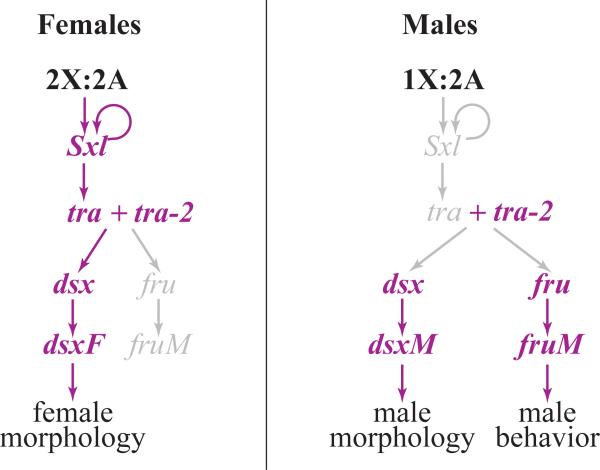Abstract
Most animals are sexually dimorphic, yet different taxa have different sex-specific traits. Despite major differences in the genetic control of sexual development among animal lineages, the Dmrt family of transcription factors has been shown to be involved in sex-specific differentiation in all animals studied so far. In recent years, the functions of Dmrt genes have been characterized in many animal groups, opening the way for a broad comparative perspective. In this review, I focus on the similarities and differences in the functions of Dmrt genes across the animal kingdom. I highlight a number of common themes in the sexual development of different taxa, discuss how Dmrt genes have acquired new roles during animal evolution, and show how they contributed to the origin of novel sex-specific traits.
Dmrt genes: a common theme amidst diversity
Sexual dimorphism (phenotypic differences between males and females of the same species) is one of the most pervasive and diverse features of animal morphology, physiology, and behavior. The demands of sexual reproduction and competition for mates have led each animal lineage to evolve its own suite of sex-specific characters. Lion manes, butterfly wings, and bird songs seem to have nothing in common beyond the fact that they differ between males and females. The molecular mechanisms responsible for sexual dimorphism are almost as diverse, ranging from cell-autonomous, splicing-based sex determination in insects to gonad-dependent endocrine control of sexual traits in mammals and other vertebrates [1,2]. For a long time, these disparities encouraged a narrow, taxon-by-taxon approach to the study of sex-specific development. While evolutionary biology has provided a universal framework for understanding the evolution of sex in all organisms, such broad approach has been slow to take hold in developmental biology.
In recent years, however, some common themes in the development of sex-specific traits in different animal lineages have started to emerge. Central to this trend has been the discovery of the doublesex/mab-3 related (Dmrt) family of transcription factors [3] (Boxes 1, 2). Dmrt genes share a common DNA-binding domain (DM domain) but otherwise show little sequence conservation, making their phylogenetic relationships obscure. Members of this ancient gene family shape sexual dimorphism in organisms as diverse as mammals, insects, and nematodes. Initially their roles seemed very different in different taxa: acting in a global alternative splicing cascade in Drosophila, making the choice between testis and ovary development in the vertebrate gonad, or cell-autonomously controlling sensory organ differentiation in Caenorhabditis elegans [4,5,6]. But upon closer examination, these differences may hide deeper similarities. It now appears that in all animals, Dmrt genes work as tissue-specific developmental regulators that integrate information about sex, position, and time to direct narrow populations of cells toward male or female fates. The most striking taxon-specific functions of Dmrt genes – such as alternative splicing in insects or primary sex determination in some vertebrates– are derived from this common ancestral function.
The molecular functions of Dmrt genes are well understood (Boxes 1, 2), and their roles in specific animal groups or developmental processes have been the subject of several recent reviews [7,8,9,10,11]. However, accumulating evidence from a variety of models opens the way for a broader comparative perspective. In this paper, I focus on the similarities and differences in the roles of Dmrt genes across the animal kingdom. The two complementary goals of this analysis are to identify common themes in the sex-specific development of different taxa and to examine lineage-specific changes in the development and evolution of sexual dimorphism.
A deeply conserved role in gonad development
Although virtually every animal lineage has evolved its own somatic sex-specific characters, the one trait most animals have in common is the presence of sexually dimorphic gonads. Despite profound differences in gonad structure and development among animal phyla, Dmrt genes are specifically expressed in the developing gonads of almost all animals: in vertebrates including mammals [4,12,13], birds [7], turtles and alligators [14,15], amphibians [9], and teleost fishes [8]; in arthropods including Drosophila [16] and diverse crustacean taxa [17,18,19]; and in different classes of mollusks [20,21]. A Dmrt gene is also present in the coral Acropora millepora, where its increased expression coincides with seasonal sexual reproduction [22]. The only known exception is C. elegans, where several Dmrt genes are involved in the sexual differentiation of somatic tissues but are dispensable for gonad development.
The main function of Dmrt genes in the gonad is to promote male-specific and repress female-specific differentiation. This function is best understood in the case of mouse Dmrt1, but the roles of other family members are also beginning to be characterized [23,24]. The vertebrate gonad is bipotential: the same embryonic cell lineages give rise to either ovaries or testes depending on the sex of the animal. Once established, sex-specific gonad differentiation must be actively maintained during embryonic and postnatal development. In mice, neither Dmrt1 nor other Dmrt genes are involved in the initial sex determination; however, Dmrt1 is essential for maintaining testis identity. Differentiated states of the testis and ovary are controlled by a genetic circuit centered on the competition between two transcription factors: Foxl2 (female-specific forkhead box L2), which promotes the female-specific granulosa and theca cell fates, and Sox9 (Sry-related box 9), which promotes the male-specific Sertoli cell fate [25,26,27]. During embryonic development, a sex-determining signal, which can be either genetic or environmental, directs gonad development down the male or female pathway. In most mammals, the Y-linked Sry (sex-determining region on the Y chromosome) gene is necessary to induce testis differentiation in males, while the absence of Sry leads to ovary development [2,26]. Dmrt1, along with Sox9, is one of the key SRY targets in mammalian testis development. In early mouse embryos, Dmrt1 is expressed in the genital ridge of both sexes before any overt signs of sex-specific differentiation [4]. Later, Dmrt1 expression declines in the ovary but is maintained in the testis, where it becomes restricted to germline and Sertoli cells [28]. Loss of DMRT1 function in Sertoli cells in the postnatal testis results in the loss of Sox9 expression and the ectopic expression of Foxl2 and other feminizing genes, leading to the transdifferentiation of Sertoli into granulosa cells [27]. DMRT1 both activates testis-specific genes such as Sox9 and Sox8 and represses ovary-specific genes including Foxl2, the Wnt4 and R-spondin-1 signaling proteins, and estrogen receptors [27,29]. In addition to these regulators, DMRT1 controls hundreds of target genes in both Sertoli and germline cells, including genes involved in cell differentiation, cell cycle control, and pluripotency [29].
doublesex (dsx) function in arthropods shows interesting parallels with the vertebrate Dmrt1. In Drosophila, male and female gonads arise from the same somatic gonad precursor (SGP) cells. In males, dsx is expressed in SGPs and is required for the recruitment of additional precursors from the mesoderm, whereas no recruitment is observed in females [16,30,31,32]. In the crustacean Daphnia magna, loss of dsx transforms testes toward ovary-like morphology; conversely, ectopic dsx expression in female embryos can induce testis development [33]. Thus, in arthropods as well as vertebrates, the function of Dmrt genes is to promote testis and repress ovary development in bipotential primordia.
This function of Dmrt genes remains conserved regardless of the mating system and the mode of sex determination. Drosophila, mammals, and birds have genetic sex determination, while in Daphnia and turtles, sex is determined by environmental factors. Dmrt1 genes promote testis development both in the male-heterogametic (XY) mammals and medaka fish, and in the female-heterogametic (ZW) birds and frogs [7,8,9,27]. Fish studies are particularly illuminating due to the diversity of sexual lifestyles in this lineage. Sex determination in fish can be either genetic or environmental, and some species are gonochoristic while others are hermaphrodites. Despite these disparities, expression of Dmrt genes correlates with testis development in all teleosts. In the gonochorists, Dmrt1 expression in the gonad is male-limited in some species and strongly male-biased in others. In hermaphroditic species, Dmrt1 expression parallels the development of testes in protogynous, and their regression in protandrous, hermaphrodites [8].
Does the shared gonad function of Dmrt genes reflect conservation or convergence? Tissue- and sex-specific expression of Dmrt genes in the somatic gonads of phyla as divergent as chordates, arthropods, and mollusks suggests that they already functioned in testis development in the common Bilaterian ancestor. Conversely, the functions of Dmrt genes in the germline of some animals are far less consistent and may reflect independent cooption (Box 3).
Somatic sexual dimorphism – integration of sex and pattern
The nature and development of sex-specific somatic traits show great diversity among animals, but once again the Dmrt genes emerge as one of the few common features. Their roles in sexual differentiation are best understood in Drosophila and C. elegans, where sex determination is largely cell-autonomous (Box 1). In flies, recent work has shown that dsx is transcribed in tightly controlled spatial patterns. Most cells of both male and female flies do not express either dsx isoform, resulting in a complex mosaic of “sex-aware” and “sex-ignorant” cells [31,32].
The central function of dsx in somatic tissues is to induce localized sex-specific differentiation by integrating information about sex, position, and time. This role is perfectly illustrated by the Drosophila sex comb – a strictly male-specific organ that develops on the first, but not the second or third, pair of legs (Figure 1 A). Spatial control is accomplished in this case by the HOX genes while the sex-specificity is controlled by alternative dsx splicing (Box 1). During larval and pupal stages, dsx is expressed in the presumptive sex comb region and is necessary for its sex-specific differentiation [34]. dsx transcription is induced in the first leg by the HOX gene Sex combs reduced (Scr), which is not expressed in the more posterior segments, and further refined by intrasegmental positional cues. Ectopic expression of dsxM in male pupae induces extra sex combs in all three legs and outside of the normal sex comb position, while a switch from male to female isoform results in feminization without affecting the spatial pattern [34].
Figure 1.
Dmrt genes integrate sex-specific, spatial, and temporal cues to induce sexually dimorphic differentiation in restricted groups of cells. A. In the D. melanogaster sex comb, dsx is controlled by the splicing-based sex determination pathway (red), the HOX gene Scr and intrasegmental positional cues (green), and presumably by ecdysone signaling that regulates the timing of metamorphosis (purple). dsx induces sex comb development by controlling cell fate decisions and several distinct morphogenetic processes [34]. B. In C. elegans, dmd-3 and mab-3 are controlled by the sex determination pathway (red), by posterior group HOX genes, Wnt signaling and GATA transcription factors (green), and by the heterochronic pathway (purple). dmd-3 and mab-3 induce male-specific tail differentiation by regulating cell-autonomous morphogenetic processes as well as signaling pathways including TGF-β (sma-3) and Rho kinase (vav-1). In both organisms, arrows indicate either direct or indirect regulation [43,46].
The role of localized dsx expression in sex-specific development can also be seen in other sexually dimorphic organs such as genitalia and central nervous system (CNS). dsx mutant flies develop both male and female genitalia, indicating that dsx acts as a switch between two alternative differentiation pathways but is not itself required to specify either [35,36,37]. In the CNS, dsx is expressed and required in discrete neuronal clusters that direct courtship song, ejaculation, and other sex-specific behaviors [38,39,40]. Here, dsx establishes sexual dimorphism by controlling neuroblast proliferation and death [40,41] and the differentiation of sex-specific neurons [38].
Unlike the insect mechanism where dsx plays active roles in both males and females, the Daphnia dsx is essential for male development but is dispensable in females. Males of Daphnia magna differ from females in having larger eyes, thoracic hooks, and longer antennae. All these structures express dsx in males but not females and are feminized by RNAi-induced dsx knockdown. Conversely, ectopic dsx expression in female embryos can masculinize these structures [33].
A similar pattern is observed in nematodes. In C. elegans, sexual differentiation is controlled by the transcription factor transformer-1 (tra-1), unrelated to the Drosophila tra [42]. Males differ from hermaphrodites in several morphological traits that require Dmrt genes for male-specific differentiation. For example, mab-23 promotes male development in the proctodeum, dmd-3 is required for male tail and sensory organ formation, and mab-3 is necessary for both proctodeum and tail development in addition to controlling gene expression in the intestine [43,44,45]. Similar to the Drosophila and Daphnia dsx, these genes are tightly regulated at the transcriptional level. All three have strictly male-limited functions and are dispensable in hermaphrodites [43,44,45].
The integration of sex and pattern by Dmrt genes in C. elegans is exemplified by dmd-3 and mab-3 (Figure 1 B) [43,46]. These genes are expressed sex-specifically in the male tail and can induce male morphogenesis when misexpressed in hermaphrodites. tra-1 is the key sex-specific regulator that limits dmd-3 and mab-3 expression to males; their spatial pattern is established in part by posterior HOX genes and localized Wnt signaling; and the timing of their activation is determined by the heterochronic pathway that controls the general progression of worm development [43,46]. dmd-3 and mab-3 then jointly activate eff-1, a gene required for cell fusion, and other effector genes that mediate male morphogenesis (Figure 1 B). Thus, dmd-3 and mab-3 integrate sexual, positional, and temporal cues to initiate a sex-specific developmental program [43,46]. This is remarkably similar to the function of dsx in the Drosophila sex comb, where it integrates sex (through alternative splicing) with spatial information (from the HOX code) and timing (through an unknown but presumably ecdysone-dependent mechanism) to promote male-specific morphogenesis (Figure 1 A) [34].
In vertebrates, although the gonad plays the central role in sexual differentiation, nongonadal cells also have an intrinsic sexual identity [7,47]. Many vertebrate Dmrts are expressed in somatic tissues including the brain [48], and there are tantalizing hints that some of them may have sex-specific functions. In Dmrt4 mutant mice, for example, a substantial proportion of males display same-sex copulatory behavior, raising the possibility that Dmrt genes act in the nervous system to control sexual behavior in vertebrates as well as in insects and nematodes [24]. However, it remains to be seen whether the vertebrate Dmrt genes play any cell-autonomous roles in the sexual differentiation of non-gonadal tissues.
Non-autonomous control of sexual dimorphism
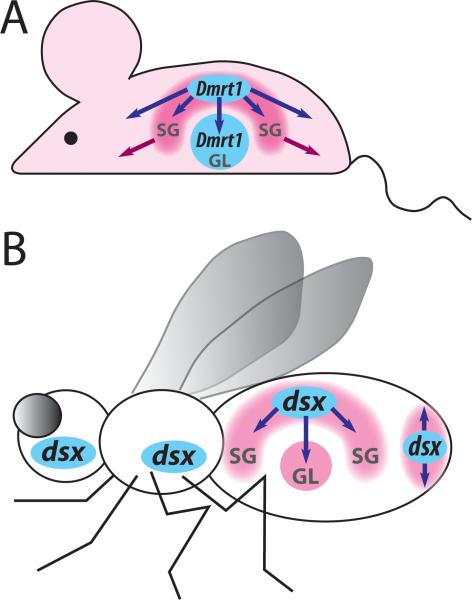
Precise spatial regulation of Dmrt genes is essential for normal development in animals as diverse as insects, vertebrates, and nematodes. In all these systems, most cells do not express any Dmrt genes. However, the relatively small populations of Dmrt-expressing cells can have a profound influence on sexual differentiation in the rest of the body. The most obvious of these roles is the endocrine control of vertebrate sexual differentiation. For example, the mammalian Sertoli cells, whose development depends on Dmrt1 expression [27,28], orchestrate the hormonal control of sex-specific development in non-gonadal tissues both by secreting sex hormones themselves and by controlling the development of other hormone-producing cell types in the gonad [2] (Figure 2 A).
Figure 2.
Cell-autonomous and non-autonomous functions of Dmrt genes in mouse and Drosophila. Tissues where Dmrt genes are expressed and act cell-autonomously are shown in blue; tissues that do not express Dmrt genes but undergo sex-specific differentiation under the non-autonomous influence of Dmrt genes are in pink; sexually monomorphic cells are in white; arrows indicate non-autonomous functions of Dmrt genes. GL, germline; SG, somatic gonad. A. In the mouse, Dmrt1 is expressed and acts cell-autonomously in germline cells and in the Sertoli cells of the somatic gonad. In addition, Dmrt1 expression in Sertoli cells controls the development of the germline and other somatic gonad cells non-autonomously. Hormones secreted by the Sertoli cells (blue arrows) and other somatic gonad cells (purple arrows) regulate sex-specific development in many non-gonadal tissues. Dmrt1 is not known to play any cell-autonomous roles in sexual differentiation outside of the gonad. B. In Drosophila, dsx is expressed in somatic gonad progenitor cells, which recruit additional cells into the gonad through cell-cell signaling. dsx is not expressed in germline cells, but its function in the somatic gonad is necessary for sex-specific germline development. dsx is expressed in a subset on non-gonadal cells, where it acts cell-autonomously to control somatic sexual differentiation. Some of these cells also induce sex-specific development in adjacent tissues by a signaling mechanism.
Dmrt genes have a widespread non-autonomous role in germline sexual dimorphism. Although mice have cell-autonomous germline sexual identity and require Dmrt1 expression in germline cells, Dmrt1 function in Sertoli cells is necessary for germline cell maintenance and meiotic progression [27,28,49] (Figure 2 A). In medaka fish, where Dmrt1 is only expressed in Sertoli cells, it is required for mitotic arrest in the primordial germ cells [50]. Drosophila, like mouse, has intrinsic germline sexual identity, but dsx is not expressed in germline cells [16]. Rather, sexual differentiation of the germline is controlled by a combination of dsx-independent intrinsic mechanisms and dsx-dependent signals from the somatic gonad [51] (Figure 2 B). In adult testes, dsxM continues to be expressed in the “hub”, a key part of the somatic niche that maintains germline stem cells [16,32].
dsx also acts non-autonomously in the Drosophila somatic gonad, where signaling from dsx-expressing gonad precursor cells is necessary to recruit additional groups of male-specific precursors from the mesoderm [16,30,31]. Similarly, in the external genitalia dsxM and dsxF organize the global morphogenesis of the entire tissue by controlling the activation of several long-range signaling pathways [36] (Figure 2 B). In these and other examples, Dmrt genes control sexual differentiation far beyond their own expression domains – in fact, some of the most dramatic aspects of sexual dimorphism are due to the non-autonomous functions of Dmrt genes.
Evolutionary takeovers of sex determination
Sex determination signals and mechanisms evolve so rapidly that the master gene rarely stays at the top of the hierarchy for very long. For example, Sry does not exist outside of mammals [52], while Sex-lethal (Sxl) (Box 1) exists but does not play a sex-determining role outside of drosophilids [10,11]. In other lineages, sex-determining mechanisms turn over even more rapidly and can differ within species or between sibling species [53,54]. In vertebrates, the gonad controls sex-specific differentiation in the rest of the body through endocrine signaling [2], although non-gonadal cells have intrinsic sexual identity as well [7,47]. Male- and female-specific steroid hormones secreted by somatic gonad cells bind to nuclear hormone receptors in other tissues to promote sexually dimorphic development. One consequence of this mechanism is that the gene located at the top of the gonad-determining hierarchy essentially becomes the main sex-determining gene.
Dmrt1 and its paralogs have taken over this role in several vertebrate lineages. One of these regulatory coups has occurred recently in the evolution of medaka fish. In the model species Oryzias latipes, Dmrt1 has undergone a duplication and one of the newly derived paralogs, called Dmrt1bY or Dmy became the dominant male-determining gene [55,56]. Other Oryzias species lack Dmy, underscoring the recent origin of this sex-determining system [57].
In the frog Xenopus laevis, sex determination is female-heterogametic (ZZ males and ZW females). X. laevis is tetraploid and so has two DMRT1 co-orthologs. Similar to medaka, one of the DMRT1 genes underwent an additional duplication and one of the duplicates (called DM-W) is located on the W chromosome [58,59]. DM-W has acquired a dominant female-determining function by repressing the transcriptional targets of DMRT1 and tilting the balance of the bipotential gonad toward ovary development [9,58,60]. As in medaka, this sex-determining role is a recent innovation: closely related species such as X. tropicalis (also known as Silurana tropicalis) lack the DM-W gene [59].
Birds are also female-heterogametic, but unlike the dominant-W mechanism of X. laevis, sex in birds appears to be controlled by Z chromosome dosage [7]. In all birds, DMRT1 is located on the Z chromosome but is absent from the W. In chicken embryos, it is expressed in the early bipotential gonad and shows higher expression in males than in females [13]. As in the mouse, DMRT1 promotes testis development by activating SOX9 and repressing female-specific genes [61]. Chicken sex is determined, at least in part, by DMRT1 dosage: the two copies present in males are sufficient to turn on the testis specification pathway while the single dose in females fails to override ovary development [7].
Thus, Dmrt1 genes have moved up the regulatory hierarchy, from downstream positions in gonad differentiation to the top sex-determining role, in at least three distantly related clades. Why Dmrt rather than, say, Sox9 or Foxl2? One possible answer may lie in the large number of target genes they control and in their ability to either activate or repress transcription [29]. These features may place structural or regulatory changes in Dmrt1 outside the buffering capacity of the finely balanced gene circuit that controls testis versus ovary decision, leading to a rapid evolutionary takeover of sex determination.
Sex-specific splicing – an evolutionary switch in regulatory activity
A comparison of Dmrt genes from different animal lineages reveals important differences in their molecular functions despite their conserved roles in sexual differentiation. The mouse DMRT1 is a bifunctional regulator, activating some direct targets and repressing others [29]. The molecular basis of this versatility is unknown, but may involve the recruitment of different cofactors (coactivators vs corepressors) to the enhancers of different DMRT-regulated genes. The nematode MAB-3 is only known to act as a transcriptional repressor [62]. The most peculiar regulatory switch, however, occurred in insects.
In Drosophila, sex-specific splicing of dsx is the central event in sexual differentiation (Box 1), as the male- and female-specific Dsx isoforms exert distinct effects on the development of sexual traits. For example, the yolk protein (yp) genes are transcribed at a basal level in dsx mutants; DsxM further represses their transcription, while DsxF causes activation above the basal level [63,64]. desaturase-F (desat-F), which encodes an enzyme involved in pheromone synthesis, is activated by DsxF but not affected by DsxM [65]. Microarray experiments also reveal genes that are repressed by DsxF and/or activated by DsxM, but it is unclear whether any of these genes are direct transcriptional targets of Dsx [66]. Recently, a large number of potential direct targets have been identified by chromatin profiling [67]. Analysis of these genes should elucidate the activator and repressor functions of the male- and female-specific Dsx proteins.
dsx is spliced sex-specifically and has distinct DsxM and DsxF regulatory activities in other dipterans, moths, beetles, and Hymenopterans [68,69,70,71]. In all these insects, tra is also spliced sex-specifically and controls the sex-specific splicing of dsx, leading to the idea that the mechanism of sex determination based on the alternative splicing of dsx is common to all insects [10,11]. However, only holometabolous insects have been studied to date. Holometabola are a monophyletic group within insects that represents only a subset of order-level diversity. In Daphnia, the closest relative of insects examined so far, neither dsx nor tra show any evidence of sex-specific splicing, and dsx does not play any role in female development. Moreover, RNAi knock-down of the Daphnia tra has no effect on sex-specific development or dsx expression [33,72]. Similarly, the nematode and vertebrate Dmrt genes are regulated sex-specifically at the transcriptional level but do not show sex-specific splicing. Thus, although more basal insects and other arthropods need to be examined, the sex-specific splicing of dsx and its active role in female sex differentiation appear to be insect-specific (or even Holometabola-specific) innovations. An intriguing possibility is that DsxM and DsxF have subdivided the regulatory activity of the ancestral Dsx, leading to a more flexible control of sex-specific development.
Origin and evolution of new sex-specific traits
One of the most fascinating features of animal evolution is the rapid turnover of sex-specific traits. Both lions and gazelles are sexually dimorphic, but the traits distinguishing males from females are clearly different between the two. This simple observation implies that new sexual characters are gained, and old ones are lost, during the evolution of any animal lineage. The molecular mechanisms of this turnover are poorly understood, but recent evidence suggests that Dmrt genes may play important roles in this process.
The role of Dmrt genes in the evolution of sexual dimorphism may be particularly prominent in organisms like insects, where the sex of most cells is determined autonomously. For any sex-specific structure to evolve, dsx must be expressed in the cells that either give rise to this structure, or induce this structure through cell-cell signaling. In tissues that already express dsx, sexual dimorphism can originate if Dsx acquires new downstream targets through cis-regulatory changes that create new, or higher affinity, Dsx binding sites [73]. If, on the other hand, the tissue is ancestrally monomorphic and does not express dsx, changes in the spatial regulation of dsx that result in its de novo expression in this tissue must be a necessary first step [34].
Recent work shows that both of these mechanisms operate during the evolution of sexual dimorphism. For example, dsx and the HOX gene Abdominal-B (Abd-B) control sex-specific pigmentation in D. melanogaster by regulating the expression of bab1 and bab2. Sexually dimorphic pigmentation is a recent evolutionary innovation: in most Drosophila species, males and females are pigmented identically. The origin of this novel pattern was caused by cis-regulatory changes in a bab enhancer, which changed the way in which the bab genes are regulated by Dsx and Abd-B [74,75]. Similarly, the evolution of sex-specific pheromone profiles in Drosophila reflects multiple gains and losses of Dsx binding sites in the enhancer of desat-F [65].
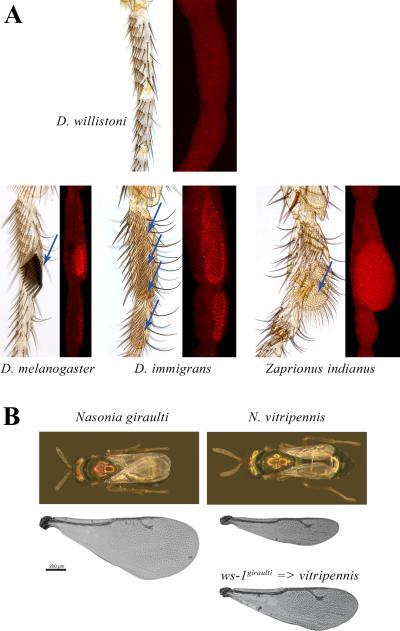
Sex comb evolution provides the clearest example of association between changes in the spatial expression of dsx and morphological diversity. The sex comb is another recent innovation restricted to a single lineage within Drosophila; outside of this lineage, males and females have identical leg bristle patterns [76]. In species that have sex combs, dsx is expressed in the presumptive sex comb region, where it is activated by Scr, while Scr expression is in turn regulated by dsx and is sexually dimorphic (Figure 3 A). However, in species that primitively lack sex combs, dsx is not expressed in the homologous region, and Scr expression is monomorphic [34,77]. Thus, sex comb evolution was associated with the origin of novel regulatory interactions between HOX and sex determination genes, leading to the gain of a new dsx expression domain. Two other, distantly related lineages in the family Drosophilidae that independently evolved new male-specific structures also show recent evolutionary gains of dsx expression in the corresponding tissues (Figure 3 A) [34].
Figure 3.
Changes in Dmrt genes are responsible for the origin and evolution of sex-specific traits. Only males are shown for all species. A. In Drosophila and related genera, the first pair of legs is sexually monomorphic in the ancestral condition, illustrated here by D. willistoni, and dsx is not expressed during the critical stages of pupal leg development. Three different lineages of Drosophilidae have independently evolved new male-specific structures: sex comb in D. melanogaster and its relatives, a long brush of tightly packed bristles in the immigrans species group, and a tarsal bulge with a round brush of fine hairs in the genus Zaprionus (arrows in Fig. 2 A). In each case, the novel morphology is associated with a novel expression domain of dsx (Dsx antibody staining in red) (Modified from [34]). B. In the wasp Nasonia, male-specific wing reduction is much more pronounced in N. vitripennis than in N. giraulti. Introgression of the ws-1 genomic region, which spans the dsx locus, from N. giraulti into N. vitripennis is sufficient to increase wing size [78] (modified from [78] with permission).
In the parasitoid wasp Nasonia, wing size and shape are sexually dimorphic in all species but to a different extent. In N. vitripennis, males have much smaller wings than females and are flightless, while in N. giraulti the dimorphism is only mild, and the males can fly (Figure 3 B). Greatly reduced male wings are a recently derived trait [78]. One of the loci responsible for the difference in male wing size between N. giraulti and N. vitripennis maps to the noncoding region immediately upstream from dsx, and the N. vitripennis allele associated with smaller wings increases dsx expression in the larval wing primordium more than two-fold (Figure 3 B) [78]. These observations in flies and wasps suggest that changes in dsx regulation may be a key mechanism that enables the evolution of new sex-specific traits in insects and other arthropods, contributing to some of the most dramatic examples of phenotypic diversification in nature.
Concluding remarks and future perspectives
Despite the endlessly diverse manifestations of sexual dimorphism and the profound differences between the mechanisms that specify it in different phyla, Dmrt genes provide a common framework for understanding the development and evolution of sex-specific traits. In all animals studied to date, members of the Dmrt family are expressed in tightly restricted spatial patterns in association with the development of sex-specific organs. Superficial differences between the roles of these genes in different phyla hide deeper similarities. In Drosophila, recent discoveries have demoted dsx from a global sex-determining gene to a tissue-specific regulator of cell differentiation. The fact that most cells in Drosophila lack dsx expression shows that only those cells that need to differentiate in a distinct male- or female-specific way are provided with the molecular machinery necessary to make the right choice. In this respect, the role of dsx in Drosophila is remarkably similar to the roles of Dmrt genes in nematodes and vertebrates. The canonical Drosophila mechanism of sex determination based on sex-specific splicing is an insect-specific innovation that was superimposed on a more ancestral mechanism based on localized transcription. Similarly, the role of Dmrt1 genes as the master sex switches in several vertebrate taxa is also a recent innovation that can ultimately be traced to their localized activation in crucial cell types in the somatic gonad.
The key function of Dmrt genes lies in the integration of spatial, temporal, and sexual information to direct male- or female-specific differentiation of a relatively small number of cell types. This function can be as minor as regulating a few enzymes or cytoskeletal molecules, or as major as controlling the deployment of long-distance morphogens or endocrine signals that induce drastically different patterns of tissue growth and differentiation. Characteristic features of Dmrt genes such as limitation to a small number of cell types, upstream co-regulation by global sex-determining pathways and local spatial activators, as well as the ability to promote sex-specific differentiation both autonomously and non-autonomously turn out to be surprisingly similar in widely diverged animal phyla.
In animals as diverse as vertebrates, arthropods, and mollusks, Dmrt genes are involved in gonad development, suggesting that they acted as selector genes that controlled testis versus ovary decision in the last Bilaterian ancestor, if not earlier. Starting from this ancestral role, changes in the transcriptional regulation of Dmrt genes could have led to their independent cooption for the development of other sex-specific structures in different lineages. In insects, this mechanism is still playing an important role in the evolution of sexual dimorphism. Every group of insects has evolved its own sex-specific structures, which often reach truly bizarre proportions but have no clear homologues in other taxa. Many of these innovations may be due to the origin of new dsx expression domains.
Many unanswered questions remain both about the functions of Dmrt genes and about their roles in evolution (Box 4). However, the last few years have laid an excellent foundation for this work. Going forward, this unique gene family will be central to our understanding of sexual dimorphism throughout the animal kingdom.
Box 1. Where it all began: doublesex and sex determination in Drosophila.
doublesex (dsx), the founding member of the Dmrt gene family, was first identified as a mutation affecting sexual differentiation in Drosophila [35]. Its name stems from the intersexual phenotype of dsx mutants: some traits are intermediate between males and females, while in other tissues both male and female structures develop in parallel [35,37].
The central role in Drosophila sex determination is played by alternative splicing (Fig. I). Drosophila has male-heterogametic sex determination where sex depends on the X:autosome ratio: XX individuals are females and XY are males. The primary genetic switch that interprets X chromosome dose is the RNA-binding protein Sex-lethal (Sxl), which regulates both its own splicing and the splicing of another RNA-binding factor, transformer (tra); as a result, only females have functional Tra protein [79,80]. dsx and an unrelated transcription factor called fruitless (fru) lie at the bottom of this splicing cascade: dsx controls sexual differentiation in most somatic tissues, while fru functions in the nervous system [37,81,82]. In females, the presence of functional Tra causes dsx to be spliced into a female-specific isoform (dsxF), while the male-specific isoform (dsxM) is produced by default [1,6].
The two dsx isoforms have opposite effects on sex differentiation: dsxM promotes the development of male-specific structures and represses female-specific structures, while dsxF promotes female-specific and represses male-specific structures [37,83,84]. DsxM and DsxF are transcription factors that share a common N-terminal DNA-binding domain, but have different C-terminal domains with distinct effects on target gene expression [64,85,86].
Until recently, only a few direct Dsx targets had been identified. These include the yolk protein genes, which are expressed in females and deposited in oocytes; bric a brac 1 (bab1) and bric a brac 2 (bab2), which encode transcription factors that control sex-specific color patterns; and desaturase-F, which encodes an enzyme involved in the synthesis of sex-specific pheromones [64,65,75]. With the growing application of chromatin profiling techniques, this paucity of data is finally at an end. A recent paper lists several dozen likely direct targets of Dsx [67], and more genomic data are on the way.
Box 1 Figure I.
Sex determination pathway in Drosophila. Active gene products and gene interactions are shown in purple and inactive in grey. Primary sex determination occurs very early in embryonic development when zygotically transcribed genes located on the X chromosome and the autosomes activate Sxl in females but fail to activate it in males. Sxl makes sex determination permanent by establishing a positive autoregulatory loop in females and controls the splicing of tra. In females, the presence of functional Sxl and Tra proteins leads to female-specific splicing of dsx and suppresses male-specific splicing of fru. In males, the absence of Sxl and Tra allows the default splicing of dsx and fru to produce male-specific isoforms.
Box 2. The Dmrt gene family.
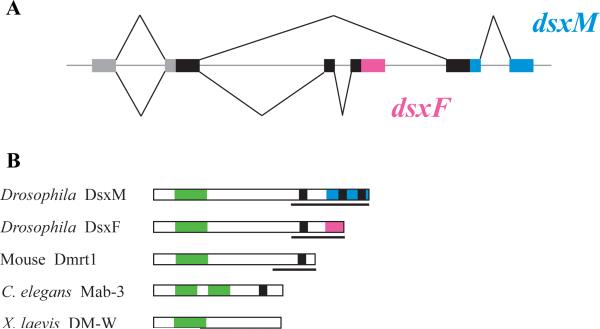
A breakthrough discovery came when the mab-3 gene of C. elegans, long known to control the development of male-specific sensory organs, was shown to encode a homologue of the Drosophila dsx [5]. A homologous gene was also identified in humans, where it was found to map to a chromosomal region associated with sex reversal [5]. Almost immediately, the mouse and chicken homologues of dsx and mab-3 were shown to have gonad-specific expression and to correlate with testis development in particular [13] (Fig. I). The DNA-binding domain common to these genes was named the DM (Dsx/MAB-3) domain, and the family was named Dmrt (doublesex/mab-3 related) genes [5,13]. Outside of the DM domain, most Dmrt genes show little or no similarity across taxa [87].
In vertebrates, which have multiple Dmrt paralogs (e. g. Dmrt1 through Dmrt7 in mice and humans), Dmrt1 plays a particularly important role in gonad development and sexual differentiation and has received the most attention, but other paralogs have also been characterized [23,24]. Orthology relationships between fish and mammalian genes are well supported, indicating that this family diversified prior to vertebrate radiation [87]. All invertebrates for which genome sequences are available also have multiple Dmrt genes; C. elegans holds the current record at 11. However, there is little sequence conservation between the vertebrate and invertebrate genes, making their relationships hard to determine. In particular, it is not clear whether dsx, Dmrt1, and mab-3 are orthologous.
The DM domain is a zinc finger DNA binding motif that interacts with DNA in the minor groove [86]. The regulatory domains of DMRT transcription factors are located at their C-termini. Different DMRT proteins, such as DsxM and DsxF as well as different vertebrate DMRT paralogs, can bind DNA as either homo- or heterodimers [88,89]. DMRT proteins have fairly high specificity compared to other minor groove-binding proteins, but much of this specificity appears to be shared across the gene family. For example, the Drosophila Dsx and the C. elegans MAB-3 bind similar DNA sequences, and dsxM can partially substitute for mab-3 in vivo [89,90].
Box 2 Figure I.
The structure of Dmrt genes and proteins. A. Genomic structure of the Drosophila dsx. Coding sequences common to dsxM and dsxF are shown by thick black bars, transcribed untranslated regions by grey bars, and introns and intergenic regions by a thin grey line; splicing patterns are indicated by thin black lines above and below the gene region. dsxM and dsxF share the 5’ exon that encodes the DNA-binding DM domain, but include mutually exclusive 3’ exons that encode different dimerization/activation domains. dsxM-specific exons are shown in blue and dsxF-specific sequence in pink. B. Protein structure of the Drosophila Dsx, mouse DMRT1, C. elegans MAB-3, and Xenopus DM-W. DM domains are shown in green and proline/serine-rich domains found in most DMRT proteins are in black. Pink and blue bars show the sex-specific domains of DsxF and DsxM, respectively. Lines under the protein schematics indicate characterized protein domains involved in transcriptional regulation. MAB-3 is unique in having two DM domains, and DM-W is unusual in lacking a C-terminal regulatory domain.
Box 3. Germline functions of Dmrt genes.
Anisogamy – a sexually dimorphic germline – is even more ancient than the gonads, predating animal multicellularity. Although gametes are probably the most sexually dimorphic cell type, in many animals (such as frogs, houseflies, or medaka fish) the germline lacks cell-autonomous sexual identity; sex-specific differentiation is imposed instead by the soma. Mice and Drosophila, by contrast, do have cell-autonomous germline sex but it is determined by mechanisms distinct from those operating in somatic cells [49]. Dmrt genes can be expressed in germline cells in both types of animals but, in contrast to the somatic gonad, their germline functions are not conserved.
In the mouse testis, Dmrt1 is expressed in germline cells and is required cell-autonomously for their survival, migration, and the suppression of pluripotency and proliferation [28]. Its two major functions in the male germline are to repress the meiotic inducer Stra8 and activate the spermatogonial differentiation factor Sohlh1 [91]. Dmrt1 function is different in the female germline, where it activates Stra8 and is required for normal meiotic prophase [92]. In the testes of teleost fish, Dmrt1 orthologs can be expressed in both somatic and germline cells (platyfish and catfish [93,94]), only in the soma (medaka, tilapia, and pufferfish [95,96,97]), or only in the germline (zebrafish and cod [98,99]). In species where Dmrt1 is expressed in both sexes, female expression is usually restricted to the germline while in males it is seen in both germline and somatic cells.
Although Drosophila, like mouse, has cell-autonomous germline sex, dsx is not expressed in the germline, and the only function of dsx in germline development in flies is non-autonomous [49,51,100]. Interestingly, a Dmrt gene identified in the crustacean Eriocheir sinensis is expressed in both somatic and germline cells of the testis [18] – a situation closer to vertebrates than to Drosophila. In contrast to the near-universal conservation of gonad function, it is not clear whether the role of Dmrt genes in germline development is ancient or has evolved independently in different lineages.
Box 4. Unanswered questions.
- How do Dmrt genes regulate their transcriptional targets? Can the Drosophila DsxM act as an activator, and DsxF as a repressor? How does mammalian DMRT1 achieve its dual function as both an activator and a repressor? What cofactors does it interact with to activate or repress transcription?
- Was the ancestral function of Dmrt genes restricted to males, or were they involved in gonad specification in both sexes? Is their function in germline cells ancestral, or were they independently recruited in germline development in some taxa?
- In vertebrates, what are the direct transcriptional regulators and targets of Dmrt1 in the gonad? How evolutionarily labile are these interactions, and what changes in these interactions allow Dmrt1 paralogs to acquire top positions in the sex-determining hierarchy?
- When and how did the splicing-based mechanism of insect sex determination evolve? What was the regulatory activity of arthropod Dsx before the evolution of sex-specific splicing? How did the newly independent DsxM and DsxF proteins acquire their distinct regulatory specificities?
- Do vertebrate Dmrt genes play cell-autonomous roles in sex-specific development outside of the gonad?
- How common is the evolution of new dsx expression domains in insects, and what role does it play in the origin of new sex-specific structures? Does a similar mechanism operate in other animal groups with cell-autonomous sex determination?
Acknowledgements
I am grateful to David Loehlin for providing Nasonia images, and to David Plachetzki, Joel Savard, David Zarkower, and three anonymous reviewers for their comments on the manuscript. Work in my laboratory is supported by NIH grant 5-R01GM082843-02.
Glossary
- Bilateria
a group of animal phyla that evolved from a common bilaterally symmetric ancestor; it includes all multicellular animals except the phyla Porifera, Placozoa, Cnidaria, and Ctenophora
- Cell-autonomous gene function
developmental mechanism where the expression of a regulatory gene in a particular cell determines the fate or functional properties of that same cell
- cis-regulatory changes
nucleotide substitutions in non-coding gene sequences that result in altered expression of that gene in the course of evolution
- Courtship song
in Drosophila males, a precisely modulated series of wing vibrations that produce species-specific sounds recognized by females
- DM domain
a DNA-binding domain common to all DMRT transcription factors
- Dmrt genes
the family of doublesex / mab-3 related transcription factors, characterized by the presence of the DNA-binding DM domain
- Ecdysone
an insect hormone that controls molting, metamorphosis, and tissue remodeling
- Gonochorists
species where each individual is either male or female, but not both
- Granulosa cells
In vertebrates, somatic ovary cells closely associated with developing female gametes; granulosa cells produce female sex hormones from a precursor secreted by the theca cells
- Hermaphrodites
species where a single individual can function as both male or female, either sequentially or at the same time
- Heterochronic pathway
the genetic pathway that controls the timing of development in C. elegans
- Non-autonomous gene function
developmental mechanism where the expression of a regulatory gene in some cells determines the fates of other cells that do not express this gene, usually through a signaling pathway
- Orthologs
genes in different species that evolved from a common ancestral gene by speciation, e. g. Dmrt1 in mouse and Dmrt1 in human
- Paralogs
genes that evolved by duplication of a common ancestral gene within a genome, e. g. Dmrt1 and Dmrt2 in mouse, or mouse Dmrt1 and human Dmrt2
- Primordial germ cells
embryonic stem cells that give rise to adult germline cells
- Proctodeum
posterior ectodermal part of the digestive tract connecting to the hindgut
- Protandrous hermaphrodites
sequential hermaphrodites where individuals function first as males, and later as females
- Protogynous hermaphrodites
sequential hermaphrodites where individuals function first as females, and later as males
- Sertoli cells
somatic testis cells associated with developing male gametes; Sertoli cells produce male sex hormones and control the development of germline cells and other hormone-producing somatic cells
- Sequential hermaphrodites
hermaphroditic species where some or all individuals change sex at some point during adult life
- Sexual dimorphism
phenotypic differences between males and females of the same species
- Theca cells
In vertebrates, somatic gonad cells associated with ovarian follicles; theca cells secrete a steroid precursor that is converted into female sex hormones by the granulosa cells
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.McKeown M. Sex differentiation: the role of alternative splicing. Curr Opin Genet Dev. 1992;2:299–303. doi: 10.1016/s0959-437x(05)80288-6. [DOI] [PubMed] [Google Scholar]
- 2.Wilhelm D, Palmer S, Koopman P. Sex determination and gonadal development in mammals. Physiol Rev. 2007;87:1–28. doi: 10.1152/physrev.00009.2006. [DOI] [PubMed] [Google Scholar]
- 3.Zarkower D. Establishing sexual dimorphism: conservation amidst diversity? Nat Rev Genet. 2001;2:175–185. doi: 10.1038/35056032. [DOI] [PubMed] [Google Scholar]
- 4.Raymond CS, Murphy MW, O'Sullivan MG, Bardwell VJ, Zarkower D. Dmrt1, a gene related to worm and fly sexual regulators, is required for mammalian testis differentiation. Genes Dev. 2000;14:2587–2595. doi: 10.1101/gad.834100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raymond CS, Shamu CE, Shen MM, Seifert KJ, Hirsch B, et al. Evidence for evolutionary conservation of sex-determining genes. Nature. 1998;391:691–695. doi: 10.1038/35618. [DOI] [PubMed] [Google Scholar]
- 6.Burtis KC, Baker BS. Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell. 1989;56:997–1010. doi: 10.1016/0092-8674(89)90633-8. [DOI] [PubMed] [Google Scholar]
- 7.Chue J, Smith CA. Sex determination and sexual differentiation in the avian model. FEBS J. 2011;278:1027–1034. doi: 10.1111/j.1742-4658.2011.08032.x. [DOI] [PubMed] [Google Scholar]
- 8.Herpin A, Schartl M. Dmrt1 genes at the crossroads: a widespread and central class of sexual development factors in fish. FEBS J. 2011;278:1010–1019. doi: 10.1111/j.1742-4658.2011.08030.x. [DOI] [PubMed] [Google Scholar]
- 9.Yoshimoto S, Ito M. A ZZ/ZW-type sex determination in Xenopus laevis. FEBS J. 2011;278:1020–1026. doi: 10.1111/j.1742-4658.2011.08031.x. [DOI] [PubMed] [Google Scholar]
- 10.Gempe T, Beye M. Function and evolution of sex determination mechanisms, genes and pathways in insects. Bioessays. 2011;33:52–60. doi: 10.1002/bies.201000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verhulst EC, van de Zande L, Beukeboom LW. Insect sex determination: it all evolves around transformer. Curr Opin Genet Dev. 2010;20:376–383. doi: 10.1016/j.gde.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 12.Kim S, Kettlewell JR, Anderson RC, Bardwell VJ, Zarkower D. Sexually dimorphic expression of multiple doublesex-related genes in the embryonic mouse gonad. Gene Expr Patterns. 2003;3:77–82. doi: 10.1016/s1567-133x(02)00071-6. [DOI] [PubMed] [Google Scholar]
- 13.Raymond CS, Kettlewell JR, Hirsch B, Bardwell VJ, Zarkower D. Expression of Dmrt1 in the genital ridge of mouse and chicken embryos suggests a role in vertebrate sexual development. Dev Biol. 1999;215:208–220. doi: 10.1006/dbio.1999.9461. [DOI] [PubMed] [Google Scholar]
- 14.Sinclair A, Smith C, Western P, McClive P. A comparative analysis of vertebrate sex determination. Novartis Found Symp. 2002;244:102–111. discussion 111-104, 203-106, 253-107. [PubMed] [Google Scholar]
- 15.Shoemaker C, Ramsey M, Queen J, Crews D. Expression of Sox9, Mis, and Dmrt1 in the gonad of a species with temperature-dependent sex determination. Dev Dyn. 2007;236:1055–1063. doi: 10.1002/dvdy.21096. [DOI] [PubMed] [Google Scholar]
- 16.Hempel LU, Oliver B. Sex-specific DoublesexM expression in subsets of Drosophila somatic gonad cells. BMC Dev Biol. 2007;7:113. doi: 10.1186/1471-213X-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato Y, Kobayashi K, Watanabe H, Iguchi T. Environmental sex determination in the branchiopod crustacean Daphnia magna: deep conservation of a Doublesex gene in the sex-determining pathway. PLoS Genet. 2011;7:e1001345. doi: 10.1371/journal.pgen.1001345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang EF, Qiu GF. A novel Dmrt gene is specifically expressed in the testis of Chinese mitten crab, Eriocheir sinensis. Dev Genes Evol. 2010;220:151–159. doi: 10.1007/s00427-010-0336-2. [DOI] [PubMed] [Google Scholar]
- 19.Farazmand A, Inanloo K, Agh N. Expression of Dmrt family genes during gonadal differentiation in two species of Artemia (Branchiopoda, Anostraca) from Urmia Lake (Iran). Crustaceana. 2010;83:1153–1165. [Google Scholar]
- 20.Klinbunga S, Amparyup P, Khamnamtong B, Hirono I, Aoki T, et al. Isolation and characterization of testis-specific DMRT1 in the tropical abalone (Haliotis asinina). Biochem Genet. 2009;47:66–79. doi: 10.1007/s10528-008-9207-1. [DOI] [PubMed] [Google Scholar]
- 21.Naimi A, Martinez AS, Specq ML, Mrac A, Diss B, et al. Identification and expression of a factor of the DM family in the oyster Crassostrea gigas. Comp Biochem Physiol A Mol Integr Physiol. 2009;152:189–196. doi: 10.1016/j.cbpa.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 22.Miller SW, Hayward DC, Bunch TA, Miller DJ, Ball EE, et al. A DM domain protein from a coral, Acropora millepora, homologous to proteins important for sex determination. Evol Dev. 2003;5:251–258. doi: 10.1046/j.1525-142x.2003.03032.x. [DOI] [PubMed] [Google Scholar]
- 23.Kim S, Namekawa SH, Niswander LM, Ward JO, Lee JT, et al. A mammal-specific Doublesex homolog associates with male sex chromatin and is required for male meiosis. PLoS Genet. 2007;3:e62. doi: 10.1371/journal.pgen.0030062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balciuniene J, Bardwell VJ, Zarkower D. Mice mutant in the DM domain gene Dmrt4 are viable and fertile but have polyovular follicles. Mol Cell Biol. 2006;26:8984–8991. doi: 10.1128/MCB.00959-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Uhlenhaut NH, Jakob S, Anlag K, Eisenberger T, Sekido R, et al. Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell. 2009;139:1130–1142. doi: 10.1016/j.cell.2009.11.021. [DOI] [PubMed] [Google Scholar]
- 26.Sekido R, Lovell-Badge R. Sex determination and SRY: down to a wink and a nudge? Trends Genet. 2009;25:19–29. doi: 10.1016/j.tig.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 27.Matson CK, Murphy MW, Sarver AL, Griswold MD, Bardwell VJ, et al. DMRT1 prevents female reprogramming in the postnatal mammalian testis. Nature. 2011;476:101–104. doi: 10.1038/nature10239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim S, Bardwell VJ, Zarkower D. Cell type-autonomous and non-autonomous requirements for Dmrt1 in postnatal testis differentiation. Dev Biol. 2007;307:314–327. doi: 10.1016/j.ydbio.2007.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murphy MW, Sarver AL, Rice D, Hatzi K, Ye K, et al. Genome-wide analysis of DNA binding and transcriptional regulation by the mammalian Doublesex homolog DMRT1 in the juvenile testis. Proc Natl Acad Sci U S A. 2010;107:13360–13365. doi: 10.1073/pnas.1006243107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DeFalco T, Camara N, Le Bras S, Van Doren M. Nonautonomous sex determination controls sexually dimorphic development of the Drosophila gonad. Dev Cell. 2008;14:275–286. doi: 10.1016/j.devcel.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Camara N, Whitworth C, Van Doren M. The creation of sexual dimorphism in the Drosophila soma. Curr Top Dev Biol. 2008;83:65–107. doi: 10.1016/S0070-2153(08)00403-1. [DOI] [PubMed] [Google Scholar]
- 32.Robinett CC, Vaughan AG, Knapp JM, Baker BS. Sex and the Single Cell. II. There Is a Time and Place for Sex. PLoS Biol. 2010;8:e1000365. doi: 10.1371/journal.pbio.1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kato M, Takehana Y, Fukuda Y, Naruse K, Sakaizumi M, et al. An autosomal locus controls sex reversal in interspecific XY hybrids of the medaka fishes. Heredity. 2011 doi: 10.1038/hdy.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tanaka K, Barmina O, Sanders LE, Arbeitman MN, Kopp A. Evolution of Sex-Specific Traits through Changes in HOX-Dependent doublesex Expression. PLoS Biol. 2011;9:e1001131. doi: 10.1371/journal.pbio.1001131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hildreth PE. Doublesex, Recessive Gene That Transforms Both Males and Females of Drosophila into Intersexes. Genetics. 1965;51:659–678. doi: 10.1093/genetics/51.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Christiansen AE, Keisman EL, Ahmad SM, Baker BS. Sex comes in from the cold: the integration of sex and pattern. Trends Genet. 2002;18:510–516. doi: 10.1016/s0168-9525(02)02769-5. [DOI] [PubMed] [Google Scholar]
- 37.Baker BS, Ridge KA. Sex and the single cell. I. On the action of major loci affecting sex determination in Drosophila melanogaster. Genetics. 1980;94:383–423. doi: 10.1093/genetics/94.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kimura K, Hachiya T, Koganezawa M, Tazawa T, Yamamoto D. Fruitless and doublesex coordinate to generate male-specific neurons that can initiate courtship. Neuron. 2008;59:759–769. doi: 10.1016/j.neuron.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 39.Rideout EJ, Billeter JC, Goodwin SF. The sex-determination genes fruitless and doublesex specify a neural substrate required for courtship song. Curr Biol. 2007;17:1473–1478. doi: 10.1016/j.cub.2007.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Billeter JC, Villella A, Allendorfer JB, Dornan AJ, Richardson M, et al. Isoform-specific control of male neuronal differentiation and behavior in Drosophila by the fruitless gene. Curr Biol. 2006;16:1063–1076. doi: 10.1016/j.cub.2006.04.039. [DOI] [PubMed] [Google Scholar]
- 41.Sanders LE, Arbeitman MN. Doublesex establishes sexual dimorphism in the Drosophila central nervous system in an isoform-dependent manner by directing cell number. Dev Biol. 2008;320:378–390. doi: 10.1016/j.ydbio.2008.05.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zarkower D. Somatic sex determination. WormBook; 2006. pp. 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mason DA, Rabinowitz JS, Portman DS. dmd-3, a doublesex-related gene regulated by tra-1, governs sex-specific morphogenesis in C. elegans. Development. 2008;135:2373–2382. doi: 10.1242/dev.017046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lints R, Emmons SW. Regulation of sex-specific differentiation and mating behavior in C. elegans by a new member of the DM domain transcription factor family. Genes Dev. 2002;16:2390–2402. doi: 10.1101/gad.1012602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yi W, Ross JM, Zarkower D. Mab-3 is a direct tra-1 target gene regulating diverse aspects of C. elegans male sexual development and behavior. Development. 2000;127:4469–4480. doi: 10.1242/dev.127.20.4469. [DOI] [PubMed] [Google Scholar]
- 46.Nelson MD, Zhou E, Kiontke K, Fradin H, Maldonado G, et al. A bow-tie genetic architecture for morphogenesis suggested by a genome-wide RNAi screen in Caenorhabditis elegans. PLoS Genet. 2011;7:e1002010. doi: 10.1371/journal.pgen.1002010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arnold AP. The end of gonad-centric sex determination in mammals. Trends Genet. 2011 doi: 10.1016/j.tig.2011.10.004. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hong CS, Park BY, Saint-Jeannet JP. The function of Dmrt genes in vertebrate development: it is not just about sex. Dev Biol. 2007;310:1–9. doi: 10.1016/j.ydbio.2007.07.035. [DOI] [PubMed] [Google Scholar]
- 49.Murray SM, Yang SY, Van Doren M. Germ cell sex determination: a collaboration between soma and germline. Curr Opin Cell Biol. 2010;22:722–729. doi: 10.1016/j.ceb.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Herpin A, Schindler D, Kraiss A, Hornung U, Winkler C, et al. Inhibition of primordial germ cell proliferation by the medaka male determining gene Dmrt I bY. BMC Dev Biol. 2007;7:99. doi: 10.1186/1471-213X-7-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Casper AL, Van Doren M. The establishment of sexual identity in the Drosophila germline. Development. 2009;136:3821–3830. doi: 10.1242/dev.042374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wallis MC, Waters PD, Graves JA. Sex determination in mammals--before and after the evolution of SRY. Cell Mol Life Sci. 2008;65:3182–3195. doi: 10.1007/s00018-008-8109-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mank JE, Avise JC. Evolutionary diversity and turn-over of sex determination in teleost fishes. Sex Dev. 2009;3:60–67. doi: 10.1159/000223071. [DOI] [PubMed] [Google Scholar]
- 54.Hediger M, Henggeler C, Meier N, Perez R, Saccone G, et al. Molecular characterization of the key switch F provides a basis for understanding the rapid divergence of the sex-determining pathway in the housefly. Genetics. 2010;184:155–170. doi: 10.1534/genetics.109.109249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nanda I, Kondo M, Hornung U, Asakawa S, Winkler C, et al. A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc Natl Acad Sci U S A. 2002;99:11778–11783. doi: 10.1073/pnas.182314699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matsuda M, Nagahama Y, Shinomiya A, Sato T, Matsuda C, et al. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature. 2002;417:559–563. doi: 10.1038/nature751. [DOI] [PubMed] [Google Scholar]
- 57.Kondo M, Nanda I, Hornung U, Asakawa S, Shimizu N, et al. Absence of the candidate male sex-determining gene dmrt1b(Y) of medaka from other fish species. Curr Biol. 2003;13:416–420. doi: 10.1016/s0960-9822(03)00125-8. [DOI] [PubMed] [Google Scholar]
- 58.Yoshimoto S, Okada E, Umemoto H, Tamura K, Uno Y, et al. A W-linked DM-domain gene, DM-W, participates in primary ovary development in Xenopus laevis. Proc Natl Acad Sci U S A. 2008;105:2469–2474. doi: 10.1073/pnas.0712244105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bewick AJ, Anderson DW, Evans BJ. Evolution of the closely related, sex-related genes DM-W and DMRT1 in African clawed frogs (Xenopus). Evolution. 2011;65:698–712. doi: 10.1111/j.1558-5646.2010.01163.x. [DOI] [PubMed] [Google Scholar]
- 60.Yoshimoto S, Ikeda N, Izutsu Y, Shiba T, Takamatsu N, et al. Opposite roles of DMRT1 and its W-linked paralogue, DM-W, in sexual dimorphism of Xenopus laevis: implications of a ZZ/ZW-type sex-determining system. Development. 2010 doi: 10.1242/dev.048751. [DOI] [PubMed] [Google Scholar]
- 61.Smith CA, Roeszler KN, Ohnesorg T, Cummins DM, Farlie PG, et al. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature. 2009;461:267–271. doi: 10.1038/nature08298. [DOI] [PubMed] [Google Scholar]
- 62.Ross JM, Kalis AK, Murphy MW, Zarkower D. The DM domain protein MAB-3 promotes sex-specific neurogenesis in C. elegans by regulating bHLH proteins. Dev Cell. 2005;8:881–892. doi: 10.1016/j.devcel.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 63.Coschigano KT, Wensink PC. Sex-specific transcriptional regulation by the male and female doublesex proteins of Drosophila. Genes Dev. 1993;7:42–54. doi: 10.1101/gad.7.1.42. [DOI] [PubMed] [Google Scholar]
- 64.Burtis KC, Coschigano KT, Baker BS, Wensink PC. The doublesex proteins of Drosophila melanogaster bind directly to a sex-specific yolk protein gene enhancer. Embo J. 1991;10:2577–2582. doi: 10.1002/j.1460-2075.1991.tb07798.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shirangi TR, Dufour HD, Williams TM, Carroll SB. Rapid evolution of sex pheromone-producing enzyme expression in Drosophila. PLoS Biol. 2009;7:e1000168. doi: 10.1371/journal.pbio.1000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Goldman TD, Arbeitman MN. Genomic and functional studies of Drosophila sex hierarchy regulated gene expression in adult head and nervous system tissues. PLoS Genet. 2007;3:e216. doi: 10.1371/journal.pgen.0030216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Luo SD, Shi GW, Baker BS. Direct targets of the D. melanogaster DSXF protein and the evolution of sexual development. Development. 2011;138:2761–2771. doi: 10.1242/dev.065227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Salvemini M, Mauro U, Lombardo F, Milano A, Zazzaro V, et al. Genomic organization and splicing evolution of the doublesex gene, a Drosophila regulator of sexual differentiation, in the dengue and yellow fever mosquito Aedes aegypti. BMC Evol Biol. 2011;11:41. doi: 10.1186/1471-2148-11-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cho S, Huang ZY, Zhang J. Sex-specific splicing of the honeybee doublesex gene reveals 300 million years of evolution at the bottom of the insect sex-determination pathway. Genetics. 2007;177:1733–1741. doi: 10.1534/genetics.107.078980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oliveira DC, Werren JH, Verhulst EC, Giebel JD, Kamping A, et al. Identification and characterization of the doublesex gene of Nasonia. Insect Mol Biol. 2009;18:315–324. doi: 10.1111/j.1365-2583.2009.00874.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Suzuki MG, Funaguma S, Kanda T, Tamura T, Shimada T. Role of the male BmDSX protein in the sexual differentiation of Bombyx mori. Evol Dev. 2005;7:58–68. doi: 10.1111/j.1525-142X.2005.05007.x. [DOI] [PubMed] [Google Scholar]
- 72.Kato Y, Kobayashi K, Oda S, Tatarazako N, Watanabe H, et al. Sequence divergence and expression of a transformer gene in the branchiopod crustacean, Daphnia magna. Genomics. 2010;95:160–165. doi: 10.1016/j.ygeno.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 73.Williams TM, Carroll SB. Genetic and molecular insights into the development and evolution of sexual dimorphism. Nat Rev Genet. 2009;10:797–804. doi: 10.1038/nrg2687. [DOI] [PubMed] [Google Scholar]
- 74.Kopp A, Duncan I, Godt D, Carroll SB. Genetic control and evolution of sexually dimorphic characters in Drosophila. Nature. 2000;408:553–559. doi: 10.1038/35046017. [DOI] [PubMed] [Google Scholar]
- 75.Williams TM, Selegue JE, Werner T, Gompel N, Kopp A, et al. The regulation and evolution of a genetic switch controlling sexually dimorphic traits in Drosophila. Cell. 2008;134:610–623. doi: 10.1016/j.cell.2008.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kopp A. Drosophila Sex Combs as a Model of Evolutionary Innovations. Evol Dev. 2011 doi: 10.1111/j.1525-142X.2011.00507.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Barmina O, Kopp A. Sex-specific expression of a HOX gene associated with rapid morphological evolution. Dev Biol. 2007;311:277–286. doi: 10.1016/j.ydbio.2007.07.030. [DOI] [PubMed] [Google Scholar]
- 78.Loehlin DW, Oliveira DC, Edwards R, Giebel JD, Clark ME, et al. Non-coding changes cause sex-specific wing size differences between closely related species of Nasonia. PLoS Genet. 2010;6:e1000821. doi: 10.1371/journal.pgen.1000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bell LR, Horabin JI, Schedl P, Cline TW. Positive autoregulation of sex-lethal by alternative splicing maintains the female determined state in Drosophila. Cell. 1991;65:229–239. doi: 10.1016/0092-8674(91)90157-t. [DOI] [PubMed] [Google Scholar]
- 80.Nagoshi RN, McKeown M, Burtis KC, Belote JM, Baker BS. The control of alternative splicing at genes regulating sexual differentiation in D. melanogaster. Cell. 1988;53:229–236. doi: 10.1016/0092-8674(88)90384-4. [DOI] [PubMed] [Google Scholar]
- 81.Manoli DS, Foss M, Villella A, Taylor BJ, Hall JC, et al. Male-specific fruitless specifies the neural substrates of Drosophila courtship behaviour. Nature. 2005;436:395–400. doi: 10.1038/nature03859. [DOI] [PubMed] [Google Scholar]
- 82.Demir E, Dickson BJ. fruitless splicing specifies male courtship behavior in Drosophila. Cell. 2005;121:785–794. doi: 10.1016/j.cell.2005.04.027. [DOI] [PubMed] [Google Scholar]
- 83.Jursnich VA, Burtis KC. A positive role in differentiation for the male doublesex protein of Drosophila. Dev Biol. 1993;155:235–249. doi: 10.1006/dbio.1993.1021. [DOI] [PubMed] [Google Scholar]
- 84.Waterbury JA, Jackson LL, Schedl P. Analysis of the doublesex female protein in Drosophila melanogaster: role on sexual differentiation and behavior and dependence on intersex. Genetics. 1999;152:1653–1667. doi: 10.1093/genetics/152.4.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Erdman SE, Burtis KC. The Drosophila doublesex proteins share a novel zinc finger related DNA binding domain. Embo J. 1993;12:527–535. doi: 10.1002/j.1460-2075.1993.tb05684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhu L, Wilken J, Phillips NB, Narendra U, Chan G, et al. Sexual dimorphism in diverse metazoans is regulated by a novel class of intertwined zinc fingers. Genes Dev. 2000;14:1750–1764. [PMC free article] [PubMed] [Google Scholar]
- 87.Volff JN, Zarkower D, Bardwell VJ, Schartl M. Evolutionary dynamics of the DM domain gene family in metazoans. J Mol Evol. 2003;57(Suppl 1):S241–249. doi: 10.1007/s00239-003-0033-0. [DOI] [PubMed] [Google Scholar]
- 88.An W, Cho S, Ishii H, Wensink PC. Sex-specific and non-sex-specific oligomerization domains in both of the doublesex transcription factors from Drosophila melanogaster. Mol Cell Biol. 1996;16:3106–3111. doi: 10.1128/mcb.16.6.3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Murphy MW, Zarkower D, Bardwell VJ. Vertebrate DM domain proteins bind similar DNA sequences and can heterodimerize on DNA. BMC Mol Biol. 2007;8:58. doi: 10.1186/1471-2199-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yi W, Zarkower D. Similarity of DNA binding and transcriptional regulation by Caenorhabditis elegans MAB-3 and Drosophila melanogaster DSX suggests conservation of sex determining mechanisms. Development. 1999;126:873–881. doi: 10.1242/dev.126.5.873. [DOI] [PubMed] [Google Scholar]
- 91.Matson CK, Murphy MW, Griswold MD, Yoshida S, Bardwell VJ, et al. The mammalian doublesex homolog DMRT1 is a transcriptional gatekeeper that controls the mitosis versus meiosis decision in male germ cells. Dev Cell. 2010;19:612–624. doi: 10.1016/j.devcel.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Krentz AD, Murphy MW, Sarver AL, Griswold MD, Bardwell VJ, et al. DMRT1 promotes oogenesis by transcriptional activation of Stra8 in the mammalian fetal ovary. Dev Biol. 2011;356:63–70. doi: 10.1016/j.ydbio.2011.05.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Veith AM, Schafer M, Kluver N, Schmidt C, Schultheis C, et al. Tissue-specific expression of dmrt genes in embryos and adults of the platyfish Xiphophorus maculatus. Zebrafish. 2006;3:325–337. doi: 10.1089/zeb.2006.3.325. [DOI] [PubMed] [Google Scholar]
- 94.Raghuveer K, Senthilkumaran B, Sudhakumari CC, Sridevi P, Rajakumar A, et al. Dimorphic expression of various transcription factor and steroidogenic enzyme genes during gonadal ontogeny in the air-breathing catfish, Clarias gariepinus. Sex Dev. 2011;5:213–223. doi: 10.1159/000328823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kobayashi T, Matsuda M, Kajiura-Kobayashi H, Suzuki A, Saito N, et al. Two DM domain genes, DMY and DMRT1, involved in testicular differentiation and development in the medaka, Oryzias latipes. Dev Dyn. 2004;231:518–526. doi: 10.1002/dvdy.20158. [DOI] [PubMed] [Google Scholar]
- 96.Kobayashi T, Kajiura-Kobayashi H, Guan G, Nagahama Y. Sexual dimorphic expression of DMRT1 and Sox9a during gonadal differentiation and hormone-induced sex reversal in the teleost fish Nile tilapia (Oreochromis niloticus). Dev Dyn. 2008;237:297–306. doi: 10.1002/dvdy.21409. [DOI] [PubMed] [Google Scholar]
- 97.Yamaguchi A, Lee KH, Fujimoto H, Kadomura K, Yasumoto S, et al. Expression of the DMRT gene and its roles in early gonadal development of the Japanese pufferfish Takifugu rubripes. Comp Biochem Physiol Part D Genomics Proteomics. 2006;1:59–68. doi: 10.1016/j.cbd.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 98.Johnsen H, Seppola M, Torgersen JS, Delghandi M, Andersen O. Sexually dimorphic expression of dmrt1 in immature and mature Atlantic cod (Gadus morhua L.). Comp Biochem Physiol B Biochem Mol Biol. 2010;156:197–205. doi: 10.1016/j.cbpb.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 99.Guo Y, Cheng H, Huang X, Gao S, Yu H, et al. Gene structure, multiple alternative splicing, and expression in gonads of zebrafish Dmrt1. Biochem Biophys Res Commun. 2005;330:950–957. doi: 10.1016/j.bbrc.2005.03.066. [DOI] [PubMed] [Google Scholar]
- 100.Hempel LU, Kalamegham R, Smith JE, 3rd, Oliver B. Drosophila germline sex determination: integration of germline autonomous cues and somatic signals. Curr Top Dev Biol. 2008;83:109–150. doi: 10.1016/S0070-2153(08)00404-3. [DOI] [PMC free article] [PubMed] [Google Scholar]