Abstract
In Arabidopsis thaliana, acyl-CoA-binding protein 3 ( ACBP3), one of six ACBPs, is unique in terms of the C-terminal location of its acyl-CoA-binding domain. It promotes autophagy-mediated leaf senescence and confers resistance to Pseudomonas syringae pv. tomato DC3000. To understand the regulation of ACBP3, a 1.7 kb 5'-flanking region of ACBP3 and its deletion derivatives were characterized using β-glucuronidase (GUS) fusions. A 374 bp minimal fragment (–151/+223) could drive GUS expression while a 1698 bp fragment (–1475/+223) conferred maximal activity. Further, histochemical analysis on transgenic Arabidopsis harbouring the largest (1698 bp) ACBP3pro::GUS fusion displayed ubiquitous expression in floral organs and vegetative tissues (vascular bundles of leaves and stems), consistent with previous results showing that extracellularly localized ACBP3 functions in plant defence. A 160 bp region (–434/–274) induced expression in extended darkness and caused down-regulation in extended light. Electrophoretic mobility shift assay (EMSA) and DNase I footprinting assay showed that the DNA-binding with one finger box (Dof-box, –341/–338) interacted specifically with leaf nuclear proteins from dark-treated Arabidopsis, while GT-1 (–406/–401) binds both dark- and light-treated Arabidopsis, suggesting that Dof and GT-1 motifs are required to mediate circadian regulation of ACBP3. Moreover, GUS staining and fluorometric measurements revealed that a 109 bp region (–543/–434) was responsive to phytohormones and pathogens. An S-box of AT-rich sequence (–516/–512) was identified to bind nuclear proteins from pathogen-infected Arabidopsis leaves, providing the basis for pathogen-inducible regulation of ACBP3 expression. Thus, three cis-responsive elements (Dof, GT-1, and the S-box) in the 5'-flanking region of ACBP3 are proven functional in the regulation of ACBP3.
Keywords: Arabidopsis ACBP3, dark/light regulation, defence response, Dof-box, GT-1 cis-acting element, S-box
Introduction
Acyl-CoA-binding proteins (ACBPs) bind to long-chain acyl-CoA esters and are ubiquitous in eukaryotes (Shoyab et al., 1986; Xiao and Chye, 2009). The highly conserved 10 kDa ACBPs from mammals and yeast protect cytosolic acyl-CoAs from cellular acyl-CoA hydrolases (Knudsen et al., 2000; Faergeman and Knudsen, 2002). Larger ACBPs are prevalent in eukaryotes, but inconsistent nomenclature has made comparison difficult (Xiao and Chye, 2011a). Besides the 10 kDa ACBP6, five larger forms (ACBP1–ACBP3) ranging from 37.5 kDa to 73.1 kDa co-exist in Arabidopsis thaliana (Xiao and Chye, 2011a). They exhibit differing binding affinities for acyl-CoA esters and are localized to various subcellular compartments, suggesting they are biologically non-redundant in vivo (Chye, 1998; Chye et al., 1999, 2000; Leung et al., 2004, 2006; Gao et al., 2009; Xiao et al., 2009, 2010; Xiao and Chye, 2011b). ACBP1 and ACBP2 share 87.4% identity, contain N-terminal transmembrane domains and C-terminal ankyrin repeats, and are targeted to the endoplasmic reticulum (ER) and plasma membrane (Chye et al., 1999; Li and Chye, 2003, 2004; Chen et al., 2010; Gao et al., 2009, 2010). In contrast, ACBP4, ACBP5, and ACBP6 are cytosolic proteins (Chen et al., 2008; Li et al., 2008; Xiao et al., 2008b). ACBP4 and ACBP5 share 81.4% homology, contain conserved kelch motifs, and bind oleoyl-CoA ester, suggesting that ACBP4 and ACBP5 are potentially oleoyl-CoA pool formers in the cytosol and facilitate oleoyl-CoA ester transfer between the plastids and the ER (Leung et al., 2004; Chen et al., 2008; Xiao et al., 2008b). Recent investigations have revealed that in addition to the differential roles in phospholipid metabolism, Arabidopsis ACBPs are involved in responses to a variety of biotic and abiotic stimuli, such as heavy metals, low temperature, oxidative stress, and pathogens (Chen et al., 2008; Li et al., 2008; Xiao et al., 2008a, 2010; Gao et al., 2009, 2010; Du et al., 2010; Xiao and Chye, 2011b).
In ACBP3, the ACB domain resides at the C-terminus unlike other Arabidopsis ACBPs (Leung et al., 2006; Xiao and Chye, 2011a). Autofluorescence-tagged ACBP3 is targeted extracellularly in tobacco Bright-Yellow-2 cells and onion epidermal cells (Leung et al., 2006). ACBP3 mRNA accumulates in vegetative rather than floral organs of mature Arabidopsis and is up-regulated in young/senescent rosettes and by dark treatment (Xiao et al., 2010). ACBP3 overexpressors displayed accelerated leaf senescence while the ACBP3 T-DNA insertional mutant and RNA interference lines were delayed (Xiao et al., 2010). ACBP3 interacts with phosphatidylethanolamine (PE) in vitro and probably regulates PE homeostasis and metabolism in vivo (Xiao et al., 2010). Given that the overexpression of ACBP3 enhanced the degradation of the autophagy-related protein 8 (ATG8) and disrupted autophagosome formation, ACBP3 through its interaction with PE may interfere with ATG8–PE complex formation and regulate autophagy-mediated leaf senescence (Xiao et al., 2010). ACBP3 expression is up-regulated by bacterial pathogen infection and treatments with pathogen elicitors, as well as by defence-related phytohormones (Xiao and Chye, 2011b). Hence it is a phospholipid-binding protein that regulates leaf senescence and defence against pathogen infection (Xiao et al., 2010; Xiao and Chye, 2011b).
Given the importance of ACBPs in abiotic and biotic stresses, the characterization of the 5′-flanking regions of Arabidopsis ACBPs was initiated. The 5′-flanking regions of ACBP3 were first selected to understand its circadian regulation and pathogen-induced expression (Xiao et al., 2010; Xiao and Chye, 2011b). Light affects transcription as well as post-transcriptional processes in plant growth and development (Green et al., 1987, 1988; Gilmartin et al., 1990; Lam and Chua, 1990), while pathogen attack is an environmental stress that triggers a variety of defence-responsive genes via transcription factors bound to specific cis-acting elements in the 5'-flanking regions (Cheong et al., 2002; Kunkel and Brooks, 2002). Here, it is shown that two motifs, Dof (DNA-binding with one finger) and GT-1, are required for dark/light regulation of ACBP3 expression, while the S-box appears to participate in regulation following infection by Pseudomonas syringae pv. tomato DC3000.
Materials and methods
Construction of the ACBP3pro::GUS fusion and its deletion derivatives
A 1698 bp (–1475/+223) 5'-flanking region of ACBP3 (AT4G24230, GenBank accession no. NM_118556, http://www.arabidopsis.org) and its six 5'-truncated derivatives were fused to the GUS (β-glucuronidase) reporter gene. PCR was performed using Arabidopsis genomic DNA as template and various primer pairs to amplify the ACBP3 5'-flanking fragments (Supplementary Fig. S1, Table S1 available at JXB online). All forward primers contain a BamHI site and the reverse primers a SmaI site. Fragments were purified and cloned into pGEMT-Easy vector (Promega). Each BamHI–SmaI fragment was subcloned to corresponding restriction sites on the binary vector pBI101.3 (Clontech) to generate a series of seven ACBP3pro::GUS fusions. The resultant plasmids were designated as constructs pAT436, pAT437, pAT438, pAT439, pAT440, pAT441, and pAT442. The cloning junctions in each resultant plasmid were verified by DNA sequence analysis.
Generation of transgenic plants
Each construct was mobilized from Escherichia coli to Agrobacterium tumefaciens strain LBA4404 by triparental mating. ACBP3pro::GUS fusions were introduced into Arabidopsis wild-type (ecotype, Columbia-0) using Agrobacterium-mediated transformation (Clough and Bent, 1998). The T0 transformants were grown to set seed in a growth chamber. Seeds were collected, surface sterilized, and then germinated on Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) supplemented with kanamycin (50 μg ml−1). Putative T1 transformants were confirmed by PCR using ACBP3 5'-flanking sequence-specific forward primers and a GUS-3' reverse primer (Supplementary Table S1 at JXB online). These PCR-confirmed seedlings were potted to yield the T2 generation. An average of 3–5 independent T2 lines per construct, all harbouring single-copy inserts that showed a simple Mendelian 3:1 segregation ratio to kanamycin, were selected. Seeds from T2 lines were germinated, and resultant T3 lines that showed 100% kanamycin-resistant segregation were deemed homozygous for further analysis.
Plant materials, growth conditions, and treatments
Seeds from Arabidopsis wild-type and ACBP3pro::GUS lines were surface sterilized and plated on MS medium followed by chilling for 4 d at 4 °C in darkness before germination. Plates were incubated in a tissue culture room at 21 °C under continuous light for 2 weeks. Seedlings were potted in soil and raised in a growth chamber with 23 °C/21 °C (day/night) cycles, plus a daylength regime of 16 h light from 06:00 to 21:00 and 8 h dark from 21:00 to 06:00 (LD). For dark or light treatment, 2- or 3-week-old LD-grown plants were incubated in constant darkness (DD) or constant light (LL). Samples were collected at the indicated time points as shown in the figures. For experiments involving phytohormone treatment, leaves from 5-week-old plants derived from pAT436 transformation were submerged in either 1 mM 1-aminocyclopropane-1-carboxylic acid (ACC; Sigma-Aldrich, St Louis, MO, USA), 100 μM methyl jasmonate (MeJA; Sigma-Aldrich), or 1 mM salicylic acid (SA; Sigma-Aldrich). Control leaves were submerged in distilled water. After 12 h of treatment, samples were harvested and stained with X-Gluc (5-bromo-4-chloro-3-indolyl-β-D-glucuronide; Invitrogen) in histochemical staining assays and analysed quantitatively for GUS activity.
Culture of pathogen and plant inoculation
Pseudomonas syringae pv. tomato DC3000 (American Type Culture Collection no. BBA-871) was cultured according to Xiao and Chye (2011b). A day before plant inoculation, a single bacterial colony from a plate of King’s B medium containing 100 μg ml−1 rifampicin was transferred to 3 ml of King’s B liquid supplemented with rifampicin (100 μg ml−1) and agitated at 28 °C overnight until mid-log growth phase. The bacteria were harvested by centrifugation at 4000 g for 7 min, and resuspended in 5 ml of 10 mM MgCl2. Rosettes from 5-week-old Arabidopsis were syringe-infiltrated with bacterial suspensions or MgCl2 (control). After inoculation, plants were placed in a growth chamber under 16 h light (23 °C)/8 h dark (21 °C) cycles. Leaves were collected 48 h and 72 h post-inoculation (Xiao and Chye, 2011b) for GUS assays and electrophoretic mobility shift assays (EMSAs).
Histochemical GUS assays
Histochemical staining for GUS (Jefferson et al., 1987) of Arabidopsis tissues was carried out by incubation in X-Gluc dissolved in standard buffer [100 mM sodium phosphate, pH 7.5, 2 mM K3Fe(CN)6, 2 mM K4Fe(CN)6, 0.1% (v/v) Triton X-100, 1 mg ml−1 X-Gluc]. Samples and control were vacuum infiltrated in GUS staining solution for 1 h, followed by 2 h to overnight incubation at 37 °C. Chlorophyll was removed with several changes of 70% ethanol. Stained samples were analysed and photographed.
Fluorometric assays of GUS activity
Arabidopsis leaves were collected and analysed for GUS activity by quantification of 4-methylumbelliferone (MUG) with substrate β-D-glucuronide (Jefferson et al., 1987). GUS activity was normalized to protein concentration as pmole of product generated per mg of total protein per minute. The protein content of tissue homogenates was quantified with Bradford reagent (Bradford, 1976) using bovine serum albumin (BSA) as a standard. An average of 3–5 lines per genotype were analysed and three independent experiments were conducted.
Preparation of nuclear protein extracts
For binding studies in dark/light regulation, 3-week-old Arabidopsis sown in soil under a 16 h light/8 h dark cycle (LD), as well as plants adapted to 48 h darkness (DD) or 48 h light (LL), were used. Leaves from LD-grown plants were harvested at 12:00. For pathogen infection, leaves from 5-week-old wild-type Arabidopsis that had been syringe-infiltrated with either bacterial strain P. syringae or 10 mM MgCl2 (control) were collected 48 h post-inoculation for nuclear protein extraction. Nuclear extracts were freshly isolated according to Maxwell et al. (2003). Following determination of protein concentration (Bradford, 1976), aliquots of nuclear extracts were stored at –80 °C until use.
Electrophoretic mobility shift assays
Probes for EMSAs were 3′-end biotin-labelled double-stranded DNA. Seven such DNA probes were prepared for dark/light regulation analysis (Supplementary Table S1 at JXB online): three pairs of Dof-box wild-type probes designated as GpI-Dof-wt, GpII-Dof-wt, and GpIII-Dof-wt, each corresponding to a cluster of six putative Dof-boxes; two Dof-box mutant probe pairs, Dof-(–341/–338)-mut and Dof-(–326/–323)-mut, which correspond to the two putative Dofs at –341/–338 and –326/–323, respectively; a GT-1-(–406/–401)-wt pair which maps to the GT-1 cis-element (–406/–401) and a GT-1-(–406/–401)-mut pair which contains the correspondingly mutated GT-1. For pathogen-related EMSAs, a wild-type probe pair [S-box-(–516/–512)-wt], and a mutant probe pair [S-box-(–516/–512)-mut] were used. All probes were 3′ end labelled with biotin using the Biotin 3′-End DNA Labeling Kit (Pierce), and unlabelled oligonucleotides were used as competitors in binding. Labelling efficiency was estimated before EMSA studies.
EMSAs were carried out using the LightShift Chemiluminescent EMSA Kit (Pierce). For investigation on dark/light regulation, crude nuclear extracts (5 μg) from LD-, DD-, or LL-treated 3-week-old Arabidopsis leaves were incubated for 30 min on ice in binding buffer [20 mM HEPES-KOH (pH 7.9), 0.5 mM dithiothreitol (DTT), 0.1 mM EDTA, 40 mM KCl, 15% glycerol (v/v)] with or without a 200-fold molar excess of specific competitor oligonucleotide to a total volume of 20 μl. Labelled DNA (20 fmol) was added to the binding mixture. For investigations on pathogen induction, binding mixtures containing nuclear extracts (4 μg), 20 fmol of binding probe, and a 50-fold molar excess of specific competitor DNA (as specified), in binding buffer (10 mM TRIS 50 mM KCl, 1 mM DTT, pH 7.5) in a 20 μl reaction volume, were incubated at room temperature for 15 min. Poly(dI/dC) (50 ng) was added as a non-specific competitor in all EMSAs. The reaction products were loaded on a 6% native polyacrylamide gel, which was run for 75 min in 0.5× TBE buffer at 100 V at 4 °C. The DNA and DNA–protein complex were fixed to a positively charged nylon membrane (Pierce Biotechnology) and were visualized by exposing the membrane to X-ray film for 2–5 min, depending on the signal intensity. All EMSAs were repeated using 2–3 independent batches of nuclear extracts to confirm the results.
Capillary electrophoresis in DNase I footprinting
Probes for DNase I footprinting assays were designed according to the ACBP3 5′-flanking sequence (Supplementary Table S1 at JXB online). The premium length probe recommended in capillary electrophoresis footprinting is ∼300 bp because too short or too long a probe will yield unwanted trace signals. The expected protection region should correspond to the middle region of the probe to enable differentiation from the unprotected region (Wilson et al., 2001; Zianni et al., 2006). Sequences (–450/–143) between the primer pair ML1171/ML1172 comprising putative cis-elements include one GT-1 cis-element (–406/–401) and six Dof-boxes (–341/–338, –326/–323, –240/–237, –231/–228, –225/–222, and –201/–198). The forward primer ML1171 was commercially synthesized and 5′ end labelled with 6-carboxyfluorescein phosphoramidate (6-FAM) (Molecular Informatrix Laboratory, Tech Dragon) while the reverse primer ML1172 was 5′ end labelled with benzofluorotrichlorocarboxy-fluorescein (NED) (Applied Biosystems). The probe was PCR amplified with construct pAT436 plasmid DNA as template. The PCR products were gel purified using the QIAquick PCR purification Kit (QIAGEN). The concentration of the probe was determined by measurement of absorbance at 260 nm.
Capillary electrophoresis DNase I footprinting experiments were carried out following Wilson et al. (2001) and Zianni et al. (2006). The 20 μl reactions consisted of 50 ng of fluorescein-labelled DNA and 10 μg of nuclear extracts from 48 h dark-treated (DD) or untreated (LD) 3-week-old Arabidopsis leaves using the binding buffer as in EMSAs. The control reaction contains an equal amount of labelled DNA and 20 μg of BSA. Binding reactions were initiated by addition of BSA or nuclear extracts followed by incubation on ice for 20 min. A 20 μl aliquot of cofactor solution (10 mM MgCl2, 5 mM CaCl2) was added together with 0.0025 Kunitz units of DNase I (Roche) to a final volume of 50 μl for each set of reactions. The protein-bound DNA was digested for 2 min at room temperature. The reactions were terminated by addition of 100 μl of EDTA (100 mM, pH 8.0) and incubation at 75 °C for 15 min. Samples were then extracted twice with an equal volume of phenol–chloroform–isoamyl alcohol (25:24:1) and DNA precipitated with 1 vol. of isopropanol at –80 °C for 1 h. After 10 min centrifugation at 14 000 rpm, the DNA pellets were washed once with absolute ethanol, after which the samples were heated for 2 min at 95 °C to remove residual ethanol. The digested DNA, at solid phase, was added to 9.5 μl of HiDi formamide (Applied Biosystems) and 0.5 μl of GeneScan-600 LIZ size standards (Applied Biosystems). The samples were analysed using the 3730 DNA Analyzer (Applied Biolosystems). Capillary electrophoresis traces were examined by Peak Scanner Software version 1.0 (Applied Biosystems) for loss of signal in the protein-containing samples and were compared with the control to identify protein-bound protected regions within the DNA fragment being examined.
Results
Sequence and deletion analyses of the ACBP3 5′-flanking region
Given that ACBP3 functions in important processes such as in senescence and defence (Xiao and Chye, 2011a, b; Xiao et al., 2010), a 1698 bp (–1475/+223) 5'-flanking region of ACBP3 was isolated and investigated. Interestingly, ACBP3 represents the only member of the six-membered Arabidopsis ACBP family that shows dark-induced expression. Results using SoftBerry PlantProm DB (http://www.softberry.com) (Shahmuradov et al., 2003) and PlantCare (http://sphinx.rug.ac.be:8080/PlantCARE/) (Rombauts et al., 1999) revealed that the putative transcription start site of ACBP3 maps 93 bp 5′ to the translation initiation codon (designated as +1 in Supplementary Fig. S1 at JXB online). The basal regulatory elements identified include a putative TATA-box (–89/–86) for RNA polymerase binding, a putative initiator element (INR: –446/–442) for binding large complex general transcription factors (Willmott et al., 1998), and two putative CAAT boxes (–372/–368 and –261/–257). Also, several putative dark/light-responsive and pathogen-inducible cis-elements were identified, including two putative nine nucleotide evening element (EE) motifs (–1047/–1039 and –1005/–997), noting that EEs mediate peak expression in late light (Harmer et al., 2000; Rawat et al., 2005); a putative S-box (–516/–512) which may be related to light regulation as in Brassica napus rbcSF1 (Nantel et al., 1991); a putative GT-1 cis-element (–406/–401) which could positively or negatively control transcription (Fisscher et al., 1994; Park et al., 2004), and six putative Dof-binding sites (–341/–338, –326/–323, –240/–237, –231/–228, –225/–222, and –201/–198) which are known to participate in diverse functions including dark regulation (Yanagisawa, 2002). In addition, the 5'-flanking region of ACBP3 harboured a putative highly conserved 7 bp P-box (–568/–562) (Wang et al., 2007).
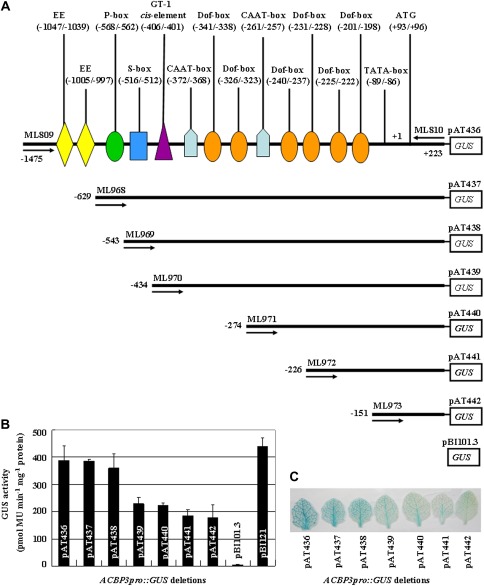
To define the minimal basal length and the maximal boundary for ACBP3 promoter activity, 5′ end deletion analysis of seven ACBP3pro::GUS fusions was used (Fig. 1A). Three to five independent transgenic Arabidopsis lines per construct were assayed for promoter strength and compared with pBI121 [Cauliflower mnosiac virus (CaMV) 35S promoter-GUS] or pBI101.3 (promoterless-GUS) transformants as positive and negative controls, respectively. When leaves from 3-week-old Arabidopsis grown under 16 h light/8 h dark cycles were analysed, no difference in GUS activities of pAT436 and pAT437 transformants was observed. In contrast, pAT438 transformants showed a 1.4- to 1.6-fold decrease in activity (Fig. 1B). A statistically significant decline in GUS expression was observed with the next four deletions (constructs pAT439, pAT440, pAT441, and pAT442). GUS expression from the largest fragment (construct pAT436) was ∼2.2-fold higher than that from the shortest (construct pAT442). Moreover, pAT436 transformants displayed approximately the same activity as pBI121 transformants (Fig. 1B). In histochemical GUS staining, a blue colour was visible in leaf tissue from all seven constructs, with the strongest from pAT436. Also, the minimal core promoter of 374 bp (–151/+223 in construct pAT442) was capable of driving GUS expression (Fig. 1C).
Fig. 1.
Analysis of GUS expression driven by deletion derivatives of the 5'-flanking region of ACBP3. (A) A schematic diagram of constructs developed by 5′ end deletion of the ACBP3 5′-flanking region (–1475/+223). Promoter fragments of various sizes were inserted into vector pBI101.3 containing the GUS reporter gene. Black bars (not to scale) indicate each truncated fragment, and predicted cis-elements are denoted on the ACBP3pro::GUS construct pAT436. Numbers on the left represent the end position of each deletion. PCR primers used for generating constructs are marked with forward or reverse arrows (the number above the arrow indicates the primer name). Putative cis-acting regulatory elements are represented by various symbols. (B) Quantitative fluorimetric measurement of GUS activity in leaf nuclear extracts from 3-week-old plants of ACBP3pro::GUS deletion constructs. Average values were obtained from experiments performed with 3–5 independent lines per construct, each line represented by 8–10 individual plants. Bars indicate the standard errors of three replicates. (C) Histochemical GUS staining of seven ACBP3pro::GUS constructs. Leaves from 3-week-old Arabidopsis transformants were stained with substrate X-gluc. The experiment was repeated three times; each test examined leaves from 8–10 individual plants per construct.
ACBP3pro::GUS is developmentally regulated
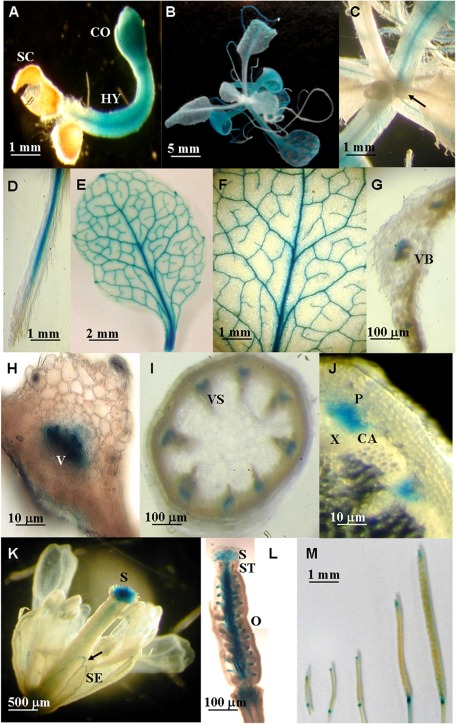
Transgenic plants harbouring the 1698 bp ACBP3pro::GUS fusion derived from construct pAT436 were used in investigations on temporal expression by GUS histochemical analysis. In the very first stage (3 d) post-seed germination, constitutive GUS activation was detected in hypocotyl and cotyledons, but not in radicle or seed coat (Fig. 2A). In 3-week-old seedlings, expression was strongest in the first pair of true leaves and declined in the second and third pairs (Fig. 2B). Interestingly, the newly emerged leaves in seedlings did not show any GUS staining (Fig. 2C, arrow). In 3-week-old plants, GUS staining was observed in root tissues excluding root hairs and tips (Fig. 2D), and in vascular bundles of intact leaves (Fig. 2E) including both major and minor veins (Fig. 2F). When hand-cut cross-sections of leaves adjacent to the petiole and stem were examined (Fig. 2G, H), GUS expression was restricted to the vascular cells in leaves and was expressed in phloem, and cambial zones of stems (Fig. 2I, J). In the open flower, ACBP3pro::GUS was detected in the stigma, style, and ovary in the pistil, and in sepals (Fig. 2K, L). Furthermore, relatively lower GUS expression was observed when siliques matured (Fig. 2M).
Fig. 2.
Spatial and temporal expression patterns of ACBP3pro::GUS fusions. Histochemical GUS staining shows expression of GUS from the ACBP3 5′-flanking region in a 3-day-old seedling (A); a 3-week-old seedling (B and C), with the arrow in C showing newly produced leaves; root (D); 32-day-old rosette leaf with the major and side veins (E and F); horizontal section of leaf (G and H); stem (I and J); fully opened flower, with the arrow showing the sepal (K); hand-section of a pistil showing the expression of ACBP3pro::GUS in stigma, style, and ovary (L); siliques from a 40-day-old transgenic plant (M). SC, seed coat; CO, cotyledons; HY, hypocotyl; VB, vascular bundle; V, vascular element; VS, vascular system; P, phloem; X, xylem; CA, cambium; SE, sepal; S, stigma; ST, style; O, ovary.
ACBP3pro::GUS is subject to circadian regulation
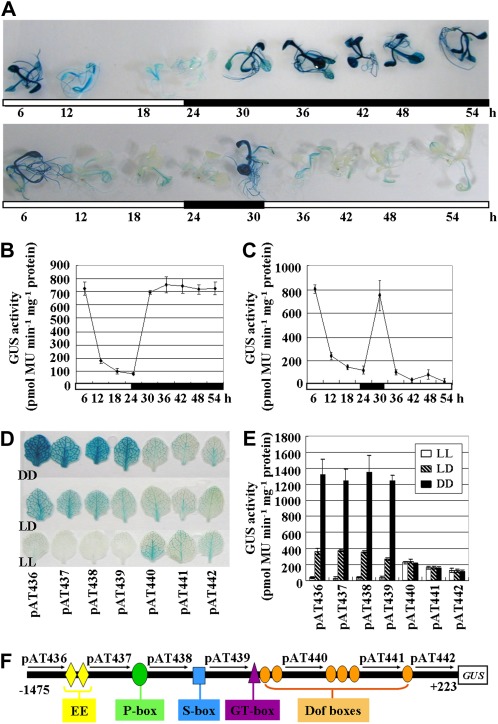
Xiao et al. (2010) had demonstrated that the ACBP3 transcript is up-regulated in constant darkness (DD) and down-regulated in constant light (LL). To identify the dark/light-responsive cis-element(s) in ACBP3 regulation, stable transgenic Arabidopsis harbouring the 1.7 kb ACBP3 5′-flanking region was tested in GUS assays. Two-week-old seedlings germinated in normal 16 h light/8 h dark cycles (LD) were subject to DD or LL and examined every 6 h for 54 h. GUS analysis indicated that seedlings germinated under LD were subject to circadian regulation (for the first 30 h). Subsequently, GUS expression gradually faded when plants were shifted to LL and vice versa for DD (Fig. 3A, bottom and top panels, respectively). GUS was expressed at very high levels, and whole seedlings were stained dark blue in DD (Fig. 3A, top panel, from 30 h to 54 h) in comparison with only light blue in vascular tissues under LL (Fig. 3A, bottom panel, from 30 h to 54 h). When samples were analysed by fluorometric GUS assays, consistent results were obtained. ACBP3pro::GUS expression decreased in the light (from 6 h to 24 h), but was rapidly elevated in darkness (from 24 h to 30 h) during a 1 d rhythm (Fig. 3B). Quantitatively, there was an ∼7-fold decrease from peak expression as recorded at 6 h in the dark versus its lowest level at 24 h in the light (Fig. 3B). GUS expression in the dark peaked again at 36 h ∼7-fold from the lowest level and remained stable for the next 18 h (Fig. 3B). Expression in DD and under LD regulation reached similar peaks, indicating the absence of any cumulative effect on ACBP3 expression (Fig. 3B). When plants were exposed to LL, GUS activity decreased 5-fold (Fig. 3C, between 30 h and 54 h). These results confirm that ACBP3 is up-regulated in darkness but is repressed by light.
Fig. 3.
Effect of dark and light on ACBP3pro::GUS expression. (A–C) GUS assays on ACBP3pro::GUS construct pAT436 under LD (the first 30 h) followed by DD [(A) top panel and (B), from 30 h to 54 h] or LL [(A) bottom panel and (C), from 30 h to 54 h] conditions. Two-week-old seedlings grown on MS medium (supplemented with 50 μg ml−1 kanamycin) in LD cycles were shifted to continuous darkness (DD) or continuous light (LL) and then harvested over a 54 h period at 6 h intervals. White and black bars indicate light and dark periods, respectively. Numbering under each bar indicates the time of treatment. (D) Leaves from 3-week-old Arabidopsis containing ACBP3pro::GUS deletions were sampled after 48 h dark treatment (DD, top row) or 48 h light exposure (LL, bottom row). Plants grown under LD cycles (middle row) were collected as a control. The experiment was repeated three times, each using leaves from 8–10 individual plants per construct. (E) Quantitative fluorimetric measurement of GUS activity in nuclear extracts from the seven ACBP3pro::GUS constructs after DD and LL treatment of LD-grown plants. Average values were obtained from experiments performed with 3–5 independent lines per construct, each line represented by 8–10 individual plants. Bars indicate the standard errors of three replicates. (F) A pictorial representation of the 1698 bp (–1475/+223) ACBP3 5′-flanking region linked to GUS. Putative cis-acting regulatory elements in the region are represented by various symbols. Numbers above the arrows indicate the corresponding construct.
To identify the relevant putative dark/light-responsive cis-element(s) in transcriptional regulation of ACBP3 expression, seeds from transformants of six truncated ACBP3 5′-flanking sequences (constructs pAT437, pAT438, pAT439, pAT440, pAT441, and pAT442), together with those of the largest 1698 bp ACBP3pro::GUS construct pAT436, were germinated and grown for 3 weeks in LD followed by shifting to either 48 h DD or LL. In histochemical GUS staining, whole leaves from transformants of constructs pAT436, pAT437, pAT438, and pAT439 were stained dark blue after dark treatment (Fig. 3D, DD). In comparison, after extended light, only weak traces of blue were evident in the veins of leaves (Fig. 3D, LL). The middle row in Fig. 3D depicts samples grown under LD cycles as a control. In contrast, DD or LL treatment did not affect GUS expression in pAT440, pAT441, and pAT442 transformants when compared with LD (Fig. 3D). Quantitative measurement of GUS activity also showed similar results (Fig. 3E). DD and LL samples were similar to untreated control (LD) for pAT440, pAT441, and pAT442 transformants, while for the four progressively longer ACBP3 5′-flanking regions (constructs pAT439, pAT438, pAT437, and pAT436), there was as a >3-fold induced GUS activity upon DD treatment and a <4-fold decrease under LL conditions in comparison with LD samples. Taken together, these results support the presence of functional cis-element(s) within a 160 bp (–434/–274) region, as delinated by constructs pAT439 and pAT440, that regulate ACBP3 expression in response to dark and light (Fig. 3F).
Role of a Dof-box and a GT-1 cis-element in regulation of ACBP3
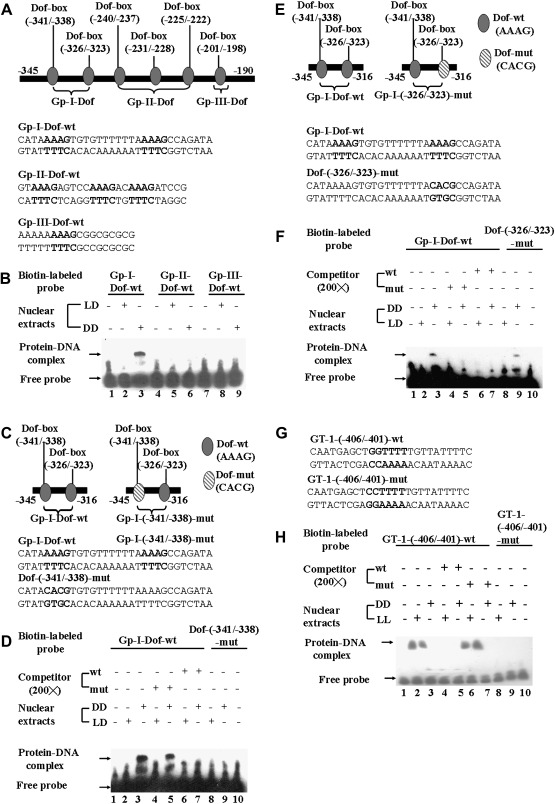
A PlantProm DB database search of the ACBP3 5′-flanking DNA sequence revealed several putative cis-elements in dark regulation including six predicted Dof-boxes (–341/–338, –326/–323, –240/–237, –231/–228, –225/–222, and –201/–198). Subsequently, EMSAs were performed using double-stranded DNA probes covering the six predicted light-responsive Dof-boxes dispersed on the 155 bp (–345/–190) ACBP3 5′-flanking region. The six Dof-boxes were initially divided into three subgroups, Gp-I-Dof (–345/–316), Gp-II-Dof (–242/–218), and Gp-III-Dof (–206/–190) (Fig. 4A). When crude nuclear extracts from leaves of 3-week-old Arabidopsis grown under LD followed by 48 h DD were tested, a strong DNA–protein binding complex was observed with the Gp-I-Dof-wt probe spanning the first two Dof-boxes (–341/–338 and –326/–323) (Fig. 4B, lane 3). The Gp-II-Dof and Gp-III-Dof probes showed no binding to either untreated (LD) or dark-treated (DD) leaf proteins (Fig. 4B, lanes 5, 6, 8, and 9). These results suggest that the light-responsive cis-element(s) are confined to the Gp-I-Dof sequence.
Fig. 4.
EMSAs on the Dof-box (–341/–338) and GT-1 cis-element (–406/–401) in vitro. (A) Schematic illustration of six putative Dof-boxes on the ACBP3 5'-flanking region. The oval-shaped Dof-boxes were artificially separated into three subgroups, and nucleotide sequences of double-stranded oligonucleotides used in EMSAs are marked below the corresponding Dof groups. The nucleotides of six Dof-boxes are shown in bold. (B) Interaction of nuclear extracts from 3-week-old Arabidopsis leaves with Gp-I-Dof-wt, Gp-II-Dof-wt, and Gp-III-Dof-wt probes, respectively. Crude nuclear extracts (5 μg) from 48 h dark-treated Arabidopsis (DD in lanes 3, 6, and 9) or LD-grown plants (LD in lanes 2, 5, and 8) were incubated with biotin end-labelled Gp-I-Dof-wt (lanes 2 and 3), Gp-II-Dof-wt (lanes 5 and 6), and Gp-III-Dof-wt (lanes 8 and 9) probes. Lanes 1, 4, and 7 are free probes without addition of crude nuclear extracts. (C) Schematic illustration of the mutated Dof-box (–341/–338) (right panel) and its corresponding location in the Gp-I–Dof (left panel) sequence. Nucleotide sequences of double-stranded oligonucleotides used in EMSAs are marked below the corresponding Dof-boxes. Mutated nucleotides in Dof-box (–341/–338) and its corresponding sequences are shown in bold. (D) Interaction of nuclear extracts from 3-week-old Arabidopsis leaves with Gp-I-Dof-wt and Dof-(–341/–338)-mut probes. Crude nuclear extracts (5 μg) from 48 h dark-treated plants (DD in lanes 3, 5, 7, and 9) or control plants (LD in lanes 2, 4, 6, and 8) were incubated with biotin end-labelled Gp-I-Dof-wt (lanes 2–7) or Dof-(–341/–338)-mut (lanes 8 and 9) probes, in the absence (lanes 2, 3, 8, and 9) or presence of a 200-fold molar excess of unlabelled competitor, Gp-I-Dof-wt (lanes 6 and 7), or Dof-(–341/–338)-mut (lanes 4 and 5). Lanes 1 and 10 are free probes without addition of crude nuclear extracts. (E) Schematic illustration of the mutated Dof-box (–326/–323) (right panel) and its corresponding location in the Gp-I–Dof (left panel) sequence. Nucleotide sequences of double-stranded oligonucleotides used in EMSAs are marked below the corresponding Dof-boxes. Mutated nucleotides in Dof-box (–326/–323) and its corresponding sequences are shown in bold. (F) Interaction of nuclear extracts from 3-week-old Arabidopsis leaves with Gp-I-Dof-wt and Dof-(–326/–323)-mut probes. Crude nuclear extracts (5 μg) from 48 h dark-treated plants (DD in lanes 3, 5, 7, and 9) or control plants (LD in lanes 2, 4, 6, and 8) were incubated with biotin end-labelled Gp-I-Dof-wt (lanes 2–7) or Dof-(–326/–323)-mut (lanes 8 and 9) probes, in the absence (lanes 2, 3, 8, and 9) or presence of a 200-fold molar excess of unlabelled competitor, Gp-I-Dof-wt (lanes 6 and 7), or Dof-(–326/–323)-mut (lanes 4 and 5). Lanes 1 and 10 are free probes without addition of crude nuclear extracts. (G) Nucleotide sequences of double-stranded oligonucleotides used in EMSAs for characterization of the GT-1 cis-element in the ACBP3 5'-flanking region. The mutated nucleotides in GT-1-(–406/–401)-mut and their corresponding sequences in GT-1-(–406/–401)-wt are shown in bold. (H) Interaction of nuclear extracts from 48 h dark-treated (DD) or 48 h light-treated (LL) 3-week-old Arabidopsis leaves with GT-1-(–406/–401)-wt and GT-1-(–406/–401)-mut probes. Crude nuclear extracts (5 μg) from dark-treated (DD in lanes 3, 5, 7, and 9) or light-treated plants (LL in lanes 2, 4, 6, and 8) were incubated with biotin end-labelled GT-1-(–406/–401)-wt (lanes 2–7) or GT-1-(–406/–401)-mut (lanes 8 and 9) probes, in the absence (lanes 2, 3, 8, and 9) or presence of a 200-fold molar excess of unlabelled competitor, GT-1-(–406/–401)-wt (lanes 4 and 5), or GT-1-(–406/–401)-mut (lanes 6 and 7). Lanes 1 and 10 are free probes without addition of crude nuclear extracts.
Subsequently, another set of EMSAs was used to distinguish between the two putative Dof-boxes (–341/–338 and –326/–323) within the Gp-I-Dof sequence in dark-induced binding to nuclear proteins. To this end, each was mutated to generate Dof-(–341/–338)-mut and Dof-(–326/–323)-mut probes (Fig. 4C, E). The results indicate that dark-treated (DD) nuclear extracts bound to Dof (–341/–338) (Fig. 4D), but not Dof (–326/–323) (Fig. 4F). The Gp-I-Dof-wt probe bound to nuclear proteins from DD leaves (Fig. 4D, lane 3), but not LD leaves (Fig. 4D, lane 2). Binding was eliminated in specific competition with a 200-fold excess of unlabelled Gp-I-Dof-wt probe (Fig. 4D, lane 7). In contrast, the corresponding Dof-(–341/–338)-mut probe showed no binding to either control (LD) (Fig. 4D, lane 8) or DD leaves (Fig. 4D, lane 9). The specificity of this binding was further demonstrated when unlabelled Dof-(–341/–338)-mut could not compete against labelled Gp-I-Dof-wt (Fig. 4D, lane 5). Its corresponding mutant probe Dof-(–326/–323)-mut bound to nuclear extracts from DD leaves (Fig. 4F, lane 9), in comparison with LD samples (Fig. 4F, lane 8). In cold competition experiments, the addition of a 200-fold molar excess of unlabelled Dof-(–326/–323)-mut interrupted the binding of the labelled Gp-I-Dof-wt probe to DD leaf extracts (Fig. 4F, lane 5). This suggests that the second putative Dof-box (–326/–323) does not function in dark regulation. EMSA studies confirmed that the first Dof-box (–341/–338) solely controls dark-induced regulation in ACBP3.
Furthermore, analysis of deletion constructs pAT439 (–434/+223) and pAT440 (–274/+223) expressed in transgenic Arabidopsis indicated a putative GT-1 cis-element between –434 and –274. EMSAs using leaf nuclear extracts from 2 d dark-adapted or 2 d light-grown 3-week-old Arabidopsis were used to investigate the role of the putative GT-1 cis-element. Formation of DNA–protein complexes (Fig. 4H, lanes 2 and 3) indicates that the GT sequence binds nuclear extracts from both LL and DD leaves. To assess GT-1 binding further, specific competition assays were included. Addition of a 200-fold molar excess of unlabelled GT-1-(–406/–401)-wt probe altered binding in dark- as well as light-treated leaves (Fig. 4H, lanes 5 and 4, respectively). The corresponding mutant probe (Fig. 4G) GT-1-(–406/–401)-mut, designed by mutation of the boxII tetramer by replacement of a crucial pair of adjacent G residues with CC (Green et al., 1987, 1988), abolished binding to GT-1 (Fig. 4H, lanes 8 and 9). A 200-fold molar excess of unlabelled GT-1-(–406/–401)-mut probe failed to compete out the labelled GT-1-(–406/–401)-wt probe, indicating that the two consecutive G residues in the GT-1 (5′-GGTTTT-3′) are essential for regulation of ACBP3 (Fig. 4H, lanes 6 and 7).
DNase I footprinting confirms that the Dof-box and GT-1 element function in dark regulation of ACBP3
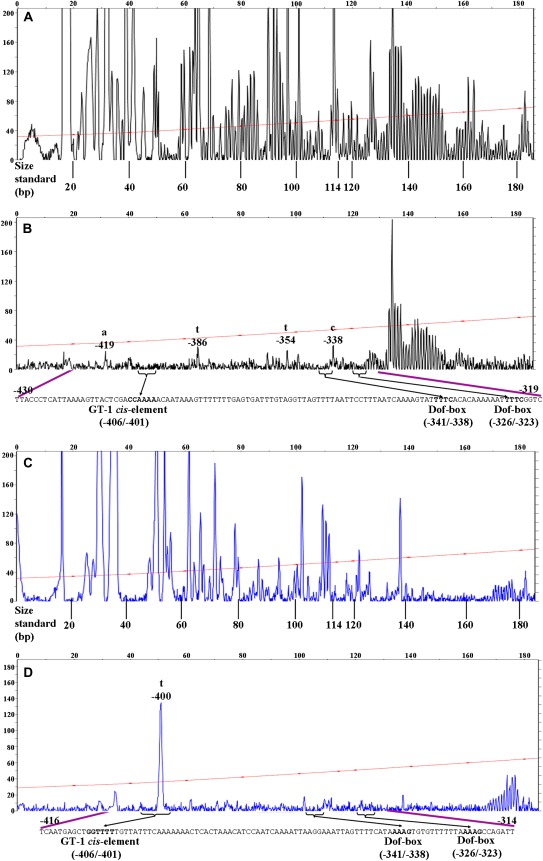
The putative cis-elements involved in dark induction were further characterized by DNase I footprinting analysis on a 307 bp region (–450/–143) spanning one GT-1 cis-element (–406/–401) and six Dof-boxes (–341/–338, –326/–323, –240/–237, –231/–228, –225/–222, and –201/–198). The 5′ end was labelled with fluorescent dyes, NED in the non-coding strand and 6-FAM in the coding strand. Analysis of the 185 bp (–450/–265) fragment displayed both protein-protected and -unprotected fragments within the sense and antisense strands. The position and extent of the protected sequences were deduced by alignment with an accompanying Genescan 600-LIZ size standard to achieve a more thorough and specific comparison of the protection pattern. This alignment is known to be very accurate, with an R2 value of ≥0.98 for each of the size standard curves (Zianni et al. 2006). The footprinting gaps in the signal of protein-containing samples are indicated by purple lines on both dye-labelled probes (Fig. 5).
Fig. 5.
In vitro DNase I footprinting analysis of the ACBP3 5′-flanking region. Digestion patterns of 5′-end NED-labelled (black peaks) ACBP3 antisense strand incubated in the absence (A) or presence (B) of nuclear extracts from dark-treated (DD) Arabidopsis leaves. The 5′-end 6-FAM-labelled (blue peaks) sense strand was also used in the same type of analysis without (C) or with (D) DD crude nuclear extracts. The purple lines identify the area that shows a significant difference in the peak pattern. The putative transcription factors of one GT-1 cis-element (–406/–401) and two Dof-boxes (–341/–338 and –326/–323) are in bold on the corresponding sequences and are shown by the horizontal brackets on traces. The numbers refer to the nucleotide sequence in the ACBP3 5'-flanking region. The LIZ-600 standard (red line with asterisks) was used in localization of the protected region, with the sizes marked below. The fluorescence intensity of DNA fragments (ordinate) is plotted against the sequence length of the fragment (abscissa).
Comparison of DNA digestion of the antisense strand pattern in the presence of dark-treated nuclear extracts (Fig. 5B) and in the absence of nuclear extracts (Fig. 5A) revealed a strongly protected area of 111 bp (–430/–319). There were several obvious hypersensitive locations on DNase I footprint analysis (Fig 5B, bases –419, –386, –354, and –338). The –430/–319 region is known to contain three transcription factor-binding sites, one GT-1 cis-element (–406/–401) and two Dof-boxes (–341/–338 and –326/–323). However, the interaction of Dof-box (–341/–338), rather than Dof-box (–326/–323), with dark-treated nuclear extracts has been ascertained in gel retardation studies. As expected, in the negative control using BSA, no protection was observed and uniform peaks appeared (Fig. 5A). The traces (blue peaks) were also examined for loss of signal in the protein-containing sample (Fig. 5D) in comparison with the BSA control from the sense strand (Fig. 5C). The probable binding location from –416 to –314 was deemed to be DNase I resistant, with one unprotected base at –400 that splits this region; this base was accessible to DNase I regardless of the presence of dark-treated nuclear extracts. As very similar peak patterns occurred from –265 to –143 when the dark-treated sample was compared with the control from both strands (data not shown), it was inferred that proteins from dark-treated leaves do not protect this region. Results from DNase I footprinting revealed one distinct foot signature region corresponding to the GT-1-binding site (5′-GGTTTT-3′) and the Dof-box-binding site (5′-AAAG-3′) on both coding and non-coding strands. This clearly illustrates specific binding, at base pair resolution, of putative transcription-regulated proteins. The finding is consistent with results from EMSAs in that the five predicted Dof-boxes (–326/–323, –240/–237, –231/–228, –225/–222, and –201/–198) lack activity in dark-induced regulation, and the Dof-box (–341/–338) and GT-1 cis-element (–406/–401) are the confirmed dark-responsive elements interacting with protein extracts from dark-treated leaves.
ACBP3pro::GUS expression is induced by pathogen and pathogen-related phytohormones
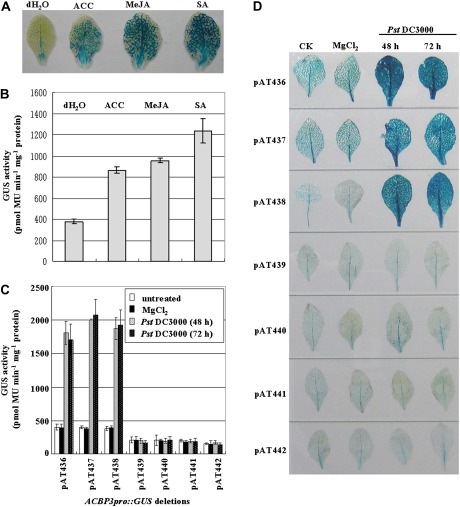
Previous analysis identified ACBP3 as playing a crucial role in plant defence (Xiao and Chye, 2011b). To identify the cis-elements of the ACBP3 5′-flanking region in phytopathogen- and phytohormone-induced regulation, GUS assays were performed on 5-week-old transgenic plants harbouring the 1698 bp ACBP3pro::GUS fusion following treatment with phytohormones, including ACC (precursor of ethylene), MeJA, and SA, the latter two being secondary messengers in signal transduction (Kunkel and Brooks, 2002). GUS was strongly expressed in leaf veins after phytohormone treatment (Fig. 6A). Exogenous application of ACC, MeJA, and SA triggered GUS activity 2.3-, 2.5-, and 3.3-fold, respectively (Fig. 6B).
Fig. 6.
ACBP3pro::GUS is inducible by phytohormones and pathogen infection. (A and B) The expression of construct pAT436 in response to ACC, MeJA, and SA treatment. Five-week-old transgenic Arabidopsis were treated with distilled water, 1 mM ACC, 100 μM MeJA, or 1 mM SA for 12 h before GUS staining assay (A) or quantitative fluorometric assays for GUS activity (B). (C and D) The expression of the ACBP3pro::GUS deletion derivatives in response to pathogen infection. Five-week-old Arabidopsis harbouring various ACBP3 5′-deletion constructs were inoculated with Pseudomonas syringae pv. tomato DC3000 or 10 mM MgCl2 (control) and then collected 48 h and 72 h after inoculation before quantitative fluorometric assays for GUS activity (C) or GUS staining assay (D). For GUS activity data, average values were obtained from experiments performed with 3–5 independent lines per construct, each line represented by 8–10 individual plants. Bars indicate the standard errors of three replicates. For histochemical GUS staining data, the experiment was repeated three times using 8–10 individual plants of each construct.
After treatment with P. syringae, transformants of the three largest constructs pAT436, pAT437, and pAT438 showed 5-fold up-regulation in comparison with the control (MgCl2) and uninfected leaves (Fig. 6C). No induction was evident in both quantitative (Fig. 6C) and qualitative (Fig. 6D) assays from the other four deletions (pAT439, pAT440, pAT441, and pAT442). In addition, a similar pattern was observed at 48 h and 72 h post-inoculation. Little GUS activity was observed at the site of inoculation, and pathogen-induced expression was detected throughout the entire leaf (Fig. 6D), presumably because the cells around the inoculation site were damaged by the injection. These findings indicate that pathogen-inducible motif(s) are located between nucleotide positions –543 and –434.
Pathogen-induced expression of ACBP3pro::GUS involves an S-box regulatory element
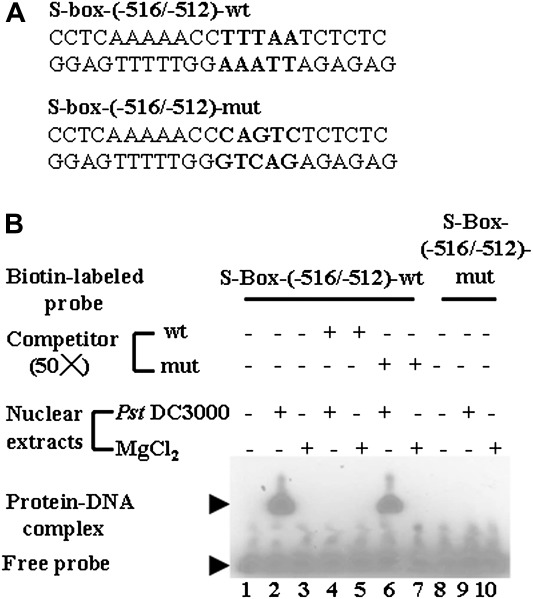
An attempt was made to identify the pathogen-inducible cis-acting element(s). Sequence analysis between nucleotides –543 and –434 identified an S-box (TTTAA) at position–516/–512 by computer program (PlantProm) DB prediction. When EMSAs with two double-stranded DNA probes, S-box-(–516/–512)-wt and its corresponding mutant S-box-(–516/–512)-mut (Fig. 7A), were used to detect interaction using pathogen-infected crude nuclear extracts, one band was observed in Fig. 7B (lane 2), indicating that the S-box-(–516/–512)-wt probe reacted with P. syringae-infected nuclear extracts but not in the mock inoculation (MgCl2). The mobility shift was completely abolished by the addition of a 50-fold molar solution of cold unlabelled S-box-(–516/–512)-wt, but not by the unlabelled S-box-(–516/–512)-mut probe (Fig. 7B, lanes 4 and 6, respectively). Subsequently, when MgCl2- or pathogen-treated nuclear extracts were incubated with the S-box mutant probe, no binding was observed (Fig. 7B, lanes 9 and 10). From these results, the S-box (TTTAA) at –516/–512 was confirmed to bind nuclear extracts from pathogen-infected leaves.
Fig. 7.
Identification of an S-box in the ACBP3 5'-flanking region. (A) Nucleotide sequences of double-stranded oligonucleotides used in EMSAs. The mutated nucleotides in S-box-(–516/–512)-mut and their corresponding sequences in S-box-(–516/–512)-wt are shown in bold. (B) Interaction of nuclear extracts from 5-week-old Arabidopsis leaves with S-box-(–516/–512)-wt and S-box-(–516/–512)-mut probes. Crude nuclear extracts (4 μg) from 48 h pathogen-infected (Pseudomonas syringae pv. tomato DC3000, lanes 2, 4, 6, and 9) or MgCl2-treated (control) Arabidopsis (MgCl2, lanes 3, 5, 7, and 10) were incubated with biotin-labelled S-box-(–516/–512)-wt (lanes 2–7) or S-box-(–516/–512)-mut (lanes 9 and 10) in the absence (lanes 2, 3, 9, and 10) or presence of a 50-fold molar excess of unlabelled competitor, S-box-(–516/–512)-wt (lanes 4 and 5), or S-box-(–516/–512)-mut (lanes 6 and 7). Lanes 1 and 8 are free probes without addition of crude nuclear extracts.
Discussion
The ACBP family of six members in Arabidopsis has been characterized using gene knock-out mutants and overexpression lines, but none of their corresponding promoters has been cloned and analysed (Xiao and Chye, 2009, 2011a). Here, analysis of a 1.7 kb 5'-flanking region of ACBP3 from Arabidopsis, representing the first ACBP promoter from plants, revealed that Dof and GT-1 activate dark-responsive regulation of ACBP3 expression. Another cis-element, the S-box, seems to be involved in transcriptional regulation of ACBP3 expression during pathogen response.
Temporal and spatial expression of ACBP3
The high activity from the ACBP3 promoter fragment (–1475/+223) in comparison with the CaMV 35S promoter indicates that this 1698 bp ACBP3 5′-flanking region confers relatively strong expression. Given its pathogen inducibility, this promoter has potential applications in driving heterologous gene expression in other higher plants. A significant increase in activity occurred between –543 and –434, suggesting the presence of enhancer(s). Given that CAAT-boxes are known to act as enhancers to potentiate transcription and to regulate transcription frequency, an adjacent putative CAAT-box (–372/–368) may be responsible for up-regulation (Fang et al., 1989).
Investigations on the spatial and temporal expression revealed that GUS is ubiquitously expressed in all vegetative tissues, more highly in young seedlings, but less in siliques. Such constitutively high expression patterns may be attributed to protection against pathogen invasion in vegetative organs, and these results are consistent with previous findings on ACBP3 mRNA induction upon pathogen infection (Xiao et al., 2010; Xiao and Chye, 2011b). High expression of GUS was observed in pistils of fully opened flowers, and such accumulation in floral organs (i.e. stigma, style, and ovary) may be related to ACBP3 function in defence against pathogens. Given that ACBP3 is a phospholipid-binding protein with secretory activity, and pollen–stigma interaction during pollination requires various lipids (Edlund et al., 2004; Sanchez et al., 2004), ACBP3 may well play a role in this process. Further investigations by lipid profiling are needed to elucidate ACBP3 function in flowers. ACBP3pro::GUS was also expressed in the vascular tissues in leaves and stems, prompting the speculation that ACBP3 may be associated with defence and long-distance lipid transport (Proels and Roitsch, 2009; Xiao et al., 2010). ACBP3pro::GUS expression in leaf veins was inducible by phytohormone treatments and pathogen infection. Given that plant defence-related genes are often expressed in the vasculature across a variety of plant species (Eyal et al., 1993; Breda et al., 1996), the accumulation of ACBP3pro::GUS in the vascular structures would strengthen its role in plant defence.
Role of the Dof-box and GT-1 in regulation of dark-inducible ACBP3 expression
GUS activities in the transformants expressing construct pAT436 conferred ACBP3 circadian regulation and dark/light responsiveness, in good agreement with results of ACBP3 mRNA expression under dark and light conditions and the role of ACBP3 in regulating dark starvation-induced leaf senescence (Xiao et al., 2010). By computational prediction, two EEs in the 5'-flanking region of ACBP3 were identified. EEs have been located in the 5′-flanking regions of many clock-controlled genes from various plants (Harmer et al., 2000). Two EEs from the 5′-flanking region of SmCP from Solanum melongena show cooperative binding activity in late light (Rawat et al., 2005), while another two have been identified in the tobacco ZGT 5′-flanking region which is subject to circadian regulation (Xu and Johnson, 2001). Recently, Wang et al. (2011) have reported that EEs are enriched in the 5′-flanking regions of 22 novel plant defence genes, and 14 of these 22 contain the EE-binding site and/or are subject to rhythmic regulation. However, the putative EEs in the ACBP3 5′-flanking region were not observed to be functional despite circadian regulation and dark induction of ACBP3. Instead, a 160 bp region containing a functional Dof-box (–341/–338) was identified, suggesting that it confers dark-induced expression of ACBP3. Wang et al. (2011) proposed that defence genes are circadian regulated and infection is anticipated at dawn, coinciding with pathogen activity in spore dispersal during the light period. Given that ACBP3 is pathogen inducible, it is not surprising that there is a link between dark induction and plant defence.
A GT-1 cis-element (–406/–401) was also functional in dark regulation. Thus the Dof and GT-1 proteins may share both redundant and non-redundant roles in dark regulation of ACBP3 expression. Given that these two binding motifs (GT-1 and Dof-box) are located within 63 bp from each other, they may cooperate as a combined ensemble and ACBP3 could possibly be transcriptionally regulated by two separate regulatory pathways, the Dof/Dof-box pathway and the GT-1/GT-1 cis-element pathway. However, further investigations are needed to explore if these motifs interact with other (positive/negative) motifs in dark regulation of ACBP3.
The Dof domain is unique to higher plants and typically consists of 52 amino acids with one C2–C2-type zinc finger motif. The binding site of an AAAG sequence or its reverse orientated sequence (CTTT) is the Dof protein recognition core (Yanagisawa, 2002). In this study, it was found that although all six putative Dof-binding domains comprise the conserved AAAG sequence, only one Dof-box (–341/–338) regulates ACBP3 expression in response to dark.
GT-1 is a well-studied cis-acting DNA element in the plant kingdom and was first identified in Pisum sativum and termed as a boxII element in the promoter of rbcS-3A (Green et al., 1987). Four to five Ts or As preceded by one or two G nucleotides at the 5' end is a common feature of the core sequence (Zhou, 1999). In contrast to the Dof-box, its high degeneracy results in only a moderate consensus [5′-G-Pu-(T/A)-A-A-(T/A)], and its diverse functions include the induction of genes subject to many environmental responses, predominantly in light and pathogen regulation (Dehesh et al., 1992; Pasquali et al., 1999). In comparison, the GT-1 cis-element in the 5′-flanking region of ACBP3 at position –406/–401 (GGTTTT) shares lower homology to those from pathogen-regulated gene promoters, including soybean SCaM-4, Catharanthus roseus cpr, bean chs, and tobacco PR-la which are induced by pathogen, salt stress, fungal elicitor, or SA (Lawton et al., 1991; Buchel et al., 1996; Cardoso et al., 1997; Park et al., 2004). This may provide a reason for the lack of pathogen responsiveness of this GT-1 cis-element. In contrast, sequence comparisons revealed that this GT-1 cis-element fits the consensus of many light-regulatory GT-binding sequences in a variety of genes, such as the paired GT-1-binding site in the rbcS-3A promoter in pea (GGTTAA and GGTAAT, boxII and boxII*, respectively), and the GT1-bx and GT2-bx in the PhyA promoter of rice (GGTTAA and GGTAAT, respectively) (nucleotide mismatches underlined) (Green et al., 1987, 1988; Kay et al., 1989; Dehesh et al., 1992). Given that all these genes are dark/light responsive, the ACBP3 5′-flanking region may recruit a similar regulatory mechanism to regulate its response to dark/light. More interestingly, it was observed that nuclear extracts isolated from dark- as well as light-treated Arabidopsis bind to the GT-1 cis-element in the 5′-flanking region of ACBP3, similar to those of pea rbcS-3A and the Arabidopsis Pc promoters (Green et al., 1987; Fisscher et al., 1994). Also, it has been reported that the increased expression of mRNA levels of GT-1a from tobacco and GT-2 protein from Arabidopsis are independent of light (Gilmartin et al., 1992; Perisic and Lam, 1992; Kuhn et al., 1993). The present findings suggest that the Dof-box (–341/–338) and the GT-1 cis-element (–406/–401) in the ACBP3 5′-flanking region regulate dark-inducible ACBP3 expression.
Control of pathogen-inducible ACBP3 expression by an S-box element
Here, it is further reported that the ACBP3 5′-flanking region is responsive to phytohormones and pathogens. EMSA results show that an S-box (–516/–512) is essential in binding pathogen-treated nuclear extracts, suggesting that it positively regulates ACBP3 during pathogen attack. The S-box is a small AT-rich motif (TTTAA) that binds to the high mobility group I (HMG I) protein of the HMG family (Lund et al., 1983). HMG I has been isolated from representatives in all eukaryotes and characterized in numerous plant species including wheat, barley, maize, and Arabidopsis (Jacobsen et al., 1990). This protein prefers to bind double-stranded DNA with six or more AT base pairs, and the functions of HMG I have been proposed to include nuclear scaffold–DNA interactions in vivo (Solomon et al., 1986). Jacobsen et al. (1990) showed that nuclear factors isolated from soybean recognize specific AT-rich sequences in nodulin promoters, suggesting that plant HMG I protein is more tightly bound to chromatin than in mammals. Similar to ACBP3, B. napus rbsSF1 is also light regulated, and an S-box that binds to leaf nuclear proteins in vitro has been identified (Nantel et al., 1991).
Taken together, the present observations have identified three cis-elements in the ACBP3 5'-flanking region, and their significance in regulation of ACBP3 expression in response to dark/light and pathogens has been documented. Subsequent investigations to isolate and characterize the transcription factors which bind to the Dof-box, GT-1, and the S-box, as well as confirmation of their potential roles in DNA–protein interactions should be carried out to better understand ACBP3 expression during dark/light and plant defence responses.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Nucleotide sequence of the ACBP3 5'-flanking region.
Table S1. Oligonucleotide primers used in this study for PCR walking, sequence analysis, EMSAs, and DNase I footprinting.
Acknowledgments
This work was supported by the University Grants Committee of the Hong Kong Special Administrative Region, China (Project no. AoE/B-07/99) and The University of Hong Kong (10208034 and 10400058). S-XZ was supported by a postgraduate studentship from The University of Hong Kong.
Glossary
Abbreviations
- ACBP
acyl-CoA-binding protein
- ACC
1-aminocyclopropane-1-carboxylic acid
- DD
continuous darkness
- Dof
DNA binding with one finger
- EE
evening element
- EMSA
electrophoretic mobility shift assay
- ER
endoplasmic reticulum
- GUS
β-glucuronidase
- HMG
high mobility group
- LD
16 h light/8 h dark cycle
- LL
continuous light
- MeJA
methyl jasmonate
- PE
phosphatidylethanolamine
- SA
salicylic acid
References
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Analytical Biochemistry. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Breda C, Sallaud C, El-Turk J, Buffard D, de Kozak I, Esnault R, Kondorosi A. Defense reaction in Medicago sativa: a gene encoding a class 10 PR protein is expressed in vascular bundles. Molecular Plant-Microbe Interactions. 1996;9:713–719. doi: 10.1094/mpmi-9-0713. [DOI] [PubMed] [Google Scholar]
- Buchel AS, Molenkamp R, Bol JF, Linthorst HJ. The PR-1a promoter contains a number of elements that bind GT-1-like nuclear factors with different affinity. Plant Molecular Biology. 1996;30:493–504. doi: 10.1007/BF00049327. [DOI] [PubMed] [Google Scholar]
- Cardoso MI, Meijer AH, Rueb S, Machado JA, Memelink J, Hoge JH. A promoter region that controls basal and elicitor-inducible expression levels of the NADPH:cytochrome P450 reductase gene (Cpr) from Catharanthus roseus binds nuclear factor GT-1. Molecular and General Genetics. 1997;256:674–681. doi: 10.1007/pl00008617. [DOI] [PubMed] [Google Scholar]
- Chen QF, Xiao S, Chye ML. Overexpression of the Arabidopsis 10-kilodalton acyl-coenzyme A-binding protein ACBP6 enhances freezing tolerance. Plant Physiology. 2008;148:304–315. doi: 10.1104/pp.108.123331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen QF, Xiao S, Qi W, Mishra G, Ma J, Wang M, Chye ML. The Arabidopsis acbp1acbp2 double mutant lacking acyl-CoA-binding proteins ACBP1 and ACBP2 is embryo lethal. New Phytologist. 2010;186:843–855. doi: 10.1111/j.1469-8137.2010.03231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong YH, Chang HS, Gupta R, Wang X, Zhu T, Luan S. Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiology. 2002;129:661–677. doi: 10.1104/pp.002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chye ML. Arabidopsis cDNA encoding a membrane-associated protein with an acyl-CoA binding domain. Plant Molecular Biology. 1998;38:827–838. doi: 10.1023/a:1006052108468. [DOI] [PubMed] [Google Scholar]
- Chye ML, Huang BQ, Zee SY. Isolation of a gene encoding Arabidopsis membrane-associated acyl-CoA binding protein and immunolocalization of its gene product. The Plant Journal. 1999;18:205–214. doi: 10.1046/j.1365-313x.1999.00443.x. [DOI] [PubMed] [Google Scholar]
- Chye ML, Li HY, Yung MH. Single amino acid substitutions at the acyl-CoA-binding domain interrupt 14[C]palmitoyl-CoA binding of ACBP2, an Arabidopsis acyl-CoA-binding protein with ankyrin repeats. Plant Molecular Biology. 2000;44:711–721. doi: 10.1023/a:1026524108095. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Dehesh K, Hung H, Tepperman JM, Quail PH. GT-2: a transcription factor with twin autonomous DNA-binding domains of closely related but different target sequence specificity. The EMBO Journal. 1992;11:4131–4144. doi: 10.1002/j.1460-2075.1992.tb05506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du ZY, Xiao S, Chen QF, Chye ML. Depletion of the membrane-associated acyl-coenzyme A-binding protein ACBP1 enhances the ability of cold acclimation in Arabidopsis. Plant Physiology. 2010;152:1585–1597. doi: 10.1104/pp.109.147066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlund AF, Swanson R, Preuss D. Pollen and stigma structure and function: the role of diversity in pollination. The Plant Cell. 2004;16(Suppl):S84–S97. doi: 10.1105/tpc.015800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyal Y, Meller Y, Lev-Yadun S, Fluhr R. A basic-type PR-1 promoter directs ethylene responsiveness, vascular and abscission zone-specific expression. The Plant Journal. 1993;4:225–234. doi: 10.1046/j.1365-313x.1993.04020225.x. [DOI] [PubMed] [Google Scholar]
- Faergeman NJ, Knudsen J. Acyl-CoA binding protein is an essential protein in mammalian cell lines. Biochemical Journal. 2002;368:679–682. doi: 10.1042/BJ20021413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang RX, Nagy F, Sivasubramaniam S, Chua NH. Multiple cis regulatory elements for maximal expression of the cauliflower mosaic virus 35S promoter in transgenic plants. The Plant Cell. 1989;1:141–150. doi: 10.1105/tpc.1.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisscher U, Weisbeek P, Smeekens S. Identification of potential regulatory elements in the far-upstream region of the Arabidopsis thaliana plastocyanin promoter. Plant Molecular Biology. 1994;26:873–886. doi: 10.1007/BF00028855. [DOI] [PubMed] [Google Scholar]
- Gao W, Li HY, Xiao S, Chye ML. Acyl-CoA-binding protein 2 binds lysophospholipase 2 and lysoPC to promote tolerance to cadmium-induced oxidative stress in transgenic Arabidopsis. The Plant Journal. 2010;62:989–1003. doi: 10.1111/j.1365-313X.2010.04209.x. [DOI] [PubMed] [Google Scholar]
- Gao W, Xiao S, Li HY, Tsao SW, Chye ML. Arabidopsis thaliana acyl-CoA-binding protein ACBP2 interacts with heavy-metal-binding farnesylated protein AtFP6. New Phytologist. 2009;181:89–102. doi: 10.1111/j.1469-8137.2008.02631.x. [DOI] [PubMed] [Google Scholar]
- Gilmartin PM, Memelink J, Hiratsuka K, Kay SA, Chua NH. Characterization of a gene encoding a DNA binding protein with specificity for a light-responsive element. The Plant Cell. 1992;4:839–849. doi: 10.1105/tpc.4.7.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmartin PM, Sarokin L, Memelink J, Chua NH. Molecular light switches for plant genes. The Plant Cell. 1990;2:369–378. doi: 10.1105/tpc.2.5.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green PJ, Kay SA, Chua NH. Sequence-specific interactions of a pea nuclear factor with light-responsive elements upstream of the rbcS-3A gene. EMBO Journal. 1987;6:2543–2549. doi: 10.1002/j.1460-2075.1987.tb02542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green PJ, Yong MH, Cuozzo M, Kano-Murakami Y, Silverstein P, Chua NH. Binding site requirements for pea nuclear protein factor GT-1 correlate with sequences required for light-dependent transcriptional activation of the rbcS-3A gene. EMBO Journal. 1988;7:4035–4044. doi: 10.1002/j.1460-2075.1988.tb03297.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- Jacobsen K, Laursen NB, Jensen EO, Marcker A, Poulsen C, Marcker KA. HMG I-like proteins from leaf and nodule nuclei interact with different AT motifs in soybean nodulin promoters. The Plant Cell. 1990;2:85–94. doi: 10.1105/tpc.2.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO Journal. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay SA, Keith B, Shinozaki K, Chye ML, Chua NH. The rice phytochrome gene: structure, autoregulated expression, and binding of GT-1 to a conserved site in the 5' upstream region. The Plant Cell. 1989;1:351–360. doi: 10.1105/tpc.1.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen J, Neergaard TB, Gaigg B, Jensen MV, Hansen JK. Role of acyl-CoA binding protein in acyl-CoA metabolism and acyl-CoA-mediated cell signaling. Journal of Nutrition. 2000;130:294S–298S. doi: 10.1093/jn/130.2.294S. [DOI] [PubMed] [Google Scholar]
- Kuhn RM, Caspar T, Dehesh K, Quail PH. DNA binding factor GT-2 from Arabidopsis. Plant Molecular Biology. 1993;23:337–348. doi: 10.1007/BF00029009. [DOI] [PubMed] [Google Scholar]
- Kunkel BN, Brooks DM. Cross talk between signaling pathways in pathogen defense. Current Opinion in Plant Biology. 2002;5:325–331. doi: 10.1016/s1369-5266(02)00275-3. [DOI] [PubMed] [Google Scholar]
- Lam E, Chua NH. GT-1 binding site confers light responsive expression in transgenic tobacco. Science. 1990;248:471–474. doi: 10.1126/science.2330508. [DOI] [PubMed] [Google Scholar]
- Lawton MA, Dean SM, Dron M, Kooter JM, Kragh KM, Harrison MJ, Yu L, Tanguay L, Dixon RA, Lamb CJ. Silencer region of a chalcone synthase promoter contains multiple binding sites for a factor, SBF-1, closely related to GT-1. Plant Molecular Biology. 1991;16:235–249. doi: 10.1007/BF00020555. [DOI] [PubMed] [Google Scholar]
- Leung KC, Li HY, Mishra G, Chye ML. ACBP4 and ACBP5, novel Arabidopsis acyl-CoA-binding proteins with kelch motifs that bind oleoyl-CoA. Plant Molecular Biology. 2004;55:297–309. doi: 10.1007/s11103-004-0642-z. [DOI] [PubMed] [Google Scholar]
- Leung KC, Li HY, Xiao S, Tse MH, Chye ML. Arabidopsis ACBP3 is an extracellularly targeted acyl-CoA-binding protein. Planta. 2006;223:871–881. doi: 10.1007/s00425-005-0139-2. [DOI] [PubMed] [Google Scholar]
- Li HY, Chye ML. Membrane localization of Arabidopsis acyl-CoA binding protein ACBP2. Plant Molecular Biology. 2003;51:483–492. doi: 10.1023/a:1022330304402. [DOI] [PubMed] [Google Scholar]
- Li HY, Chye ML. Arabidopsis acyl-CoA-binding protein ACBP2 interacts with an ethylene-responsive element-binding protein, AtEBP, via its ankyrin repeats. Plant Molecular Biology. 2004;54:233–243. doi: 10.1023/B:PLAN.0000028790.75090.ab. [DOI] [PubMed] [Google Scholar]
- Li HY, Xiao S, Chye ML. Ethylene- and pathogen-inducible Arabidopsis acyl-CoA-binding protein 4 interacts with an ethylene-responsive element binding protein. Journal of Experimental Botany. 2008;59:3997–4006. doi: 10.1093/jxb/ern241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund T, Holtlund J, Fredriksen M, Laland SG. On the presence of two new high mobility group-like proteins in HeLa S3 cells. FEBS Letters. 1983;152:163–167. doi: 10.1016/0014-5793(83)80370-6. [DOI] [PubMed] [Google Scholar]
- Maxwell BB, Andersson CR, Poole DS, Kay SA, Chory J. HY5, Circadian Clock-Associated 1, and a cis-element, DET1 dark response element, mediate DET1 regulation of chlorophyll a/b-binding protein 2 expression. Plant Physiology. 2003;133:1565–1577. doi: 10.1104/pp.103.025114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologia Plantarum. 1962;15:473–497. [Google Scholar]
- Nantel AM, Lafleur F, Boivin R, Baszczynski CL, Bellemare G. Promoter for a Brassica napus ribulose bisphosphate carboxylase/oxygenase small subunit gene binds multiple nuclear factors and contains a negative-strand open reading frame encoding a putative transmembrane protein. Plant Molecular Biology. 1991;16:955–966. doi: 10.1007/BF00016068. [DOI] [PubMed] [Google Scholar]
- Park HC, Kim ML, Kang YH, et al. Pathogen- and NaCl-induced expression of the SCaM-4 promoter is mediated in part by a GT-1 box that interacts with a GT-1-like transcription factor. Plant Physiology. 2004;135:2150–2161. doi: 10.1104/pp.104.041442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquali G, Erven AS, Ouwerkerk PB, Menke FL, Memelink J. The promoter of the strictosidine synthase gene from periwinkle confers elicitor-inducible expression in transgenic tobacco and binds nuclear factors GT-1 and GBF. Plant Molecular Biology. 1999;39:1299–1310. doi: 10.1023/a:1006177414456. [DOI] [PubMed] [Google Scholar]
- Perisic O, Lam E. A tobacco DNA binding protein that interacts with a light-responsive box II element. The Plant Cell. 1992;4:831–838. doi: 10.1105/tpc.4.7.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proels RK, Roitsch T. Extracellular invertase LIN6 of tomato: a pivotal enzyme for integration of metabolic, hormonal, and stress signals is regulated by a diurnal rhythm. Journal of Experimental Botany. 2009;60:1555–1567. doi: 10.1093/jxb/erp027. [DOI] [PubMed] [Google Scholar]
- Rawat R, Xu ZF, Yao KM, Chye ML. Identification of cis-elements for ethylene and circadian regulation of the Solanum melongena gene encoding cysteine proteinase. Plant Molecular Biology. 2005;57:629–643. doi: 10.1007/s11103-005-0954-7. [DOI] [PubMed] [Google Scholar]
- Rombauts S, Déhais P, Van Montagu M, Rouzé P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Research. 1999;27:295–296. doi: 10.1093/nar/27.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez AM, Bosch M, Bots M, Nieuwland J, Feron R, Mariani C. Pistil factors controlling pollination. The Plant Cell. 2004;16(Suppl):S98–S106. doi: 10.1105/tpc.017806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahmuradov IA, Gammerman AJ, Hancock JM, Bramley PM, Solovyev VV. PlantProm: a database of plant promoter sequences. Nucleic Acids Research. 2003;31:114–117. doi: 10.1093/nar/gkg041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoyab M, Gentry LE, Marquardt H, Todaro GJ. Isolation and characterization of a putative endogenous benzodiazepineoid (endozepine) from bovine and human brain. Journal of Biological Chemistry. 1986;261:11968–11973. [PubMed] [Google Scholar]
- Solomon MJ, Strauss F, Varshavsky A. A mammalian high mobility group protein recognizes any stretch of six A·T base pairs in duplex DNA. Proceedings of the National Academy of Sciences, USA. 1986;83:1276–1280. doi: 10.1073/pnas.83.5.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HW, Zhang B, Hao YJ, Huang J, Tian AG, Liao Y, Zhang JS, Chen SY. The soybean Dof-type transcription factor genes, GmDof4 and GmDof11, enhance lipid content in the seeds of transgenic Arabidopsis plants. The Plant Journal. 2007;52:716–729. doi: 10.1111/j.1365-313X.2007.03268.x. [DOI] [PubMed] [Google Scholar]
- Wang W, Barnaby JY, Tada Y, Li H, Tör M, Caldelari D, Lee DU, Fu XD, Dong X. Timing of plant immune responses by a central circadian regulator. Nature. 2011;470:110–114. doi: 10.1038/nature09766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmott RL, Rushton PJ, Hooley R, Lazarus CM. DNase1 footprints suggest the involvement of at least three types of transcription factors in the regulation of alpha-Amy2/A by gibberellin. Plant Molecular Biology. 1998;38:817–825. doi: 10.1023/a:1006084104041. [DOI] [PubMed] [Google Scholar]
- Wilson DO, Johnson P, McCord BR. Nonradiochemical DNase I footprinting by capillary electrophoresis. Electrophoresis. 2001;22:1979–1986. doi: 10.1002/1522-2683(200106)22:10<1979::AID-ELPS1979>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Xiao S, Chen QF, Chye ML. Expression of ACBP4 and ACBP5 proteins is modulated by light in Arabidopsis. Plant Signaling and Behavior. 2009;4:1063–1065. doi: 10.4161/psb.4.11.9718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Chye ML. An Arabidopsis family of six acyl-CoA-binding proteins has three cytosolic members. Plant Physiology and Biochemistry. 2009;47:479–484. doi: 10.1016/j.plaphy.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Xiao S, Chye ML. New roles for acyl-CoA-binding proteins (ACBPs) in plant development, stress responses and lipid metabolism. Progress in Lipid Research. 2011a;50:141–151. doi: 10.1016/j.plipres.2010.11.002. [DOI] [PubMed] [Google Scholar]
- Xiao S, Chye ML. Overexpression of Arabidopsis ACBP3 enhances NPR1-dependent plant resistance to Pseudomonas syringe pv. tomato DC3000. Plant Physiology. 2011b;156:2069–2081. doi: 10.1104/pp.111.176933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Gao W, Chen QF, Chan SW, Zheng SX, Ma J, Wang M, Welti R, Chye ML. Overexpression of Arabidopsis acyl-CoA binding protein ACBP3 promotes starvation-induced and age-dependent leaf senescence. The Plant Cell. 2010;22:1463–1482. doi: 10.1105/tpc.110.075333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Gao W, Chen QF, Ramalingam S, Chye ML. Overexpression of membrane-associated acyl-CoA-binding protein ACBP1 enhances lead tolerance in Arabidopsis. The Plant Journal. 2008a;54:141–151. doi: 10.1111/j.1365-313X.2008.03402.x. [DOI] [PubMed] [Google Scholar]
- Xiao S, Li HY, Zhang JP, Chan SW, Chye ML. Arabidopsis acyl-CoA-binding proteins ACBP4 and ACBP5 are subcellularly localized to the cytosol and ACBP4 depletion affects membrane lipid composition. Plant Molecular Biology. 2008b;68:571–583. doi: 10.1007/s11103-008-9392-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Johnson CH. A clock- and light-regulated gene that links the circadian oscillator to LHCB gene expression. The Plant Cell. 2001;13:1411–1425. doi: 10.1105/tpc.13.6.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S. The Dof family of plant transcription factors. Trends in Plant Science. 2002;7:555–560. doi: 10.1016/s1360-1385(02)02362-2. [DOI] [PubMed] [Google Scholar]
- Zhou DX. Regulatory mechanism of plant gene transcription by GT-elements and GT-factors. Trends in Plant Science. 1999;4:210–214. doi: 10.1016/s1360-1385(99)01418-1. [DOI] [PubMed] [Google Scholar]
- Zianni M, Tessanne K, Merighi M, Laguna R, Tabita FR. Identification of the DNA bases of a DNase I footprint by the use of dye primer sequencing on an automated capillary DNA analysis instrument. Journal of Biomolecular Techniques. 2006;17:103–113. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.