS on chromosome V and the prenyltransferase on chromosome VIII allowed the isolation of several stable intermediates in amount sufficient for full NMR characterization. Many of the intermediates isolated share structural similarities to the proposed structures from the earlier labeling work and also intermediates isolated from wild type strain of Penicillium sp. LaBioMi-02423 and provide direct support for the pathway proposed by the Simpson group.
In summary we have shown that genes from two different gene clusters on two different chromosomes are required to make for dehydroaustinol and austinol in A. nidulans. One cluster contains the PKS gene and the other cluster contains the prenyltransferase gene. By creating a series of deletants and analyzing the metabolite profiles using LC/MS, fourteen genes were identified to be involved in austinol/dehydroaustinol biosynthesis. Based on the intermediates and bioinformatics study, we have proposed a biosynthetic pathway for austinol/dehydroaustinol.
MATERIALS AND METHODS
Strains and molecular genetic manipulations
A. nidulans strains used in this study are listed in Table S1 in the supplemental material. All the deletant strains were generated by replacing each gene with the Aspergillus fumigatus pyrG gene in the A. nidulans strain LO2026. The construction of fusion PCR products, protoplast production, and transformation were carried out as described previously.19 For the construction of the fusion PCR fragments, two 1,000-bp fragments of genomic A. nidulans DNA, upstream and downstream of the targeted gene, were amplified by PCR. Primers used in this study are listed in Table S2 in the supplemental material. Fusion PCR was set up with the two amplified flanking sequences and the A. fumigatus pyrG selectable marker cassette as the template DNA. The three fragments were fused into a single molecule and amplified with two nested primers. Diagnostic PCR analyses of the deletion mutants and their parent strain, LO2026, were performed with the external primers used in the first round of PCR. The difference in the size between the gene replaced by the selective marker and the native gene allowed the determination of correct gene replacement. In addition, the correctness of the gene replacements was further confirmed using external primers along with primers located inside the marker gene. Diagnostic PCR with multiple primer sets allowed us to determine reliably if correct gene targeting had occurred. Although this technique does not allow us to determine if additional heterologous integrations have occurred, extra insertions rarely happen (<2%) with the strains used.19 At least two transformants per gene were selected and their metabolite profiles were examined. The probability that two random extra insertions would occur and would give the same altered metabolite profile is vanishingly small.
Fermentation and LC/MS analysis
The A. nidulans control strain LO2026 and deletion strains were cultivated at 37°C on solid YAG (complete medium; 5 g of yeast extract/liter, 15 g of agar/liter, and 20 g of D-glucose/liter supplemented with 1.0-ml/liter of a trace element solution and required supplements) at 1 × 107 spores per 10 cm diameter plate (with 18 ml of medium per plate). After 3 days, agar was chopped into small pieces and the material was extracted with 50 ml of methanol (MeOH) followed by 50 ml of 1:1 CH2Cl2-MeOH, each with 1 h of sonication. The extract was evaporated in vacuo to yield a residue, which was suspended in H2O (25 ml), and this suspension was then partitioned with ethyl acetate (EtOAc) twice. The combined EtOAc layer was evaporated in vacuo, redissolved in 1ml DMSO, and then diluted with 4 volumes of MeOH. 4ul of this dilute crude extract was injected for high-performance liquid chromatography (HPLC)-photodiode array detection-mass spectrometry (MS) analysis.
Liquid chromatography-MS (LC-MS) was carried out using a ThermoFinnigan LCQ advantage ion trap mass spectrometer with a reverse-phase C18 column (2.1 by 100 mm with a 3 µm particle size; Alltech Prevail) at a flow rate of 125 µl/min. The solvent gradient for HPLC was 95% acetonitrile (MeCN)-H2O (solvent B) in 5% MeCN-H2O (solvent A), both containing 0.05% formic acid, as follows: 0% solvent B from 0 to 5 min, 0 to 100% solvent B from 5 to 35 min, 100% solvent B from 35 to 40 min, 100 to 0% solvent B from 40 to 45 min, and reequilibration with 0% solvent B from 45 to 50 min. The positive mode was used for the detection of austinol/dehydroaustinol and their intermediates. Negative ion electrospray ionization (ESI) was used for the detection of compound 3. Conditions for MS included a capillary voltage of 5.0 kV, a sheath gas flow rate at 60 arbitrary units, an auxiliary gas flow rate at 10 arbitrary units, and an ion transfer capillary temperature of 350°C.
Isolation, identification of secondary metabolites and compound spectral data
For structure elucidation, each A. nidulans deletant was cultivated at 37 °C on 30 YAG plates at 2.25 × 107 spores per 15-cm plate (~55 ml of medium per plate). The desired intermediates from crude extracts of each deletant strain were isolated with flash chromatography and reverse phase HPLC. (Please see supplemental information for details). Melting points were determined with a Yanagimoto micromelting point apparatus and are uncorrected. IR spectra were recorded on a GlobalWorks Cary 14 Spectrophotometer. Optical rotations were measured on a JASCO P-200 digital polarimeter. NMR spectra were collected on a Varian Mercury Plus 400 spectrometer. High-resolution mass spectrometry was performed on Agilent 6210 time of flight LC–MS. The X-ray crystallographic data were collected using Bruker KAPPA APEX II diffractometer in the X-ray crystallography laboratory of the California Institute of Technology.
3,5-Dimethylorsellinic acid
Yellowish white solid, mp 182 – 185 °C; IRmaxZnSe 3448, 1685, 1208, 1142 cm−1; For UV-Vis and ESIMS spectra, see Figure S1; 1H NMR (acetone-d6, 400 MHz) 2.10 (3H, s), 2.14 (3H, s), 2.50 (3H, s); 13C NMR (acetone-d6, 100 MHz) 8.8 (q), 12.4 (q), 19.0 (q), 106.2 (s), 108.9 (s), 116.4 (s), 138.6 (s), 159.0 (s), 161.6 (s), 175.3 (s). 1H and 13C NMR data were in good agreement with the published data.42
Protoaustinoid A
Yellowish gum; For UV-Vis and ESIMS spectra, see Figure S1; For 1H and 13C NMR data, see Table S3.
Preaustinoid A3
Colorless solid, mp 209 – 212 °C; [α]D24 +250.4° (CHCl3, c 0.9); IRmaxZnSe 3371, 1733, 1707, 1646, 1374, 1297, 1230, 1110, 1004, 923 cm−1; For UV-Vis and ESIMS spectra, see Figure S1; For 1H and 13C NMR data, see Table S9 and S10. 1H and 13C NMR data were in good agreement with the published data.23
Preaustinoid A4
Colorless needles, mp 167–171 °C; [α]D24 +469.5° (CHCl3, c 1.7); IRmaxZnSe 3383, 1759, 1715, 1654, 1376, 1282, 1239, 1156, 1109, 984, 911 cm−1; For UV-Vis and ESIMS spectra, see Figure S1; For 1H and 13C NMR data, see Table S4; HRESIMS, [M + H]+ m/z found 457.2227; calc. for C26H32O7: 457.2221.
Isoaustinone
Colorless solid, mp >300 °C; [α]D24 +462.6° (CHCl3, c 0.3); IRmaxZnSe 3490, 1752, 1708, 1375, 1285, 1236, 1159, 1114, 1083, 1053, 981, 925 cm−1; For UV-Vis and ESIMS spectra, see Figure S1; For 1H and 13C NMR data, see Table S9 and S10. 1H and 13C NMR data were in good agreement with the published data.23
11 β-Hydroxyisoaustinone
Colorless needles, mp 225 – 229 °C; [α]D24 +292.2° (CHCl3, c 0.1); IRmaxZnSe 3490, 3378, 1764, 1750, 1691, 1312, 1241, 1177, 1118, 1086, 1037, 983, 927 cm−1; For UV-Vis and ESIMS spectra, see Figure S1; For 1H and 13C NMR data, see Table S5; HRESIMS, [M + H]+ m/z found 443.2069; calc. for C25H30O7: 443.2064.
Austinolide
Colorless needles, mp 259 – 263 °C; [α]D24 +436.3° (CHCl3, c 1.0); IR nmaxZnSe 3458, 1780, 1717, 1375, 1293, 1218, 1179, 1117, 1075, 979, 910 cm−1; For UV-Vis and ESIMS spectra, see Figure S1; For 1H and 13C NMR data, see Table S9 and S10. 1H and 13C NMR data were in good agreement with the published data.23
Preaustinoid A5
Colorless amorphous solid, mp unmeasurable; [α]D24 +162.0° (CHCl3, c 0.6); IRmaxZnSe 3426, 1760, 1715, 1374, 1265, 1235, 1110, 1048, 973, 908 cm−1; For UV-Vis and ESIMS spectra, see Figure S1; For 1H and 13C NMR data, see Table S6; HRESIMS, [M + H]+ m/z found 457.2228; calc. for C26H32O7: 457.2221.
Austinoneol A
Colorless needles, mp 219 – 224 °C; [α]D24 +366.8° (CHCl3, c 0.9); IRmaxZnSe 3403, 1755, 1721, 1711, 1661, 1393, 1375, 1278, 1223, 1112, 1060, 974, 906 cm−1; For UV-Vis and ESIMS spectra, see Figure S1; For 1H and 13C NMR data, see Table S9 and S10. 1H and 13C NMR data were in good agreement with the published data.43
(5'R)-Isoaustinone
Colorless plates, mp 234–237 °C; [α]D24 +428.7° (CHCl3, c 0.3); IRmaxZnSe 3382, 1755, 1712, 1375, 1285, 1201, 1113, 1066, 1003, 915 cm−1; ; For UV-Vis and ESIMS spectra, see Figure S1; For 1H and 13C NMR data, see Table S7; HRESIMS, [M + H]+ m/z found 427.2120; calc. for C25H30O6: 427.2115.
Neoaustinone
Yellowish gum; [α]D24 +357.1° (CHCl3, c 0.2); IRmaxZnSe 3373, 1760, 1711, 1657, 1380, 1312, 1196, 1112, 1071, 1017, 979 cm−1; For UV-Vis and ESIMS spectra, see Figure S1; For 1H and 13C NMR data, see Table S8; HRESIMS, [M + H]+ m/z found 443.2070; calc. for C25H30O6: 443.2064.
X-ray Crystallographic Analysis of (5'R)-Isoaustinone
A colorless crystal of 12 with dimensions 1.21 × 0.21 × 0.19 mm3 was selected for X-ray analysis. Structure analysis was performed using SHELXS-97 program. The compound crystallized in the monoclinic space group I2, with a = 16.9929(9) Å, b = 7.3879(4) Å, c = 17.1390(9) Å, = 95.180(3)°, volume = 2142.9(2) Å3, Z = 4, ρcalc = 1.322 Mg/m3, λ = 0.71073 Å. Intensity data were collected on a Bruker instrument with an APEX II detector and crystals were mounted on a glass fiber using Paratone oil then placed on the diffractometer under a nitrogen stream at 100K. A total of 30291 data points were collected in the range 2.39° ≤ θ ≤ 31.83°, yielded 7321 unique reflections. Crystallographic data have been deposited at the CCDC, 12 Union Road, Cambridge CB2 1EZ, UK and copies can be obtained on request, free of charge, by quoting the publication citation and the deposition number 773039.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by grant PO1GM084077 from the National Institute of General Medical Sciences. We thank an anonymous reviewer for pointing out the PKS-like sequence between AN11205.4 and AN9256.4.
Footnotes
SUPPORTING INFORMATION
A. nidulans strains and primers used in this study, detailed structural characterization, NMR spectral data , purification methods and results of diagnostic PCR. This information is available free of charge via the internet at http://pubs.acs.org/.
REFERENCES
- 1.Hoffmeister D, Keller NP. Nat. Prod. Rep. 2007;24:393–416. doi: 10.1039/b603084j. [DOI] [PubMed] [Google Scholar]
- 2.Geris R, Simpson TJ. Nat. Prod. Rep. 2009;26:1063–1094. doi: 10.1039/b820413f. [DOI] [PubMed] [Google Scholar]
- 3.Mcintyre CR, Simpson TJ. Chem. Comm. 1981;20:1043–1044. [Google Scholar]
- 4.Chen JW, Luo YL, Hwang MJ, Peng FC, Ling KH. J. Biol. Chem. 1999;274:34916–34923. doi: 10.1074/jbc.274.49.34916. [DOI] [PubMed] [Google Scholar]
- 5.Omura S, Tomoda H, Kim YK, Nishida H. Jpn. J. Antibiot. 1993;46:1168–1169. doi: 10.7164/antibiotics.46.1168. [DOI] [PubMed] [Google Scholar]
- 6.Sunazuka T, Hirose T, Omura S. Accounts Chem. Res. 2008;41:302–314. doi: 10.1021/ar6000044. [DOI] [PubMed] [Google Scholar]
- 7.Simpson TJ, Stenzel DJ, Bartlett AJ, O’Brien E, Holker JSE. J. Chem. Soc. Perk. T. 1. 1982:2687–2692. [Google Scholar]
- 8.Márquez-Fernández O, Trigos A, Ramos-Balderas JL, Viniegra-González G, Deising HB, Aguirre J. Eukaryot. Cell. 2007;6:710–720. doi: 10.1128/EC.00362-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szewczyk E, Chiang YM, Oakley CE, Davidson AD, Wang CCC, Oakley BR. Appl. Environ. Microb. 2008;74:7607–7612. doi: 10.1128/AEM.01743-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mclntyre CR, Simpson TJ, Stenzel DJ, Bartlett AJ, Brien EO, Holkerb JSE. J. Chem. Soc. Chem. Comm. 1982:781–782. [Google Scholar]
- 11.Simpson TJ, Stenzel DJ, Moore RN, Trimble LA, Vederas JC. J. Chem. Soc. Chem. Comm. 1984:1242–1243. [Google Scholar]
- 12.Ahmed SA, Scott FE, Stenzel DJ, Simpson TJ, Moore RN, Trimble LA, Arai K, Vederas JC. J. Chem. Soc. Perk. T. 1. 1989:807–816. [Google Scholar]
- 13.Itoh T, Tokunaga K, Matsuda Y, Fujii I, Abe I, Ebizuka Y, Kushiro T. Nat. Chem. 2010;2:858–864. doi: 10.1038/nchem.764. [DOI] [PubMed] [Google Scholar]
- 14.Sanchez JF, Entwistle R, Hung JH, Yaegashi J, Jain S, Chiang YM, Wang CCC, Oakley BR. J. Am. Chem. Soc. 2011;133:4010–4017. doi: 10.1021/ja1096682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nielsen ML, Nielsen JB, Rank C, Klejnstrup ML, Holm DK, Brogaard KH, Hansen BG, Frisvad JC, Larsen TO, Mortensen UH. FEMS microbiol. lett. 2011;321:157–166. doi: 10.1111/j.1574-6968.2011.02327.x. [DOI] [PubMed] [Google Scholar]
- 16.Bok JW, Chiang YM, Szewczyk E, Reyes-domingez Y, Davidson AD, Sanchez JF, Lo HC, Watanabe K, Strauss J, Oakley BR, Wang CCC, Keller NP. Nat. Chem. Biol. 2009;5:462–464. doi: 10.1038/nchembio.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanchez JF, Chiang YM, Szewczyk E, Davidson AD, Ahuja M, Oakley CE, Bok JW, Keller N, Oakley BR, Wang CCC. Mol. biosyst. 2010;6:587–593. doi: 10.1039/b904541d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiang YM, Szewczyk E, Davidson AD, Keller N, Oakley BR, Wang CCC. J. Am. Chem. Soc. 2009;131:2965–2970. doi: 10.1021/ja8088185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nayak T, Szewczyk E, Oakley CE, Osmani A, Ukil L, Murray SL, Hynes MJ, Osmani SA, Oakley BR. Genetics. 2006;172:1557–1566. doi: 10.1534/genetics.105.052563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiang YM, Szewczyk E, Nayak T, Davidson AD, Sanchez JF, Lo HC, Ho WY, Simityan H, Kuo E, Praseuth A, Watanabe K, Oakley BR, Wang CCC. Chem. Biol. 2008;15:527–532. doi: 10.1016/j.chembiol.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melzer M, Heide L. Biochem. Biophys. Acta. 1994;1212:93–102. doi: 10.1016/0005-2760(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 22.Brown DW, Yu JH, Kelkar HS, Fernandes M, Nesbitt TC, Keller NP, Adams TH, Leonard TJ. P. Natl. Acad. Sci. USA. 1996;93:1418–1422. doi: 10.1073/pnas.93.4.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fill TP, Pereira GK, Geris dos Santos RM, Rodrigues-Fo EZ. Naturforsch. 2007;62b:1035–1044. [Google Scholar]
- 24.Kroken S, Glass NL, Taylor JW, Yoder OC, Turgeon BG. Natl. Acad. Sci. USA. 2003;100:15670–15675. doi: 10.1073/pnas.2532165100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yazaki K, Kunihisa M, Fujisaki T, Sato F. J. Biol. Chem. 2002;277:6240–6246. doi: 10.1074/jbc.M106387200. [DOI] [PubMed] [Google Scholar]
- 26.Heide L. Curr. Opin. Chem. Biol. 2009;13:171–179. doi: 10.1016/j.cbpa.2009.02.020. [DOI] [PubMed] [Google Scholar]
- 27.Saikia S, Parker EJ, Koulman A, Scott B. FEBS Lett. 2006;580:1625–1630. doi: 10.1016/j.febslet.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 28.Thoma R, Schulz-Gasch T, D’Arcy B, Benz J, Aebi J, Dehmlow H, Hennig M, Stihle M, Ruf A. Nature. 2004;432:118–122. doi: 10.1038/nature02993. [DOI] [PubMed] [Google Scholar]
- 29.Iwaki H, Wang S, Grosse S, Lertvorachon J, Yang J, Konishi Y, Hasegawa Y, Lau PCK. Appl. Environ. Microb. 2006;72:2707–2720. doi: 10.1128/AEM.72.4.2707-2720.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fraaije M, Kamerbeek NM, Berkel WJH, Janssen DB. FEBS Lett. 2002;518:43–47. doi: 10.1016/s0014-5793(02)02623-6. [DOI] [PubMed] [Google Scholar]
- 31.Cary JW, Wright M, Bhatnagar D, Lee R, Chu FS. Appl. Environ. Microb. 1996;62:360–366. doi: 10.1128/aem.62.2.360-366.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bömke C, Rojas MC, Gong F, Hedden P, Tudzynski B. Appl. Environ. Microb. 2008;74:5325–5339. doi: 10.1128/AEM.00694-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayashi K, Schoonbeek H, Waard MAD. Appl. Environ. Microb. 2002;68:4996–5004. doi: 10.1128/AEM.68.10.4996-5004.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang PK, Yu J, Yu JH. Fungal. Genet. Biol. 2004;41:911–920. doi: 10.1016/j.fgb.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 35.Pitkin JW, Panaccione DG, Walton JD. Microbiol-SGM. 1996;142:1557–1565. doi: 10.1099/13500872-142-6-1557. [DOI] [PubMed] [Google Scholar]
- 36.Alexander NJ, McCormick SP, Hohn TM. Mol. Gen. Genet. 1999;261:977–984. doi: 10.1007/s004380051046. [DOI] [PubMed] [Google Scholar]
- 37.Callahan TM, Rose MS, Meade MJ, Ehrenshaft M, Upchurch RG. Mol. .Plant. Microbe. In. 1999;12:901–910. doi: 10.1094/MPMI.1999.12.10.901. [DOI] [PubMed] [Google Scholar]
- 38.Osbourn A. Trends. Genet. 2010;26:449–457. doi: 10.1016/j.tig.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 39.Slot JC, Rokas A. Curr. Biol. 2011;21:134–139. doi: 10.1016/j.cub.2010.12.020. [DOI] [PubMed] [Google Scholar]
- 40.Walton JD. Fungal. Genet. Biol. 2000;30:167–171. doi: 10.1006/fgbi.2000.1224. [DOI] [PubMed] [Google Scholar]
- 41.Inderbitzin P, Asvarak T, Turgeon BG. Mol .Plant. Microbe. In. 2010;23:458–472. doi: 10.1094/MPMI-23-4-0458. [DOI] [PubMed] [Google Scholar]
- 42.Hirota A, Morimitsu Y, Hiroshi H. Biosci. Biotech. Biochem. 1997;61:647–650. [Google Scholar]
- 43.Santos RMGD, Rodrigues-Filho E. J. Brazil. Chem. Soc. 2003;14:722–727. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.