Abstract
Small DNA tumour viruses have evolved a number of mechanisms to drive nondividing cells into S phase. Virally encoded oncoproteins such as adenovirus E1A and human papillomavirus (HPV) E7 can bindan array of cellular proteins to override proliferation arrest. The DNA methyltransferase Dnmt1 is the major mammalian enzyme responsible for maintaining CpG methylation patterns in the cell following replication. One of the hallmarks of tumour cells is disrupted DNA methylation patterns, highlighting the importance of the proper regulation of DNA methyltransferases in normal cell proliferation. Here, we show that adenovirus 5 E1A and HPV-16 E7 associate in vitro and in vivo with the DNA methyltransferase Dnmt1. Consistent with this interaction, we find that E1A and E7 can purify DNA methyltransferase activity from nuclear extracts. These associations are direct and mediated by the extreme N-terminus of E1A andthe CR3 zinc-finger domain of E7. Furthermore, we findthat a point mutant at leucine 20 of E1A, a residue known to be critical for its transformation functions, is unable to bind Dnmt1 and DNA methyltransferase activity. Finally, both E1A and E7 can stimulate the methyltransferase activity of Dnmt1 in vitro. Our results provide the first indication that viral oncoproteins bind and regulate Dnmt1 enzymatic activity. These observations open up the possibility that this association may be usedto control cellular proliferation pathways andsuggest a new mechanism by which small DNA tumour viruses can steer cells through the cell cycle.
Keywords: E1A, E7, Dnmt1, DNA methyltransferase, viral oncoproteins
Small DNA tumour viruses such as adenovirus and papillomaviruses require viral and cellular host proteins to replicate their genomes. The adenovirus oncoprotein E1A possesses multiple functions, and can affect gene transcription, induce DNA synthesis, immortalize cells and inhibit differentiation (Frisch and Mymryk, 2002). E1A associates with a large number of cellular proteins in infected or transformed cells, including the coactivators and histone acetyltransferases CBP/p300, the tumour-suppressor Rb, as well as sequence-specific transcription factors. E1A has several conserved regions which mediate most of its functions, designated conserved region 1 (CR1, residues 40–80), CR2 (121–139) and CR3 (140–188). The immortalization and transformation functions of E1A are mediated by CR1 and CR2, and these regions correspond with the interaction sites of Rb and CBP/p300 (Frisch and Mymryk, 2002).
Human papillomaviruses (HPVs) are another group of small DNA tumour viruses, which are of particular importance as infection with the high risk human types (such as HPV16 and 18) can lead to anogenital cancers. E7 is the major transforming protein of human papillomaviruses and shares sequence homology with E1A. E7 targets the Rb family to overcome proliferation arrest, by sequestering Rb from E2F complexes (McCance, 2005).
DNA methyltransferases are responsible for the methylation of cytosine in mammals and play a role in gene silencing. Dnmt1 is the main ‘maintenance’ methyltransferase, restoring the methylation pattern on newly replicated DNA (Fuks, 2005). Aberrant methylation patterns are increasingly being recognized as an important and frequent event in cancers (Robertson, 2001; Jones and Baylin, 2002). Tumours and transformed cell lines exhibit abnormal methylation of CpG islands and frequent inactivation of tumour suppressor genes. Several lines of evidence point to an increase in Dnmt1 activity being an important step in carcinogenesis. Overexpression of Dnmt1 in mouse fibroblasts leads to transformation (Wu et al., 1993). Dnmt1 has been reported to be required for maintaining cellular transformation by fos (Ordway et al., 2005) and a number of studies have reported elevated mRNA levels and Dnmt1 activity in cancers (Robertson, 2001). Although it remains to be established how increased levels of Dnmt1 may contribute to hypermethylation of CpG islands and tumour development, inhibition of Dnmt1 has been shown to suppress tumorigenesis in vitro and in vivo (Laird et al., 1995). These studies point to a role for increases in Dnmt1 activity playing an important part in CpG methylation-mediated cellular transformation.
Viral regulation of Dnmt1 has been reported. SV40 large T antigen upregulates Dnmt1 RNA and protein levels post-transcriptionally, leading to a 5- to 15-fold increase in methyltransferase activity and an increase in genomic methylation (Slack et al., 1999), and Dnmt1 acts as a downstream effector of the oncogenic programme of large T antigen. Similarly, HIV-1 infection leads to the upregulation of Dnmt1 expression and activity, resulting in global DNA hypermethylation with subsequent methylation of the IFN-γ promoter (Mikovits et al., 1998). Furthermore, an increase in DNA methylation at a particular promoter has been observed in cells infected with a range of tumorigenic viruses, such as HTLV-1, EBV and SV-40 (de Bustros et al., 1988). However, none of these previous studies have demonstrated a direct association between particular viral proteins and Dnmt1.
In the present study, we sought to determine whether viral oncoproteins can target Dnmt1 directly. We find that adenovirus 5 E1A and HPV-16 E7 bind to Dnmt1 and precipitate DNA methyltransferase activity. These interactions are direct and map to the extreme N-terminus of E1A and to the zinc-finger CR3 region of E7. We also find that leucine 20 of E1A, known to play a key role in the ability of the virus to transform cells, is required for the association of E1A with Dnmt1 as well as with DNA methyltransferase activity. Finally, both E1A and E7 upregulate the methyltransferase activity of Dnmt1 in vitro. These results reveal that Dnmt1 is a novel cellular target for E1A and E7 and uncover a novel mechanism whereby E1A and E7 may transform cells.
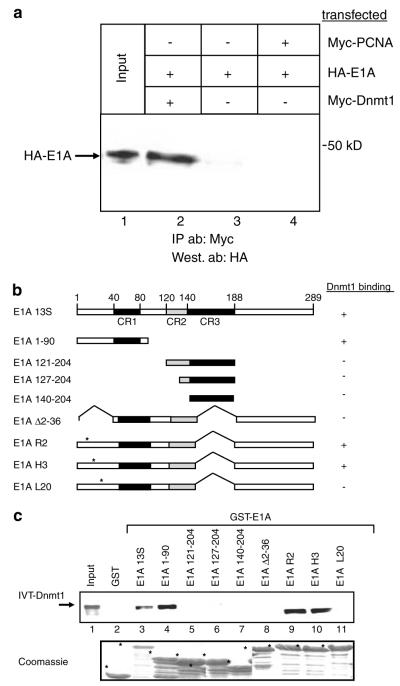
To investigate whether adenovirus E1A interacts with Dnmt1, a co-immunoprecipitation approach was employed. HA-tagged Ad5 E1A 12S (HA-E1A) and Myc-tagged, full-length Dnmt1 (Myc-Dnmt1) were transfected into 293T cells. Cells were lysed, immunoprecipitation was carried out using an anti-Myc antibody, and precipitates were subjected to Western blot analysis with an anti-HA antibody. Figure 1 (lane 2) indicates that E1A associates specifically with Dnmt1 in vivo.
Figure 1.
E1A interacts with Dnmt1 through its extreme N-terminal region. (a) E1A co-immunoprecipitates with Dnmt1 from transfected cell extracts. 293T cells were transfected with HA-Ad5 E1A (expressing HA-tagged E1A 12S) alone or together with either Myc-Dnmt1 (expressing Myc-tagged full-length Dnmt1, Myc-Dnmt1) or pMyc-PCNA (Myc-tagged PCNA, Myc-PCNA) as a control, as indicated. After 48 h, cells were lysed in IPH buffer (Brenner et al., 2005) and whole-cell extracts were then precipitated with anti-Myc antibody (9E10, Roche) and the presence of E1A in the immunoprecipitates was visualized using anti-HA antibody (3F10, Roche), as described (Vire et al., 2006). (b) Schematic representation of E1A with the deletions and point mutants used in GST-pulldowns. (c) In vitro translated (IVT) Dnmt1 was incubated with the indicated GST fusions. The reactions were subjected to GST-pulldown assays as described (Deplus et al., 2002). Bottom panel: Coomassie-stained gel showing the input of GST proteins used.
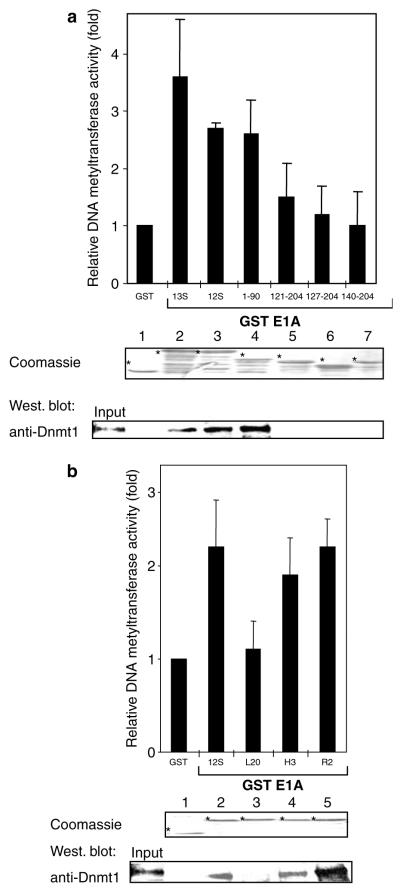
We next wished to define the sequences within E1A that are required for the association with Dnmt1. glutathione S-transferase (GST)-pulldown experiments indicate that the E1A N-terminus (amino acids 1–90) is needed for the association with in vitro translated Dnmt1, whereas E1A CR2 or CR3 are not (Figure 1c, lanes 3–7). Of note, Dnmt1 is known to associate with Rb (Pradhan and Kim, 2002) and Rb itself is a target of E1A (Whyte et al., 1988). We observed, however, that the major Rb binding site within CR2 (the LXCXE motif) does not seem to contribute to the association of E1A with Dnmt1 (Figure 1c, compare lanes 5 and 6). We further found that the extreme N-terminus of E1A (amino acids 2–36), which is involved in transformation, cell growth control and transcriptional regulation (Wang et al., 1993; Sang and Giordano, 1997; Boyd et al., 2002), is essential for the interaction with Dnmt1 (Figure 1c, lane 8). The use of well-defined single point mutants within this region of E1A shows that mutation of leucine 20 specifically disrupts the binding to Dnmt1 (Figure 1c, lanes 9–11). Amino acid 20 is critical for immortalization as well as transformation (Wang et al., 1993; Sang and Giordano, 1997; Boyd et al., 2002). These results were confirmed by means of an in vitro DNA methyltransferase enzymatic assay. In this assay, liquid scintillation counting is used to monitor the incorporation of [3H]-S-adenosyl-l-methionine into a synthetic 33-bp hemimethylated oligonucleotide. As shown in Figure 2a we found that GST-E1A associates specifically, through residues 1–90, with DNA methyltransferase activity from nuclear extracts. Moreover, mutation of leucine 20 of E1A strongly impairs association with DNA methyltransferase activity (Figure 2b).
Figure 2.
E1A associates with DNA methyltransferase activity and leucine 20 is important for enzymatic binding. (a) E1A precipitates DNA methyltransferase activity through residues 1–90. Equivalent amounts of GST or GST-E1A fusion proteins bound to Sepharose beads were incubated with HeLa nuclear extract, washed and assayed for DNA methyltransferase activity as described (Fuks et al., 2000). Activity is shown as c.p.m. of S-adenosyl-l-(methyl-3H)-methionine incorporated into a synthetic 33-bp hemimethylated oligonucleotide substrate (upper strand: GAT meCG-meC meCGA TGmeC GmeCG AAT meCGmeC GAT meCGA TGmeC GAT; lower strand: ATC GCA TCG ATC GCG ATT CGC GCA TCG GCG ATC). Values are normalized to background controls lacking substrate. (b) Point mutation within leucine 20 of E1A strongly impaired association with DNA methyltransferase activity. For panels (a) and (b): middle panels show Coomassie-stained gel with the input of GST proteins; bottom panels represent Western-blotting following the enzymatic assays using anti-Dnmt1 (Pradhan and Kim, 2002). The results from (a) and (b) are the average of at least three independent experiments with s.d.
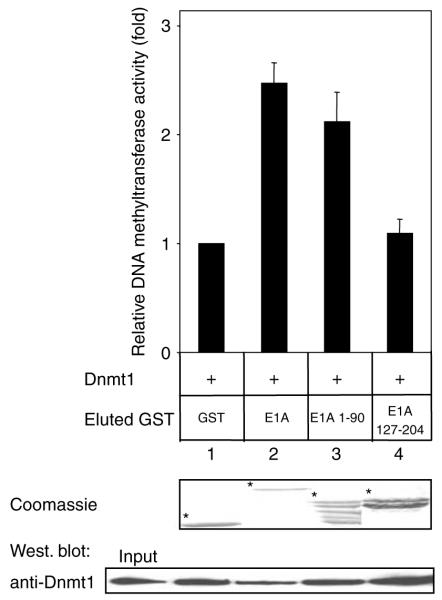
E1A interacts directly with several histone acetyltransferases, and it has been reported that E1A has the ability to regulate the acetyltransferase activity of CBP/ p300 both positively (Ait-Si-Ali et al., 1998) and negatively (Chakravarti et al., 1999). This prompted us to determine whether the association of E1A with Dnmt1 could modulate its enzymatic activity. To this end, baculovirus-expressed Dnmt1 was preincubated with GST-E1A and the reactions were subjected to DNA methyltransferase assays. As depicted in Figure 3, incubation of Dnmt1 with GST-E1A significantly stimulates DNA methyltransferase activity as compared to GST alone (lanes 1 and 2). Region 1–90 of E1A upregulates DNA methyltransferase activity as efficiently as full-length E1A, whereas another region of E1A (residues 127–204) is unable to do so (Figure 3). Western blotting of Dnmt1 following DNA methyltransferase assays shows that the observed enzymatic stimulation is not simply due to stabilizing the Dnmt1 protein (Figure 3, bottom panel). Thus, E1A can stimulate the methyltransferase activity of Dnmt1 and residues 1–90 of E1A are essential to mediate this effect.
Figure 3.
E1A upregulates the methyltransferase activity of Dnmt1. We expressed and purified baculovirus human Dnmt1 as described previously (Pradhan et al., 1999). Baculovirus-expressed Dnmt1 was preincubated for 10 min on ice with increasing concentrations (1 and 5 μg) of eluted and the indicated dialysed GST proteins. Activity is shown as c.p.m. of S-adenosly-l-(methyl-3H)-methionine incorporated into a hemimethylated oligonucleotide substrate. Bottom panel: Western blotting following the enzymatic assays using anti-Dnmt1 (Pradhan and Kim, 2002) indicates that the E1A-mediated enzymatic stimulations (lanes 2 and 3) are not simply due to stabilizing Dnmt1. The results are the average of at least three independent experiments with s.d.
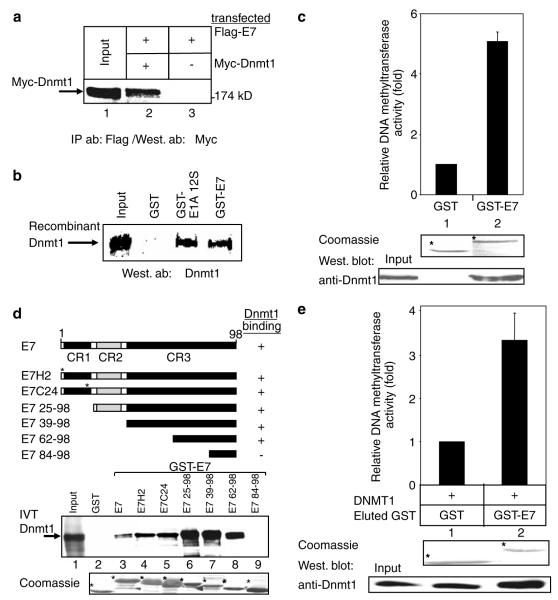
As HPV E7 shares sequence homology to E1A, it was of interest to determine whether E7 could also target Dnmt1. Figure 4a indicates that Dnmt1 and HPV-16 E7 interact when they are coexpressed in mammalian cells. The contacts between Dnmt1 and E7, as well as E1A, are direct since GST-E7 and GST-E1A produced in Escherichia coli bind efficiently to baculovirus-expressed Dnmt1 (Figure 4b). Next, we performed DNA methyltransferase assays to determine whether E7 could purify DNA methyltransferase activity from nuclear extracts. As shown in Figure 4c, GST-E7 precipitates significant levels of DNA methyltransferase activity compared to GST alone. Deletion analysis in GST-pulldowns indicates that the association of E7 with Dnmt1 is mediated by residues 62–84 encompassing the CR3 zinc-finger region (Figure 4d), which is known to contribute to E7 transformation functions (McCance, 2005). Deletion of the CR1 and CR2 regions as well as point mutations of residue at position 2 or within the Rb-binding motif of E7 (residue C24) did not alter the interaction with Dnmt1 (Figure 4d). Interestingly, constructs lacking the CR1 region appear to bind more strongly than wild-type E1A. We speculate that this may be due to conformation changes within the truncated E1A proteins, allowing the binding region within the C-terminus greater access to Dnmt1.
Figure 4.
HPV-16 E7 associates with Dnmt1 in vitro and in vivo and can upregulate its DNA methyltransferase activity. (a) HPV16 E7 co-immunoprecipitates with Dnmt1. 293 T cell were transfected with pcDNA3-E7-F(Flag-E7), Myc-Dnmt1 and empty expression vectors as indicated. Cell extracts were precipitated with anti-Flag (M2, Kodak) and the presence of Myc-Dnmt1 in the immunoprecipitates was visualized by Western blot analysis using anti-Myc (3F10, Roche). (b) Dnmt1 binds E7 and E1A directly. The indicated GST proteins were incubated with baculovirus-expressed Dnmt1, followed by Western blotting using anti-Dnmt1. (c) E7 associates with DNA methyltransferase activity. Equivalent amounts of GST or GST-E7 were incubated with HeLa nuclear extract and assayed for DNA methyltransferase activity as described previously (Fuks et al., 2000). Middel panel: Coomassie-stained gel shows the input of GST fusion proteins used. Bottom panel: Western analysis using anti-Dnmt1 following enzymatic assays. Error bars represent s.d. (d) The indicated GST-E7 fusion proteins were tested in a GST-pulldown assay for binding to in vitro translated and 35S-radiolabelled full-length Dnmt1 (IVT-Dnmt1). (e) E7 stimulates DNA methyltransferase activity. Baculovirus-expressed Dnmt1 was preincubated as described in Figure 3 with eluted and dialysed GST protein alone or GST-E7. Samples were assayed for DNA methyltransferase activity as described previously (Fuks et al., 2000). The results show the average of three independent experiments with s.d.
Using a similar approach as for E1A, the effect of HPV-16 E7 on the DNA methyltransferase activity of Dnmt1 was investigated. As shown in Figure 4e, GST-E7 significantly upregulates the enzymatic activity of baculovirus-expressed Dnmt1 while equivalent amounts of GST-alone have no effect on Dnmt1 activity. Following DNA methyltransferase assays, we performed Western blotting of Dnmt1. Interestingly, Dnmt1 appears to be partially stabilized by E7, suggesting a possible mechanism for how E7 stimulates the enzymatic activity of the protein (Figure 4e, bottom panel). Thus, in addition to E1A, E7 also targets Dnmt1, and this interaction appears to involve the C-terminal CR3 zinc-finger domain of E7. Furthermore, E7 also upregulates the enzymatic activity of Dnmt1, and this targeting thus represents a conserved feature of these viral oncoproteins.
In summary, we have shown that the viral oncoproteins E1A and E7 bind directly to Dnmt1 using sequences involved in their transformation functions. In addition, we found that E1A as well as E7 can upregulate the DNA methyltransferase activity of Dnmt1. The molecular basis underlying the stimulation of Dnmt1’s enzymatic activity by E1A and E7 remains to be elucidated. One mechanism could be related to the stimulation of the methyltransferase activity of Dnmt3a by Dnmt3L, a regulator of imprinting. In that situation, recent biochemical studies suggest that Dnmt3L induces a conformational change in Dnmt3a that opens the active site of the enzyme and promotes binding of DNA and the S-adenosyl-l-methionine (Adomet). In other words, Dnmt3L seems to act as a substrate exchange factor that accelerates DNA and Adomet binding to Dnmt3a, thereby stimulating its enzymatic activity (Gowher et al., 2005).
The stimulation of Dnmt1 activity shown in the present study may mimic the increase in DNA methyltransferase activity as a result of elevated levels of Dnmt1 observed with other viral proteins and that observed in some cancers. Our data, however, represent a much more direct link between viral proteins and Dnmt1, where the activity of the enzyme is stimulated directly. This increased activity may lead to similar cellular consequences of aberrant methylation of the genome followed by cellular transformation as a result of tumour suppressor gene silencing.
Acknowledgements
We thank B Moran for the kind gift of E1A constructs. WAB was supported by a scholarship from the National Research Foundation of South Africa. LB was supported by the FNRS and the ‘Fondation pour la Recherche Médicale’. SP was supported by NEB. FF is a ‘Chercheur Qualifié du FNRS’. This work was funded by a programme grant from the Cancer Research Campaign to TK and by grants from the ‘Fédération Belge contre le Cancer’, the FNRS, ‘FB Assurances’, and ‘ARC de la Communauté Française de Belgique’ to YdL and FF.
References
- Ait-Si-Ali S, Ramirez S, Barre FX, Dkhissi F, Magnaghi-Jaulin L, Girault JA, et al. Histone acetyltransferase activity of CBP is controlled by cycle-dependent kinases and oncoprotein E1A. Nature. 1998;396:184–186. doi: 10.1038/24190. [DOI] [PubMed] [Google Scholar]
- Boyd JM, Loewenstein PM, Tang QQ, Yu L, Green M. Adenovirus E1A N-terminal amino acid sequence requirements for repression of transcription in vitro and in vivo correlate with those required for E1A interference with TBP-TATA complex formation. J Virol. 2002;76:1461–1474. doi: 10.1128/JVI.76.3.1461-1474.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner C, Deplus R, Didelot C, Loriot A, Vire E, De Smet C, et al. Myc represses transcription through recruitment of DNA methyltransferase corepressor. Embo J. 2005;24:336–346. doi: 10.1038/sj.emboj.7600509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti D, Ogryzko V, Kao HY, Nash A, Chen H, Nakatani Y, et al. A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell. 1999;96:393–403. doi: 10.1016/s0092-8674(00)80552-8. [DOI] [PubMed] [Google Scholar]
- de Bustros A, Nelkin BD, Silverman A, Ehrlich G, Poiesz B, Baylin SB. The short arm of chromosome 11 is a ‘hot spot’ for hypermethylation in human neoplasia. Proc Natl Acad Sci USA. 1988;85:5693–5697. doi: 10.1073/pnas.85.15.5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplus R, Brenner C, Burgers WA, Putmans P, Kouzarides T, de Launoit Y, et al. Dnmt3L is a transcriptional repressor that recruits histone deacetylase. Nucleic Acids Res. 2002;30:3831–3838. doi: 10.1093/nar/gkf509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch SM, Mymryk JS. Adenovirus-5 E1A: paradox and paradigm. Nat Rev Mol Cell Biol. 2002;3:441–452. doi: 10.1038/nrm827. [DOI] [PubMed] [Google Scholar]
- Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- Fuks F. DNA methylation and histone modifications: teaming up to silence genes. Curr Opin Genet Dev. 2005;15:490–495. doi: 10.1016/j.gde.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Gowher H, Liebert K, Hermann A, Xu G, Jeltsch A. Mechanism of stimulation of catalytic activity of Dnmt3A and Dnmt3B DNA-(cytosine-C5)-methyltransferases by Dnmt3L. J Biol Chem. 2005;280:13341–13348. doi: 10.1074/jbc.M413412200. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Laird PW, Jackson-Grusby L, Fazeli A, Dickinson SL, Jung WE, Li E, et al. Suppression of intestinal neoplasia by DNA hypomethylation. Cell. 1995;81:197–205. doi: 10.1016/0092-8674(95)90329-1. [DOI] [PubMed] [Google Scholar]
- McCance DJ. Transcriptional regulation by human papillomaviruses. Curr Opin Genet Dev. 2005;15:515–519. doi: 10.1016/j.gde.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Mikovits JA, Young HA, Vertino P, Issa JP, Pitha PM, Turcoski-Corrales S, et al. Infection with humanimmunodeficiency virus type 1 upregulates DNA methyltransferase, resulting in de novo methylation of the gamma interferon (IFN-gamma) promoter and subsequent downregulation of IFN-gamma production. Mol Cell Biol. 1998;18:5166–5177. doi: 10.1128/mcb.18.9.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordway JM, Fenster SD, Ruan H, Curran T. A transcriptome map of cellular transformation by the fos oncogene. Mol Cancer. 2005;4:19. doi: 10.1186/1476-4598-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan S, Bacolla A, Wells RD, Roberts RJ. Recombinant human DNA (cytosine-5) methyltransferase I. Expression, purification, and comparison of de novo and maintenance methylation. J Biol Chem. 1999;274:33002–33010. doi: 10.1074/jbc.274.46.33002. [DOI] [PubMed] [Google Scholar]
- Pradhan S, Kim GD. The retinoblastoma gene product interacts with maintenance human DNA (cytosine-5) methyltransferase and modulates its activity. EMBO J. 2002;21:779–788. doi: 10.1093/emboj/21.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson KD. DNA methylation, methyltransferases, and cancer. Oncogene. 2001;20:3139–3155. doi: 10.1038/sj.onc.1204341. [DOI] [PubMed] [Google Scholar]
- Sang N, Giordano A. Extreme N terminus of E1A oncoprotein specifically associates with a new set of cellular proteins. J Cell Physiol. 1997;170:182–191. doi: 10.1002/(SICI)1097-4652(199702)170:2<182::AID-JCP10>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Slack A, Cervoni N, Pinard M, Szyf M. DNA methyltransferase is a downstream effector of cellular transformation triggered by simian virus 40 large T antigen. J Biol Chem. 1999;274:10105–10112. doi: 10.1074/jbc.274.15.10105. [DOI] [PubMed] [Google Scholar]
- Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- Wang HG, Yaciuk P, Ricciardi RP, Green M, Yokoyama K, Moran E. The E1A products of oncogenic adenovirus serotype 12 include amino-terminally modified forms able to bind the retinoblastoma protein but not p300. J Virol. 1993;67:4804–4813. doi: 10.1128/jvi.67.8.4804-4813.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte P, Buchkovich KJ, Horowitz JM, Friend SH, Raybuck M, Weinberg RA, et al. Association between an oncogene and an anti-oncogene: the adenovirus E1A proteins bind to the retinoblastoma gene product. Nature. 1988;334:124–129. doi: 10.1038/334124a0. [DOI] [PubMed] [Google Scholar]
- Wu J, Issa JP, Herman J, Bassett DE, Jr, Nelkin BD, Baylin SB. Expression of an exogenous eukaryotic DNA methyltransferase gene induces transformation of NIH 3T3 cells. Proc Natl Acad Sci USA. 1993;90:8891–8895. doi: 10.1073/pnas.90.19.8891. [DOI] [PMC free article] [PubMed] [Google Scholar]