Abstract
The molecular chaperone HSP90 maintains the activity and stability of a diverse set of “client” proteins that play key roles in normal and disease biology. Around 20 HSP90 inhibitors that deplete the oncogenic clientele have entered clinical trials for cancer. However, the full extent of the HSP90-dependent proteome, which encompasses not only clients but also proteins modulated by downstream transcriptional responses, is still incompletely characterized and poorly understood. Earlier large-scale efforts to define the HSP90 proteome have been valuable but are incomplete because of limited technical sensitivity. Here, we discuss previous large-scale surveys of proteome perturbations induced by HSP90 inhibitors in light of a significant new study using state-of-the-art stable isotope labeling by amino acids (SILAC) technology combined with more sensitive high-resolution mass spectrometry (MS) that extends the catalog of proteomic changes in inhibitor-treated cancer cells. Among wide-ranging changes, major functional responses include downregulation of protein kinase activity and the DNA damage response alongside upregulation of the protein degradation machinery. Despite this improved proteomic coverage, there was surprisingly little overlap with previous studies. This may be due in part to technical issues but is likely also due to the variability of the HSP90 proteome with the inhibitor conditions used, the cancer cell type and the genetic status of client proteins. We suggest future proteomic studies to address these factors, to help distinguish client protein components from indirect transcriptional components and to address other key questions in fundamental and translational HSP90 research. Such studies should also reveal new biomarkers for patient selection and novel targets for therapeutic intervention.
Key words: HSP90, HSP90 proteome, HSP90 inhibitors, HSP90 biomarkers, cancer
Introduction
Heat-shock protein 90 (HSP90) is an extraordinarily versatile molecular chaperone with important roles in healthy cell protein homeostasis (proteostasis) and also in the pathology of many diseases.1,2 These include cancer, Alzheimer disease, Parkinson disease and prion disease as well as viral and protozoan infections. Pharmacologic HSP90 inhibitors are in clinical development for cancer treatment. In an informative new article in Molecular Cell Proteomics, Sharma et al. present a quantitative proteomics study designed to map global cellular changes in protein levels upon treatment with a pharmacological inhibitor of HSP90. Using state-of-the-art stable isotope labeling by amino acids (SILAC) technology combined with high-resolution mass spectrometry (MS), Sharma et al. have been able to provide us with a much more detailed picture than was available previously of how the cellular proteome responds to HSP90 inhibition.
Summarizing their main findings, in addition to significantly expanding the “HSP90 proteome,” Sharma et al. also demonstrate, in the most sensitive and systematic analysis to date, that HSP90 inhibition preferentially affects kinases and the DNA damage response. Furthermore, a follow-up global analysis of protein phosphorylation reveals that a much greater proportion of the phosphoproteome is decreased than is increased in response to HSP90 inhibitor treatment. In this Perspective, we highlight the advances made by the Sharma et al. study over earlier proteome-wide surveys of the effects of HSP90 inhibition. In light of this, we discuss topical issues in the HSP90 field, placing particular emphasis on therapeutic insights that might be obtained through further analysis of the HSP90-dependent proteome. Finally, we propose how global genomic and proteomic approaches can be used in the future to address important remaining questions in basic and therapeutic research on HSP90 and its pharmacological inhibitors.
Basic Research and Drug Development Interests
Over and above its importance to fundamental molecular, cellular and whole-organism research, HSP90 has attracted much recent interest in the field of cancer drug development, with some 20 inhibitors currently in clinical trials.4,5 The basis for this interest is HSP90's ability to facilitate both the activation and stabilization, through physical interaction, of a wide range of “client” proteins, many of which are involved in oncogenesis and malignant progression.1 Inhibition of HSP90 leads to the loss of this physical interaction and to ubiquitination and degradation of clients via the 26S proteasome (Fig. 1A). To date, over 200 proteins have been identified as clients of HSP90 (www.picard.ch/downloads/downloads.htm). However, the proteome-modulating scope of HSP90 inhibition is not limited to depletion of client proteins: since many clients are actively involved in a variety of signal transduction pathways, the loss of their activity ultimately leads to changes in gene expression programs controlled by the signaling output of those pathways; in addition, HSP90 inhibition causes activation of the heat shock response mediated by heat shock factor 1 (HSF1), leading to upregulation of many cytoprotective proteins (see later). Especially given the emerging clinical finding that cancers respond differently to HSP90 inhibitors depending on their molecular background,1 unbiased genome- and proteome-wide global approaches should help in the identification of molecular biomarkers to enable selection of patients most likely to respond to treatment with HSP90 inhibitors, as well as being of fundamental research value.6
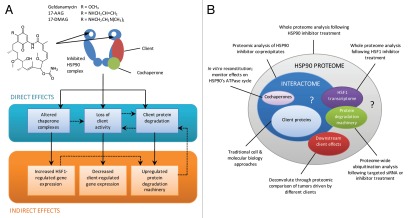
Figure 1.
(A and B) The proteomic response to pharmacological inhibition of HSP90. The molecular chaperone HSP90 functions in the cell as a dimer and interacts with regulatory cochaperones and client proteins. Most pharmacologic HSP90 inhibitors, including all of those currently in clinical trials, bind to the N-terminal nucleotide binding site, thereby inhibiting the essential ATPase activity, interrupting the chaperone cycle and blocking the function of the chaperone. The consequences may be separated into direct effects, e.g., those caused by disruption of the active HSP90 complex, as defined by the interactome, and indirect effects, e.g., downstream alterations to transcriptional programs. Note that inhibitor-induced alteration of HSP90 complexes, such as disruption of client and HSF1 interactions, are believed to lead to many of the indirect transcriptional effects. There is evidence for the interdependence of many aspects of the HSP90 proteome, as indicated by dotted arrows in (A) and overlapping circles in (B).
Previous Proteomic Studies
For the majority of the proteins known to interact physically with HSP90, either as client proteins or cochaperones, the evidence for this binding was obtained from conventional small-scale biochemical studies in which they were detected as either co-purifying in a complex with HSP90 or pulled-down with it in immunoprecipitation assays. Putative protein clients have also been detected by observing their depletion following the treatment of cells with HSP90 inhibitors, typically using western blotting. Hartson and Matts7 have recently reviewed more than 23 studies in which higher-throughput, unbiased survey approaches have been taken to detect proteins interacting directly or indirectly with HSP90. These studies have commonly involved genetic, chemical genetic and physical interaction screens performed in yeast8–14 as well as large-scale analyses, commonly utilizing affinity purification of HSP90, of proteins present in HSP90 complexes.7,15–20
Only a few efforts have been made previously toward characterizing the global cellular effects of HSP90 inhibition in mammalian cells. The first published report by Clarke et al.21 used mRNA expression profiling, by cDNA microarray, to characterize in an unbiased fashion the effects in a panel of human colon cancer cell lines of treatment with the first clinically evaluated HSP90 inhibitor, the geldanamycin analog 17-AAG (tanespimicin). Among the effects seen were the upregulation of several heat-shock proteins and molecular chaperones;21 this HSF1-mediated induction of the heat shock response—via release of HSP90-mediated inhibition of the transcription factor—is now considered a bona fide component of the molecular signature of HSP90 inhibition and is used alongside client protein depletion to demonstrate target engagement in clinical trials.22,23 Moreover, since many of the heat-shock proteins are anti-apoptotic, this response limits the effectiveness of HSP90 inhibitors.24 Clarke et al. also observed altered expression at the mRNA level of genes encoding keratin 8, keratin 18 and caveolin-1, the last of which was confirmed at the protein level.21
More recent large-scale unbiased profiling campaigns have incorporated advances in quantitative proteomics to determine alterations at the protein level. The benefit of such approaches is that not only do they provide information on functional protein expression, but they also take into account posttranslational changes; for example, protein degradation and protein phosphorylation can be cataloged. The first such study by Maloney et al.25—which monitored global changes in both mRNA and protein levels induced in human ovarian cancer cells by treatment with 17-AAG—was notable in its use of an inactive analog of 17-AAG, together with the chemically distinct HSP90 inhibitor radicicol, employed as control compounds in order to discount off-target expression changes. Proteomic analysis was done by two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) followed by protein identification through matrix-assisted laser desorption/ionization mass spectrometry (MALDI MS) of excised spots. The profiling studies revealed that around 3% of the total gene transcripts examined and 4% of the detectable proteins were responsive to 17-AAG treatment. Highlighted in the proteomic analysis were the induction of heat-shock proteins and the HSP90 ATPase-activating protein AHA1. Of further interest and novelty were changes in expression of chromatin-associated proteins. Expression of heterochromatin protein 1 HnRNP3 (hnRNP 2H9A), a member of the heterogeneous nuclear ribonucleoprotein family, was increased, while levels of histone acetyltransferase-1 (HAT-1) and the protein arginine methyl transferase PRMT5 were reduced by 17-AAG treatment. The key protein changes were confirmed by western blots and were accompanied by a decrease in cellular protein histone acetylation, and therapeutic antagonism was reported between 17-AAG and a histone deacetylase inhibitor.25 Binding of PRMT5 to HSP90 was shown by co-immunoprecipitation, suggesting PRMT5 as a possible new HSP90 client.25
In the same year, Schumacher et al. published their results of a proteomic analysis—using liquid chromatography tandem mass spectrometry (LC-MS/MS) in combination with cleavable isotope-coded affinity tag (cICAT™) labeling—of geldanamycin-treated ALK-positive human anaplastic large-cell lymphoma cells. Of the 176 cICAT-labeled peptides, 119 were differentially expressed by 1.5-fold or greater 12 h after geldanamycin treatment, with 59% of these being downregulated. Ingenuity pathway analysis of differentially expressed proteins revealed deregulation of a variety of pathways, including integrin, estrogen receptor, MAPK, JAK/STAT, PPAR and NFκB signaling as well as ubiquitin-mediated proteolytic and cell cycle-regulatory pathways. Interestingly, some previously identified HSP90 client proteins and interactors were found to be downregulated in the proteomic analysis and validated by western blotting, including NPM-ALK, STIP1, IKKκ, MAK and MCPH1.
Another LC-MS/MS study, this time using iTRAQ (isobaric tag for relative and absolute quantitation)-labeled peptides, tracked proteomic changes in two human pancreatic cancer cell lines treated with the more water-soluble geldanamycin analog IPI-504.27 The investigators identified 20 downregulated and 42 upregulated proteins common to both cell lines. Analysis of these proteins in the human protein database revealed most downregulated proteins to be involved in macromolecule metabolism, cell localization, cell growth and maintenance and transport, whereas the upregulated proteins were mostly involved in cellular metabolism and cell organization and biogenesis.
A recent global HSP90 inhibitor study was reported by Yao et al. in human retinal pigment epithelial cells treated with 17-AAG. They performed MALDI-MS on proteins separated by 2D-PAGE, in a manner similar to Maloney et al.25 and identified 87 proteins with altered expression. Ingenuity pathway analysis of these proteins revealed a strong effect on oxidative stress pathways. Additionally, canonical pathway analysis found glycolysis/gluconeogenesis networks to be deregulated.
Each of these large-scale surveys has provided extensive and important new information on the HSP90 inhibition proteome. However, although the Maloney et al. study identified the protein arginine methyl transferase PRMT5 as a potential new client protein,25 none of the global profiling campaigns were able to identify key established clients of HSP90. This was presumably because the proteomic methods used tend to detect only the most abundant proteins, whereas the majority of well-categorized clients of HSP90 are signaling proteins—exemplified by protein kinases—that are generally expressed at low levels in cells.
Expanding the HSP90 Proteome Using SILAC Technology
In order to address the sensitivity issue, Sharma et al. combined SILAC technology and high resolution LC-MS/MS to more fully catalog the HSP90 proteome and in particular to determine systematically and in an unbiased manner the proteins and pathways affected by the HSP90 inhibitor 17-DMAG (alvespimicin, another clinically trialed drug closely related to 17-AAG) in the HeLa human cervical cancer cell line. From a technical point of view, their data exhibit a high level of consistency, with 85% of protein hits being found in at least 3 of their 5 biological replicates. Of the 6,000 proteins that could be accurately quantified, 724 (12.4%) showed a decreased abundance, while 97 (1.7%) exhibited an increased abundance, using a cut-off of at least a 1.5-fold change compared with untreated controls. As well as obtaining substantially more proteomic coverage than any previous HSP90 inhibition study, this new analysis also provides the clearest and most extensive demonstration to date that blockade of the molecular chaperone leads to significantly more down- than upregulation of cellular proteins, most likely due to the now well-established destabilization of client proteins and their subsequent proteasomal degradation.
Consistent with that hypothesis and endorsing the technology used, among the downregulated proteins identified by Sharma et al. are numerous previously identified bona fide protein kinase clients of HSP90, including ARAF, AKT, CDK4, MET and PDK1. Furthermore, using a bioinformatic process known as 1D annotation enrichment analysis, the authors identified “protein kinase activity” as one of the most significantly downregulated functional categories (p < 10−13). In agreement with this, Sharma et al. performed a global analysis of 4,000 protein phosphorylation sites, 34% of which were found to be downregulated more than 2-fold, as opposed to only 6% that were upregulated upon HSP90 inhibition. Interestingly, the overall level of kinase downregulation in the total proteome was fairly modest (indicated by a moderately negative difference value in their 1D annotation enrichment analysis). In addition to the overall analytical sensitivity limitation mentioned earlier, this provides a further explanation for why protein kinases were was not identified by previous, less-sensitive proteome analysis methods. Also of interest was that more than half of the downregulated protein phosphorylation events involved “proline-directed” motifs, indicating important effects on mitogen-activated or cyclin-dependent kinases.
Comparisons and Interpretation
Despite the large number of inhibitor-modulated hits obtained by Sharma et al., surprisingly few of the proteins identified by previous proteomic surveys25–28 were reproduced in this new study. Comparison of the proteins the levels of which were changed by more than 1.5-fold revealed that only 8 of the 23 detected by Maloney et al.,25 9 of the 97 found by Schumacher et al.,26 5 of the 34 from Song et al.27 and 3 of the 87 from Yao et al.28 were also present in the Sharma et al. catalog. No proteins were common to more than 3 of the 5 studies. In fact, many of the common hits were affected in opposite ways in the different studies. Such discordance was also found in three genome-wide analyses of HSP90s “interactome” in yeast.29 While the limited overlap and disparities observed both in yeast and in human cells might be caused at least in part by technical differences between the various studies, such as the use of different inhibitors and cell types; it might also indicate the varying nature of the HSP90-dependent proteome in different biological contexts.
It is possible that such biological variability represents the different ways in which a cell can respond to HSP90 inhibition, depending on its molecular circuitry. On the other hand, despite the few hits shared by the analyses of Schumacher et al. and Sharma et al., both studies found significant upregulation of proteins involved in protein degradation and the 26S proteasome. It is well established that HSP90 inhibition leads to the ubiquitin-dependent proteasomal degradation of HSP90 client proteins. Indeed, one analysis of the ubiquitinated proteome following concomitant HSP90 and proteasome inhibition identified numerous known HSP90 interactors.30 However, with few exceptions, the degradative molecular components involved are as yet poorly characterized. An interesting possibility is that different clients are acted upon by different degradative proteins, e.g., E3 ubiquitin ligases.31 Thus, variation in the expression or role of the various elements of the protein degradative machinery between the distinct model systems studied could contribute to differential impacts on HSP90's clientele following chaperone inhibition.
Another key contributory factor in cancer cells is likely to be the molecular status of its HSP90 clients, in particular whether any oncoprotein clients are mutated or overexpressed. It is now clear that many amplified or mutated oncoproteins are more dependent on HSP90 than the wild-type equivalents, as in the case of BRAF.32,33 Extrapolating this thinking to the clinic is helping to explain why patients with tumors that are driven by particular genetically activated oncoprotein clients of HSP90—e.g., amplified and overexpressed ERBB2 in many breast cancers and the ELM4-ALK fusion protein in a proportion of non-small cell lung cancers—are able to respond especially well to HSP90 inhibitors.1 Moreover, whereas previous research has focused on the oncoprotein clients, an improved understanding of which parts of the degradation machinery are involved with which particular HSP90 clients might enable us to predict even better how patients will respond to HSP90 inhibitors and would provide a basis for potential combinatorial therapeutic strategies involving specific proteasomal components as additional drug targets. In support of such an approach, bortezomib is already approved in multiple myeloma, and further proteasome inhibitors are in development.34,35 Furthermore, combinations of bortezomib with HSP90 inhibitors are showing clinical promise in multiple myeloma, a cancer in which buffering of proteotoxic stress is especially critical for survival.1,5
A further important aspect brought to light by genome-wide surveys is the extensive functional diversity of proteins affected by HSP90 inhibition. Although the Sharma et al. analysis is the first of its kind to confirm at the whole-proteome level what was already known through individual candidate studies to be a high preponderance of kinases within the proteins affected by HSP90 inhibition (www.picard.ch/downloads/downloads.htm), the multitude of other affected protein classes lends further weight to the developing view that HSP90 should not be regarded only as a chaperone of signaling proteins, as originally thought,36 but rather has substantially more far-reaching cellular roles, including, for example, glucose metabolism, ribonucleoprotein (RNP) assembly and the DNA damage response.7 Indeed, aside from “protein kinase activity,” Sharma et al. identified “DNA metabolic processes,” including proteins involved in DNA repair, as the other most significantly downregulated functional category from their bioinformatic analysis, an observation that is consistent with the preclinical findings that HSP90 inhibitors sensitize cancer cells to radiation through inhibition of base excision repair.37
Whether these various far-reaching effects exemplified above can all be explained as downstream consequences of client protein signaling will remain an area for future research. One must be very careful when interpreting mechanistically and categorizing the changes in expression seen in large-scale proteomic surveys of drug effects. For example, in the present case, it is difficult to determine whether the changes seen are direct or indirect consequences of HSP90 inhibition, and to see in what way these effects are inter-related. As a case in point, the upregulated levels of components of the proteasomal pathway observed by Sharma et al. are likely caused, at least in part, by inhibitor-induced destabilization of HSP90 clients, which include many protein kinases. Therefore, rather than classifying kinases and proteasomal proteins as distinct components of the response to HSP90 inhibition, they could alternatively be considered conceptually as intrinsically linked, albeit opposing, parts of the HSP90-dependent proteome. Furthermore, many of the proteomic changes observed, including upregulation of the protein ubiquitination and degradation machinery, are likely to be mediated through activation of HSF1,38 as further emphasized by the observation that about a quarter of the proteins that were at least 1.5-fold upregulated in the Sharma et al. analysis were identified by the authors as the products of HSF1-dependent genes.
The challenge in classifying different parts of the HSP90 inhibitor response highlights a general limitation of the global proteomic approach when used in isolation: that is, its inability to distinguish direct relationships, e.g., interactions of HSP90 with cochaperones and clients, from indirect ones, e.g., transcriptional effects mediated by client proteins and HSF1 (Fig. 1A and B). One recent study used MS analysis of proteins chemically precipitating with the purine-class HSP90 inhibitor PU-H71 to specifically identify proteins in HSP90 complexes that were being inhibited.20 Such large-scale analysis of HSP90 complexes could be employed alongside more global cellular proteomic approaches to help define the immediate HSP90 “interactome” within the more general HSP90-dependent proteome.
Further Questions and Future Challenges
While the significant extension of the HSP90 proteome afforded by Sharma et al. will be welcomed as a valuable resource by the HSP90 research community, as always with such global surveys, the results leave open or create many questions that must be answered by further research. Some additional, specific follow-up proteomic experiments are recommended by the present authors: drug concentration-response studies, particularly using 17-DMAG concentrations lower than the 50 µM employed by Sharma et al., since the average peak concentrations achieved at the recommended Phase II dose was only 2.6 µM;39 time-dependence studies, especially looking at earlier responses than those seen at the single 24 h time-point used by Sharma et al.; confirmation of the effects seen with 17-DMAG—the development of which is now discontinued—using alternative HSP90 inhibitors with different chemical scaffolds and that currently show promise in the clinic;1,4,5 alongside these alternative chemotypes, use of inactive analogs as negative controls to help exclude off-target effects,40 as employed by Maloney et al.;25 and, finally, studies to compare normal responses in multiple cancer and normal non-tumorigenic cell lines. This is a large body of work, and is important to help establish the universality or otherwise of the proteome-wide effects reported.
Importantly, confirmation of new protein hits is necessary, especially given the common lack of reproducibility and validation data seen in such proteomic surveys, potentially due to labeling errors in databases.7,41,42
The biological and clinical significance of the effects on particular biological pathways and candidate proteins identified by Sharma and colleagues will also need to be explored further (e.g., see Fig. 2). Potentially contributing to the combinatorial therapeutic benefits of HSP90 inhibition are effects observed on the WNT receptor tyrosine kinase ROR2 and on ADAMTS1, which is involved in angiogenesis, as well as those seen on proteins involved in sphingolipid metabolism. Possible disadvantageous effects requiring further work include those on the tumor suppressor LRIG1, which acts as a feedback negative regulator of signaling by receptor tyrosine kinases, through a mechanism that involves enhancement of receptor ubiquitination and accelerated intracellular degradation, and on TM4SF1, which is a member of the transmembrane 4 or tetraspanin family and is involved in angiogenesis, invasion and metastasis.
Figure 2.
Statistically significant pathways found to be modulated in the Sharma et al. analysis, ordered from most significant (top) to least significant (bottom). The ‘s’ score (between -1 and 1) indicates where the center of the distribution of values for the protein category is located relative to the overall distribution of values. Negative ‘s’ scores (indicating downregulation) are shown in green; positive ‘s’ scores (indicating upregulation) are shown in red.
Future applications of the sensitive proteomic analytical technique used by Sharma et al. should allow researchers to address several of the key questions that have been cause for much speculation in the HSP90 field. For example, the complete cataloging of the HSP90 clientele might shed light on the molecular factors that confer dependence of a given protein on chaperoning by HSP90—probably the most frustrating and perplexing fundamental question for the HSP90 community.
An additional key question that could be addressed by proteomic technology is the role of the different HSP90 family members. Although Sharma et al. provide some indirect evidence to suggest a predominant role for the protein changes induced by 17-DMAG being mediated through the cytosolic HSP90α and β isoforms with a lesser involvement for the endoplasmic reticulum family member GRP94, there is now a need to investigate these isoforms in more detail together with the mitochondrial member TRAP1.43–46 Indeed, proteomic technology could certainly illuminate the as yet poorly defined differences and similarities in the fundamental biological functions of the four family members in healthy and diseased cells, as well as their relative contributions as targets of the many inhibitors that are currently undergoing development.
Furthermore, by comparing differences in the HSP90-dependent proteome between cancer cells and normal cells, it might be possible to determine the precise biochemical basis for the tumor selectivity of HSP90 inhibitors that is apparently associated with the differential make-up of HSP90 complexes in cancer vs. normal cells, first reported over 8 y ago47 but still not fully understood at the fundamental molecular level. We could envisage using sensitive proteomic profiling to obtain a much more detailed picture of HSP90 inhibitor effects in anatomically and genetically different cancer cell types, potentially providing additional important biomarkers for prediction of sensitivity and enabling patient selection as part of modern personalized treatment.48
Yet another key area of current interest that could be probed in more detail by large-scale proteomics is the role of the HSF1-regulated heat shock program in cancer cells, especially given the recent finding that high nuclear levels of HSF1 are associated with poor prognosis in a cohort study of almost 2,000 breast cancer patients.49 Direct comparison of proteomic changes caused by HSF1 inhibition or knockdown with those associated with HSP90 inhibition or, alternatively, proteomic analysis of cells genetically lacking HSF1, such as Hsf1-/- mouse embryonic fibroblasts, treated with an HSP90 inhibitor could identify in more precise detail what proportion of the proteomic response to HSP90 inhibition is due to activation of HSF1, and could thereby help provide a rationale for combinatorial therapies targeting both HSP90 and HSF1.
Concluding Remarks
Thus, while it is fascinating for us to observe the ever-expanding proteome of the HSP90 molecular chaperone, there remains much mechanistic research to be performed—both to interpret the impressive catalog of HSP90s proteomic associations (direct and indirect) and also to exploit the plethora of emerging new findings for therapeutic intervention in diseases in which pathogenic alterations in the proteostasis network are involved. A final comment is to encourage—even in diseases where pathogenic proteostasis is the main driver—the integration of proteomic approaches with other large-scale interrogations, including genome sequencing, transcriptomics, epigenomics and metabolomics, so that the maximal value and understanding can be realized.
Acknowledgments
The authors thank members of the Workman lab and many colleagues and collaborators for helpful discussions. Paul Workman is a Cancer Research UK Life Fellow [grant number C309/A8992]. Paul Workman and Paul Clarke acknowledge grant funding from Cancer Research UK [grant number C309/A8274]. Rahul Samant was supported by a PhD studentship from Cancer Research UK [grant number C309/A8140]. The authors acknowledge funding from the NHS to the NIHR Biomedical Research Centre.
Note Added in Proof
A very recent paper (Wu Z, et al. Systematic identification of the HSP90 regulated proteome. Mol Cell Proteomics 2012; PMID: 22337586) described another large-scale analysis of proteome-wide changes in four human cancer cell lines following exposure to geldanamycin, as measured by SILAC and quantitative MS. The authors showed that, of 288 identified protein kinases, 98 were downregulated and 17 were upregulated in response to HSP90 inhibition, including, they suggested, >50 kinases not formerly known to be regulated by HSP90. The upregulated kinases included several involved in cell cycle control (e.g., (AURA, PBK, PKAb, NEK2, ERK3, TTk, PKR) and two involved in stress responses (OSR1, MST3). Interestingly, protein turnover studies using pulsed SILAC indicated that, for a significant number of proteins the rate of protein depletion correlated with the protein half-life in untreated cells, with protein kinases showing significantly faster turnover than other proteins. The proteomic response to geldanamycin was shown to be similar to that for the alternative purine chemotype and clinical developmental HSP90 inhibitor PU-H71. Application of HSP90 immunoprecipitation and pulldowns with immobilized geldanamycin indicated that several kinases (AXL, DDR1, TRIO) and other signalling proteins (BIRC6, ISG15, FLII) were HSP90 client proteins. Although differences in response were seen between the four different cancer cell lines, the most significant common gene ontology categories affected were, not surprisingly, the unfolded protein response for upregulated proteins and kinase activity for depleted proteins, followed by cell cycle and response to DNA damage. Of note, of the proteins responding to HSP90 inhibition in the Wu et al. study, ∼500 were reported to be in common with those observed by Sharma et al.
Disclosure of Potential Conflict of Interest
All the authors are or were employees or students of the Institute of Cancer Research, which has a commercial interest in HSP90 inhibitors and operates a reward to inventors scheme. Paul Workman received research funding from Vernalis for the discovery of HSP90 inhibitors, and intellectual property for this program was licensed to Vernalis Ltd., and Novartis. He has also been involved in a research collaboration with AstraZeneca in the area of stress and chaperone pathways. Paul Workman has been a consultant to Novartis, is a founder of Chroma Therapeutics and is scientific advisory board member of Astex Pharmaceuticals and Nextech Invest.
References
- 1.Neckers L, Workman P. Hsp90 molecular chaperone inhibitors: are we there yet? Clin Cancer Res. 2012;18:64–76. doi: 10.1158/1078-0432.CCR-11-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 3.Sharma K, Vabulas RM, Macek B, Pinkert S, Cox J, Mann M, et al. Quantitative proteomics reveals that Hsp90 inhibition preferentially targets kinases and the DNA damage response. Mol Cell Proteomics. 2011 doi: 10.1074/mcp.M111.014654. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jhaveri K, Taldone T, Modi S, Chiosis G. Advances in the clinical development of heat-shock protein 90 (Hsp90) inhibitors in cancers. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbamcr.2011.10.008. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Travers J, Sharp S, Workman P. HSP90 inhibition: two-pronged exploitation of cancer dependencies. Drug Discov Today. 2011 doi: 10.1016/j.drudis.2011.12.021. In press. [DOI] [PubMed] [Google Scholar]
- 6.da Silva VC, Ramos CH. The network interaction of the human cytosolic 90 kDa heat-shock protein Hsp90: A target for cancer therapeutics. J Proteomics. 2012 doi: 10.1016/j.jprot.2011.12.028. In press. [DOI] [PubMed] [Google Scholar]
- 7.Hartson SD, Matts RL. Approaches for defining the Hsp90-dependent proteome. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbamcr.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kimura Y, Yahara I, Lindquist S. Role of the protein chaperone YDJ1 in establishing Hsp90-mediated signal transduction pathways. Science. 1995;268:1362–1365. doi: 10.1126/science.7761857. [DOI] [PubMed] [Google Scholar]
- 9.Nathan DF, Vos MH, Lindquist S. Identification of SSF1, CNS1 and HCH1 as multicopy suppressors of a Saccharomyces cerevisiae Hsp90 loss-of-function mutation. Proc Natl Acad Sci USA. 1999;96:1409–1414. doi: 10.1073/pnas.96.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao R, Davey M, Hsu YC, Kaplanek P, Tong A, Parsons AB, et al. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell. 2005;120:715–727. doi: 10.1016/j.cell.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 11.Millson SH, Truman AW, King V, Prodromou C, Pearl LH, Piper PW. A two-hybrid screen of the yeast proteome for Hsp90 interactors uncovers a novel Hsp90 chaperone requirement in the activity of a stress-activated mitogen-activated protein kinase, Slt2p (Mpk1p) Eukaryot Cell. 2005;4:849–860. doi: 10.1128/EC.4.5.849-60.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao R, Houry WA. Molecular interaction network of the Hsp90 chaperone system. Adv Exp Med Biol. 2007;594:27–36. doi: 10.1007/978-0-387-39975-1_3. [DOI] [PubMed] [Google Scholar]
- 13.McClellan AJ, Xia Y, Deutschbauer AM, Davis RW, Gerstein M, Frydman J. Diverse cellular functions of the Hsp90 molecular chaperone uncovered using systems approaches. Cell. 2007;131:121–135. doi: 10.1016/j.cell.2007.07.036. [DOI] [PubMed] [Google Scholar]
- 14.Echeverría PC, Forafonov F, Pandey DP, Mühlebach G, Picard D. Detection of changes in gene regulatory patterns, elicited by perturbations of the Hsp90 molecular chaperone complex, by visualizing multiple experiments with an animation. BioData Min. 2011;4:15. doi: 10.1186/1756-0381-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falsone SF, Gesslbauer B, Tirk F, Piccinini AM, Kungl AJ. A proteomic snapshot of the human heat-shock protein 90 interactome. FEBS Lett. 2005;579:6350–6354. doi: 10.1016/j.febslet.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 16.Te J, Jia L, Rogers J, Miller A, Hartson SD. Novel subunits of the mammalian Hsp90 signal transduction chaperone. J Proteome Res. 2007;6:1963–1973. doi: 10.1021/pr060595i. [DOI] [PubMed] [Google Scholar]
- 17.Gong Y, Kakihara Y, Krogan N, Greenblatt J, Emili A, Zhang Z, et al. An atlas of chaperone-protein interactions in Saccharomyces cerevisiae: implications to protein folding pathways in the cell. Mol Syst Biol. 2009;5:275. doi: 10.1038/msb.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsaytler PA, Krijgsveld J, Goerdayal SS, Rüdiger S, Egmond MR. Novel Hsp90 partners discovered using complementary proteomic approaches. Cell Stress Chaperones. 2009;14:629–638. doi: 10.1007/s12192-009-0115-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gano JJ, Simon JA. A proteomic investigation of ligand-dependent HSP90 complexes reveals CHORDC1 as a novel ADP-dependent HSP90-interacting protein. Mol Cell Proteomics. 2010;9:255–270. doi: 10.1074/mcp.M900261-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moulick K, Ahn JH, Zong H, Rodina A, Cerchietti L, Gomes DaGama EM, et al. Affinity-based proteomics reveal cancer-specific networks coordinated by Hsp90. Nat Chem Biol. 2011;7:818–826. doi: 10.1038/nchembio.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clarke PA, Hostein I, Banerji U, Stefano FD, Maloney A, Walton M, et al. Gene expression profiling of human colon cancer cells following inhibition of signal transduction by 17-allylamino-17-demethoxygeldanamycin, an inhibitor of the hsp90 molecular chaperone. Oncogene. 2000;19:4125–4133. doi: 10.1038/sj.onc.1203753. [DOI] [PubMed] [Google Scholar]
- 22.Banerji U, O'Donnell A, Scurr M, Pacey S, Stapleton S, Asad Y, et al. Phase I pharmacokinetic and pharmacodynamic study of 17-allylamino, 17-demethoxygeldanamycin in patients with advanced malignancies. J Clin Oncol. 2005;23:4152–4161. doi: 10.1200/JCO.2005.00.612. [DOI] [PubMed] [Google Scholar]
- 23.Maloney A, Clarke PA, Workman P. Genes and proteins governing the cellular sensitivity to HSP90 inhibitors: a mechanistic perspective. Curr Cancer Drug Targets. 2003;3:331–341. doi: 10.2174/1568009033481822. [DOI] [PubMed] [Google Scholar]
- 24.Powers MV, Clarke PA, Workman P. Dual targeting of HSC70 and HSP72 inhibits HSP90 function and induces tumor-specific apoptosis. Cancer Cell. 2008;14:250–262. doi: 10.1016/j.ccr.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 25.Maloney A, Clarke PA, Naaby-Hansen S, Stein R, Koopman JO, Akpan A, et al. Gene and protein expression profiling of human ovarian cancer cells treated with the heat-shock protein 90 inhibitor 17-allylamino-17-demethoxygeldanamycin. Cancer Res. 2007;67:3239–3253. doi: 10.1158/0008-5472.CAN-06-2968. [DOI] [PubMed] [Google Scholar]
- 26.Schumacher JA, Crockett DK, Elenitoba-Johnson KS, Lim MS. Proteome-wide changes induced by the Hsp90 inhibitor, geldanamycin in anaplastic large cell lymphoma cells. Proteomics. 2007;7:2603–2616. doi: 10.1002/pmic.200700108. [DOI] [PubMed] [Google Scholar]
- 27.Song D, Chaerkady R, Tan AC, García-García E, Nalli A, Suárez-Gauthier A, et al. Antitumor activity and molecular effects of the novel heat-shock protein 90 inhibitor, IPI-504, in pancreatic cancer. Mol Cancer Ther. 2008;7:3275–3284. doi: 10.1158/1535-7163.MCT-08-0508. [DOI] [PubMed] [Google Scholar]
- 28.Yao JQ, Liu QH, Chen X, Yang Q, Xu ZY, Hu F, et al. Hsp90 inhibitor 17-allylamino-17-demethoxygeldanamycin inhibits the proliferation of ARPE-19 cells. J Biomed Sci. 2010;17:30. doi: 10.1186/1423-0127-17-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dezwaan DC, Freeman BC. HSP90: the Rosetta stone for cellular protein dynamics? Cell Cycle. 2008;7:1006–1012. doi: 10.4161/cc.7.8.5723. [DOI] [PubMed] [Google Scholar]
- 30.Falsone SF, Gesslbauer B, Rek A, Kungl AJ. A proteomic approach towards the Hsp90-dependent ubiquitinylated proteome. Proteomics. 2007;7:2375–2383. doi: 10.1002/pmic.200600996. [DOI] [PubMed] [Google Scholar]
- 31.Xu W, Marcu M, Yuan X, Mimnaugh E, Patterson C, Neckers L. Chaperone-dependent E3 ubiquitin ligase CHIP mediates a degradative pathway for c-ErbB2/Neu. Proc Natl Acad Sci USA. 2002;99:12847–12852. doi: 10.1073/pnas.202365899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.da Rocha Dias S, Friedlos F, Light Y, Springer C, Workman P, Marais R. Activated B-RAF is an Hsp90 client protein that is targeted by the anticancer drug 17-allylamino-17-demethoxygeldanamycin. Cancer Res. 2005;65:10686–10691. doi: 10.1158/0008-5472.CAN-05-2632. [DOI] [PubMed] [Google Scholar]
- 33.Grbovic OM, Basso AD, Sawai A, Ye Q, Friedlander P, Solit D, et al. V600E B-Raf requires the Hsp90 chaperone for stability and is degraded in response to Hsp90 inhibitors. Proc Natl Acad Sci USA. 2006;103:57–62. doi: 10.1073/pnas.0609973103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neznanov N, Komarov AP, Neznanova L, Stanhope-Baker P, Gudkov AV. Proteotoxic stress targeted therapy (PSTT): induction of protein misfolding enhances the antitumor effect of the proteasome inhibitor bortezomib. Oncotarget. 2011;2:209–221. doi: 10.18632/oncotarget.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Molineaux SM. Molecular pathways: targeting proteasomal protein degradation in cancer. Clin Cancer Res. 2012;18:15–20. doi: 10.1158/1078-0432.CCR-11-0853. [DOI] [PubMed] [Google Scholar]
- 36.Richter K, Buchner J. Hsp90: chaperoning signal transduction. J Cell Physiol. 2001;188:281–290. doi: 10.1002/jcp.1131. [DOI] [PubMed] [Google Scholar]
- 37.Camphausen K, Tofilon PJ. Inhibition of Hsp90: a multitarget approach to radiosensitization. Clin Cancer Res. 2007;13:4326–4330. doi: 10.1158/1078-0432.CCR-07-0632. [DOI] [PubMed] [Google Scholar]
- 38.Page TJ, Sikder D, Yang L, Pluta L, Wolfinger RD, Kodadek T, et al. Genome-wide analysis of human HSF1 signaling reveals a transcriptional program linked to cellular adaptation and survival. Mol Biosyst. 2006;2:627–639. doi: 10.1039/b606129j. [DOI] [PubMed] [Google Scholar]
- 39.Pacey S, Wilson RH, Walton M, Eatock MM, Hardcastle A, Zetterlund A, et al. A phase I study of the heat-shock protein 90 inhibitor alvespimycin (17-DMAG) given intravenously to patients with advanced solid tumors. Clin Cancer Res. 2011;17:1561–1570. doi: 10.1158/1078-0432.CCR-10-1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Workman P, Collins I. Probing the probes: fitness factors for small molecule tools. Chem Biol. 2010;17:561–577. doi: 10.1016/j.chembiol.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bell AW, Deutsch EW, Au CE, Kearney RE, Beavis R, Sechi S, et al. HUPO Test Sample Working Group A HUPO test sample study reveals common problems in mass spectrometry-based proteomics. Nat Methods. 2009;6:423–430. doi: 10.1038/nmeth.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams AJ, Ekins S. The long term cost of inferior database quality. Drug Discov Today. 2012 http://www.drugdiscoverytoday.com/view/22579/the-long-term-cost-of-inferior-database-quality/ [Google Scholar]
- 43.Sreedhar AS, Kalmár E, Csermely P, Shen YF. Hsp90 isoforms: functions, expression and clinical importance. FEBS Lett. 2004;562:11–15. doi: 10.1016/S0014-5793(04)00229-7. [DOI] [PubMed] [Google Scholar]
- 44.Eletto D, Dersh D, Argon Y. GRP94 in ER quality control and stress responses. Semin Cell Dev Biol. 2010;21:479–485. doi: 10.1016/j.semcdb.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Costantino E, Maddalena F, Calise S, Piscazzi A, Tirino V, Fersini A, et al. TRAP1, a novel mitochondrial chaperone responsible for multi-drug resistance and protection from apoptotis in human colorectal carcinoma cells. Cancer Lett. 2009;279:39–46. doi: 10.1016/j.canlet.2009.01.018. [DOI] [PubMed] [Google Scholar]
- 46.Landriscina M, Amoroso MR, Piscazzi A, Esposito F. Heat-shock proteins, cell survival and drug resistance: the mitochondrial chaperone TRAP1, a potential novel target for ovarian cancer therapy. Gynecol Oncol. 2010;117:177–182. doi: 10.1016/j.ygyno.2009.10.078. [DOI] [PubMed] [Google Scholar]
- 47.Kamal A, Thao L, Sensintaffar J, Zhang L, Boehm MF, Fritz LC, et al. A high-affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors. Nature. 2003;425:407–410. doi: 10.1038/nature01913. [DOI] [PubMed] [Google Scholar]
- 48.Yap TA, Workman P. Exploiting the cancer genome: strategies for the discovery and clinical development of targeted molecular therapeutics. Annu Rev Pharmacol Toxicol. 2012;52:549–573. doi: 10.1146/annurev-pharmtox-010611-134532. [DOI] [PubMed] [Google Scholar]
- 49.Santagata S, Hu R, Lin NU, Mendillo ML, Collins LC, Hankinson SE, et al. High levels of nuclear heat-shock factor 1 (HSF1) are associated with poor prognosis in breast cancer. Proc Natl Acad Sci USA. 2011;108:18378–18383. doi: 10.1073/pnas.1115031108. [DOI] [PMC free article] [PubMed] [Google Scholar]