Abstract
Dimethylation of histone H3 arginine 2 (H3R2me2) maintains transcriptional silencing by inhibiting Set-1 mediated trimethylation of H3K4. Here we demonstrate that arginine 2 is also monomethylated (H3R2me1) in yeast but that its functional characteristics are distinct from H3R2me2: (a) H3R2me1 does not inhibit H3K4 methylation, (b) it is present throughout the coding region of genes and (c) it correlates with active transcription. Collectively, these results indicate that different H3R2 methylated states have defined roles in gene expression.
Keywords: chromatin modifications, histone, arginine methylation, H3R2me1
Covalent post-translational modifications of histones play a fundamental role in chromatin structure and function 1. Histone arginine methylation is one such modification that has been linked to transcriptional regulation 2. Arginines are methylated on the terminal guanidino nitrogens and can exist in three different methylation states; monomethylated (me1), symmetrically dimethylated (me2s) or asymmetrically dimethylated (me2a) 3. Studies in mammalian and yeast cells have demonstrated that histone arginine methylation can influence both gene activation and repression 4-7. For example, we have recently shown that asymmetric dimethylation of arginine 2 of H3 (H3R2me2a) in yeast contributes to transcriptional repression by inhibiting trimethylation of H3K4. Specifically, H3R2me2a inhibits H3K4me3 by blocking the PHD domain of Set1 complex component Spp1 from binding to methylated H3K4 and therefore, abrogating H3K4 trimethylation by the Set1 methyltransferase 8. Similarly in mammals, H3R2me2a, catalyzed by PRMT6, blocks binding of the WD40 domain of WDR5 to histone H3 thus inhibitng H3K4 trimethylation mediated by MLL1 9-11. Overall, these studies described a function for dimethyl-arginines in transcriptional regulation. However, the role of mono-methyl arginine still remains unexplored, even though this modification is detected in vivo on mammalian histones 6.
Using an antibody against the monomethylated form of H3R2 we show that this modification (H3R2me1) occurs on yeast histone H3 (Supplementary Fig. 1a). The antibody recognizes only arginine 2 on histone H3 as mutation of arginine to alanine (H3R2A) or glutamine (H3R2Q) abolishes the signal (Supplementary Fig. 1b). Furthermore, we demonstrate the specificity of this antibody towards the monomethylated version of H3R2 by dot blot and peptide-competition analyses (Supplementary Fig. 1c and d).
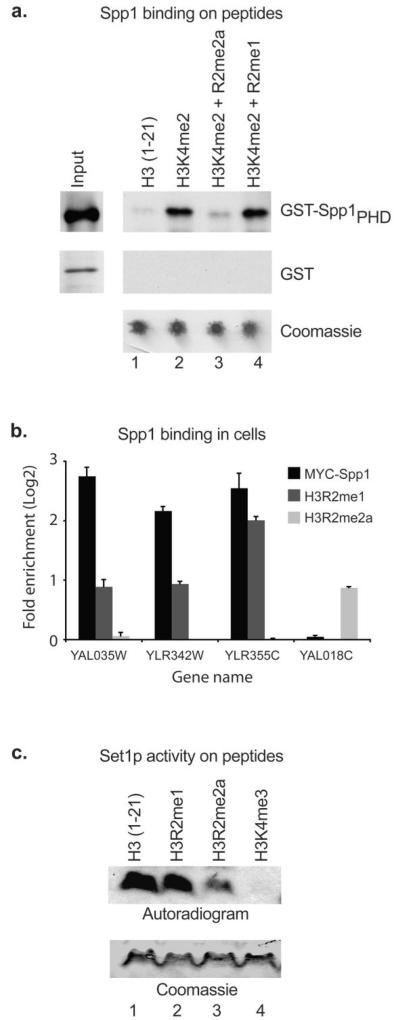
To determine whether H3R2me1 functions in a similar manner to H3R2me2a we first examined the effect of this methyl H3R2 state on the recruitment of Spp1 to H3K4 in vitro and in vivo. Figure 1A shows that in pull-down assays H3R2me1 does not block the interaction of the Spp1 PHD-finger with dimethylated H3K4 peptides (Fig 1A, lane 4). Consistently, ChIP assays show that in vivo Spp1 occupancy on the 5′-end of genes can coincide with H3R2me1 enrichment but not with the presence of H3R2me2a (Fig. 1b). We next sought to determine the effect of H3R2me1 on the ability of Set1 to trimethylate H3K4. In agreement with the above results, figure 1c shows that the purified Set1 complex is able to methylate an H3R2me1 peptide almost as well as an unmodified peptide. The specificity of the Set1 activity is demonstrated by the fact that an H3K4me3 peptide is not methylated while the presence of H3R2me2a reduces this activity (Fig. 1c, lanes 3 and 4). These results show that, H3R2me1 does not abrogate H3K4 trimethylation (unlike H3R2me2a) and suggest that H3R2 monomethylation has a distinct function from asymmetric dimethylation.
Figure 1. H3R2me1 does not block Set1 complex activity towards H3K4.
(a) Pull-down assays using synthesized peptides and recombinant GST-Spp1PHD or GST-only as a negative control. Equal loading of peptides is monitored by coomassie staining. (b) Chromatin immunoprecipitation (ChIP) analysis of logarithmically growing yeast cells using antibodies towards Myc-Spp1, H3R2me1 and H3R2m2a. Error bars represent s.e.m for duplicate experiments. (c) In vitro methyltransferase assays using purified Set1 complex and synthesized peptides. Equal amounts of peptides were used in the methyltransferase reactions as shown by coomassie staining.
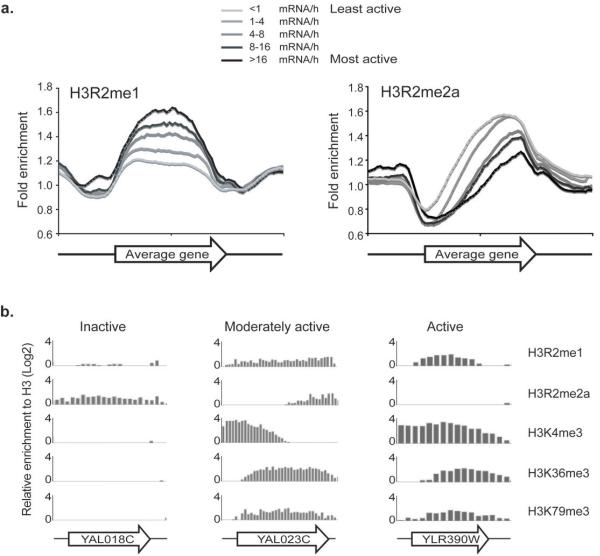
Having determined that H3R2me1 has functional characteristics distinct from H3R2me2a, we attempted to determine the role of H3R2me1 in transcription. We used a high-resolution genome-wide ChIP-chip analysis to determine the location of H3R2me1 occupancy and its relationship to gene expression (Supplementary Methods). We found that H3R2me1 is localized mainly within transcriptional units and is present in 85% of yeast genes (Supplementary Fig. 2). To determine the relationship of H3R2me1 to gene expression we first divided 5065 genes into five groups according to their transcriptional rate as previously determined 12 and then examined the average enrichment of H3R2me1 for each gene group (Fig. 2a). Average gene profiles of H3R2me1 indicated that this modification is present evenly throughout the entire coding region of transcriptionally active genes. The enrichment of H3R2me1 correlated with levels of transcriptional activity since the most active genes were most enriched in this modification (Fig. 2a, left panel). In contrast, analysis of previously reported data of H3R2me2a showed that this mark is present on the 3′-end of genes and correlates with transcriptional repression (Fig. 2a, right panel and ref. 8). Additionally, H3R2me2a covers inactive genes entirely (Fig. 2b, left panel). We next divided all genes into three differentially expressed categories (inactive, moderately active and active) in order to investigate the relationship of H3R2me1 with other histone methyl marks. Figure 2b shows that H3R2me1 overlaps to some extend with the active lysine methyl marks H3K4me3, K36me3, and K79me3 on representative moderately transcribed (YAL023C) and active (YLR390W) genes. Unlike H3R2me2a, the monomethylated form of this residue overlaps with H3K4me3 at the 5′end of moderately active genes (Fig. 2b, middle panel). Thus, together these results indicate that H3R2me1 coincides with all other active modifications on yeast genes and its correlation to transcription is opposite to that of H3R2me2a.
Figure 2. H3R2me1 associates with transcriptional activation.
(a) Composite profiles of ChIP-chip experiments. (b) ChIP-chip analysis compares the distribution of various histone methyl marks across three differentially expressed genes. The H3R2me2a and H3K4me3 data have been previously described 8.
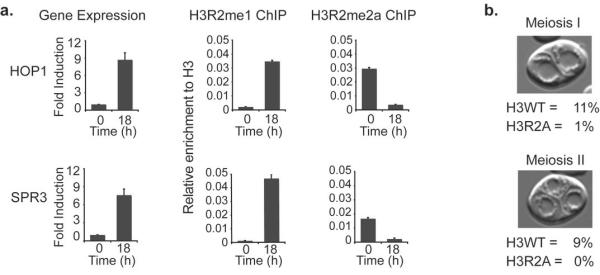
The above analyses suggested that H3R2me1 and H3R2me2a might have opposite roles in transcription. Therefore we next asked whether H3R2me1 and H3R2me2a are dynamically exchanged on nucleosomes upon induction of gene expression. To test this, we used the sporulation pathway as a model system of inducible gene activity. ChIP analysis of cells grown in rich media (repressed condition) showed high enrichment of H3R2me2a at sporulation genes (Hop1 and Spr3) as expected, while H3R2me1 was not detected at all on the same nucleosomes (Fig. 3a, time 0 hrs). Most significantly, shifting the cells in sporulation media which induced activation of these genes (Fig. 3a, left panels) completely reversed the levels of the two modifications at those same nucleosomes. H3R2me1 was robustly enriched but there were no traces of H3R2me2a (Fig. 3a, time 18 hrs). These results confirm that the monomethyl state of H3R2 associates with transcriptional activation.
Figure 3. H3R2 is necessary for sporulation.
(a) Gene expression analysis and ChIP experiments during induction of sporulation. Gene expression levels were normalised to a gene, HSD1, whose transcription remains unchanged before and after sporulation. Error bars represent s.e.m for duplicate experiments. (b) Sporulation assays of H3WT and H3R2A strains.
Since methylation at H3R2 associated with sporulation genes we next sought to determine whether H3R2 is necessary for sporulation in yeast. We grew cells that expressed either wild-type histone H3 (H3WT) or the mutant H3R2A in sporulation media for 7 days and then using microscopy we counted the number of cells that have undergone meiosis I or meiosis II. Mutation of H3R2 to alanine resulted in a severe defect of sporulation as only 1% of cells managed to undergo meiotic nuclear divisions, as opposed to 20% of H3WT cells (Fig. 3b). This result is consistent with the dynamic regulation of H3R2 methylation on sporulation genes and suggests that H3R2 plays an important role in the early stages of the sporulation process in yeast.
In summary, this report unveils for the first time a role for a mono-methyl arginine state in transcription. The results presented here provide evidence that H3R2me1 is a methylation state that occurs in vivo on yeast nucleosomes. The presence of H3R2me1 correlates with transcriptional activity, which is opposite to the relationship of H3R2me2a with gene expression. Although both H3R2 methyl-states are enriched within the coding region of genes their distribution is also different: mono-methylation is enriched throughout the coding region of active genes whereas di-methylation is enriched throughout inactive genes and towards the 3′-end of active genes. The distinct distribution patterns of mono- and di-methylation are a strong indicator that these two modifications are associated with different functions.
The functional difference amongst these H3R2 methyl states is probably conserved in higher eukaryotes. Recent findings show that PRMT6, the predominant H3R2 methyltransferase, catalyzes preferentially asymmetric dimethylation implying the existence of a distinct enzyme that carries out monomethylation 10,13. The need for two separate enzymes to catalyze these modifications suggests that possibly these two methyl states function differently. The identity of an enzyme that would catalyze exclusively H3R2me1 in mammals or yeast remains elusive. Combinatorial deletions of the three putative yeast arginine methyltransferases (Rmt1, Rmt2, Hsl7) and individual deletions of 35 other yeast methyltransferases do not affect the levels of this modification (data not shown and Supplementary Table 1).
The precise function of H3R2me1 in the process of transcriptional activation remains to be resolved. There are at least two distinct mechanisms by which H3R2me1 can function in gene expression. In one model, the mono-methyl state is a “passive” mark on chromatin which is used to identify actively transcribed regions that need to be silenced. In this model, the mono-methyl mark is deposited on active genes in order to allow subsequent di-methylation and consequent repression of transcription. In a second model, mono-methylation dictates a function (such as the recruitment of a protein) that is necessary for genes to become or remain active. Such a model would be analogous to the recruitment of chromatin effectors by specific lysine methylation states 14,15. Future studies will aim to decipher the molecular mechanism employed by H3R2me1 during gene expression.
Supplementary Material
Acknowledgements
We thank members of the T.K. laboratory for helpful discussions and M. Gilchrist for help with depositing genomic data. This work was supported by postdoctoral fellowship grants to A.K. from the European Molecular Biology Organization (EMBO) and Marie Curie. The T.K. laboratory is funded by grants from Cancer Research UK (CRUK) and the 6th Research Framework Program of the European Union (Epitron and Heroic). The microarray data sets are available from GEO (Gene Expression Omnibus) under accession number GSE14453.
References
- 1.Kouzarides T. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Wysocka J, Allis CD, Coonrod S. Front Biosci. 2006;11:344–55. doi: 10.2741/1802. [DOI] [PubMed] [Google Scholar]
- 3.Turner BM. Nat Struct Mol Biol. 2005;12:110–2. doi: 10.1038/nsmb0205-110. [DOI] [PubMed] [Google Scholar]
- 4.Bauer UM, Daujat S, Nielsen SJ, Nightingale K, Kouzarides T. EMBO Rep. 2002;3:39–44. doi: 10.1093/embo-reports/kvf013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma H, et al. Curr Biol. 2001;11:1981–5. doi: 10.1016/s0960-9822(01)00600-5. [DOI] [PubMed] [Google Scholar]
- 6.Strahl BD, et al. Curr Biol. 2001;11:996–1000. doi: 10.1016/s0960-9822(01)00294-9. [DOI] [PubMed] [Google Scholar]
- 7.Wang H, et al. Science. 2001;293:853–7. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- 8.Kirmizis A, et al. Nature. 2007;449:928–32. doi: 10.1038/nature06160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guccione E, et al. Nature. 2007;449:933–7. doi: 10.1038/nature06166. [DOI] [PubMed] [Google Scholar]
- 10.Hyllus D, et al. Genes Dev. 2007;21:3369–80. doi: 10.1101/gad.447007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iberg AN, et al. J Biol Chem. 2007 [Google Scholar]
- 12.Holstege FC, et al. Cell. 1998;95:717–28. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 13.Lakowski TM, Frankel A. J Biol Chem. 2008 doi: 10.1074/jbc.M710176200. [DOI] [PubMed] [Google Scholar]
- 14.Ruthenburg AJ, Allis CD, Wysocka J. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 15.Sims RJ, 3rd, Reinberg D. Genes Dev. 2006;20:2779–86. doi: 10.1101/gad.1468206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.