Abstract
Induction of gene expression in yeast and human cells involves changes in histone modifications associated with promoters. Here we identify a histone H3 endopeptidase activity in S. cerevisiae that may regulate these events. The endopeptidase cleaves H3 after alanine 21, generating a histone lacking the first 21 residues and displays a preference for H3 tails carrying repressive modifications. In vivo, the H3 N-terminus is clipped, specifically within the promoter of genes following the induction of transcription. H3 clipping precedes the process of histone eviction seen when genes become fully active. A truncated H3 product is not generated in yeast carrying a mutation of the endopeptidase recognition site (H3 Q19L20->AA) and the gene induction is defective in these cells. These findings identify clipping of H3 tails as a novel modification of promoter-bound nucleosomes, which may result in the localised clearing of repressive signals during the induction of gene expression.
Keywords: Histone H3 cleavage, protease activity, nucleosome eviction, transcription activation
In all eukaryotes DNA is packed into chromatin, whose fundamental subunit is the nucleosome. Chromatin enables the compaction of the genome in the limited nuclear space but also represent a physical barrier to DNA replication, repair and transcription. Nucleosomes are evicted at many yeast promoters during gene activation, to allow access to the transcriptional machinery, and reassembled on the DNA template upon transcription repression (1-5). ATP-dependent chromatin remodelling complexes are involved in this process and a wave of histone acetylation precedes nucleosome removal from promoters of induced genes (6-8). Thus, post-translational histone modifications affect nucleosome dynamics.
There is evidence that histone turnover is regulated by proteolytic activities. For example, an H2A-specific protease activity has been described during granulocyte differentiation (9), which removes a pentadecapeptide from the carboxyl terminus of H2a cutting between V114 and L115. The resulting H2A: H2B dimer has a reduced affinity for the H3:H4 tetramer destabilizing the whole nucleosome. This function may contribute to a more “open chromatin” which facilitates transcription or replication. In the parasite Chlamydia trachomatis, chromatin decondensation occurring during the early life cycle is accompanied by the C-terminal proteolysis of the histone H1-like Hc1 protein by the EUO protease, eliminating its DNA interacting domain (10). In addition, Tetrahymena transcriptional inactive micronucleus and transcriptional active macronucleus differ on their histone complement. Macronuclear linker histone H1 is missing in the micronucleus, which contains α, β, γ and δ H1-like forms produced by proteolytic cleavage of a precursor (11) and a form of H3 that lacks the first 6 residues (12). Also the acetylated N-terminus of histone H4 (up to amino acid 21) seems to be proteolytically removed from the macronuclear genome during conjugation in Tetrahymena (13). Finally, there is also evidence that in S. cerevisiae a shorter version of histone H3 binds to Spt6 (14).
Results
Identification of a histone H3 endopeptidase activity in S. cerevisiae
While assaying yeast protein complexes for their capacity to demethylate histone H3, we noticed an endopeptidase activity in our nuclei preparations which cleaves the exogenously provided substrate (calf H3). The activity was low in the nuclei of cells growing exponentially in rich medium but increased in cells grown into stationary phase or shifted to sporulation medium (Figure 1A). In an attempt to biochemically purify this activity (see Materials and Methods) we found that it was retained on sepharose-based matrices (Figure 1B). To identify the site of cleavage on calf H3, we analyzed the reactions by mass spectrometry. Matrix-assisted laser desorption/ionization (MALDI) on the substrate (calf H3) identified a broad peak representing H3 carrying combinations of post-translational modifications (Supplementary Fig. 1, dark blue circle). When reactions were performed in the presence of the endopeptidase activity from stationary cells, two additional peaks were detected (Supplementary Fig.1, red and light blue circles). Electrospray sequencing of the smaller peak revealed products corresponding to the first 21 amino acids of calf H3 carrying various modifications (Supplementary Fig. S2). The N-terminal tail of H3 is sufficient for endopeptidase recognition as peptides spanning amino acids 1 to 30 are also cleaved after alanine 21 (Supplementary Fig. S3). To establish whether flanking residues represent a recognition site for the enzyme, full-length H3 was mutated at position 19 and 20 from QL to AA, expressed in E. coli and used as a substrate for the endopeptidase activity. Figure 1C shows that the wild type full length H3 is cleaved whereas the QL to AA mutant is resistant to the endopeptidase activity.
Figure 1.
A histone H3 endopeptidase activity in S. cerevisiae.
(A) A H3 endopeptidase activity is present in the yeast nuclei.
Nuclear extracts from early exponential, sporulation or stationary phase cultures were assayed for endopeptidase activity on recombinant H3. The reactions were stained with Ponceau (15 μl reaction, to visualize the products) and analyzed by western blot with anti C-terminal H3 antibody (5 μl reaction, to avoid signal saturation). The clipped H3 product is highlighted.
(B) The H3 endopeptidase activity is enriched upon nutrient starvation.
Extracts from early exponential, sporulation or stationary phase cultures, purified on sepharose beads, were assayed for endopeptidase activity on calf H3. The reactions were analyzed by western blot with anti C-terminal H3 antibody. The clipped H3 product is highlighted.
(C) The Q19L20A21 is the recognition sequence for the H3 endoppetidase.
Extract from yeast cell on stationary phase was pull down on Sepharose beads and assayed against recombinant wild type H3 and recombinant Q19L20->AA mutant H3.The reactions were analysed by western blot with anti C-terminal H3 antibody.
(D) Activity against different histones.
Stationary phase pull down on Sepharose beads was assayed against identical amounts of calf histones. The reactions were analysed by western blot with antibodies specific for each histone. The clipped H3 product is highlighted.
Next we analyzed whether other histones could be substrate for the endopeptidase. We used calf thymus H2A, H2B and H4 in the reactions and analyzed them with antibodies specific to each histone. Faster migrating bands were only detected in reactions performed on H3, indicating that the endopeptidase is specific for this histone (Figure 1D).
Modifications on the H3 tail affect clipping
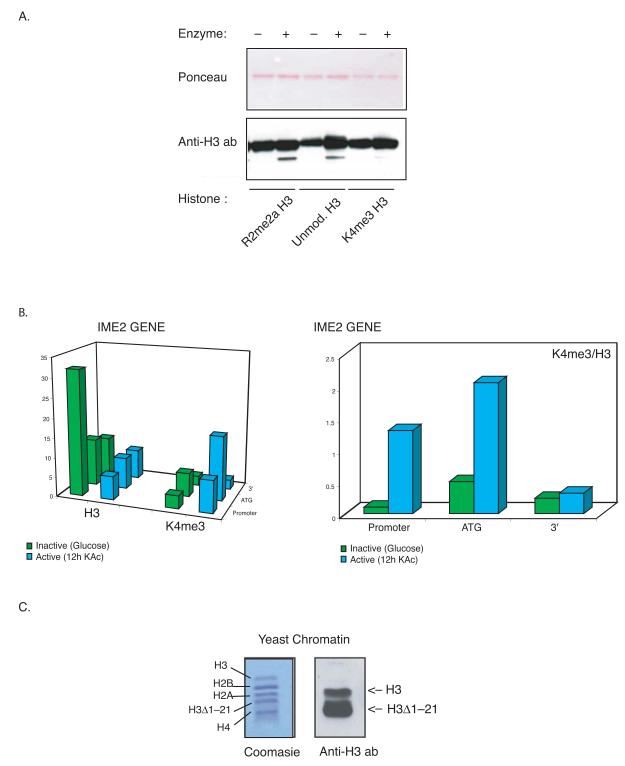
The electrospray analysis on cleaved calf thymus H3 showed clipped tail peptides carrying various modifications (Supplementary Fig. 4-6). Interestingly, peptides containing methylated K4 were absent among the most prominent N-terminal cleavage products. In contrast, K9 di- and tri methylated products could be detected. To test directly whether post-translational modifications alter the activity of the H3 clipping enzyme, we assayed this activity on full length modified H3. The H3 substrate was generated by ligating recombinant H3 spanning residues 32 to 135 to synthetic peptides (residues 1-31) containing trimethylated K4 (an activatory mark) or asymmetrical dimethyl R2 (a repressive mark, see Material and Methods). This procedure enables us to use a fully modified population of histones carrying a single post-translational mark. As shown in Figure 2A, the endopeptidase activity purified from a stationary culture was able to cut the unmodified H3 as well as R2 dimethylated H3. However the endopeptidase activity was reduced when H3 trimethylated at K4 was the substrate, suggesting that methylation of K4 is inhibitory to the H3 clipping activity. Chromatin Inmunoprecipitation analysis of inducible genes support this view. Figure 2B shows that IME2 induction is accompanied by a strong reduction in nucleosome occupancy at the promoter of the gene (left panel, H3 green and blue bars). This reduction is not observed on K4me3 H3 at the same location. In fact, the actual ratio of K4me3 with respect to the H3 content increases significantly from the repressed to the active transcription state of the gene (right panel). This behaviour was observed on other genes when induced (HOP1, SPR3, data not shown) and suggests that the K4 trimethylated H3 nucleomes are not removed from the promoters.
Figure 2.
The histone H3 endopeptidase is a serine protease with preference for non-activating marks.
(A) Histone H3 tails carrying non activatory marks are preferentially clipped.
Histone H3 modified at different residues (see Materials and Methods) was used as substrate for the endopeptidase activity pulled down from stationary phase culture. The reactions were analyzed by western blot with anti C-terminal H3 antibody.
(B) K4 me3 H3 are not evicted from promoters upon gene induction.
Chromatin immunoprecipitation experiments were performed in yeast cells cultured in either glucose (green bars) or sporulation medium for 12 hours (blue bars) using anti K4 me3 H3 antibody and anti C-terminal H3 antibody. The precipitated DNA was analyzed by quantitative PCR using primers specific to the indicated positions within the IME2 gene. The diagrams represent relative fluorescent units normalized to an intergenic region on chromosome V (see Material and Methods).
(C) H3Δ1-21 occurs in vivo in S. cerevisiae.
Yeast chromatin was prepared from early stationary phase culture and analyzed by Coomasie (left panel) or western blot (right panel). The full length and clipped H3 (H3Δ1-21) are highlighted.
The clipped H3 product can be detected the in vivo in chromatin prepared from yeast grown to stationary phase (Figure 2C). Edman degradation sequencing verified that this band between H2A and H4 corresponds to a version of H3 starting at amino acid 22 (data not shown). This result confirms that a histone H3 proteolytic product (1-21Δ H3) occurs in vivo in yeast.
The H3 endopeptidase is a serine protease
In order to determine the identity of the H3 endopeptidase, extracts from cells grown in stationary phase were subjected to chromatography onto sepharose beads and then eluted with 2M NaCl (Supplementary Fig. 7). SDS-PAGE analysis of the eluted active sample shows very few proteins present at the detection level of our Silver-staining (Supplementary Fig. 7). The proteins identified in this fraction by mass spectrometry did not match to any known or putative protease. Repeated attempts to identify the H3 endopeptidase by mass spectrometry have failed, possibly due to very low abundance of the enzyme in our preparations or difficulties in yielding peptides from this protein that can be subjected to sequencing.
As an alternative approach to determine the identity of the enzyme, we performed the cleavage reactions in the presence of inhibitors specific for different families of proteases (Figure 3). This screen shows that the serine protease inhibitors PMSF and leupeptin inhibit the activity to different extends (Figure 3A, lanes 5 and 9). Given that some cysteine proteases can also be inhibited by these compounds, we tested more cysteine protease inhibitors but none of them blocked the activity (Figure 3B). Furthermore, Figure 3A shows that pefacbloc SC plus, which reacts only with the nucleophilic serine in the catalytic site of proteases, can robustly inhibit the H3 endopeptidase activity (lane 12). Together, these results strongly suggest that the activity purified from yeast extracts is a serine protease.
Figure 3.
The H3 endopeptidase is a serine protease.
(A) Inhibition of the H3 endopeptidase activity.
The endopeptidase activity on recombinant H3 was tested in the presence of different proteases inhibitors. The reactions were analysed by Western Blot with anti C-terminal H3 antibody.
(B) Protease inhibitor profile of the H3 endopeptidase activity.
In S. cerevisiae there are 24 serine proteases, 21 of which are not essential for viability (supplementary Table 1). To determine if anyone of these serine proteases is responsible for the endopeptidase activity, we assayed yeast strains deleted in the genes encoding each of these proteases. Unfortunately, none of the deletion mutants lost the ability to cleave H3. These results suggest that the H3 endopeptidase is distantly related to the currently defined serine proteases or that redundant enzymes can perform this cleavage.
H3 tail clipping precedes H3 eviction from induced promoters
We next sought to establish an assay for the detection of H3 clipping on chromatin. To do this we used strains expressing a N-terminal myc tagged H3 allele (5). This strategy allows us to monitor the H3 N-terminus using a myc antibody and H3 C-terminus using a C-teminal H3 antibody. We studied genes induced during sporulation and stationary phase, since the endopeptidase activity is enhanced under these conditions. Figure 4A (green bars) shows that during exponential growth similar amounts of H3 N- and C-termini are detected at the promoter of the early meiotic gene IME1. However, 2 hours after induction of sporulation we observed a loss of the N-terminus relative to the H3 core (Figure 4A, red bars). After 12 hours of induction, histone H3 appears to be displaced from the IME1 promoter since a loss of both the N- and C-terminal H3 signals is observed (Figure 4A, blue bars). Tail-loss and histone H3 displacement were not observed within the open reading frame (ORF) of IME1 (Figure 4A). Analysis of a stationary induced gene ACH1 showed a very similar pattern of tail-loss followed by H3 displacement in the promoter, suggesting that tail-loss might be a promoter specific event.
Figure 4.
Histone tails are removed prior core depletion from promoters.
(A) Histone H3 tails are removed specifically from promoters. Chromatin immunoprecipitation experiments were performed in yeast cells cultured in either glucose (green bars) or sporulation medium (read and blue bars) using anti-myc antibody to detect the N-terminus of histone H3 (N) and anti C-terminal H3 antibody to detect the H3 core (C). The precipitated DNA was analyzed by quantitative PCR using primers specific to the indicated positions within the gene. The diagrams represent relative fluorescent units normalized to an intergenic region on chromosome V (see Material and Methods).
(B) Histone H3 tails are removed on induction of the sporulation.
Left panel: Chromatin immunoprecipitation experiments were performed in yeast cells cultured in either glucose (green bars) or sporulation medium (read and blue bars) using anti-myc antibody to detect the N-terminus of histone H3 (N) and anti C-terminal H3 to detect the H3 core (C). The precipitated DNA was analyzed by quantitative PCR using primers specific to the indicated promoters. The diagrams represent relative fluorescent units normalized to an intergenic region on chromosome V. Right panel: RT-PCR analysis was performed on the same cultures described (left panel), using primers specific to the indicated genes. The expression level of each gene was normalized to the RNA levels of RTG2.
(C) Histone H3 tails are removed on the transition to stationary phase.
Left panel: Chromatin immunoprecipitation experiments were performed in yeast cells cultured in glucose to OD600nm: 0.6 (green bars), OD600nm: 5 (read bars), OD600nm: 11 (blue bars) using anti-myc antibody to detect the N-terminus of histone H3 (N) and anti C-terminal H3 to detect the H3 core (C). The precipitated DNA was analyzed by quantitative PCR using primers specific to the indicated genes. The diagrams represent relative fluorescent units normalized to an intergenic region on chromosome V. Right panel: RT-PCR analysis was performed on the same cultures described (left panel), using primers specific to the indicated genes. The expression level of each gene was normalized to the RNA levels of RTG2.
To confirm this hypothesis we analyzed promoters of several sporulation (SPO11, HOP1, IME1) and stationary-phase (HSP26, HSP12, ACH1) induced genes (Figure 3B and 3C, left panels). Generally, we observe that upon gene induction H3 N-terminal tail loss precedes displacement of the histone core (Figure 4B and 4C, left panels). Gene expression analysis shows that reduction of H3 tail coincides with low accumulation of transcript, whereas displacement of core H3 associates with high levels of transcription (Figure 4B and 4C, right panels and Supplementary Fig. 8 for HSP12). In contrast, SPR3, which is induced much later in the sporulation course, does not show H3 tail-loss or displacement (Figure 4B). These results indicate that loss of the H3 N-terminal tail is an early event in the induction of gene expression that precedes the previously observed process of H3 displacement (1-4, 8).
Abrogation of H3 tail clipping impairs gene induction
To test if the H3 tail-loss is important for gene induction we constructed a yeast strain harbouring the mutation in the histone H3 cleavage site (Q19L20->AA H3). Analysis of the nuclear fraction from yeast grown to stationary phase shows the presence of full length and cleaved H3 in the wild type strain while only full length is present in the mutant (Figure 5A). This mutant strain, which is unable to cleave H3 tails, was then tested for changes in the expression of genes activated on stationary phase (HSP26 and HSP12) and sporulation (IME2). Figure 5B shows that the mutant exhibits impaired induction of these genes. In contrast RTG2, a gene which is known not to change transcription under the tested conditions, does not show any difference between WT and mutant strains. These findings suggest that proteolytic cleavage of the H3 tail at gene promoters contributes to proper induction of gene expression.
Figure 5.
Histone H3 Q19L20->AA mutation abrogates tail loss and cause transcription defects.
(A) The H3 Q19L20->AA yeast strain does not loose H3 tails.
Nuclear fractions from wild type and Q19L20->AA H3 yeast strains cultured in glucose to OD600nm: 7 were analyzed by western blot with anti C-terminal H3 antibody. The clipped H3 product is highlighted.
(B) Gene induction is defective in the H3 Q19L20->AA yeast strain.
RT-PCR analysis was performed on cultures of wild type and Q19L20->AA H3 grown on rich medium to OD600nm: 0.6 and OD600nm: 7, using primers specific to the indicated genes. The histogram bar represent folds induction for each strain and gene, calculated as the ration of [induced mRNA: non induced mRNA].
Discussion
Collectively these results identify a protease activity in S. cerevisiae which cleaves histone H3 after alanine 21 and has a recognition site Q19L20A21. This activity is induced under conditions of nutrient deprivation (stationary phase) and sporulation. Genes activated under these conditions are sensitive to H3 tail clipping and as a result, the levels of expression from these genes is compromised. The endopeptidase is also present at low levels on cells growing exponentially in rich medium, so we can not exclude the possibility that H3 tail clipping also controls the activity of genes induced under these conditions.
Our results indicate that the endopeptidase activity is regulated in some way during the activation of different pathways leading to gene expression. This regulation could come from transcriptional induction of the enzyme or by the activation of a pre-existing inactive enzyme. In the latter case, the switch in activity could reflect a covalent modification of the enzyme or (as is common for proteases) the conversion of the enzyme from an inactive pre-protease to proteolytically-cleaved active form.
The endopeptidase activity is sensitive to the modification status of the H3 tail. The presence of H3K4me3, an active mark, is inhibitory for clipping whereas the repressive H3R2me2 mark is not. This suggests that during activation, H3 tails methylated at H3K4 may be protected from being clipped. The selectivity for the clipping of tails with repressive marks is consistent with the idea that clipping may be necessary for the removal of transcription inhibitors at the onset of gene expression. Repressive protein complexes bound to the H3 tail at promoter regions may thus be removed to allow activator protein complexes to take over during the induction process. Thus, H3 tail clipping can be considered as a histone modification that allows “mass clearance” of repressive marks and proteins at promoter, a process that may be a necessary prelude to gene activation.
Our data also suggest another possible mechanism by which clipping may regulate gene expression. Eviction of nucleosomes is known to occur at promoters during gene induction and we observe that H3 clipping takes place just prior to nucleosome eviction. It is therefore possible that a clipped H3 tail may mark which nucleosomes are to be displaced prior to gene induction. The inability of the endopeptidase to clip H3K4 methylated tails is consistent with this model. During gene activation, H3K4 methylation is maintained on promoters despite the decrease in nucleosome occupancy (our data and 15) suggesting that the nucleosomes targeted for eviction are not methylated at H3K4. Together these observations are compatible with the model whereby H3 tails trimethylated at H3K4 are protected against clipping and are therefore resistant to eviction.
If clipping is directly linked to the process of eviction, the tail-less H3 may have a passive role, by occluding repressors of this process or it may have an active role by recruiting a protein complex necessary for the process. The generation of a novel N-terminus beginning at residue 22 may allow the recruitment of complexes which are normally unable to bind full length H3.
Another important question involves the targeting of the endopeptidase to the promoter of genes, to allow for the specificity of nucleosomal H3 clipping. One possibility is that there are specific mechanisms to deliver the enzyme to the correct region, for example by a set of DNA binding transcription factors or cofactors that recognise the promoter. In an alternative scenario, the enzyme may already be present at the promoter of genes in an inactive state, becoming active only following the stimulation of the pathway that regulates gene expression. Unmasking the endopeptidase activity of an already localized enzyme would provide a very rapid response to gene induction.
Recently an enzyme has been identified in mouse ES cells, Cathepsin L, that has the ability to cleave H3 after residue 21 and progressively remove several residues between amino acid 21 and 27 (16). This mammalian H3 clipping activity is stimulated during a specific period in the differentiation of ES cells. The existence of a mammalian enzyme (16) and a yeast enzyme (this paper) that clips H3 are consistent with the fact that the Q19L20A21 recognition site in the tail of H3 is conserved from yeast to man.
Although both Cathepsin L and the yeast enzyme have similar endopeptidase activity, (which cleaves H3 after residue 21), the data presented here suggest that the enzymatic activity in yeast is distinct from that of Cathepsin L. Firstly Cathepsin L, unlike the yeast activity, possesses an intrinsic aminopeptidase activity in addition to being an endopeptidase. Consequently Cathepsin L can generate several H3 forms with distinct N-termini whereas the yeast enzyme only generates H3 with an N-terminus at alanine 22. Secondly, Cathepsin L is a cystein protease and as such its activity is sensitive to cystein protease inhibitors. Our analysis of the yeast enzyme indicate that it is not sensitive to several cystein protease inhibitors, including cathepsin L I inhibitor (Z-Phe-Phe-FMK) that has been shown to inhibit Cathepsin L (16). Thirdly, inhibitors of serine proteases inhibit the yeast H3 endopeptidase. Taken together these studies indicate that the yeast enzyme is unlikely to be a homologue of Cathepsin L. Thus, there may be several classes of enzymes that can clip H3. Perhaps the serine protease family identified in yeast may have a more general role in transcription, given its involvement in several transcription pathways. In contrast Cathepsin L may represent a family of enzymes that have appeared late in evolution to regulate specific processes during cell differentiation.
Together the results presented here identify an enzyme in yeast that has the capacity to clip the H3 tail. Removal of the first 21 residues of H3 is shown to be a process necessary for the correct induction of gene expression. This modification of H3 may have consequences not only for transcription, but for many other events involving chromatin. The identification of the yeast endopeptidase will shed light on these processes and the ultimate biological consequences of H3 tail clipping.
Materials and methods
Yeast strains
BY4741 (Open Biosystems): MATa, ura3Δ0, leu2Δ0, his3Δ1, met15Δ0; BY4743 (Open Biosystems); JHY6: MATa, ura3-52, lys2-801, ade2-101, trp1–289, his3Δ1, leu2–3,112, Δhhf2-hht2, Δhhf1-hht1, pMS333[URA3- HHT2-HHF2]; JHY7: MATa/α, ura3-52, lys2-801, ade2-101, trp–289, his3Δ1, leu2–3,112, Δhhf2-hht2, Δhhf1-hht1, pMS333[URA3- HHT2-HHF2]. JHY7 was constructed by expressing the HO endonuclease in JHY6 to change its mating type, followed by mating to JHY6. Mutations of H3Q19L20 ->AA were introduced into the HHT2 gene in plasmids pMR206 (TRP1-HHT2-HHF2) and PNOY439 (CEN6 ARS4 TRP1-myc-HHT2-HHF2) using the QuickChange (Stratagene) site directed mutagenesis kit and verified by sequencing. Mutant and wild-type control plasmids were introduced into strain JHY6 to make the strains JHY6-H3WT and JHY6-H3 Q19L20->AA. The strain JHY6 was a gift from S. Berger.
Plasmids
PNOY439: CEN6 ARS4 TRP1-myc-HHT2-HHF2 M. Nomura
PNOY440: CEN6 ARS4 TRP1-myc-HHT2 Q19A L20A -HHF2 This study
PET21 b(+)-H3.1 This study
PET21 b(+)-H3.1 Q19A L20A This study
Protein purification and endopeptidase reactions
We initially detected an H3 endopeptidase activity from all protein complexes purified from cells grown in stationary phase. This activity was found to be associated with the sepharose component of the affinity matrix and was independent of its functional groups. We therefore used underivatized sepharose CL-4B (Sigma) for subsequent purifications.
Yeast cell were grown in rich YPAD medium. For mitotic exponential preparations cells were grown at 30C to OD600nm=0.8; for meiotic preparations cells were grown at 30C to OD600nm =0.8, collected by centrifugation, resuspended in 2% potassium acetate and incubated at 25C for additional 3h; for stationary preparations cells were grown in YPAD medium at 30C to OD600nm = 5-11 as specified in each experiment. Cells were collected by centrifugation and spheroblast prepared by treatment with Zymolyase 20T as described (17). Spheroblasts were lyzed on 20mM Tris-HCl ph 8.5; 200mM KCl; 25mM EDTA; 1% (v v−1) Triton X-100. Proteins were pulled down from each extract by chromatography on sepharose (200 microliter beads per 5g cells). Beads were washed with 40 ml lysis buffer and 40ml of the same buffer lacking Triton. Beads were resuspended in 400 microlitres of lysis buffer without Triton. Endopeptidase reactions were performed by incubating 25 microliters of beads bound proteins with 1-2 micrograms of histone H3 1h at 30C with shaking (recombinant H3.1, New England Biolabs; calf thymus H3, Roche). Assays on other histones (Calf thymus H2A, H2B and H4, Roche) were performed as described for calf H3. Proteins were separated by SDS-PAGE, stained by Ponceau and analyzed by Western Blot.
Western Blot
Western blot of total yeasts extracts were performed by standard procedures. Transfer to nitrocellulose membranes were made on carbonate buffer (21.1g l−1 NaHCO3, 18.35g l−1 Na2CO3, pH: 9.5) at 40mA for 70 min. The membranes were blocked o/n at 4 C in TBS+7% (v v-1) Tween-20+ 5% (w v−1) BSA; primary antibody for 3 hours at room temperature and secondary 1h at room temperature.
Antibodies
Histone H3: Ab1791, AbCam. Western Blot 1:10000; ChIP: 2 microliters ml-1 chromatin.
H2A: ab13923. Western Blot 1:1000
H2B: ab 1790. Western Blot 1:1000.
H4: A. Verrault. Western Blot 1:1000
Myc: M4439, Sigma. ChIP: 3 microliters ml-1 chromatin.
K36 trimethyl: ab 9050, AbCam. Western Blot 1:1000
K79 trimethyl: ab 7766, AbCam. Western Blot 1:1000
K56 Acetyl: 07-677, Upstate/Millipore. Western Blot 1:1000
Mass Spectrometry and MALDI analysis
0.5ul of incubation sample was mixed with 1.5ul of matrix solution (10mg ml−1 α-cyano-4-hydroxycinnamic acid in 50% (v v−1) aq. acetonitrile containing 0.1% (v v-1) trifluoroacetic acid) and dried onto a maldi target plate. Each sample spot was washed with 5ul of 0.2% heptafluorobutyric acid in water, dried, and analysed on a Waters TofSpec2E maldi mass spectrometer. Data was collected using a 500MHz detector in reflectron mode, averaging 50-200 laser pulses according to signal strength. Calibration was 3-point between matrix ions and 1-31thioester peptide (1+ and 2+ charge states). The mass of 1-31thioester was checked independently as correct using internal standards of substance P and oxidised bovine insulin B chain (not shown). Calibration and m z −1determination was carried out from centroid data.
Protease Inhibitors
Pepstatin A (P5318, Sigma); Iodoacetamide (I1149, Sigma); Pefabloc plus (cat. No. 11873601001, Roche); Iodoacetic acid (I4386, Sigma); Leupeptin (L2884 Sigma); Bestatin (B8385, Sigma); Calpain Inhibitor 1 (A6185, Sigma); PMSF (P7626, Sigma); Z-Phe-Phe-Fluomethyl Ketone (C9109, Sigma).
The inhibitors were incubated with the proteins for 15 min at 30C prior addition of the substrate.
Chromatin Immunoprecipitation “ChIP”
Chromatin prepared according to (17), was sonicated to produce fragmentsof 400-500 bp (determined for each experiment). Chromatin inmunoprecipitation was performed as described (17). Real time PCR analysis was performed on an ABI PRISM 7000 sequence detection system using SYBR Green (Applied Biosystems). Standard curves for each primer set were calculated from amplification of wild-type genomic DNA diluted in 1:10, 1:100. 1:1000, 1:10000, and 1:100000. After each run, a dissociation curve was performed to ensure that no primer dimmers contaminated the quantification and that the product had the expected melting temperature. Each PCR reaction was performed in duplicate and the signal intensity value for each sample was calculated from the average of the 2 experiments. Relative fluorescent intensities for the ChIP experiments were calculated as follows: [(Ab signal X :Ab signal Y)-(IgG signal X : IgG signal Y)], where Ab is the antibody of interest, IgG is the negative control antibody, H3 is the histone H3 antibody, X is the locus of interest and Y is the intergenic region on Chromosome-V which was used as an internal reference. Each experiment was repeated between 2 and 4 times, from independent cultures and using different batches for each antibody. Differences in antibody lot numbers resulted in variability of up to 20% between experiments. However, after internal normalization (to intergenic region in chromosome V) the variability was less than 10% of the value for each experiment shown. Control ChIPs were done with anti-myc antibodies from untagged strain. The Primers used for PCR analysis are available on request.
Quantitative reverse-transcription PCR
Total yeast RNA was prepared from 3 × 107 cells of each indicated growth condition using the RNeasy Mini kit (QIAGEN) according to the manufacturer’s protocol. To ensure complete removal of contaminating DNA from the RNA preparations, the TURBO DNA-free kit (Ambion) was used. First strand cDNA synthesis was achieved using SuperScript II reverse transcriptase (cat.18080, Invitrogen) with a primer cocktail, containing 50 uM oligo(dT) (Ambion) and 50 ng random hexamers (Invitrogen), as described in the manufacturer’s instructions. The cDNA samples were then used as templates for real-time PCR (see above).
Histone H3 tail ligations
Recombinant modified histone H3 was prepared following a described procedure (18).
Yeast nuclei preparation
according to (19).
Yeast chromatin preparation
Native chromatin was purified from yeast cells grown in rich YPD medium to early stationary phase (OD=2.5) following a described procedure (20). After microccocal nuclease digestion, the chromatin samples were concentrated and fractionated by gel filtration (Superose 6, Amersham Biosciences) in high-salt buffer. Histones were separated in SDS-PAGE gel and analyzed by western blot.
Supplementary Material
Acknowledgments
We thank Blerta Xhemalce for constructive discussions; Len Packman for excellent assistance with peptide and calf H3 sequencing and Alain Verreault for the gift of H3 and H4 antibodies. We are most grateful to Mark Dickman; Michiel Vermeulen & Matthias Mann; Hediye Erdjument-Bromage & Paul Tempst; Sarah Tully & Benjamin F. Cravatt and Sew Y. Peak-Chew for their attempts to identify the H3 endopeptidase by mass spectrometry.
References
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet. 2004;8:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- 3.Korber P, Luckenbach T, Blaschke D, H^rz W. Evidence for histone eviction in trans upon induction of the yeast PHO5 promoter. Mol Cell Biol. 2004;24:10965–10974. doi: 10.1128/MCB.24.24.10965-10974.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernstein BE, Liu CL, Humphrey EL, Perlstein EO, Schreiber SL. Global nucleosome occupancy in yeast. Genome Biol. 2004;5:R62. doi: 10.1186/gb-2004-5-9-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schermer UJ, Korber P, H^rz W. Histones are incorporated in trans during reassembly of the yeast PHO5 promoter. Mol Cell. 2005;19:279–85. doi: 10.1016/j.molcel.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 6.Schwabish MA, Struhl K. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol Cell. 2006;22:415–422. doi: 10.1016/j.molcel.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 7.English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK. Structural basis for the histone chaperone activity of Asf1. Cell. 2006;127:495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reinke H, H^rz W. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol Cell. 2003;11:1599–607. doi: 10.1016/s1097-2765(03)00186-2. [DOI] [PubMed] [Google Scholar]
- 9.Elia MC, Moudrianakis EN. Regulation of H2a-specific proteolysis by the histone H3:H4 tetramer. J Biol Chem. 1988;263:9958–64. [PubMed] [Google Scholar]
- 10.Kaul R, Hoang A, Yau P, Bradbury EM, Wenman WM. The chlamydial EUO gene encodes a histone H1-specific protease. J Bacteriol. 1997;179:5928–34. doi: 10.1128/jb.179.18.5928-5934.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allis CD, Allen RL, Wiggins JC, Chicoine LG, Richman R. Proteolytic processing of h1-like histones in chromatin: a physiologically and developmentally regulated event in Tetrahymena micronuclei. J Cell Biol. 1984;99:1669–77. doi: 10.1083/jcb.99.5.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allis CD, Bowen JK, Abraham GN, Glover CV, Gorovsky MA. Proteolytic processing of histone H3 in chromatin: a physiologically regulated event in Tetrahymena micronuclei. Cell. 1980;20:55–64. doi: 10.1016/0092-8674(80)90234-2. [DOI] [PubMed] [Google Scholar]
- 13.Lin R, Cook RG, Allis CD. Proteolytic removal of core histone amino termini and dephosphorylation of histone H1 correlate with the formation of condensed chromatin and transcriptional silencing during Tetrahymena macronuclear development. Genes Dev. 1991;5:1601–1610. doi: 10.1101/gad.5.9.1601. [DOI] [PubMed] [Google Scholar]
- 14.Bortvin A, Winston F. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science. 1996;272:1473–1476. doi: 10.1126/science.272.5267.1473. [DOI] [PubMed] [Google Scholar]
- 15.Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, Rando OJ. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 2005;3:328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duncan EM, Muratore-Schroeder TL, Cook RG, Garcia BA, Shabanowitz J, Hunt DF, Allis CD. Cathepsin L proteolytically processes Histone H3 diuring embryonic stem cell differentiation. Cell. 2008;135:284–294. doi: 10.1016/j.cell.2008.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 18.Shogren-Knaak MA, Peterson CL. Creating designer histones by native chemical ligation. Methods Enzymol. 2004;375:62–76. doi: 10.1016/s0076-6879(03)75004-6. [DOI] [PubMed] [Google Scholar]
- 19.Kizer KO, Xiao T, Strahl BD. Accelerated nuclei preparation and methods for analysis of histone modifications in yeast. Methods. 2006;40:296–302. doi: 10.1016/j.ymeth.2006.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altaf M, Utley RT, Lacoste N, Tan S, Briggs SD, CÙtÈ J. Interplay of chromatin modifiers on a short basic patch of histone H4 tail defines the boundary of telomeric heterochromatin. Mol Cell. 2007;28:1002–1014. doi: 10.1016/j.molcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.