Abstract
Endothelial cell dysfunction is a critical component of ocular diseases such as age-related macular degeneration and diabetic retinopathy. An important limitation in endothelial cell research is the difficulty in achieving efficient transfection of these cells. A new polymer library was synthesized and utilized to find polymeric nanoparticles that can transfect macrovascular (human umbilical vein, HUVECs) and microvascular (human retinal, HRECs) endothelial cells. Nanoparticles were synthesized that can achieve transfection efficiency of up to 85% for HRECs and 65% for HUVECs. These nanoparticle systems enable high levels of expression while avoiding problems associated with viral gene delivery. The polymeric nanoparticles also show cell-specific behavior, with a high correlation between microvascular and macrovascular transfection (R2 = 0.81), but low correlation between retinal endothelial and retinal epithelial transfection (R2 = 0.21). These polymeric nanoparticles may be used in vitro as experimental tools and potentially in vivo to target and treat vascular-specific diseases.
Keywords: Non-viral gene delivery, nanoparticles, ocular diseases, human retinal endothelial cells
Background
Endothelial cells play important roles in various ocular diseases, such as age-related macular degeneration, diabetic retinopathy, and retinoblastoma.1 The dysregulation and subsequent angiogenic proliferation of these ocular microvascular endothelial cells represents the key step in most retinal causes of blindness. Regulation of this angiogenesis by anti-angiogenic drugs such as ranibizumab is now the first-line therapy and there is continued interest in more effective anti-angiogenic therapies.2 Gene delivery is one such alternative method for delivery of antiangiogenic factors to endothelial cells, such as endostatin, angiostatin, and vascular endothelial growth factor-binding protein.3, 4 In addition, gene therapy can be used to correct specific genetic deficiencies within the endothelial cell population. In this strategy, therapeutic genes that can either add or block a function are delivered to a targeted cell population. Additionally, such gene delivery methods can also be very useful in the study of cellular biology and disease.
Non-viral transfection of human retinal endothelial cells (HRECs) remains a challenge as does transfection of many other cell types. For example, one recent report using lipid coated magnetic nanoparticles achieved only ~5% transfection efficacy.5 Leading commercial reagents, such as Lipofectamine 2000, can improve the transfection of HRECs. One study found that this approach could lead to 42%–67% knockdown of a target receptor’s surface expression following plasmid transfection.6 Macrovascular (human umbilical vein endothelial cells, HUVECs) are also generally difficult to transfect as a lead polymer (polyethylenimine) plus magnetofection yielded transfection efficacy of only 39% positive cells.7 New nanomedicines are needed to further increase the effectiveness of non-viral gene delivery.
Certain poly(β-amino esters) (PBAEs) have recently shown good transfection efficacy to a variety of cell types, including hard to transfect cell types like human mammary epithelium in 2D and 3D,8 human brain cancer cells,9 and HUVECs.10 Particular polymers formulations have shown selectivity in terms of transfecting brain cancer cells as compared to normal astrocytes.9 In addition, polymer end-group modification has been suggested as a tool to tune transfection efficacy.10–12 The objectives of this study were to investigate the endothelial and retinal cell-type specificities of PBAE-based nanoparticles and also to identify novel nanoparticles that can achieve high transfection of human endothelial cells with minimal toxicity. A new PBAE combinatorial polymer library was synthesized and evaluated to discover nanoparticles that can transfect either macrovascular (HUVECs)or microvascular(HRECs) human endothelial cells or both.
Methods
Materials
All chemicals and solutions were used as received unless otherwise indicated. Monomers and vendors used for synthesis are the following: from Acros Organics [1-(3-aminopropyl)pyrrolidine (E8)], Alfa Aesar [3-amino-1-propanol (S3), 4-amino-1-butanol (S4), 5-amino-1-pentanol (S5), 1,4-butanediol diacrylate (B4), 1,6-hexanedioldiacrylate (B6), 1-(3-aminopropyl)-4-methylpiperazine (E7)], Fluka[2-(3-aminopropylamino)ethanol (E6)], Monomer-Polymer and Dajac Laboratories [1,3-propanediol diacrylate (B3), 1,5-pentanedioldiacrylate (B5)], Sigma-Aldrich [1,3-diaminopropane (E1), 2,2-Dimethyl-1,3-propanediamine (E2), cystaminedihydrochloride (E10), 2-(1H-imidazol-4-yl)ethanamine (E12)], and TCI America [1,3-diaminopentane (E3), 2-methyl-1,5-diaminopentane (E4), (PEO)4-bis-amine (E5)]. Anhydrous DMSO and 3 M sodium acetate buffer were purchased from Sigma Aldrich. Sodium acetate buffer was diluted to 25 mM and filtered through 0.2 μm filter. pCMV-eGFP DNA was purchased from Aldevron. PBS, 0.25% trypsin-EDTA, Fu-Gene HD, and Lipofectamine 2000 were purchased from Invitrogen. CellTiter 96® AQueous One MTS assay was purchased from Promega. 96-well TCP and non-TCP round-bottom plates were purchased from Sarstedt. Human Umbilical Vein Endothelial Cells (HUVECs) and EGM-2 Bullet Kit and Reagent Pack were purchased from Lonza. Human retinal endothelial cells (HRECs) were obtained from Cell Systems and cultured in EGM2-MV (from Lonza), as previously described.13, 14 The immortalized human RPE cell line, ARPE-19, was obtained from the Dr. James Handa.15
Synthesis of Poly(beta-amino ester) (PBAE)
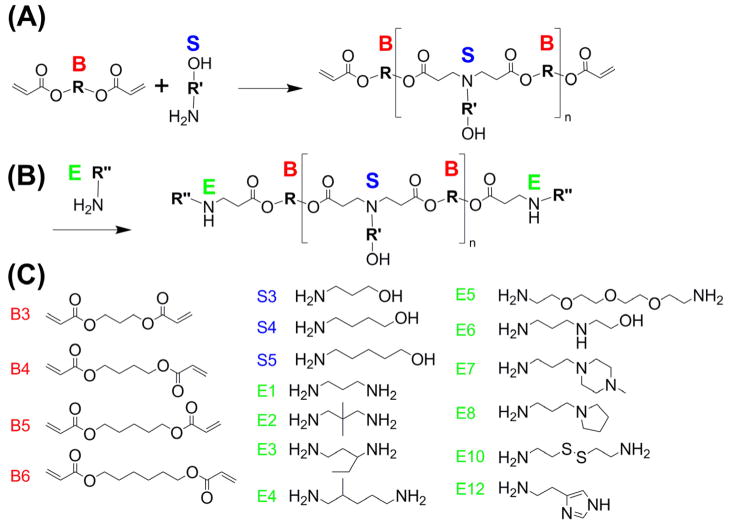
The structures of these polymers were chosen so that there are ester linkages to ensure degradability of the polymers and amine groups to ensure the ability to bind DNA and form nanoparticles. The base polymers were synthesized by mixing diacrylates, labeled as ‘B#’ in the text, and amino alcohols, ‘S#’, at a molar ratio of 1.2:1 in glass scintillating vials with teflon stir bars, forming ‘B#-S#’ base polymers. The reaction was run at 90°C for 24 hours. The base polymer was then dissolved at 167mg/mL in DMSO. In the last step of the reaction, 480 μL containing 80 mg of the base polymer and 320 μL of 0.5 M end-capping amine, ‘E#’, were mixed in a 1.5 mL eppendorf tubes in a shaker for 1 hour. Completed polymers were aliquoted into smaller volumes and stored at 4°C. The completed polymers are designated as ‘B#-S#-E#’; for example, B3-S5-E1 is an end-modified PBAE formed from the B3-S5 base polymer which is then end-modified with the end-capping amine E1. Polymer molecular weights were typically ~10 kDa as we previously have reported and among these polymers we have found that molecular weight is not correlated to transfection efficacy.16 We have recently published the polymer characterization of these synthetic biomaterials including 1H-NMR and Gel Permeation Chromatography of each polymer.16
Nanoparticle Sizing
DNA was diluted in 25 mM sodium acetate buffer to 0.01 mg/mL. Polymer was diluted in sodium acetate buffer to either 30 or 60 times that concentration (30 and 60 weight to weight, w/w, ratios), added to the DNA solution and incubated at room temperature for 10 minutes. Nanoparticles were then diluted 20 fold into PBS and loaded into NS500 nanosight tracking analysis (NTA) system. Nanosight videos were captured for 60 seconds and then analyzed used the NTA software, version 2.2.
Transmission Electron Microscopy (TEM) imaging
Nanoparticles were prepared the same way as for sizing by NTA. 10 uL of sample was dropped onto carbon coated copper grids and left to dry in chemical hood for 2 hours. Unstained TEM imaging was then performed using the Philips CM120 system. Nanoparticle sizing was performed using ImageJ software.
Green Fluorescent Protein (GFP) transfection
HUVECs and HRECs were seeded at 2,500–5,000 cells/well onto 96-well plates and allowed to adhere overnight, in 100 μL of the appropriate media per well (EGM-2 for HUVECs and EGM-2 MV for HRECs). Immediately before transfection, media in plates were replaced with 10% supplemented FBS HUVEC media, 100 μL per well. For RPE transfections, ARPE-19 cells were seeded at 15,000 cells/well and allowed to grow to confluence over 3 days, and then transfected in DMEM with 10% supplemented FBS. Polymers and pCMV-eGFP DNA were diluted in 25mM sodium acetate buffer. The final DNA concentration of the mixture was 0.03 mg/mL, with PBAEs at either at 0.9 or 1.8 mg/mL (30 w/w and 60 w/w ratios, respectively). Solution was mixed, incubated for 10 minutes at room temperature and then 20μL were added per well. FuGene HD and Lipofectamine 2000 were screened to find best formulation by following the manufacturer’s guidelines. FuGene HD-DNA ratio of 4-1 was used, with 10 μL added per well, while Lipofectamine 2000-DNA ratio of 1.5-1 was used, with 10 μL added per well. Plates then incubated for 4 hours, after which nanoparticle loaded media was removed and fresh EGM-2 and EGM-2 MV media were added, 100 μL/well. Each experimental condition was evaluated in quadruplicate. Duplicate plates were made for each experimental group to use one each for cell viability and transfection efficiency measurements.
Cell metabolism/viability
Twenty-four hours after transfection, designated plates were used for the CellTiter 96® AQueous One MTS assay. 10 μL of the aliquoted assay solution was added per well. Plates were incubated for 1–4 hours, after which absorbance at 490 nm was measured using the BioTekSynergy 2 Plate Reader. Absorbance measurements were corrected from background media signal and normalized by untreated groups. Each experimental condition was evaluated in quadruplicate.
Flow cytometry
Forty-eight hours after transfection, flow cytometry was performed using the Accuri C6 flow cytometer with IntelliCyt high-throughput attachment. Cells were washed with 1x PBS and trypsinized with 30 μL/well of 0.25% trypsin-EDTA. 170 μL FACS buffer (2% FBS, 1x PBS) was added to cells and 200 μL/well transferred to 96-well round-bottom plates. The plates were centrifuged at 1000 rpm for 5 minutes, after which 170 μL/well of the supernatant was removed. The pellets were re-suspended in the remaining 30μL of buffer. Plates were then placed onto the IntelliCyt high-throughput attachment and HyperCyt software was to acquire and process data. FlowJo was used to analyze the flow cytometry data.
Results
Polymer and Nanoparticle Synthesis
A polymer library was synthesized for gene delivery, such that a large diversity of structures could be investigated, each with small differential structural changes from each other. The structures of these polymers were chosen so that there are ester linkages to ensure degradability of the polymers and amine groups to ensure the ability to bind DNA and form nanoparticles. These polymers were synthesized from a pool of 4 acrylate monomers, 3 side chain monomers, and 12 end-chain capping molecules as described in the methods. The synthetic polymers are referred to by a ‘B’ number, a ‘S’ number, and an ‘E’ number, each referring to their constituent monomers. The number following the ‘B’ or the ‘S’ corresponds to the number of carbons between functional groups in the diacrylate or amino-alcohol monomers; thus ‘B4-S5’ is a polymer formed from a base diacrylate with 4 carbons between each acrylate group, and an amino-alcohol with 5 carbons between the amine and the alcohol functional groups.
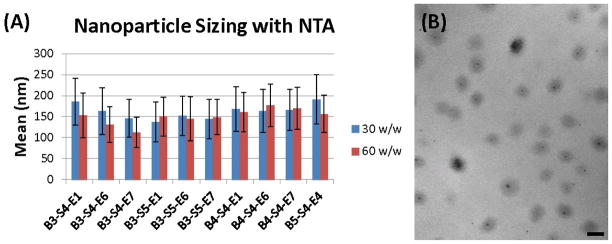
Nanoparticles are formed through electrostatic self-assembly, due to attractive interactions between cationic polymers and anionic DNA. We use relatively high polymer to DNA weight ratios (w/w) such that the polymers encapsulate 100% of the DNA as we have recently described.17 Representative polymers of interest were chosen for sizing measurements. Even though polymer structure varied, nanoparticle size was found to be similar with these different types of polymers (Figure 2A). Hydrodynamic diameter of the nanoparticles was approximately 150 nm, with a range of 110–190 nm in the particle distributions that was dependent on polymer to DNA weight-to-weight (w/w) formulation ratio. The average diameter of a nanoparticle formed from B3-S5-E1 at 60 w/w as measured by TEM was 147 nm (Figure 2B), which closely matches the results found from NTA. There was no correlation between the sizes of nanoparticles that transfected the cells well (B3-S5-E# and B4-S4-E# series) as compared to those that transfected the cells less well (B3-S4-E# series). Therefore, we hypothesize that chemical structure, not nanoparticle physical properties, is the driver of the differences seen in transfection efficacy.
Figure 2.
(A) Hydrodynamic diameter by nanoparticle tracking analysis. (B) Transmission Electron Microscopyof nanoparticles formed from B3-S5-E1 (60 w/w); scale bar is 200nm.
Nanoparticle-mediated Gene Delivery
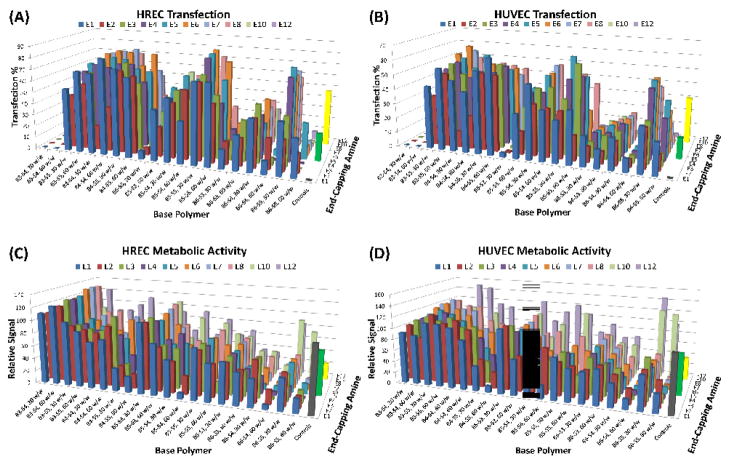
Leading polymeric nanoparticles were found to be very effective for gene delivery to human microvascular and macrovascular endothelial cells. Significantly, transfection efficiencies of up to 85% for HRECs and 65% for HUVECs were observed as opposed to 49% (HREC) and 32% (HUVEC) with a leading commercially available reagent, Lipofectamine 2000 (Figures 3A, and 3B). Lipofectamine 2000 was also more toxic than the PBAE nanoparticles (Figures 3C and 3D). The polymeric nanoparticles were found to display a wide range of transfection efficacies with both HRECs and HUVECs, which was dependent on polymer structure. Both the choice of base polymer and end-capping groups had a large effect on the transfection efficacy. The B4, B5, and B6 diacrylates differ by one carbon, yet there are significant differences in toxicity and transfection efficacy. A similar effect was observed when changing the side-chain amines. For example, B3-S4-based polymeric nanoparticles had very low transfection (< 10%), but a single carbon added to each of the side-chain monomers (B3-S5-based polymeric nanoparticles) resulted in highly effective gene delivery for both HRECs (up to 76%) and HUVECs (up to 64%). The end-group of the polymer made a big difference to endothelial cell gene delivery as well, where the best formulations usually contained E5, E6, and E7.
Figure 3.
(A) HUVEC and (B) HREC transfection percentage. (C) HREC and (D) HUVEC metabolic activity relative to untreated control. Controls: Gray=untreated, Green=FuGene HD, Yellow=Lipofectamine 2000.
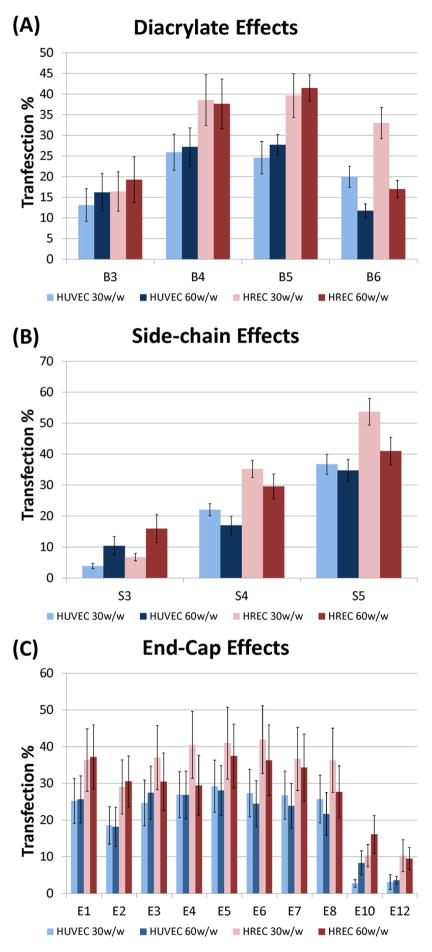
In order to better isolate the effects from changing each component of the polymer, the data were averaged together across two of the three polymer elements that compose each synthetic polymer. In general, B4 and B5 based polymers had the highest gene delivery efficacy (Figure 4A) and reveal an optimal base diacrylate length. On the other hand, we observe an increasing monotonic trend with increasing length for the various side-chain amines used in this experiment, with S5 based polymers performing best (Figure 4B). Unlike with the structure of the base polymers, a clear trend in the structure of the end-group with transfection efficacy is not observed (Figure 4C). However, there are some end-capping amines, such as E6, which dramatically improve delivery, and others, such as E12, that are much less effective. These results motivate further studies on how polymer structure affects endothelial cell gene delivery.
Figure 4.
Panel of monomer effects on transfection %. Collapsing (averaging) two of three types of monomers attempt to isolate effects of each monomer change. (A) Diacrylate base, (B) Side-chain amine base, (C) End-group amine.
Endothelial Cell-specific Gene Delivery
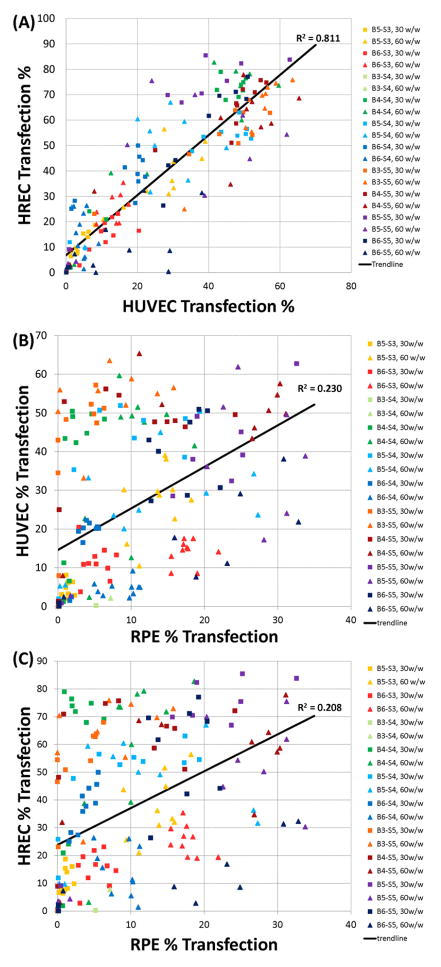
Comparing the transfection efficiencies between the different cell types and tissues revealed interesting trends (Figure 5). In this study, the HREC and HUVEC transfection profiles were positively correlated (linear regression, R2= 0.81), with greater transfection efficiencies for HRECs overall. Similar viability profiles were also observed between HUVECs and HRECs. On the other hand, using the same set of nanoparticles, there is very little correlation between the transfection efficacies for gene delivery to HRECs as compared to retinal pigment epithelium (RPEs, linear regression, R2 = 0.21), or HUVECs as compared to RPEs (linear regression, R2 = 0.23). Polymers that performed better on the endothelial cell types as compared the regression line include B4-S4 and B3-S5, as well as some B4-S5-based polymers (upper left hand corner of Figures 5B and 5C). As a group, these polymers were synthesized by diacrylate monomers that were less hydrophobic and contained 3 or 4 carbons between diacrylate monomer groups. On the other hand, there are some polymers that work better on the epithelial cells, such as B6-S5 (lower right hand corner of Figures 5B and 5C). This polymer is more hydrophobic than the polymers that tend to be better at transfecting endothelial cells and this polymer contains 6 carbons between its diacrylate monomer groups. The polymers that work well on both endothelial and epithelial cell types include B5-S5 based-polymers (upper right hand corner of Figures 5B and 5C), which have an intermediate level of hydrophobicity and 5 carbons between its diacrylate monomer groups. Strikingly, these structural changes are quite subtle and they do not influence nanoparticle size (Figure 2).
Figure 5.
(A) There is a strong correlation between transfection of HUVEC and HRECs and (B) a much weaker correlation when compared to transfection of RPEs.
Discussion
Synthesis and testing of a PBAE library resulted in the identification of polymeric nanoparticles that can transfect human endothelial cells, HRECs and HUVECs, with high efficiency as compared to current commercially available standards. The best polymers (both good transfection and viability), B3-S5-E# and B4-S4-E# series, are new PBAE structures that were able transfect at least as well as the commercially available reagents and in some cases with less toxicity. Additionally, Small molecular changes to polymer backbone, side chain or end-group structure dramatically changed transfection efficiency. The best polymers were ones that contained B4 and B5, and S4 and S5, with a few different E# groups. The most hydrophilic polymers, such as B3-S4, were not good at transfecting the cells, while their toxicity was also minimal. On the other hand, the most hydrophobic polymers, such as the B6-S5 series, were usually relatively toxic to the endothelial cells, subsequently also reducing the transfection efficiency. This trend is mirrored when looking at the effects of the different diacrylate monomers used (Figure 4A), where B4 and S5 performed best. While among certain structures, high hydrophobicity correlated to increased cytotoxicity and reduced transfection, overall, when all the data is evaluated together, there is not a strong correlation between transfection efficiency and viability (R2 =0.06, data not shown).
Interestingly, the gene delivery efficacy across the entire polymer library was highly correlated between transfection of HREC and HUVEC cells – nanoparticle formulations that performed well for HRECs also performed well for HUVECs (R2 = 0.81). This is dramatically different when compared to the performance of the same nanoparticles with retinal pigment epithelium, indicating that formulations could potentially be found which selectively transfect one ophthalmic cell type and not the other, as can be observed in Figures 5B and 5C. To our knowledge this phenomenon, that specific ocular cell types can be targeted directly by fine-tuning of the polymer structure that composes nanoparticles, is striking and surprising. This finding also suggests that biomaterial-mediated targeting from this class of materials may be able to be combined with other methods of gene delivery targeting such as ligand or coating-mediated cell uptake18–20 and transcriptional targeting21, 22 to further improve specificity and efficacy to only one specific type of cell.
In particular, we show that the polymers in the middle of the hydrophobicity scale for this PBAE library worked very well for the endothelial cells, but much less so for the epithelial cells. On the other hand, those that were more hydrophobic, such as the B6-S5, worked well for the epithelial cells. Taken together, these correlations indicate that there may be natural cell type specificity for polymeric nanoparticles of this class.
Safe and effective gene delivery vehicles can have a large impact on biological studies and for the treatment of diseases. For example, researchers have developed a system for gene delivery of sFLT using adeno-associated virus (AAV) to bind VEGF in monkeys as a potential treatment for age-related macular degeneration.23 However, viral gene therapy has limitations that may preclude many clinical applications due to carrying capacity constraints and safety concerns.24 In contrast, our study reveals premiere non-viral nanoparticles that are enabling technologies for transfection of endothelial cells in vitro and are promising for use in vivo as delivery vehicles for genetic nanomedicines. We also show that polymer structure itself, may enable cell-specific non-viral gene delivery.
Figure 1.
(A) Polymers were synthesized by reacting a diacrylate, ‘B#’, with an amino alcohol, ‘S#’; (B) These were then reacted with an end-group, ‘E#’; (C) Monomer structures.
Acknowledgments
The authors thank the TEDCO MSCRF(2009-MSCRFE-0098-00), the NIH (R21CA152473), and MSTP (to JCS) for support in part of this work. EJD is supported by a Career Development Award from Research to Prevent Blindness.
The authors thank Corey Bishop for help with TEM imaging and Dr. James Handa’s lab (JHMI-Wilmer Eye Institute) for ARPE-19 cells.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jo DH, Kim JH, Kim JH. How to Overcome Retinal Neuropathy: The Fight against Angiogenesis-related Blindness. Archives of Pharmacal Research. 2010;33:1557–1565. doi: 10.1007/s12272-010-1007-6. [DOI] [PubMed] [Google Scholar]
- 2.Bhise NS, Shmueli RB, Sunshine JC, Tzeng SY, Green JJ. Drug delivery strategies for therapeutic angiogenesis and antiangiogenesis. Expert Opinion on Drug Delivery. 2011;8:485–504. doi: 10.1517/17425247.2011.558082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adhim Z, Lin X, Huang W, Morishita N, Nakamura T, Yasui H, et al. E10A, an adenovirus-carrying endostatin gene, dramatically increased the tumor drug concentration of metronomic chemotherapy with low-dose cisplatin in a xenograft mouse model for head and neck squamous-cell carcinoma. Cancer Gene Ther. 2011 doi: 10.1038/cgt.2011.79. [DOI] [PubMed] [Google Scholar]
- 4.Campochiaro PA. Gene transfer for ocular neovascularization and macular edema. Gene Ther. 2011 doi: 10.1038/gt.2011.164. [DOI] [PubMed] [Google Scholar]
- 5.Prow T, Smith JN, Grebe R, Salazar JH, Wang N, Kotov N, et al. Construction, gene delivery, and expression of DNA tethered nanoparticles. Mol Vis. 2006;12:606–15. [PubMed] [Google Scholar]
- 6.Spoerri PE, Afzal A, Li Calzi S, Shaw LC, Cai J, Pan H, et al. Effects of VEGFR-1, VEGFR-2, and IGF-IR hammerhead ribozymes on glucose-mediated tight junction expression in cultured human retinal endothelial cells. Mol Vis. 2006;12:32–42. [PubMed] [Google Scholar]
- 7.Krotz F, Sohn HY, Gloe T, Plank C, Pohl U. Magnetofection potentiates gene delivery to cultured endothelial cells. Journal of Vascular Research. 2003;40:425–34. doi: 10.1159/000073901. [DOI] [PubMed] [Google Scholar]
- 8.Bhise NS, Gray RS, Sunshine JC, Htet S, Ewald AJ, Green JJ. The relationship between terminal functionalization and molecular weight of a gene delivery polymer and transfection efficacy in mammary epithelial 2-D cultures and 3-D organotypic cultures. Biomaterials. 2010;31:8088–96. doi: 10.1016/j.biomaterials.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tzeng SY, Guerrero-Cazares H, Martinez EE, Sunshine JC, Quinones-Hinojosa A, Green JJ. Non-viral gene delivery nanoparticles based on Poly(beta-amino esters) for treatment of glioblastoma. Biomaterials. 2011;32:5402–5410. doi: 10.1016/j.biomaterials.2011.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sunshine J, Green JJ, Mahon KP, Yang F, Eltoukhy AA, Nguyen DN, et al. Small-Molecule End-Groups of Linear Polymer Determine Cell-Type Gene-Delivery Efficacy. Advanced Materials. 2009;21:4947–4951. doi: 10.1002/adma.200901718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green JJ, Shi J, Chiu E, Leshchiner ES, Langer R, Anderson DG. Biodegradable polymeric vectors for gene delivery to human endothelial cells. Bioconjugate Chemistry. 2006;17:1162–1169. doi: 10.1021/bc0600968. [DOI] [PubMed] [Google Scholar]
- 12.Green JJ, Langer R, Anderson DG. A combinatorial polymer library approach yields insight into nonviral gene delivery. Accounts of Chemical Research. 2008;41:749–759. doi: 10.1021/ar7002336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maiti D, Xu Z, Duh EJ. Vascular endothelial growth factor induces MEF2C and MEF2-dependent activity in endothelial cells. Invest Ophthalmol Vis Sci. 2008;49:3640–8. doi: 10.1167/iovs.08-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Z, Yu Y, Duh EJ. Vascular endothelial growth factor upregulates expression of ADAMTS1 in endothelial cells through protein kinase C signaling. Invest Ophthalmol Vis Sci. 2006;47:4059–66. doi: 10.1167/iovs.05-1528. [DOI] [PubMed] [Google Scholar]
- 15.Handa JT, Reiser KM, Matsunaga H, Hjelmeland LM. The advanced glycation endproduct pentosidine induces the expression of PDGF-B in human retinal pigment epithelial cells. Exp Eye Res. 1998;66:411–9. doi: 10.1006/exer.1997.0442. [DOI] [PubMed] [Google Scholar]
- 16.Sunshine JC, Akanda MI, Li D, Kozielski KL, Green JJ. Effects of Base Polymer Hydrophobicity and End-Group Modification on Polymeric Gene Delivery. Biomacromolecules. 2011 doi: 10.1021/bm200807s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhise NS, Shmueli RB, Gonzalez J, Green JJ. A Novel Assay for Quantifying the Number of Plasmids Encapsulated by Polymer Nanoparticles. Small. 2011 doi: 10.1002/smll.201101718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Green JJ, Chiu E, Leshchiner ES, Shi J, Langer R, Anderson DG. Electrostatic ligand coatings of nanoparticles enable ligand-specific gene delivery to human primary cells. Nano Letters. 2007;7:874–9. doi: 10.1021/nl062395b. [DOI] [PubMed] [Google Scholar]
- 19.Harris TJ, Green JJ, Fung PW, Langer R, Anderson DG, Bhatia SN. Tissue-specific gene delivery via nanoparticle coating. Biomaterials. 2010;31:998–1006. doi: 10.1016/j.biomaterials.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shmueli RB, Anderson DG, Green JJ. Electrostatic surface modifications to improve gene delivery. Expert Opin Drug Deliv. 2010;7:535–50. doi: 10.1517/17425241003603653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang YH, Zugates GT, Peng W, Holtz D, Dunton C, Green JJ, et al. Nanoparticle-delivered suicide gene therapy effectively reduces ovarian tumor burden in mice. Cancer Research. 2009;69:6184–91. doi: 10.1158/0008-5472.CAN-09-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Showalter SL, Huang YH, Witkiewicz A, Costantino CL, Yeo CJ, Green JJ, et al. Nanoparticulate delivery of diphtheria toxin DNA effectively kills Mesothelin expressing pancreatic cancer cells. Cancer biology & therapy. 2008;7:1584–90. doi: 10.4161/cbt.7.10.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacLachlan TK, Lukason M, Collins M, Munger R, Isenberger E, Rogers C, et al. Preclinical Safety Evaluation of AAV2-sFLT01-A Gene Therapy for Age-related Macular Degeneration. Molecular Therapy. 2011;19:326–334. doi: 10.1038/mt.2010.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nature reviews Genetics. 2003;4:346–58. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]