Abstract
GLAST is the predominant glutamate transporter in the cerebellum and contributes substantially to glutamate transport in forebrain. This astroglial glutamate transporter quickly binds and clears synaptically released glutamate and is principally responsible for ensuring that synaptic glutamate concentrations remain low. This process is associated with a significant energetic cost. Compartmentalization of GLAST with mitochondria and proteins involved in energy metabolism could provide energetic support for glutamate transport. Therefore, we performed immunoprecipitation and co-localization experiments to determine if GLAST might co-compartmentalize with proteins involved in energy metabolism. GLAST was immunoprecipitated from rat cerebellum and subunits of the Na+/K+ ATPase, glycolytic enzymes, and mitochondrial proteins were detected. GLAST co-localized with mitochondria in cerebellar tissue. GLAST also co-localized with mitochondria in fine processes of astrocytes in organotypic hippocampal slice cultures. From these data, we hypothesized that mitochondria participate in a macromolecular complex with GLAST to support oxidative metabolism of transported glutamate. To determine the functional metabolic role of this complex, we measured CO2 production from radiolabeled glutamate in cultured astrocytes and compared it to overall glutamate uptake. Within 15 minutes, 9% of transported glutamate was converted to CO2. This CO2 production was blocked by inhibitors of glutamate transport and glutamate dehydrogenase, but not by an inhibitor of glutamine synthetase. Our data support a model in which GLAST exists in a macromolecular complex that allows transported glutamate to be metabolized in mitochondria to support energy production.
Keywords: Mitochondria, astrocyte, EAAT1, Na+/K+ ATPase, cerebellum
1. Introduction
Glutamate is the major excitatory neurotransmitter in the central nervous system. Synaptic glutamate must be kept at low levels (approximately 25 nM) because excessive extracellular glutamate is excitotoxic (Choi, 1992; Frandsen et al., 1989; Herman and Jahr, 2007). A family of plasma membrane glutamate transporters is responsible for clearing extracellular glutamate (Danbolt, 2001). The astrocytic glutamate transporters, GLAST and GLT-1, are enriched in perisynaptic astrocyte processes and are responsible for the vast majority of glutamate uptake in the central nervous system (Chaudhry et al., 1995; Danbolt, 2001; Robinson, 1998; Sheldon and Robinson, 2007). GLT-1 is the predominant transporter in the forebrain, whereas GLAST is the predominant transporter in the cerebellum.
Glutamate transporters use the Na+-electrochemical gradient maintained by the Na+/K+ ATPase to drive glutamate clearance. This process is energetically costly because 3 Na+ ions must be co-transported with each glutamate molecule, glutamate is transported against a steep gradient, and transport occurs in spatially restricted processes. The Na+/K+ ATPase is physically and functionally coupled to glutamate transporters (Rose et al., 2009). Our laboratory recently demonstrated that GLT-1 exists in a macromolecular complex that includes the Na+/K+ ATPase, most of the enzymes involved in glycolysis, and mitochondria (Genda et al., 2011). This work lead us to hypothesize that these proteins/organelles may exist in a complex with glutamate transporters in order to maintain the Na+-electrochemical gradient necessary to drive glutamate uptake.
Classically, it is believed that once transported into astrocytes, glutamate is converted to glutamine by glutamine synthetase (Norenberg and Martinez-Hernandez, 1979; Palmada and Centelles, 1998; Waniewski and Martin, 1986). Glutamine is then transported back into the presynaptic neuron and converted to glutamate, creating a glutamate-glutamine cycle. However, alternative metabolic pathways for glutamate are also described (Schousboe et al., 1993; Westergaard et al., 1995). Glutamate can be converted to alpha-ketoglutarate by transaminases or by glutamate dehydrogenase (Plaitakis et al., 2011). Alpha-ketoglutarate is a tricarboxylic acid (TCA) cycle intermediate involved in oxidative phosphorylation. Thus, the glutamate carbon backbone can be broken down to produce ATP and CO2. This pathway would be energetically favorable because oxidation of glutamate could potentially offset some of the energetic cost of glutamate transport. In contrast, conversion of glutamate to glutamine requires additional energy in the form of ATP.
We hypothesize that a macromolecular complex exists between GLAST, mitochondria, glycolytic enzymes, and the Na+/K+ ATPase. We performed coimmunoprecipitations to determine if mitochondrial proteins, glycolytic enzymes, and the Na+/K+ ATPase might be physically coupled to GLAST. We performed immunofluorescence experiments to determine whether GLAST co-localizes with mitochondria in vivo. We transfected astrocytes in hippocampal slice cultures with fluorescently labeled GLAST and a fluorescently labeled mitochondrial protein to determine whether co-localization of GLAST and mitochondria can occur within individual astrocytic processes. We hypothesize that this complex might exist to metabolically support glutamate transport within the spatially restricted space of these processes. To determine the metabolic role of a complex involving both GLAST and mitochondria, we adapted an assay to measure CO2 produced in cultured astrocytes.
2. Methods
2.1. Immunoprecipitation
Immunoprecipitations of GLAST were performed using Pierce crosslink immunoprecipitation columns (Thermo Scientific, Rockford, IL). Antibody (10-15 μg) was cross-linked to the column resin per manufacturer's instructions. Cerebellar tissue was harvested from adult male Sprague-Dawley rats after euthanasia by decapitation, to avoid potential effects of anesthetic agents, and homogenized in immunoprecipitation buffer (150 mM NaCl, 1 mM EDTA, 100 mM Tris HCl, pH 7.4, 1% Triton-X-100, and 1% sodium deoxycholate) plus protease and phosphatase inhibitors (1 μg/mL leupeptin, 260 μM PMSF, 1 μg/mL aprotinin, 1 mM iodoacetamide, 10 mM NaF, and 1 mM sodium orthovanadate) (18.5 mL/g wet weight). Homogenates were rotated on a shaker for 1 hr at 4°C and then cleared of cellular debris by centrifugation at 13,000 × g for 30 min at 4°C. One mL of the supernatant was precleared with 40 μL protein A control resin (Thermo Scientific, Rockford, IL) at 4°C for 1 hr. After analyses of protein (bicinchoninic acid protein assay kit Thermo Scientific, Rockford, IL), an aliquot of the resulting supernatant containing 750 μg of total protein was applied to the column and rotated overnight at 4°C. Columns were washed with manufacturer supplied buffers: IP Lysis/Wash Buffer (0.025 M Tris, 0.15 M NaCl, 0.001 M EDTA, 1% NP40, 5% glycerol, pH 7.4), then a neutral pH Conditioning Buffer (Thermo Scientific, Rockford, IL). Antigens were eluted using 35 μL of a low pH (2.8) elution buffer per manufacturer's instructions (Thermo Scientific, Rockford, IL). After elution, 35 μL of SDS-PAGE loading buffer was incubated with the eluate for 30 min at 25°C prior to gel loading.
Immunoprecipitations of UQCRC2 were performed by incubating 15 μg mouse anti-UQCRC2 antibody (AbCam, Cambridge, MA) with precleared cerebellar tissue (prepared as described above) overnight at 4°C. This suspension was then incubated with protein A agarose beads (Invitrogen, Grand Island, NY) for 2 hr at 4°C. After centrifugation,beads were washed 3 times, and proteins were eluted with SDS-PAGE loading buffer at 25°C for 45 min.
2.2. Western Blotting
Proteins (including rainbow molecular weight marker, Amersham) were resolved using 8% or 10% SDS-polyacrylamide gels and transferred to Immobilon FL polyvinylidene fluoride membranes (Millipore, Bedford, MA). After blocking for 1 hr at 25°C in TBS (50 mM Tris, pH 8.0, 150 mM NaCl) containing 5% nonfat dry milk, membranes were probed with the appropriate primary antibody. Mouse anti-β1 subunit of the Na+/K+ ATPase (1:200), rabbit anti-GLAST(1:200), and goat anti-NADH ubiquinone oxidoreductase complex 1 (NDUFS1) (1:200) were from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse anti-hexokinase (1:1000), mouse anti-α1 subunit of the Na+/K+ ATPase (1:2000), mouse anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1:200), and rabbit anti-α2 subunit of the Na+/K+ ATPase (1:1000) were from Millipore (Temecula, CA). Mouse anti-ubiquinol-cytochrome-c reductase complex core protein 2 (UQCRC2) (1:5000) was from Abcam (Cambridge, MA). Membranes were washed in TBS-T (50 mM Tris, pH 8.0, 150 mM NaCl, 0.1% Tween 20) and then incubated with fluorescently conjugated anti-rabbit, anti-mouse, or anti-goat antibodies (1:10,000 LiCor Biosciences, Lincoln, NE). Blots were scanned using an Odyssey Infrared Imager (LiCor Biosciences, Lincoln, NE).
2.3. Immunofluorescence
Adult rats (10-12 weeks) were anesthetized with isofluorane and transcardially perfused with ice-cold PBS followed by 4% paraformaldehyde in phosphate buffered saline (PBS), pH 7.4. Brains were post-fixed overnight (4°C), equilibrated in 30% sucrose, flash frozen in isopentane (-50°C), and stored at -80°C. After cutting coronal sections (40 μm) on a microtome, free-floating sections were subject to antigen retrieval (10 mM citrate buffer pH 8, 80°C, 30 min). Sections were blocked in PBS containing 5% normal serum and 0.4% Triton X-100 for 1 hr and then incubated with rabbit anti-EAAT1 (Abcam, 1:100) and/or mouse anti-UQCRC2 (Abcam, 1:100) overnight at 4°C. After 3 rinses, they were incubated in secondary antibody (either Alexa Fluor 488 or 633; 1:400) overnight at 4°C. Sections were mounted on pre-coated slides, cover-slipped with a DNA counterstain, 4',6-diamidino-2-phenylindole (DAPI), and stored at 4°C until analysis. Controls for each experiment included incubations to confirm the species specificity of secondary antibodies and to confirm that signal was dependent on the presence of primary antibodies. Sections were visualized on a Fluoview 1000 confocal microscope (Olympus, Center Valley, PA) equipped with a PlanApo 60x objective (numerical aperture = 1.4). All images were collected in sequential scan mode to minimize cross-contamination of fluorophores.
2.4. Preparation of cDNA constructs
Monomeric red fluorescent protein (mRFP) was a gift from Dr. Roger Tsien (University of California at San Diego)(Campbell et al., 2002). mRFP-C3 was obtained by replacing eGFP with mRFP in pEGFP-C3 (Clontech, Mountain View, CA) (see Genda et al 2011). mRFP-GLAST was generated in two steps. The coding region of GLAST was amplified from GLAST (pCDNA 3.1+) adding restriction sites for Bgl II and Sal I. The PCR product was gel purified and ligated into pCR2.1 using a Topo TA cloning kit (Invitrogen, Grand Island, NY). The coding sequence was restricted from pCR2.1 using Bgl II and Sal I and subsequently ligated into pmRFP-C3. Mito-EGFP encodes a fusion of EGFP with the mitochondrial targeting sequence of subunit VIII of cytochrome c oxidase and was a generous gift from Dr. Stanley Thayer (University of Minnesota, Minneapolis, MN) (Wang et al., 2003). The resultant fusion construct was sequenced to confirm identity.
2.5. Hippocampal slice culture
Hippocampal slices (300 μm) were prepared from rat pups between 6 and 10 days of age using a McIlwain tissue chopper (Brinkman Instruments, Westbury, NY) and placed in Millicell Culture Inserts (0.4 μm pore size) (Millipore, Bedford, MA) in six-well plates as previously described (Kayser et al., 2006). Slices were maintained in a humidified incubator with 5% CO2 at 37°C in 1 mL of medium containing: 50% Neurobasal medium, 25% Hank's buffered saline solution, 25% horse serum, supplemented with 10 mM HEPES, 36 mM glucose, 2 mM glutamine, penicillin (10 U/mL) and streptomycin (100 μg/mL), pH 7.2-7.3. After two days in vitro, hippocampal slice cultures were transduced using a Helios Gene Gun (Bio-Rad, Hercules, CA) (McAllister, 2004). cDNAs (10 μg) were combined with 10 μg of 1.0 μm gold particles in a solution with 0.02 mg/mL polyvinylpyrrolidone 20 (PVP-20), 0.05 M spermidine, and 1 M CaCl2. This suspension was used to coat Teflon tubing. Tubing was cut into cartridges and loaded into the Gene Gun, and gold particles were shot with high-pressure helium (100-120 psi) into cultured slices in inserts sitting on warmed agarose slabs. Under these conditions, astrocytes are selectively transduced (Benediktsson et al., 2005; Genda et al., 2011).
Two days post-transduction, slices were fixed on the membrane inserts in a solution of 4% paraformaldehyde in PBS for 15 min at room temperature. Tissues were rinsed in PBS and gently removed from their membrane support with a spatula. Some tissues were processed as free-floating sections for subsequent immunostaining. These sections were extracted overnight in a solution of PBS containing 1% Triton-X-100 at 4°C. Sections were blocked in 5% goat serum for 1 hr at room temperature and incubated with rabbit anti-glial fibrillary acidic protein (GFAP) polyclonal antibody (1:300; Sigma, St. Louis, MO) overnight at 4°C in PBS containing 1% Triton-X-100 and 5% goat serum. Sections were rinsed 3 × 15 min in PBS and incubated overnight with secondary antibody (goat anti-rabbit Alexa Fluor 350; Invitrogen, Grand Island, NY). Slices were rinsed 3 × 15 min in PBS before mounting on pre-coated slides (Superfrost Plus; Fisher, Pittsburgh, PA).
Images were collected using an Olympus Fluoview 1000 (Center Valley, PA) laser scanning confocal microscope that was equipped with 405, 488, 546, and 633 nm laser lines. All images were collected in sequential scan mode to limit cross-contamination of fluorescence. For 3D reconstruction of cells in slices, 40-80 optical sections were taken at an interval of 0.5 μm in the z-direction using a 40x UPlanFL objective (numerical aperture = 1.3).
2.6. Puncta Analysis
Co-clustering of RFP-GLAST with mito-EGFP was conducted as described previously (Genda et al., 2011). Briefly, images were background corrected and filtered using a Gaussian filter (σ = 1). Mitochondria and GLAST were identified by locally thresholding each channel (r = 8). The resulting images were converted into binary images. We measured the length of each punctum and the length of each process. Co-clustering of puncta was defined as >1 pixel (0.155 μm per pixel) of overlap between the two channels and expressed as a percentage of the puncta that overlapped with at least one punctum of the opposite type.
2.7. Intensity correlation/intensity co-localization quotient analysis
Single optical sections were background corrected and filtered using a Gaussian filter (σ = 1). All image analysis was conducted using NIH ImageJ software (http://rsb.info.nih.gov/ij/). Images were automatically thresholded and the Intensity Co-localization Quotient (ICQ) values calculated using the intensity correlation analysis (ICA) plug-in to the McMaster Biophotonics Facility ImageJ collection. The ICQs were calculated as described by Li et al., 2004; results were compared to zero using a one-sample t test.
2.8. Astrocyte cultures
Astrocyte cultures were prepared from cortex of P0-P3 Sprague Dawley rats as previously described (Garlin et al., 1995). After dissection and removal of meninges, cortices were trypsinized for 20 min at 37°C in 5% CO2 and washed in Hanks’ buffered saline solution. The tissue was triturated in warm media (10% heat-inactivated fetal bovine serum, 10% Ham's F-12 medium, and 80% Dulbecco's modified Eagles’ medium with 0.24% penicillin/streptomycin), and cells were plated onto T75 flasks (BD Falcon, Franklin Lakes, NJ). To remove A2B5 positive cells, cultures were incubated with anti-A2B5 antibody and rabbit complement for 45 min. Cultures were then replated onto 12-well plates (BD Falcon, Franklin Lakes, NJ).
2.9. Glutamate Uptake
Astrocytic sodium-dependent transport activity was measured in triplicate. Plates were placed in a 37°C water bath. Cells were rinsed twice with 1 mL of warm artificial cerebrospinal fluid (ACSF) (130mM NaCl, 3mM KCl, 1.25mM NaH2PO4, 26mM NaHCO3, 10mM dextrose, 1mM MgCl2, 2mM CaCl2, bubbled with 95% O2, 5% CO2 mixture for at least 30 min) and then incubated with 1 μM L-[3H]-glutamate for 5 or 15 min. After halting uptake of radioactive glutamate with 3 washes with ice-cold ACSF, the cells were solubilized in 1 mL of 0.1 N sodium hydroxide, and samples were taken for analysis of radioactivity using a Beckman LS 6500 scintillation counter (Beckman Coulter, Indianapolis, IN). Uptake was defined as the difference in radioactivity accumulated in the presence and absence of excess (10 mM) non-radioactive glutamate. The amount of radioactivity observed in the presence of excess glutamate was < 1.5% of that observed in the absence of excess glutamate (mean of 3 experiments).
2.10. CO2 measurements
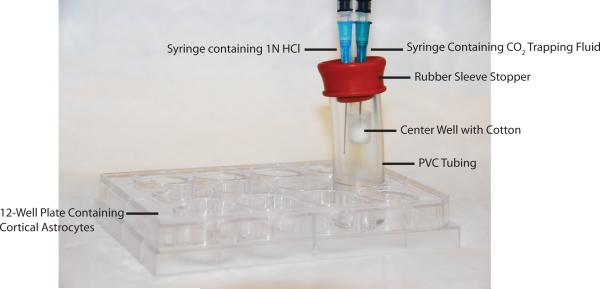
Production of CO2 from an astrocyte monolayer was measured using an adaptation of methods previously described (MacDonnell and Greengard, 1975; Yu et al., 1982). CO2 measurements were performed in duplicate in 12 well plates (BD Falcon, Franklin Lakes, NJ) at 37°C in a water bath. Wells were washed 3 times with ACSF bubbled with 5% CO2, 95% O2. After addition of 1 mL ACSF containing 1 μM L-[1-14C]- glutamate, they were sealed with polyvinyl chloride tubing and a rubber sleeve stopper (see Figure 1). After 5 or 15 minutes, the reaction was stopped by injection of 1N HCl (500 μL). In order to trap 14CO2, Carbosorb-E base (300 μL)(Perkin-Elmer, Waltham, MA) was injected into a cup (center well, Kimble Chase, Vineland, NJ) suspended above the well, and the wells remained sealed for an additional hour to allow all CO2 to be trapped. Cups were removed, placed in vials containing scintillation fluid (Permafluor E+,Perkin-Elmer, Waltham, MA), and radioactivity was analyzed using a scintillation counter. Enzyme dependent CO2 production was defined as the difference in radioactivity accumulated in the presence and absence of excess (10 mM) non-radioactive glutamate.
Figure 1. Device used to capture CO2.
Astrocytes were cultured in 12-well plates. ACSF containing L-[1-14C]-glutamate was added to the well, and the well was immediately plugged with PVC tubing and a stopper. A center well containing cotton was suspended by the stopper. The reaction was stopped by injection of HCl through a 1.5 inch needle, and then CO2 was trapped by injection of a scintillation fluid-compatible base into the center well with a 1 inch needle.
3. Results
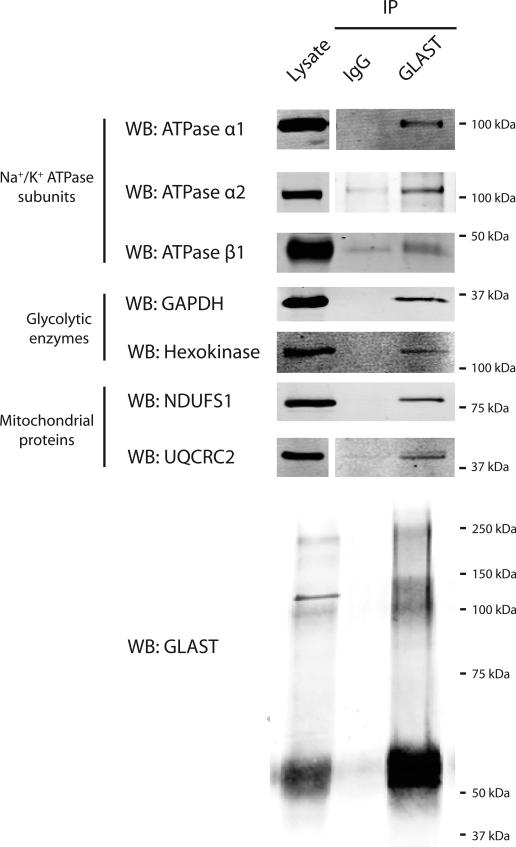
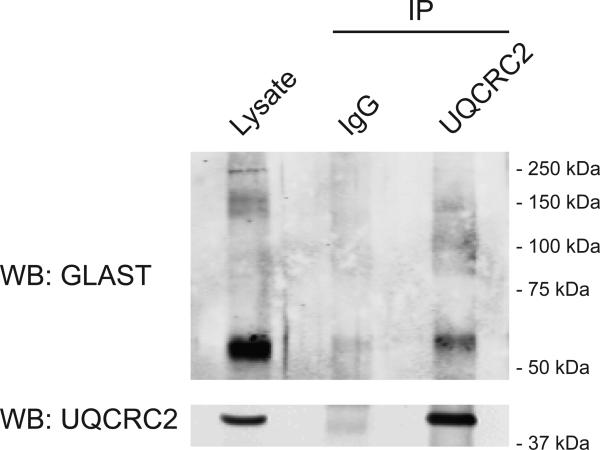
To determine whether GLAST interacts with proteins involved in energy metabolism, we immunoprecipitated GLAST from rat cerebellum and performed Western blot analysis for candidate interacting partners. Immunoprecipitates of GLAST from rat cerebellum contained subunits of the Na+/K+ ATPase, glycolytic enzymes, and mitochondrial proteins (Figure 2). Specifically, we detected the α1, α2, and β1 subunits of the Na+/K+ ATPase in GLAST immunoprecipitations. The glycolytic enzymes GAPDH, and hexokinase were also detected. We also detected the inner mitochondrial membrane proteins NDUFS1 and UQCRC2 in the GLAST immunoprecipitates. All of the co-immunoprecipitated proteins were enriched in the GLAST immunoprecipitates compared to IgG controls. To address the possibility that GLAST antibody nonspecifically immunoprecipitates these proteins in the absence of GLAST protein, we performed immunoprecipitations using antibody against UQCRC2, and tested for the presence of GLAST. Immunoprecipitates of UQCRC2 were enriched in GLAST compared to IgG controls.
Figure 2. Immunoprecipitation of GLAST from cerebellar lysates.
Anti-GLAST antibody or IgG was used for immunoprecipitations from rat cerebellar lysates (750 μg protein). In all cases, the lysates were analyzed in the same Western blots. Some blots were cropped. Immunoprecipitation of GLAST was confirmed in every immunoprecipitation and one example is presented. Data are representative of at least 3 independent experiments.
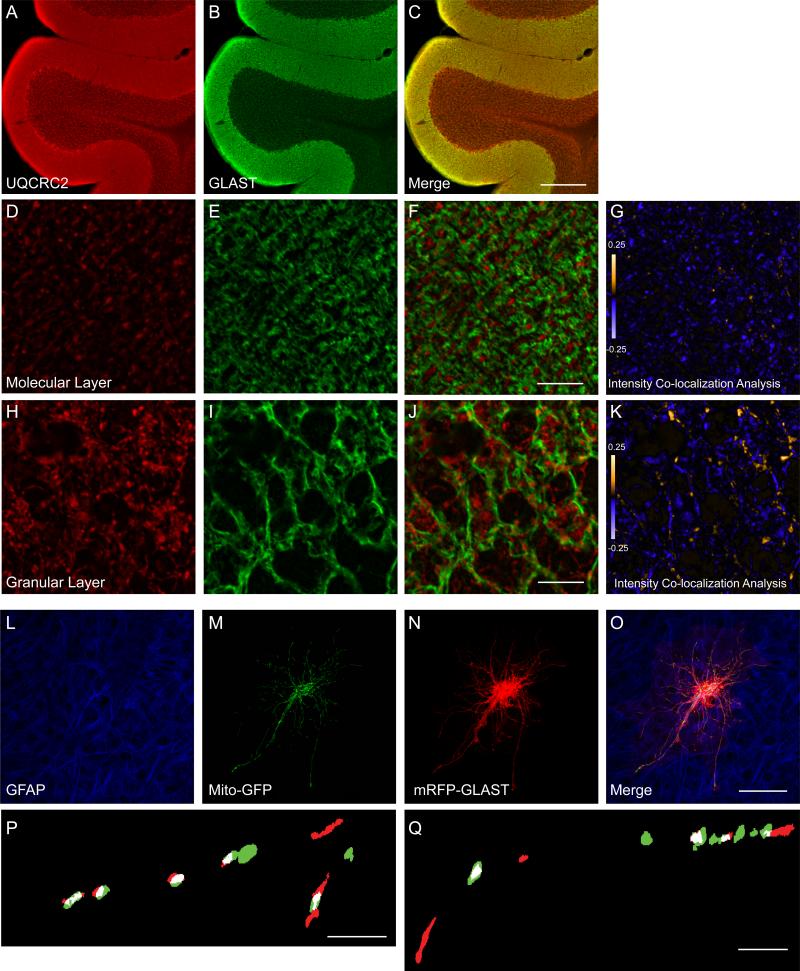
As detection of inner mitochondrial membrane proteins in an immunoprecipitation of a plasma membrane protein could be explained by post-solubilization aggregation, we tested for co-localization of GLAST and the mitochondrial protein, UQCRC2, in cerebellar tissue. If two proteins are part of the same complex, then their staining intensities should co-vary. Conversely if the proteins are part of different complexes or structures, then their staining patterns should display segregated staining. We calculated Intensity Co-localization Quotients (ICQ) for GLAST and UQCRC2 in both the molecular and granular layers of the cerebellum (Genda et al., 2011; Li et al., 2004). This measure describes the extent of correlation of the staining intensities for two proteins. ICQ values are distributed between -0.5 and +0.5. Positive ICQ values indicate that the relative intensities are correlated and consistent with co-localization. Conversely, negative ICQ values indicate that the proteins are segregated. ICQ values not significantly different from zero indicate random staining. We found no evidence of co-localization in the molecular layer of the cerebellum (ICQ = -0.02 ± 0.03; mean ± S.E.M, n = 9), but there was significant co-variance of signal for GLAST and UQCRC2 in the granular layer of cerebellum (ICQ = 0.1 ± 0.02, p < 0.001, n = 8) (Figure 4A-K).
Figure 4. Co-localization of GLAST with mitochondria in cerebellum (A-K) and in biolistically transduced astrocytes in organotypic hippocampal slice cultures (L- Q).
A-C. Representative images from adult rat cerebellum immunostained with antibodies against UQCRC2 (A; red) and GLAST (B; green). C. Merged image of GLAST and UQCRC2 immunofluorescence. Scale bar = 300 μm. D-K. Higher magnification views of the molecular layer (D, E) and granular layer (H, I) of the cerebellum immunostained for UQCRC2 (red; D, H) and GLAST (green; E, I) and merged (F, J). Scale bar = 10 μm. G, K. Pseudocolor representatations of PDM values for the image pairs (D:E and H:I), where pixel intensity is equal to the PDM value at that location. Images with positive PDM value are displayed in yellow, while those with negative values are displayed in violet. L-Q. Co-localization of mitochondria and GLAST in the processes of astrocytes in slice culture. Representative images of astrocytes in slice culture immunostained for glial fibrillary acidic protein (L; GFAP; blue) and transfected with the cDNAs encoding Mito-GFP (M, green) and mRFP-GLAST (N, red) and a merged image (O). P, Q. Binary, higher magnification view of astrocyte processes. Overlapping pixels are indicated in white.
While immunofluorescence allowed us to determine the extent of overlap between GLAST and UQCRC2 in native tissue, the resolution (~200nm) is not sufficient to determine whether the overlap is a result of co-localization within individual astrocytic processes. To address this concern, astrocytes in hippocampal slice cultures were biolistically transduced with mitochondrially targeted EGFP (Mito-EGFP) and an RFPGLAST fusion protein (mRFP-GLAST) (Figure 4L-Q). Slices were immunolabeled for glial fibrillary acidic protein (GFAP), a commonly used marker for astrocytic lineage (Figure 4L). All transduced cells co-localized with GFAP. The length of each process (52 ± 4 μm, n = 33 from 10 different cells analyzed from 4 independent experiments), the sizes (lengths) of GLAST puncta (1.5 ± 0.1 μm; n = 399), and the lengths of the mitochondria (1.9 ± 0.1 μm; n = 294) were measured. The average densities of GLAST and mitochondria (i.e., the portion of the process covered by either particle) were similar, 0.37 ± 0.17 and 0.34 ± 0.19, respectively. GLAST puncta were frequently overlapped by mitochondria and vice versa. Across all processes 52.5 ± 3.3% of GLAST puncta were overlapped by mitochondria, and 67.6 ± 3.5% of mitochondria were overlapped by GLAST puncta.
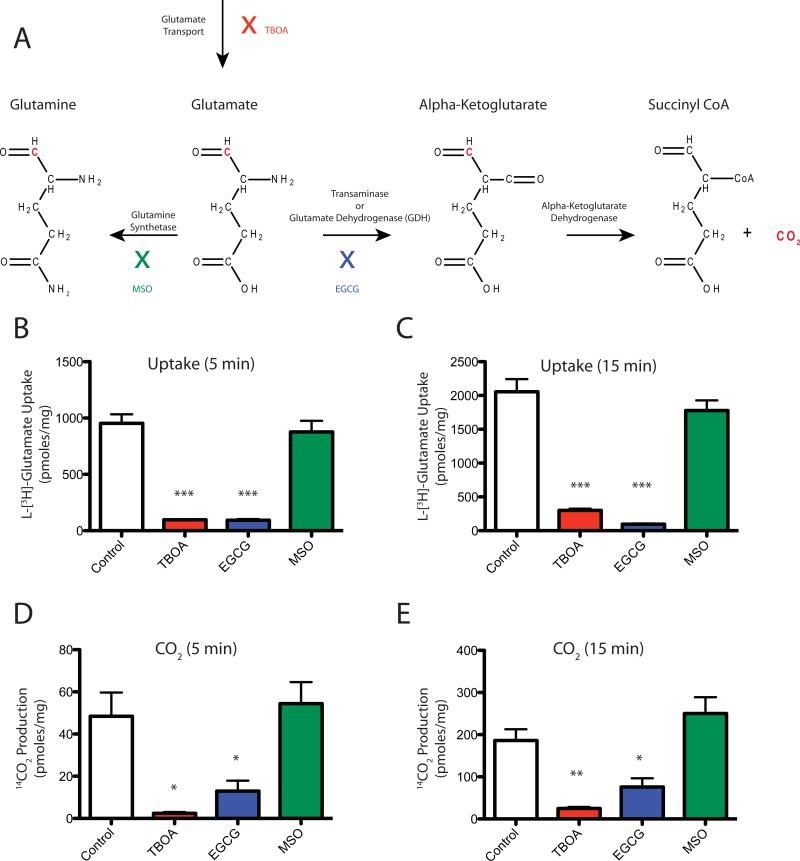
Co-compartmentalization of GLAST with the Na+/K+ ATPase, glycolytic enzymes and mitochondria could have several functional consequences. Mitochondria, glycolytic enzymes, and the Na+/K+ ATPase can provide energy (ATP and Na+ gradient) to drive uptake. Mitochondria could also buffer Na+ and H+ that are co-transported with glutamate. Finally, mitochondria provide an alternate route for glutamate metabolism via oxidation (see Figure 5A). As primary astrocyte cultures only express the glutamate transporter GLAST (Schlag et al., 1998; Swanson et al., 1997) and express no glutamic acid decarboxylase, this is an ideal model system in which to examine the metabolic fate of glutamate transported by GLAST. Earlier studies have documented both conversion of glutamate to glutamine and glutamate oxidation in astrocytes. In order to determine the percentage of transported glutamate that is converted to CO2, the amount of glutamate accumulated in astrocytes and the amount of CO2 generated from L-[1-14C] glutamate were measured in parallel (Figure 5B-E). After 5 minutes, 5 ± 0.9% of transported glutamate was converted to CO2. After 15 minutes, the percentage of glutamate converted to CO2 was significantly greater (9 ± 0.7%; p < 0.05). Both glutamate transport and CO2 production were completely blocked by the general inhibitor of Na+-dependent transport systems, DL-threo-β-benzyloxyaspartic acid (TBOA, 1 mM). To test whether these measures are dependent upon the enzyme glutamate dehydrogenase, which catalyzes the conversion of glutamate to alpha-ketoglutarate, the glutamate dehydrogenase inhibitor epigallocatechin gallate (EGCG, 1 mM) was used (Li et al., 2006). Both glutamate transport and CO2 production were inhibited by EGCG. The glutamine synthetase inhibitor methionine sulfoximine (MSO, 5 mM) inhibited neither glutamate uptake nor CO2 production.
Figure 5. Stoichiometry of CO2 production from L-[1-14C] glutamate in cortical astrocyte cultures.
Glutamate uptake and CO2 production were measured in cortical astrocyte cultures at 5 or 15 minutes in the absence or presence of TBOA (1 mM), EGCG (1 mM), or MSO (5 mM). A. Potential Glutamate Pathways: Glutamate that has been transported into astrocytes can be converted into glutamine by glutamine synthetase or converted to alpha-ketoglutarate by transaminase or glutamate dehydrogenase. Alpha-ketoglutarate can then be converted into succinyl CoA and CO2 by alpha ketoglutarate dehydrogenase. Radiolabeled carbon in the number 1 position of glutamate is depicted in red. TBOA inhibits glutamate transporters (red). EGCG inhibits glutamate dehydrogenase (blue). MSO inhibits glutamine synthetase (green). B. Glutamate uptake at 5 minutes incubation. C. Glutamate uptake at 15 minutes incubation. D. CO2 production at 5 minutes incubation. E. CO2 production at 15 minutes incubation. Note: None of the measures in B-E were normalized to time. Data are the mean ± SEM of 3 independent observations and were compared by ANOVA. * p < 0.05; ** p < 0.01; *** p < 0.001.
4. Discussion
It is increasingly apparent that proteins and organelles compartmentalize within cells to support specific functions (de Brito and Scorrano, 2008; Garcia-Perez et al., 2008; Sheng, 2001). We found that subunits of the Na+/K+ ATPase, glycolytic enzymes and mitochondrial proteins co-immunoprecipitate with GLAST. We also found that GLAST co-localizes with mitochondria in cerebellum and in fine processes of astrocytes in hippocampal slice cultures. To the best of our knowledge, prior to this work, only 6 GLAST interacting proteins have been identified: NHERF-1, NHERF-2, ezrin, septin 2, GFAP, and the α2 subunit of the Na+/K+ ATPase (Kinoshita et al., 2004; Lee et al., 2007; Ritter et al., 2011; Rose et al., 2009; Sullivan et al., 2007). In this study, we identified 6 additional proteins that co-immunoprecipitate with GLAST. Consistent with the interaction between the Na+/K+ ATPase and GLAST, we found that in addition to the α2 subunit, the α1 and β1 subunits of the ATPase also co-immunoprecipitate with GLAST. Additionally, we demonstrate that mitochondrial proteins, UQCRC2 and NDUFS1, and glycolytic enzymes, hexokinase and GAPDH, co-immunoprecipitate with GLAST. We recently demonstrated that GLT-1 is part of a similar complex (Genda et al., 2011).
While we find it unlikely that co-immunoprecipitations paired with co-localization are the result of a post-solubilization aggregation, we cannot rule out this possibility. Another potential confounding factor could be the presence of GLAST in mitochondrial membranes rather than the plasma membranes of astrocytes. GLAST has been detected in mitochondria of cardiac myocytes (Ralphe et al., 2005; Ralphe et al., 2004). In those studies, the authors observed complete overlap of GLAST with mitochondria. It is possible that the high metabolic demands of cardiac myocytes result in close coupling of GLAST to mitochondria in this system. In brain, several different studies have failed to detect GLAST in mitochondria using electron microscopy (Chaudhry et al., 1995; Lehre et al., 1995; Rinholm et al., 2007). In the present study, we detected incomplete overlap of GLAST puncta with mitochondria (fluorescence for the mitochondrial marker frequently extends beyond GLAST fluorescence within a single mitochondrion, see Figure 4P-Q for examples), indicating that this overlap is not due to targeting of GLAST to mitochondria. Furthermore, the fact that there was no significant co-localization between GLAST and UQCRC2 in the molecular layer of the cerebellum provides additional evidence that GLAST is not targeted to mitochondria.
In order for the inner mitochondrial membrane proteins UQCRC2 and NDUFS1 to interact with GLAST, there must be a scaffold of several proteins that link them. Linkages of proteins that span multiple membranes are not unprecedented. Hexokinase is constitutively associated with mitochondria in the brain (Sui and Wilson, 1997). Hexokinase interacts with the outer mitochondrial membrane protein voltage-dependent anion channel (VDAC) (Abu-Hamad et al., 2008; Sui and Wilson, 1997). VDAC, in turn, interacts with the adenine nucleotide translocase (ANT) which resides in the inner mitochondrial membrane (Beutner et al., 1998; Crompton et al., 1998). ANT interacts with prohibitin in the inner mitochondrial matrix (Osman et al., 2009). Therefore it is possible that these proteins can be connected through a series of linking proteins. This will need to be tested in future studies.
Granule cells are the only glutamatergic cells in the cerebellum. Their cell bodies are located in the granular layer, and they project to the molecular layer where GLAST is most abundant. Both GLAST and UQCRC2 were present at higher levels in the molecular layer than in the granular layer, yet we only detected significant co-localization in the granular layer. The lack of co-localization in the molecular layer may simply be attributed to the fact that both proteins were significantly more abundant in this region, and co-variance may be less likely detected in regions of high expression. It is also possible that this differential co-localization reflects an intrinsic mechanism to regulate formation of the complex. Alternatively, this may suggest that transport energetics are supported differently in these two areas. In the molecular layer, glutamate can easily be bound and buffered by GLAST because it is abundantly expressed by Bergmann glia, and thus may not need energetic support to quickly transport glutamate. In the granular layer which is densely populated with neuronal cell bodies, quick transport may be necessary to remove glutamate because the buffering capacity is decreased. Despite its low expression level in the granular layer, GLAST is primarily responsible for limiting the time course of synaptic transmission at granule cell synapses (Overstreet et al., 1999). Alternatively, the difference in co-localization could be attributable to regional heterogeneity in subpopulations of mitochondria. Mitochondria can differ in levels of alpha-ketoglutarate dehydrogenase (Waagepetersen et al., 2006), and mitochondria in astrocytes of the granular layer contain more glutamate-dehydrogenase reactive particles than mitochondria in astrocytes of the molecular layer (Rothe et al., 1994). Thus, it is possible that GLAST preferentially associates with mitochondria in the granular layer that express higher levels of enzymes required for glutamate flux through the TCA cycle. In any case, our data suggest that there are regional differences in the coupling of glutamate transport to this complex.
After transport into astrocytes, glutamate can be converted into glutamine by glutamine synthetase or converted to alpha-ketoglutarate by transaminase or glutamate dehydrogenase (Palmada and Centelles, 1998; Schousboe et al., 1993; Waniewski and Martin, 1986; Westergaard et al., 1995). Alpha-ketoglutarate can then be converted into succinyl CoA and CO2 by alpha-ketoglutarate dehydrogenase (Figure 5A). In this study we measured the stoichiomentry of CO2 produced from transported glutamate. Previous studies using astrocytes (Farinelli and Nicklas, 1992; McKenna et al., 1996; Rao and Murthy, 1993; Skytt et al., 2010; Sonnewald et al., 1997; Waniewski and Martin, 1986; Yu et al., 1982), synaptosomes (Yudkoff et al., 1994), and brain slices (El Hage et al., 2011) have examined the catabolism of glutamate and its conversion to CO2 with conflicting results. Some studies found a relatively small portion of glutamate flux through the tricarboxylic acid (TCA) cycle, whereas others found a large portion of glutamate entering the TCA cycle. The pathway by which glutamate is metabolized appears to depend on glutamate concentration, with high concentrations of glutamate favoring its flux through the TCA cycle (McKenna, 2007; McKenna et al., 1996; Sonnewald et al., 1997). We used a relatively low concentration of glutamate (1 μM), which would favor low TCA cycle flux and thus relatively low levels of CO2 production. Another variable that could influence CO2 production is incubation time. In behaving rats, the rate of glutamate oxidation increased greatly at about 45 minutes in hippocampus (Zielke et al., 1998). In the current study, the percentage of glutamate converted to CO2 nearly doubled between 5 and 15 min. It is possible that CO2 production might increase further at longer incubation times; however, glutamate uptake would cease to be linear.
Several studies have also used pharmacological agents to study flux of glutamate through metabolic pathways. Yu et al. used aminooxyacetic acid to block transaminase and found no inhibition of CO2 production, suggesting that glutamate dehydrogenase is responsible, consistent with our EGCG results (Yu et al., 1982). We believe ours is the first study using EGCG to block glutamate dehydrogenase while measuring CO2 production. EGCG did block CO2; however, it also blocked uptake. Our experiments did not test the mechanism through which EGCG blocked CO2 production. Since uptake was blocked, the simplest explanation for the reduction in CO2 is a decrease in intracellular glutamate as a precursor for CO2. The decrease in glutamate uptake could be a result of EGCG acting on targets besides glutamate dehydrogenase, or it might suggest that flux of glutamate through this pathway provides energy to support glutamate uptake. In future studies it will be important to determine the mechanism through which EGCG inhibits glutamate uptake (e.g. competitive vs. noncompetitive inhibitor). It will also be interesting to determine the effects of new pharmacological agents that have been identified to inhibit glutamate dehydrogenase (Li et al., 2011).
Glutamate uptake is energetically demanding, and the fate of transported glutamate could determine the energetic cost of this process. Glutamate that is oxidized through the TCA cycle results in the production of ATP, whereas conversion to glutamine requires expenditure of ATP. Glutamate uptake into astrocytes may additionally affect metabolism through the stimulation of glycolysis (Bittner et al., 2011; Pellerin and Magistretti, 1994) and glucose transport (Loaiza et al., 2003). Compartmentalization of GLAST with glycolytic enzymes could potentially support the stimulation of glycolysis. In addition to supporting energetic demand, mitochondria may localize near GLAST and the Na+/K+ ATPase to buffer changes in ion concentrations. Mitochondria near GLAST can sense and buffer Na+ (Bernardinelli et al., 2006), and mitochondria in astrocytes buffer protons during glutamate uptake (Azarias et al., 2011).
It is possible that formation of this complex is regulated. One potential regulator could be extracellular glutamate. There are several lines of evidence to support this hypothesis. Application of glutamate to astrocytes causes clustering of GLT-1 (Nakagawa et al., 2008) and moves mitochondria near the plasma membrane (Kolikova et al., 2006). Glutamate also increases glutamate uptake (Munir et al., 2000) and relative CO2 production in astrocytes (McKenna, 2007; McKenna et al., 1996). The complex could also be regulated by activity of the Na+/K+ ATPase. ATPase inhibitors alter distribution of GLAST (Nguyen et al., 2010), which could potentially disrupt this complex. Determining the factors that influence this complex could provide therapeutic targets for developing treatments for diseases at the crossroads of energetics and excitotoxicity such as Alzheimer's disease, Parkinson's disease, and amyotrophic lateral sclerosis.
Based on our findings, we propose a model in which GLAST compartmentalizes with mitochondria, glycolytic enzymes, and the Na+/K+ ATPase. We propose that the complex of GLAST with the Na+/K+ ATPase, glycolytic enzymes, and mitochondrial proteins exists to support local glutamate uptake. Glycolytic enzymes and mitochondria provide ATP to fuel the ATPase, the ATPase provides the sodium gradient to support glutamate uptake, and GLAST transports glutamate, which stimulates glycolysis and provides a TCA cycle substrate in mitochondria.
Figure 3. Immunoprecipitation of UQCRC2 from cerebellar lysates.
Anti-UQCRC2 antibody or IgG was used for immunoprecipitations from rat cerebellar lysates (500 μg protein). Lysates were analyzed in the same Western blots. Data are representative of 2 independent experiments. Note: GLAST monomers migrate at about 60 kDa.
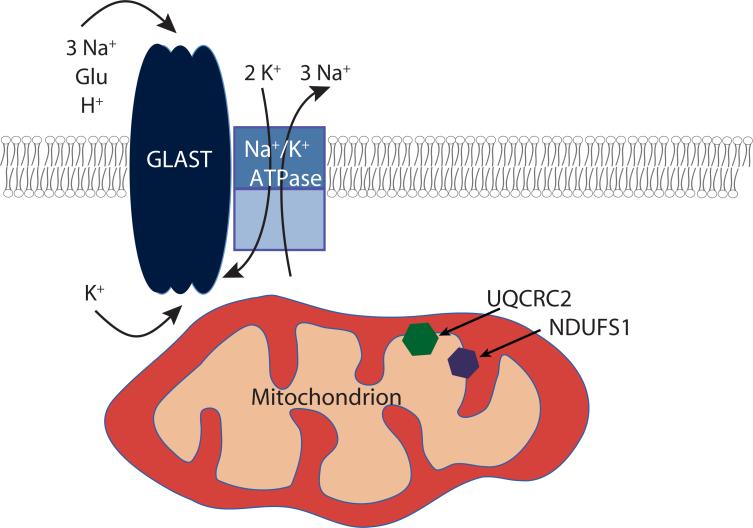
Figure 6. Schematic of a potential macromolecular complex for GLAST.
GLAST and the Na+/K+ ATPase are in the plasma membrane, and mitochondria containing UQCRC2 and NDUFS1 in the inner membrane co-localize with GLAST near the plasma membrane.
Acknowledgments
This work was supported by the Analytical Neurochemistry Core of the Institutional Intellectual and Developmental Disabilities Research Center (P30HD26979). D.E.B was supported by an Institutional Research and Academic Career Development Award (K12GM081259). J.G.J. was partially supported by a Training Grant in Neurodevelopmental Disabilities (T32NS007413). M.M.M was supported by the University of Pennsylvania Postbaccalaureate Research Education Program (R25GM071745). We thank the members of the Robinson laboratory for advice regarding this research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abu-Hamad S, Zaid H, Israelson A, Nahon E, Shoshan-Barmatz V. Hexokinase-I protection against apoptotic cell death is mediated via interaction with the voltage-dependent anion channel-1: mapping the site of binding. J. Biol. Chem. 2008;283:13482–13490. doi: 10.1074/jbc.M708216200. [DOI] [PubMed] [Google Scholar]
- Azarias G, Perreten H, Lengacher S, Poburko D, Demaurex N, Magistretti PJ, Chatton JY. Glutamate transport decreases mitochondrial pH and modulates oxidative metabolism in astrocytes. J. Neurosci. 2011;31:3550–3559. doi: 10.1523/JNEUROSCI.4378-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benediktsson AM, Schachtele SJ, Green SH, Dailey ME. Ballistic labeling and dynamic imaging of astrocytes in organotypic hippocampal slice cultures. J Neurosci Methods. 2005;141:41–53. doi: 10.1016/j.jneumeth.2004.05.013. [DOI] [PubMed] [Google Scholar]
- Bernardinelli Y, Azarias G, Chatton JY. In situ fluorescence imaging of glutamate-evoked mitochondrial Na+ responses in astrocytes. Glia. 2006;54:460–470. doi: 10.1002/glia.20387. [DOI] [PubMed] [Google Scholar]
- Beutner G, Ruck A, Riede B, Brdiczka D. Complexes between porin, hexokinase, mitochondrial creatine kinase and adenylate translocator display properties of the permeability transition pore. Implication for regulation of permeability transition by the kinases. Biochim. Biophys. Acta. 1998;1368:7–18. doi: 10.1016/s0005-2736(97)00175-2. [DOI] [PubMed] [Google Scholar]
- Bittner CX, Valdebenito R, Ruminot I, Loaiza A, Larenas V, Sotelo-Hitschfeld T, Moldenhauer H, San Martin A, Gutierrez R, Zambrano M, Barros LF. Fast and reversible stimulation of astrocytic glycolysis by K+ and a delayed and persistent effect of glutamate. J. Neurosci. 2011;31:4709–4713. doi: 10.1523/JNEUROSCI.5311-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. U. S. A. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry FA, Lehre KP, van Lookeren Campagne M, Ottersen OP, Danbolt NC, Storm-Mathisen J. Glutamate transporters in glial plasma membranes: highly differentiated localizations revealed by quantitative ultrastructural immunocytochemistry. Neuron. 1995;15:711–720. doi: 10.1016/0896-6273(95)90158-2. [DOI] [PubMed] [Google Scholar]
- Choi DW. Excitotoxic cell death. J Neurobiol. 1992;23:1261–1276. doi: 10.1002/neu.480230915. [DOI] [PubMed] [Google Scholar]
- Crompton M, Virji S, Ward JM. Cyclophilin-D binds strongly to complexes of the voltage-dependent anion channel and the adenine nucleotide translocase to form the permeability transition pore. Eur. J. Biochem. 1998;258:729–735. doi: 10.1046/j.1432-1327.1998.2580729.x. [DOI] [PubMed] [Google Scholar]
- Danbolt NC. Glutamate uptake. Prog Neurobiol. 2001;65:1–105. doi: 10.1016/s0301-0082(00)00067-8. [DOI] [PubMed] [Google Scholar]
- de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456:605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- El Hage M, Conjard-Duplany A, Baverel G, Martin G. Metabolic fate of a high concentration of glutamine and glutamate in rat brain slices: a (1)(3)C NMR study. Neurochem. Int. 2011;58:896–903. doi: 10.1016/j.neuint.2011.02.018. [DOI] [PubMed] [Google Scholar]
- Farinelli SE, Nicklas WJ. Glutamate metabolism in rat cortical astrocyte cultures. J. Neurochem. 1992;58:1905–1915. doi: 10.1111/j.1471-4159.1992.tb10068.x. [DOI] [PubMed] [Google Scholar]
- Frandsen A, Drejer J, Schousboe A. Direct evidence that excitotoxicity in cultured neurons is mediated via N-methyl-D-aspartate (NMDA) as well as non-NMDA receptors. J. Neurochem. 1989;53:297–299. doi: 10.1111/j.1471-4159.1989.tb07327.x. [DOI] [PubMed] [Google Scholar]
- Garcia-Perez C, Hajnoczky G, Csordas G. Physical coupling supports the local Ca2+ transfer between sarcoplasmic reticulum subdomains and the mitochondria in heart muscle. J. Biol. Chem. 2008;283:32771–32780. doi: 10.1074/jbc.M803385200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garlin AB, Sinor AD, Sinor JD, Jee SH, Grinspan JB, Robinson MB. Pharmacology of sodium-dependent high-affinity L-[3H]glutamate transport in glial cultures. J. Neurochem. 1995;64:2572–2580. doi: 10.1046/j.1471-4159.1995.64062572.x. [DOI] [PubMed] [Google Scholar]
- Genda EN, Jackson JG, Sheldon AL, Locke SF, Greco TM, O'Donnell JC, Spruce LA, Xiao R, Guo W, Putt M, Seeholzer S, Ischiropoulos H, Robinson MB. Co-compartmentalization of the Astroglial Glutamate Transporter, GLT-1, with Glycolytic Enzymes and Mitochondria. J. Neurosci. 2011;31:18275–18288. doi: 10.1523/JNEUROSCI.3305-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman MA, Jahr CE. Extracellular glutamate concentration in hippocampal slice. J. Neurosci. 2007;27:9736–9741. doi: 10.1523/JNEUROSCI.3009-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser MS, McClelland AC, Hughes EG, Dalva MB. Intracellular and trans-synaptic regulation of glutamatergic synaptogenesis by EphB receptors. J. Neurosci. 2006;26:12152–12164. doi: 10.1523/JNEUROSCI.3072-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita N, Kimura K, Matsumoto N, Watanabe M, Fukaya M, Ide C. Mammalian septin Sept2 modulates the activity of GLAST, a glutamate transporter in astrocytes. Genes Cells. 2004;9:1–14. doi: 10.1111/j.1356-9597.2004.00696.x. [DOI] [PubMed] [Google Scholar]
- Kolikova J, Afzalov R, Giniatullina A, Surin A, Giniatullin R, Khiroug L. Calcium-dependent trapping of mitochondria near plasma membrane in stimulated astrocytes. Brain Cell Biol. 2006;35:75–86. doi: 10.1007/s11068-006-9000-1. [DOI] [PubMed] [Google Scholar]
- Lee A, Rayfield A, Hryciw DH, Ma TA, Wang D, Pow D, Broer S, Yun C, Poronnik P. Na+-H+ exchanger regulatory factor 1 is a PDZ scaffold for the astroglial glutamate transporter GLAST. Glia. 2007;55:119–129. doi: 10.1002/glia.20439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehre KP, Levy LM, Ottersen OP, Storm-Mathisen J, Danbolt NC. Differential expression of two glial glutamate transporters in the rat brain: quantitative and immunocytochemical observations. J Neurosci. 1995;15:1835–1853. doi: 10.1523/JNEUROSCI.15-03-01835.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Allen A, Kwagh J, Doliba NM, Qin W, Najafi H, Collins HW, Matschinsky FM, Stanley CA, Smith TJ. Green tea polyphenols modulate insulin secretion by inhibiting glutamate dehydrogenase. J. Biol. Chem. 2006;281:10214–10221. doi: 10.1074/jbc.M512792200. [DOI] [PubMed] [Google Scholar]
- Li M, Li C, Allen A, Stanley CA, Smith TJ. The structure and allosteric regulation of mammalian glutamate dehydrogenase. Arch Biochem Biophys. 2011 doi: 10.1016/j.abb.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Lau A, Morris TJ, Guo L, Fordyce CB, Stanley EF. A syntaxin 1, Galpha(o), and N-type calcium channel complex at a presynaptic nerve terminal: analysis by quantitative immunocolocalization. J. Neurosci. 2004;24:4070–4081. doi: 10.1523/JNEUROSCI.0346-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loaiza A, Porras OH, Barros LF. Glutamate triggers rapid glucose transport stimulation in astrocytes as evidenced by real-time confocal microscopy. J. Neurosci. 2003;23:7337–7342. doi: 10.1523/JNEUROSCI.23-19-07337.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonnell P, Greengard O. The distribution of glutamate decarboxylase in rat tissues; isotopic vs fluorimetric assays. J. Neurochem. 1975;24:615–618. [PubMed] [Google Scholar]
- McAllister AK. Biolistic transfection of cultured organotypic brain slices. Methods Mol Biol. 2004;245:197–206. doi: 10.1385/1-59259-649-5:197. [DOI] [PubMed] [Google Scholar]
- McKenna MC. The glutamate-glutamine cycle is not stoichiometric: fates of glutamate in brain. J. Neurosci. Res. 2007;85:3347–3358. doi: 10.1002/jnr.21444. [DOI] [PubMed] [Google Scholar]
- McKenna MC, Sonnewald U, Huang X, Stevenson J, Zielke HR. Exogenous glutamate concentration regulates the metabolic fate of glutamate in astrocytes. J. Neurochem. 1996;66:386–393. doi: 10.1046/j.1471-4159.1996.66010386.x. [DOI] [PubMed] [Google Scholar]
- Munir M, Correale DM, Robinson MB. Substrate-induced up-regulation of Na(+)-dependent glutamate transport activity. Neurochem Int. 2000;37:147–162. doi: 10.1016/s0197-0186(00)00018-8. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Otsubo Y, Yatani Y, Shirakawa H, Kaneko S. Mechanisms of substrate transport-induced clustering of a glial glutamate transporter GLT-1 in astroglial-neuronal cultures. The European journal of neuroscience. 2008;28:1719–1730. doi: 10.1111/j.1460-9568.2008.06494.x. [DOI] [PubMed] [Google Scholar]
- Nguyen KT, Buljan V, Else PL, Pow DV, Balcar VJ. Cardiac glycosides ouabain and digoxin interfere with the regulation of glutamate transporter GLAST in astrocytes cultured from neonatal rat brain. Neurochem. Res. 2010;35:2062–2069. doi: 10.1007/s11064-010-0274-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norenberg MD, Martinez-Hernandez A. Fine structural localization of glutamine synthetase in astrocytes of rat brain. Brain Res. 1979;161:303–310. doi: 10.1016/0006-8993(79)90071-4. [DOI] [PubMed] [Google Scholar]
- Osman C, Merkwirth C, Langer T. Prohibitins and the functional compartmentalization of mitochondrial membranes. J. Cell Sci. 2009;122:3823–3830. doi: 10.1242/jcs.037655. [DOI] [PubMed] [Google Scholar]
- Overstreet LS, Kinney GA, Liu YB, Billups D, Slater NT. Glutamate transporters contribute to the time course of synaptic transmission in cerebellar granule cells. J Neurosci. 1999;19:9663–9673. doi: 10.1523/JNEUROSCI.19-21-09663.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmada M, Centelles JJ. Excitatory amino acid neurotransmission. Pathways for metabolism, storage and reuptake of glutamate in brain. Front Biosci. 1998;3:d701–718. doi: 10.2741/a314. [DOI] [PubMed] [Google Scholar]
- Pellerin L, Magistretti PJ. Glutamate uptake into astrocytes stimulates aerobic glycolysis: a mechanism coupling neuronal activity to glucose utilization. Proc. Natl. Acad. Sci. U. S. A. 1994;91:10625–10629. doi: 10.1073/pnas.91.22.10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaitakis A, Latsoudis H, Spanaki C. The human GLUD2 glutamate dehydrogenase and its regulation in health and disease. Neurochem. Int. 2011;59:495–509. doi: 10.1016/j.neuint.2011.03.015. [DOI] [PubMed] [Google Scholar]
- Ralphe JC, Bedell K, Segar JL, Scholz TD. Correlation between myocardial malate/aspartate shuttle activity and EAAT1 protein expression in hyper- and hypothyroidism. Am J Physiol Heart Circ Physiol. 2005;288:H2521–2526. doi: 10.1152/ajpheart.00991.2004. [DOI] [PubMed] [Google Scholar]
- Ralphe JC, Segar JL, Schutte BC, Scholz TD. Localization and function of the brain excitatory amino acid transporter type 1 in cardiac mitochondria. J Mol Cell Cardiol. 2004;37:33–41. doi: 10.1016/j.yjmcc.2004.04.008. [DOI] [PubMed] [Google Scholar]
- Rao VL, Murthy CR. Uptake and metabolism of glutamate and aspartate by astroglial and neuronal preparations of rat cerebellum. Neurochem. Res. 1993;18:647–654. doi: 10.1007/BF00966777. [DOI] [PubMed] [Google Scholar]
- Rinholm JE, Slettalokken G, Marcaggi P, Skare O, Storm-Mathisen J, Bergersen LH. Subcellular localization of the glutamate transporters GLAST and GLT at the neuromuscular junction in rodents. Neuroscience. 2007;145:579–591. doi: 10.1016/j.neuroscience.2006.12.041. [DOI] [PubMed] [Google Scholar]
- Ritter SL, Asay MJ, Paquet M, Paavola KJ, Reiff RE, Yun CC, Hall RA. GLAST stability and activity are enhanced by interaction with the PDZ scaffold NHERF-2. Neurosci. Lett. 2011;487:3–7. doi: 10.1016/j.neulet.2010.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MB. The family of sodium-dependent glutamate transporters: a focus on the GLT-1/EAAT2 subtype. Neurochem. Int. 1998;33:479–491. doi: 10.1016/s0197-0186(98)00055-2. [DOI] [PubMed] [Google Scholar]
- Rose EM, Koo JC, Antflick JE, Ahmed SM, Angers S, Hampson DR. Glutamate transporter coupling to Na,K-ATPase. J. Neurosci. 2009;29:8143–8155. doi: 10.1523/JNEUROSCI.1081-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothe F, Brosz M, Storm-Mathisen J. Quantitative ultrastructural localization of glutamate dehydrogenase in the rat cerebellar cortex. Neuroscience. 1994;62:1133–1146. doi: 10.1016/0306-4522(94)90349-2. [DOI] [PubMed] [Google Scholar]
- Schlag BD, Vondrasek JR, Munir M, Kalandadze A, Zelenaia OA, Rothstein JD, Robinson MB. Regulation of the glial Na+-dependent glutamate transporters by cyclic AMP analogs and neurons. Mol Pharmacol. 1998;53:355–369. doi: 10.1124/mol.53.3.355. [DOI] [PubMed] [Google Scholar]
- Schousboe A, Westergaard N, Sonnewald U, Petersen SB, Huang R, Peng L, Hertz L. Glutamate and glutamine metabolism and compartmentation in astrocytes. Dev Neurosci. 1993;15:359–366. doi: 10.1159/000111356. [DOI] [PubMed] [Google Scholar]
- Sheldon AL, Robinson MB. The role of glutamate transporters in neurodegenerative diseases and potential opportunities for intervention. Neurochem. Int. 2007;51:333–355. doi: 10.1016/j.neuint.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M. Molecular organization of the postsynaptic specialization. Proc Natl Acad Sci U S A. 2001;98:7058–7061. doi: 10.1073/pnas.111146298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skytt DM, Madsen KK, Pajecka K, Schousboe A, Waagepetersen HS. Characterization of primary and secondary cultures of astrocytes prepared from mouse cerebral cortex. Neurochem. Res. 2010;35:2043–2052. doi: 10.1007/s11064-010-0329-6. [DOI] [PubMed] [Google Scholar]
- Sonnewald U, Westergaard N, Schousboe A. Glutamate transport and metabolism in astrocytes. Glia. 1997;21:56–63. doi: 10.1002/(sici)1098-1136(199709)21:1<56::aid-glia6>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Sui D, Wilson JE. Structural determinants for the intracellular localization of the isozymes of mammalian hexokinase: intracellular localization of fusion constructs incorporating structural elements from the hexokinase isozymes and the green fluorescent protein. Arch Biochem Biophys. 1997;345:111–125. doi: 10.1006/abbi.1997.0241. [DOI] [PubMed] [Google Scholar]
- Sullivan SM, Lee A, Bjorkman ST, Miller SM, Sullivan RK, Poronnik P, Colditz PB, Pow DV. Cytoskeletal anchoring of GLAST determines susceptibility to brain damage: an identified role for GFAP. J Biol Chem. 2007;282:29414–29423. doi: 10.1074/jbc.M704152200. [DOI] [PubMed] [Google Scholar]
- Swanson RA, Liu J, Miller JW, Rothstein JD, Farrell K, Stein BA, Longuemare MC. Neuronal regulation of glutamate transporter subtype expression in astrocytes. J. Neurosci. 1997;17:932–940. doi: 10.1523/JNEUROSCI.17-03-00932.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waagepetersen HS, Hansen GH, Fenger K, Lindsay JG, Gibson G, Schousboe A. Cellular mitochondrial heterogeneity in cultured astrocytes as demonstrated by immunogold labeling of alpha-ketoglutarate dehydrogenase. Glia. 2006;53:225–231. doi: 10.1002/glia.20276. [DOI] [PubMed] [Google Scholar]
- Wang GJ, Jackson JG, Thayer SA. Altered distribution of mitochondria impairs calcium homeostasis in rat hippocampal neurons in culture. J. Neurochem. 2003;87:85–94. doi: 10.1046/j.1471-4159.2003.01970.x. [DOI] [PubMed] [Google Scholar]
- Waniewski RA, Martin DL. Exogenous glutamate is metabolized to glutamine and exported by rat primary astrocyte cultures. J. Neurochem. 1986;47:304–313. doi: 10.1111/j.1471-4159.1986.tb02863.x. [DOI] [PubMed] [Google Scholar]
- Westergaard N, Sonnewald U, Schousboe A. Metabolic trafficking between neurons and astrocytes: the glutamate/glutamine cycle revisited. Dev Neurosci. 1995;17:203–211. doi: 10.1159/000111288. [DOI] [PubMed] [Google Scholar]
- Yu AC, Schousboe A, Hertz L. Metabolic fate of 14C-labeled glutamate in astrocytes in primary cultures. J. Neurochem. 1982;39:954–960. doi: 10.1111/j.1471-4159.1982.tb11482.x. [DOI] [PubMed] [Google Scholar]
- Yudkoff M, Nelson D, Daikhin Y, Erecinska M. Tricarboxylic acid cycle in rat brain synaptosomes. Fluxes and interactions with aspartate aminotransferase and malate/aspartate shuttle. J. Biol. Chem. 1994;269:27414–27420. [PubMed] [Google Scholar]
- Zielke HR, Collins RM, Jr., Baab PJ, Huang Y, Zielke CL, Tildon JT. Compartmentation of [14C]glutamate and [14C]glutamine oxidative metabolism in the rat hippocampus as determined by microdialysis. J. Neurochem. 1998;71:1315–1320. doi: 10.1046/j.1471-4159.1998.71031315.x. [DOI] [PubMed] [Google Scholar]