Abstract
Abscisic acid (ABA) plays important roles during tomato fruit ripening. To study the regulation of carotenoid biosynthesis by ABA, the SlNCED1 gene encoding 9-cis-epoxycarotenoid dioxygenase (NCED), a key enzyme in the ABA biosynthesis, was suppressed in tomato plants by transformation with an RNA interference (RNAi) construct driven by a fruit-specific E8 promoter. ABA accumulation and SlNCED1 transcript levels in the transgenic fruit were down-regulated to between 20–50% of that in control fruit. This significant reduction in NCED activity led to the carbon that normally channels to free ABA as well as the ABA metabolite accumulation during ripening to be partially blocked. Therefore, this ‘backlogged’ carbon transformed into the carotenoid pathway in the RNAi lines resulted in increased assimilation and accumulation of upstream compounds in the pathway, chiefly lycopene and β-carotene. Fruit of all RNAi lines displayed deep red coloration compared with the pink colour of control fruit. The decrease in endogenous ABA in these transgenics resulted in an increase in ethylene, by increasing the transcription of genes related to the synthesis of ethylene during ripening. In conclusion, ABA potentially regulated the degree of pigmentation and carotenoid composition during ripening and could control, at least in part, ethylene production and action in climacteric tomato fruit.
Keywords: ABA, β-carotene, carotenoid biosynthesis, ethylene, fruit ripening, lycopene, SlNCED1-RNAi, tomato
Introduction
Tomato fruit are enriched in lycopene and β-carotene, both of which are highly beneficial for human health (Giuliano et al., 1993; Crozier et al., 1997; Fraser and Bramley, 2004). Therefore, obtaining fruit with higher lycopene and β-carotene levels has been a major goal in tomato breeding. In order to achieve this goal, it is necessary to understand the regulation of carotenoid biosynthesis and to develop related methodologies to enhance it (Adams-Phillips et al., 2004; Giovannoni, 2004). To date, the carotenoid biosynthesis pathway in plants has been clarified at the molecular level, and genes that encode the key enzymes have been cloned (Bramley et al., 1992; Corona et al., 1996; Cunningham and Gantt, 1998; Hirschberg, 2001; Fraser and Bramley, 2004; Romer and Fraser, 2005; DellaPenna and Pogson, 2006). The biosynthetic pathway begins with the formation of phytoene from two molecules of geranylgeranyl diphosphate (GGPP) in the central isoprenoid pathway. Four desaturation steps give rise to lycopene; cyclizations at both ends of the lycopene molecule produce α- or β-carotene, which undergo hydroxylation at C3 and C3′ to form the xanthophylls, lutein and zeaxanthin, respectively. Zeaxanthin epoxidase produces violaxanthin, which is the substrate for neoxanthin synthase. As the latter occurs in the light-harvesting complex in the 9-cis configuration, an isomerization step, probably catalysed by an, as yet unknown, enzyme, is hypothesized. 9-cis violaxanthin and possibly also 9-cis-neoxanthin undergo oxidative cleavage to give cis-xanthoxin.
In the cytosol, abscisic acid (ABA) is formed from xanthoxin via ABA-aldehyde by two oxidation reactions. The content of lycopene in a mature fruit depends on the differential expression of related biosynthesis enzyme genes at the breaker stage (Fraser et al., 1999; Ronen et al., 2000; Davuluri et al., 2005). The red pigmentation of ripening fruit is one of the most recognizable features of the cultivated tomato, and the major carotenoids that accumulate in ripening tomato are lycopene (70–90%) and β-carotene (5–40%), conferring red and orange colours, respectively (Ronen et al., 2000). To enhance coloration in tomato fruit, many attempts have been made (Mehta et al., 2002; Davidovich-Rikanati et al., 2007) using genetic engineering, for example, to transfer genes encoding carotenoid biosynthesis enzymes (Romer et al., 2000; Fraser et al., 2002) or related transcription factors (Bovy et al., 2002) to increase carotenoid content. However, these methods have generally resulted in the increase of only one or a few metabolites, and not in increased flux through the entire carotenoid pathway. It is possible that manipulating a multi-functional regulator gene, rather than a structural gene or a regulatory gene operating in a single branch of the biosynthesis pathway, may result in better effects. Recently, carotenoid and flavonoid levels were shown to be significantly increased with the use of a fruit-specific promoter to drive the DET1-RNAi (RNA interference) vector in tomato transgenic lines (Davuluri et al., 2005). In addition, when two transcription factors from snapdragon have been expressed in tomato, the fruit was shown to accumulate anthocyanins at levels substantially higher than that in previously reported efforts to engineer anthocyanin accumulation in tomato (Niggeweg et al., 2004; Butelli et al., 2008).
ABA, as a ubiquitous plant hormone, has been implicated in a myriad of physiological and developmental processes in many organisms (Leng et al., 2009; Zhang et al., 2009a, b; Ren et al., 2010, 2011; Sun et al., 2010, 2011), but its biological regulation on carotenoid biosynthesis during fruit ripening is still unclear (Buesa et al., 1994; Tung et al., 2008; Boyd et al., 2009; Prasad et al., 2010). As ABA-deficient mutants generally do not undergo normal growth and fruit development (Thompson et al., 1999, 2004), it was necessary to use an alternative method for studying the regulation of ABA in carotenoid biosynthesis. Therefore, a key step in ABA biosynthesis, 9-cis-epoxycarotenoid dioxygenase (NCED), was targeted for inhibition via RNAi in tomato fruit. Four independent transgenic plants were evaluated after greenhouse cultivation for three successive generations.
Materials and methods
Plant material
The selfed lines of tomato plants (Solanum lycopersicum cv. Jia Bao) were grown in a climate-controlled greenhouse at 25/18 °C (day/night) with natural light. The SlNCED1-RNAi transgenic plants contain four independent transgenic lines (SlNCED1-RNAi-9, SlNCED1-RNAi-11, SlNCED1-RNAi-18, and SlNCED1-RNAi-24). Each independent transgenic line included 10–15 plants coming from tissue culture. Thirty selfed non-transgenic plants coming from tissue culture were used as controls. Tomato plants of three generations were used for experiment. The plants of first generation (T1) were from tissue culture and those of the second generation (T2), and third generation (T3) came from seedlings. The ripening stages of tomato fruits were divided according to days after flowering (DAF) and fruit colour (Giovannoni, 2004). During development, fruits were sampled on the following days after anthesis (DAA) (stages): 20 d (IG, immature green), 35 d (MG, mature green), 38 d (B, breaker), 40 d (T, turning), 42 d (P, pink), 43 d (HR, harvest red), and 45 d (OR, over red). Fifteen fruits were harvested randomly at each stage from each transgenic line and control plants and then divided into three groups (five for each) as three replications. After harvest, each group of fruits was weighed and their ethylene production was investigated. Later, all fruits were dissected into three parts (peel, pulp, and seed) and sampled. All the samples were immediately frozen with liquid nitrogen, powdered, mixed, and stored at –80 °C until further use. Data are expressed as mean ±SD (standard deviation, n=3).
Construction of SlNCED1-RNAi vector and plant transformation
In order to evaluate the role of ABA during maturation and ripening accurately, it is important for the RNAi transgenic tomato plants and their fruit to be as normal and healthy as their control cognates. To harness the positive effects of suppressing the SlNCED1 gene in fruit without the collateral negative effects on plant growth, an attempt was made to inhibit SlNCED1 mRNA accumulation using a pCAM-RNAi construct in which the RNAi fusion gene is driven by the fruit-specific E8 promoter (Deikman et al., 1992). The PCR primers used for generating this construct are shown in Table 1. The E8::antisense SlNCED1::GUS::sense SlNCED1::Nos-terminator fusion gene was cloned into pCAMBIA1305.1 (Invitrogen, USA). The pCAM-RNAi recombinant plasmid thus obtained was transferred into E. coli DH5α (Tiangen, Beijing, China) and amplified. The pCAM-RNAi plasmid was subsequently introduced into Agrobacterium tumefaciens LBA4404 (Tiangen, Beijing, China) using a freeze–thaw method. Subsequently, the SlNCED1-RNAi construct was transformed into the selfed line of tomato plants via the A. tumefaciens LBA4404-mediated method (see Supplementary Figs S1 and S2 at JXB online). Four independent transgenic lines (SlNCED1-RNAi-9, SlNCED1-RNAi-11, SlNCED1-RNAi-18, and SlNCED1-RNAi-24) were evaluated after greenhouse cultivation for three successive generations.
Table 1.
Specific primers used for SlNCED1-RNAi construction
| Name | Oligonucleotides | Length (bp) |
| E8-prom-f (BstXI) | 5′-CTAGAAGGAATTTCACGAAATCGGCCCTT-3′ | 1086 |
| E8-prom-r (XbaI) | 5′-AAAAATCTCAATATGAGGA TGCCATATTT-3′ | |
| Nos-f (HindIII) | 5′-GAATTTCCCCGATCGTTCAAACATTTGGC-3′ | 265 |
| Nos-r (SalI) | 5′-CCGATCTAGTAACATAGATGACACCGCGC-3′ | |
| NCED-sense-f (EcoRI) | 5′-TGGGTCGCCCTGTTTTCCCTAAAGCCATT-3′ | 427 |
| NCED-sense-r (HindIII) | 5′-TCATGCATCATTGTTGGGTCTTCAACTGG-3′ | |
| NCED-anti-f (BamHI) | 5′-TGGGTCGCCCTGTTTTCCCTAAAGCCATT-3′ | 427 |
| NCED-anti-r (XbaI) | 5′-TCATGCATCATTGTTGGGTCTTCAACTGG-3′ | |
| GUS-f (PstI) | 5′-TGGTCAGTCCCTTATGTTACGTCCTGTAG-3′ | 1879 |
| GUS-r (EcoRI) | 5′-GGTAGCAATTCCCGAGGCTGTAGCCGACG-3′ | |
| SlNCED2-3′ RACE-F | 5′-CTATTGTTGTTCTATGCCCGTGGAG-3′ | |
| SlNCED2-3′ RACE-R | 5′-AATCCTTCAAGTTTTCTCCAGACGG-3′ |
Antisense and sense SlNCED1 fragments were amplified from tomato fruit cDNA. The E8 promoter was amplified from tomato genomic DNA. The β-glucuronidase (GUS) fragment and the Nos-terminator were amplified from a plasmid including GUS and Nos sequences.
RNA extraction, reverse transcription-PCR (RT-PCR), and sequencing
Total RNA was extracted from 1 g of flesh tissue of at least three individual fruit at each developmental stage analysed using the hot borate method (Wan and Wilkins, 1994). Synthesis of the first-strand cDNA from 2 μg of total RNA was conducted using a Moloney murine leukemia virus reverse transcriptase (Takara, Dalian, China). The cDNA of tomato flesh was used as a template for amplifying genes with degenerate or specific primers (Tables 1, 2).
Table 2.
Specific primer sequences used for real-time quantitative PCR analysis
| Target gene | Forward primer (5′-3′) | Reverse primer (5′-3′) |
| SlSAND | TTGCTTGGAGGAACAGACG | GCAAACAGAACCCCTGAATC |
| SlNCED1 | AGGCAACAGTGAAACTTCCATCAAG | TCCATTAAAGAGGATATTACCGGGGAC |
| SlNCED2 | TGGTTTTCATGGGACATTCATTAGC | ATCTCCCTTCTCAACTCCCTATTCC |
| SlACS2 | CTACGCAGCCACTGTCTTTGAC | TGATTCCGACTCTAAATCCTGGTAA |
| SlACS4 | TTGCGACGAAATATATGCTGCT | CACTCGAAATCCTGGAAAACCT |
| SlACO1 | ACTATCCACCATGTCCTAAGCCCG | TCTGTTTGTGCAATTACTCTGTGCAGC |
| SlETR1 | ATGGATGAGAATGGTGTTAGCAGGA | CACAATAAGTGGCCTACCGTGACGT |
| SlETR3 | AAGGGAACCACTGTCACGTTTGTAG | TTAATGTTCTTTGTCACACCAATGTCC |
| SlETR4 | TGAGAATTCGGAAGTTTGGTAGCCA | ACTCCACTCCTATAAGGCACCGTCA |
| SlETR6 | TTGATCATCGGTTTAGCTGCAATTACT | GTCTATTGTAAACGTTACCGTCATGGC |
| SlCTR1 | ACATTTGGATTATGTCAGGCTTGCA | TTGCTCAAACAATGGTTCAAAGAGG |
| SlCTR3 | CTGCAATCATGGATATGCTGAGACCA | CGGAACAAAAGCCTGAAGTAAACAA |
| SlCTR4 | TTATTGAAGCTTGCTGGGTGAATGA | GCCAAATACCAAGTGCAAATGTTTC |
| SlEIN2 | AAGTTCTTGGTGATGTCAGTTCCCC | TCTACTATGCCCTGAAGACGGTTGAG |
| SlERF2 | GAAAAGGGCTCCTCAGAGAGCATAT | TTTCTTCTGCCAATTCAAGTGACGA |
| SlCYP707A1 | CCCAGAGTTCTTTCCTGATCCACAA | GAATGCCACTACCAGATCCTACCAC |
| SlCYP707A2 | TCGAAAAAGGATACAATTCGATGCC | CTGCAATTTGTTCGTCAGTGAGTCC |
| SlGGPS | TGTCTTGCTGCCTGTGAACTTGTTGG | TACACTTTATGATTCGTCGGCTTTCC |
| SlPDS | TGGTAGCGAATCAATGGGTCATAAGT | ACCTGCACCAGCAATAACAATCTCC |
| SlPSY1 | GGAGAAGATGCCAGAAGAGGAAGAGT | ACAAGACCAAAGATGCCCATACAGG |
| SlPSY2 | CGTTGTGGCGAAGTATGTGCAGAGTA | TCCAGCCTTGCCTCCCACCTATCTAA |
| SlZDS | TGCATTGGCATCTCCAGATGATTACT | ATGGGTCTTTACCAGGTCCTTCACG |
| SlBcyc | GTTCTGAAAGAAGTCATTCGGGTAATG | CATGCCAATAACGAGGTTCTAAGTCA |
Quantitative real-time PCR analysis
Total RNA was isolated from tomato samples using the hot borate method (Wan and Wilkins, 1994). Genomic DNA was eliminated using an RNase Free DNase I kit (Takara, Dalian, China) according to the manufacturer’s recommendations. The quality and quantity of every RNA sample was assessed by agarose gel electrophoresis. The cDNA was synthesized from the total RNA using the PrimeScript™ RT reagent kit (Takara, Dalian, China) according to the manufacturer’s instructions. Primers used for real-time PCR, designed using Primer 5 software, are listed in Table 1. The SAND gene (SGN-U316474) encoding the SAND protein was selected as an internal control gene according to Exposito-Rodriguez et al. (2008), and the stability of its expression was tested in preliminary studies shown in Supplementary Fig. S6 at JXB online. All primer pairs were tested by PCR. A single product of correct size for each gene was confirmed by agarose gel electrophoresis and double-strand sequencing (Invitrogen, Beijing, China). The amplified fragment of each gene was subcloned into the pMD18-T vector (Takara, Dalian, China) and used to generate standard curves by serial dilution. The real-time PCR was conducted using a Rotor-Gene 3000 system (Corbett Research, Australia) with SYBR Premix Ex Taq™ (Takara, Dalian, China). Each 20 μl reaction contained 0.8 μl of primer mix (containing 4 μM of each forward and reverse primer), 1.5 μl cDNA template, 10 μl SYBR Premix Ex Taq™ (2×) mix, and 7.7 μl water. Reactions were carried out under the following conditions: 95 °C/30 s (1 cycle); 95 °C/15 s, 60 °C/20 s; 72 °C/15 s (40 cycles). Relative fold expression changes were calculated using the relative two standard curves method with Rotor-Gene 6.1.81 software.
Histochemical GUS assay for transgenic tomato plants
The GUS assay was performed according to the procedure described by Inaba et al. (2007). The assay buffer contained basic phosphate buffer, anhydrous methanol and X-Gluc. After incubation in the tissue culture room for 24 h at 25 °C, these tissues were placed into a 5 ml centrifuge tube containing excess assay buffer (3 ml). The centrifuge tubes were incubated for 24 h at 37 °C.
siRNA Northern blot analysis
SiRNA Northern blot analysis was performed according to the Hamilton and Baulcombe (1999) method. Small RNA (less than 100 bp) was extracted from 10 g of flesh using the miRcute miRNA isolation kit (TIANGEN, Beijing). The siRNA was fractionated in polyacrylamide-urea gel, and blotted onto a nylon membrane (Hybond N+, Amersham Biosciences, UK). The membrane was then hybridized with the DIG-labelled SlNCED1 cDNA fragment probe (primers are shown in Table 1) in high SDS buffer [7% (w/v) SDS, 5× SSC, 50 mM sodium phosphate, pH 7.0, 2% (w/v) blocking reagent, and 0.1% N-lauroylsarcosine] containing 50% (v/v) formamide overnight at 42 °C. After hybridization, membranes was washed twice at 37 °C in 2× SSC and 0.1% (w/v) SDS for 15 min and twice at 55 °C in 0.1× SSC and 0.1% (w/v) SDS for 30 min. The membranes were then subjected to immunological detection according to the manufacturer’s instructions using CDP-Star™ as a chemiluminescent substrate for alkaline phosphatase (Roche Diagnostics).
Extraction, separation, identification, and quantification of pigments
Tomato fruit from both control and RNAi plants were harvested at different stages. About 0.5 g of tomato pulp or peel was extracted with 50 ml of petroleum benzene plus acetone (1:0.5 v/v) until the extracts were colourless. The extracts were combined and centrifuged at 10 000 g for 20 min. The supernatant liquid was eluted through a Sep-Pak C18 cartridge (Waters, Milford, MA, USA) to remove polar compounds, extracted with petroleum benzene and stored at –20 °C for later use.
Saponification
Petroleum benzene (50 ml) containing carotenoid was mixed with 30% KOH–methanol (20 ml) in a brown bottle in the dark for 12 h. The saponified solution was transferred to the separating funnel, which was washed several times to remove the methanol and other impurities. After using a vacuum drier to remove the petroleum benzene layer, the condensed extract was dissolved in ethyl alcohol:acetonitrile (7:3, v/v), and finally filtered through a 0.4 μm filter.
Chromatography
The analysis of carotenoids was conducted using high performance liquid chromatography (HPLC, Agilent 1200, New York), equipped with an Eclipse XDB-C18 column (New York). The mobile phase was ethyl alcohol:acetonitrile (7:3, v/v). The current velocity was 0.6 ml min−1, and the wavelength was 450 nm. Peak area of the compound was determined according to the spectral characteristics. Quantification of carotenoids was performed based on the concentration curve of standard compounds.
Identification
Identification of the carotenoids was carried out by HPLC through the combined use of the retention time, UV-visible absorption spectrum obtained and co-injection with carotenoid standards.
Quantification
The standard compounds lutein (DHI, Batch No:Lut-112) and zeaxanthin (DHI, Batch No:Zea-120) were purchased from ‘DHI Water and Environment’ (The International Agency for 14C Determination, Hoersholm, Denmark). Lycopene (Wako: 125-04341) and β-carotene (Wako: 035-05531) were purchased from Wako Pure Chemical Industries, Ltd., Osaka (see Supplementary Figs S5–S7 at JXB online).
The concentrations of the carotenoids of the tomato extracts after saponification were determined according to the procedure of Hart and Scott (1995). Calibration curves were built with a minimum of five concentration levels of each carotenoid standard, and the straight line equations and their coefficients of correlation (ranging from 0.9990 to 0.9999) were calculated.
Recovery of standard addition
Recoveries were completed by standard addition before saponification for each sample analysed and used to correct carotenoid levels after HPLC analysis.
The results of recovery of standard addition (RA) were acquired by the method of extraction and determination of carotenoid compounds mentioned above.
where QA(S), QA(O), QA(O+S) are, respectively, the quantity of A added (spiked value), the quantity of A in the original sample, and the quantity of A recovered from the spiked sample.
Recovery determination
The sample with or without added standard sample was determined by HPLC, and recovery values of carotenoid compounds was calculated. Recovery of each type of carotenoid substance was above 85%, which showed that the method was accurate and reliable. Data are the mean ±SE.
Determination of ethylene production
Ethylene production from the fruit was measured by enclosing three fruits in 1.0 l airtight containers for 2 h at 20 °C, withdrawing 1 ml of the headspace gas, and injecting it into a gas chromatograph (model Agilent, 6890N, USA) fitted with a flame ionization detector and an activated alumina column. Flesh tissues from each fruit were frozen in liquid nitrogen and stored at –80 °C until use.
Determination of ABA content
For ABA extraction, 1 g of flesh was ground in a mortar and homogenized in extraction solution (80% methanol, v/v). Extracts were centrifuged at 10 000 g for 20 min. The supernatant was eluted through a Sep-Pak C18 cartridge (Waters, Milford, MA, USA) to remove polar compounds and then stored at –20 °C until use.
Quantification of ABA in pulp and peel of control and transgenic fruit was performed by indirect enzyme-linked immunosorbent assay (ELISA) as reported previously (Zhang et al., 2009a, b). The ELISA procedures were conducted according to the instructions provided by the manufacturer (China Agricultural University, Beijing, China) and reading the assay plates using the Thermo Electron (Labsystems) Multiskan MK3 (PIONEER Co., Beijing).
Results
Expression of SlNCED genes and changes of ABA accumulation in tomato fruit
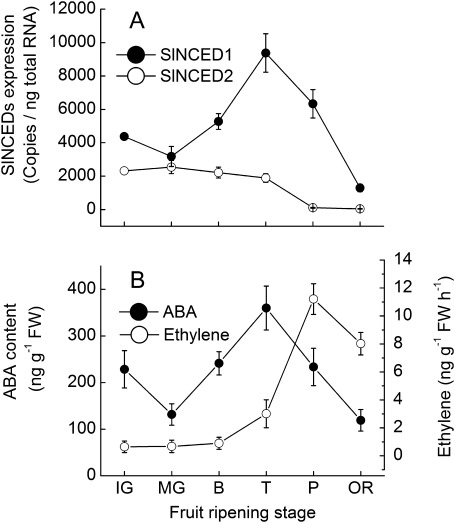
Two NCED genes (SlNCED1 and SlNCED2) were analysed from cDNA of tomato fruit. Expression of these genes in fruit displayed rather different patterns. The expression level of SlNCED1 (Z97215) (Burbidge et al., 1997) declined from the immature stage to the mature green stage and then rapidly increased to a peak at the turning stage (Fig. 1A). This fluctuation in SlNCED1 expression correlated with ABA content, which peaked prior to ethylene production during fruit ripening (Fig. 1B). Compared with SlNCED1, the expression of the SlNCED2 (EU912387) was high at the immature stage and then declined continually through all stages of fruit development (Fig. 1A). Because the decline in SlNCED1 expression correlated with that of ABA accumulation during fruit ripening, this gene was targeted by RNAi in subsequent experiments.
Fig. 1.
Expressions of ABA biosynthesis genes and changes of ABA content and ethylene production during tomato fruit development. The ABA and ethylene were measured in fruit from plants grown in the greenhouse. Tomato fruit were sampled on the following DAA (stages): 20 d (immature green), 35 d (mature green), 38 d (breaker), 40 d (turning), 42 days (pink), and 45 d (over red). Fifteen fruits were harvested randomly at each stage from control plants and then divided into three groups (five for each) as three replications. The SAND gene (SGN-U316474) is shown as an internal loading control. Data are means ±SD (n=3).
Effect of SlNCED1-RNAi on expression of SlNCED genes and changes of ABA accumulation in tomato fruit
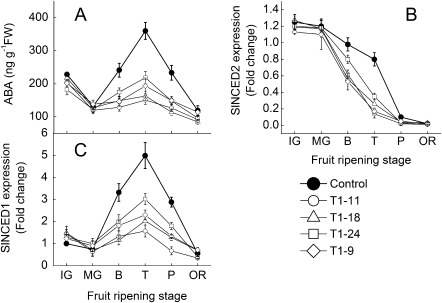
To investigate the role of ABA during fruit development, a SlNCED1-RNAi construct was employed to transform tomato plants (see Supplementary Figs S1–S4 at JXB online). Four independent RNAi plants and their segregating lines were evaluated after greenhouse cultivation for three successive generations. Tomato plants from all four transgenic lines appeared normal and as healthy as control plants (Fig. 3A–H). However, the ABA levels and expression of SlNCED1 and SlNCED2 in fruit of all transgenic lines were down-regulated by 20–50% (Fig. 2) than those in the control fruit.
Fig. 3.
Ripening in RNAi transgenic tomato fruit with suppressed SlNCED1 gene expression. Tomato fruit of transgenic and control fruit were harvested in the different stages cultivated in normal greenhouse conditions. Data include four independent transgenic lines of the T1 generation. (A) Flower of SLNCED1-RNAi line 11. (B–H) Fruit development and ripening of SlNCED1-RNAi line 11 (30, 35, 38, 40, 42, and 43 DAA). (I–K) Fruit of SlNCED1-RNAi lines 24, 9, and 18, respectively (43 DAA). (L–N) Control fruit (38, 40, and 43 DAA, respectively). (O) Greenhouse environment of SlNCED1-RNAi plants.
Fig. 2.
Expressions of ABA biosynthesis genes and changes of ABA content during fruit development in RNAi transgenic and control fruit. Data include four transgenic lines of the T1 generation. SAND expression is shown as an internal loading control. Fifteen fruits were harvested randomly at each stage from each transgenic line and control plants and then divided into three groups (five for each) as three replications. Data are means ±SD (n=3). IG, immature green; MG, mature green; B, breaker; T, turning; P, pink; OR, over red.
Ripening in transgenic tomato fruit with suppressed SlNCED1 gene expression
As Fig. 3 shows, the colour of the transgenic fruit was a deep red, while that of the typical control fruit was pink (Fig. 3H–K, N). Throughout the ripening processes, the transgenic fruit were different from control fruit in the coloration pattern. During the breaker stage, degreening of control fruit started from the fruit apex and then gradually extended toward the lower part; then the fruit apex became pink first followed by the other parts of the fruit (Fig. 3L, M, N). Compared with the control, the transgenic fruit showed a uniform degreening process. The fruit surface started to turn yellow only when the green colour almost disappeared, and then the red colour appeared after the whole fruit became yellow (Fig. 3C, D, E). Compared with other transgenic fruit, the transgenic line 24 (Fig. 3I) had a higher level of β-carotene (Fig. 5D, I) and a relatively lower level of lycopene (Fig. 5C, H); therefore, fruits from this transgenic line were yellower than those of other transgenic lines. The seeds of all the transgenic fruit were considerably fewer in number. The trait changes induced by SlNCED1-RNAi in transgenic fruit could be stably transmitted to the next generation.
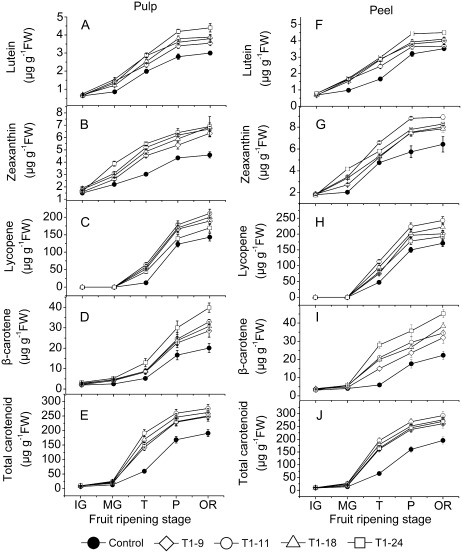
Fig. 5.
Comparison of carotenoid contents in the pulp and peel between control and transgenic tomato fruit. Data include four independent transgenic lines (T9, T11, T18, and T24 of the T1 generation). IG, immature green; MG, mature green; T, turning; P, pink; OR, over red. Fifteen fruits were harvested randomly at each stage from each transgenic line and control plants and then divided into three groups (five for each) as three replications. Data are means ±SD (n=3). The significance of differences between control and transgenic fruit was determined using Student’s t test (P ≤0.05).
Carotenoid levels are up-regulated in transgenic fruit
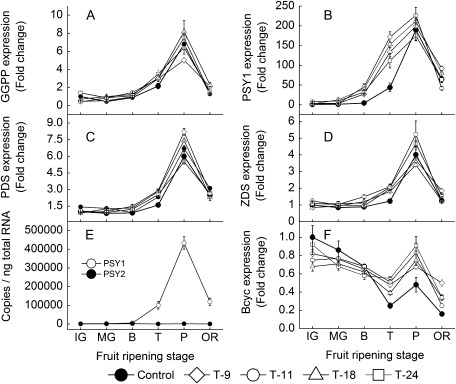
As shown in Fig. 4, analyses of absolute expression levels confirmed that SlPSY1, which encodes phytoene synthetase, was more highly expressed than SlPSY2 in tomato fruit (Fig. 4E). Analysis of relative expression levels showed that the transcription levels of SlPSY1 in the transgenic fruit were significantly up-regulated compared with that of the control fruit (Fig. 4B). Meanwhile, SlGGPP, SlPds, and SlZds, which encode geranylgeranyl diphosphate synthetase, phytoene dehydrogenase, and ζ-carotene dehydrogenase, respectively, were slightly up-regulated in the transgenics during the turning stage, but their expression levels were slightly lower than in the control fruit during the mature stage (Fig. 4A, C, D). By contrast, expression of SlBcyc, which encodes lycopene β-cyclase, was not different in the immature and mature green stages in the transgenics, while it was slightly up-regulated during the turning and mature stages compared with the control (Fig. 4F).
Fig. 4.
Expression analysis of genes encoding enzymes involving in the synthesis and catabolism of carotenoids during development in four transgenic lines of the T1 generation: (A–D, F) Relative quantitiative real-time PCR analysis. (E) Absolute quantitative real-time PCR analysis. Fifteen fruits were harvested randomly at each stage from each transgenic line and control plants and then divided into three groups (five for each) as three replications. SAND expression is shown as an internal loading control. Data are means ±SD (n=3). IG, immature green; MG, mature green; B, breaker; T, turning; P, pink; OR, over red.
To quantitate the carotenoid accumulation profiles precisely, the carotenoid composition and content of fruit from SlNCED1-RNAi lines and control were analysed by HPLC (Fig. 5A–J). The carotenoid compositions were nearly the same in all fruit of both types of plants and included lutein, zeaxanthin, β-cryptoxanthin, lycopene, β-carotene, and the fatty acid esters of lycopene and β-carotene. The transgenic fruit contained approximately 30–45% and 34–50% higher levels of total carotenoids in both pulp and peel than the control fruit (Fig. 5E, J); meanwhile, the levels of lycopene and β-carotene were enhanced by up to 18–48% and 42–98% in pulp (Fig. 5C, D) or 12–42% and 43–94% in the peel (Fig. 5H, I).
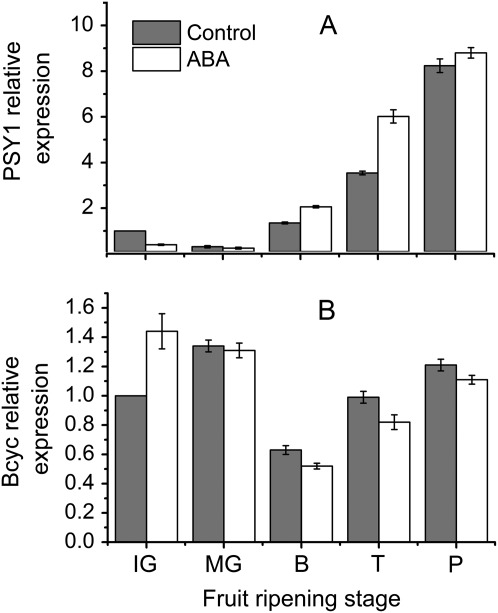
Effect of exogenous ABA application on the gene expressions of SlPSY1 and SlBcyc in control tomato fruit
Exogenous ABA markedly inhibited SlPSY1 expression (Fig. 6A) but promoted SlBcyc expression during the immature developmental stage (Fig. 6B). The exogenous ABA significantly promoted SlPSY1 expression in the turning stage, while no difference was observed between control and ABA treated fruit in the mature green stage (Fig. 6A). The expression level of SlBcyc increased after the breaker stage, but was slightly down-regulated by exogenous ABA (Fig. 6B).
Fig. 6.
Effect of exogenous ABA application on the expression of PSY1 and Bcyc genes in the tomato fruit. Fruits were harvested and then divided into two groups in every sampling time, and treated with 0.1 mM ABA for 10 min, and remain a group was as control fruits (distilled water for 10 min). After 24 h, all fruit were sampled, frozen with liquid nitrogen, powdered, mixed, and stored at –80 °C for further use. The above data are the means of three replicates including five fruits at each sampling time. The transcript levels of SAND are shown as an internal loading control. Data are means ±SD (n=3). IG, immature green; MG, mature green; T, turning; P, pink; OR, over red.
Expression of genes involved in both synthesis and signalling of ethylene in transgenic and control fruit
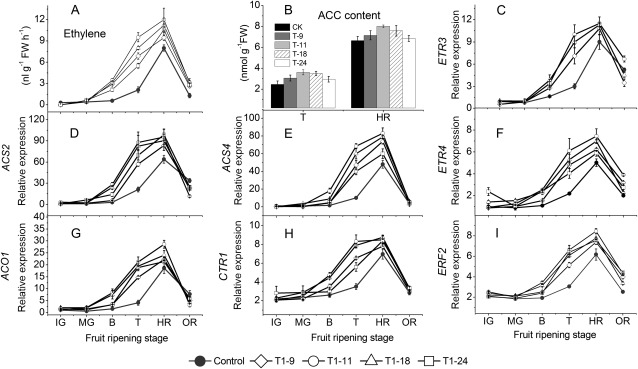
In the transgenic fruit, ethylene production was higher than control fruit during the breaker and turning stages (Fig. 7A). The relative quantitative real-time PCR analysis showed that the expressions of SlETR3 and SlETR4 (ethylene response) as well as that of SlCTR1 (constitutive triple response) and SlERF2 were consistently only slightly up-regulated (Fig. 7C, F, H, I), while the relative transcript levels of SlACS2, SlACS4, and SlACO1 were all markedly up-regulated compared with those in the non-transgenic control fruit at the breaker and turning stages (Fig. 7D, E, G). However, all these transcripts showed no significant differences in expression in the mature stage, and were coincident with ethylene production (Fig. 7A) and 1-aminocyclopropane-1-carboxylic acid (ACC) content (Fig. 7B).
Fig. 7.
Expression of genes related to synthesis and signalling of ethylene and changes in ethylene and ACC contents in control and transgenic tomato fruit. Changes in the relative transcript level profiles for SlACSs, SlACO1, SlETRs, SlCTRs, and SlERF2 genes in control and transgenic tomatoes, including four transgenic lines of three successive generation during development. Relative mRNA expression is expressed as a fold change of control fruit at 20 DAA. SAND was used as the reference gene. Fifteen fruits were harvested randomly at each stage from each transgenic line and control plants and then divided into three groups (five for each) as three replications. Data are means ±SD (n=3). IG, immature green; MG, mature green; T, turning; P, pink; OR, over red.
Discussion
SlNCED1-RNAi channels more carbon flux into carotenoid production
In this study, a key step in ABA biosynthesis, NCED, was targeted for inhibition via RNAi in tomato fruit. The observed phenotypic effects were interesting with respect to the degree of pigmentation and carotenoid composition in the SlNCED1 RNAi lines relative to the controls. The levels of lycopene and β-carotene were enhanced due to the up-regulation of phytoene synthase (SlPSY1), a rate-limiting enzyme (Fraser et al., 2002), and lycopene-β-cyclase (SlBcyc) expression when ABA levels were significantly reduced as fruit developed in the four SlNCED1 RNAi lines compared with the control fruit. The regulatory mechanisms of carotenoid biosynthesis in our study are apparently much different from those in other tomato mutant of high-pigment 3 (hp3) reported by Galpaz et al. (2008). Compared with the SlNCED1-RNAi lines, the tomato hp3 mutant contained an altered gene for zeaxanthin epoxidase (Zep), which catalyses the conversion of zeaxanthin to violaxanthin that takes place in the plastid through the intermediate antheraxanthin (Marin et al., 1996). The violaxanthin is the substrate for neoxanthin synthase. The neoxathin occurs in the light-harvesting complex in the 9-cis configuration, as an isomerization step. The 9-cis violaxanthin and 9′-cis neoxanthin are two necessary components of the light-harvesting complexes (LHC) in high plants. Therefore, the deficiency of 9-cis violaxanthin and 9′-cis neoxanthin influenced light-harvesting and plastid development. Similarly, suppressing an endogenous photomorphogenesis regulatory gene, DET1, could increase the carotenoid content in tomato (Davuluri et al., 2005). In addition, the deficiency of 9-cis violaxanthin and 9′-cis neoxanthin, which are the substrates for ABA biosynthesis, resulted in a decrease in ABA content.
On the other hand, the tomato hp3 mutant is not fruit-specific, that is, the growth and development of the entire plant was influenced by the altered ZEP gene. The metabolism of carotenoids changed from the seeding stage and throughout all of the leaf, flower, and fruit. The hp3 plant could not produce normal fruits and the fruit size was only one-half of that in control plants. The carotenoid content is increased in the tomato hp3 mutant due to the increase and enlargement of the plastid compartments. Compared with the tomato hp3 mutant, it was first verified that SlNCED1 is a major gene for ABA biosynthesis in tomato fruit, which was then suppressed using RNAi driven by the E8 promoter. Given that the E8 promoter is ethylene-induced, transgenic plants developed like normal plants up until fruit development to the breaker stage, at which stage the trace ethylene was released and RNAi silencing was activated. Then the transgenic fruits showed a different phenotype compared with the control. As shown in Fig. 3, the colour of the control fruit turned from green to white, and then transformed to pink; meanwhile, the transgenic fruit turned to yellow after degreening and then transformed to orange and then to deep red.
An alternate explanation of why the transgenic fruit contents more carotenoids could be that the carbon flux, which normally channels to free ABA and ABA metabolite accumulation during ripening, is partially blocked by a significant reduction in NCED activity, so this ‘backlogged’ carbon transformed into the carotenoid pathway in the RNAi lines resulting in the increased biosynthesis and accumulation of upstream compounds in the pathway, chiefly lycopene and β-carotene. In other words, the increased lycopene and β-carotene levels are due to an increased up-regulation of SlPSY1 channelling more carbon into carotenoid production or due to an inhibition of carbon flux to ABA and its metabolites.
Generally, manipulation of the levels of ABA, which regulates the expression of many genes involved in entire metabolic pathways, provides an effective tool for engineering high levels of metabolites, for example, an increase in the lycopene and carotene content in fruit. Consequently, inhibiting SlNCED1 in transgenic tomatoes resulted in immediate increases of lycopene and β-carotene. The promotion of ethylene biosynthesis and the regulation of fruit ripening by ABA were achieved through the accommodation of several metabolic pathways (Spanu et al., 1994). The results shown in Fig. 6 indicate that ABA could directly regulate the expressions of SlPSY1 and SlBcyc, thereby affecting the red pigmentation of ripening fruit. However, the biological effect of exogenous ABA relies on the fruit development process (Fig. 6). For example, the ABA effects on up-regulating PSY1 gene expression was more significant at breaker and turning stage; by contrast, when treated at the pink stage the ABA effect was less significant; furthermore, when exogenous ABA was applied to fruit at the mature green stage, there was almost no effect (Fig. 6). In addition, ABA may also have indirectly regulated the level of carotenoid via the regulation of ethylene release (Fig. 7).
ABA controls, at least in part, ethylene production and effects in tomato fruit
Intriguingly, both ABA and ethylene levels began to increase after the mature green stage, with ABA preceding and, based on transcriptional analyses, at least partially controlling the production and effects of ethylene (Figs 1B, 7A). Before endogenous ABA reached its maximum level in the turning stage, it could inhibit ethylene production and sensibility by regulating the biosynthesis and signalling of ethylene. However, this effect gradually weakened, but could be promoted by exogenous ABA treatment during the progress of fruit ripening (data not shown). The SlNCED1-RNAi fruit provided direct evidence for the inhibition of ethylene by endogenous ABA. When endogenous ABA was down-regulated by SlNCED1-RNAi, ethylene production in the transgenic fruit increased by advancing transcription of ACSs and ACO1, and ethylene perception was increased by advancing transcription of CTR1, ETRs, and ERF2 during the turning stage (Fig. 7H, C, F, I). As the ABA signalling pathway participates in ethylene signalling (Tieman et al., 2001; Pech et al., 2008; Rodriguez et al., 2010; Sun et al., 2010), the inhibition of SlNCED1 could affect the level of ethylene and its signalling pathway (Fig. 7D, E, G). Although assessing the physiological effects of inhibited endogenous ABA is complex, as it has pleiotropic effects involving cross-talk with other signalling pathways, the data clearly show that inhibition of SlNCED1 affected both the synthesis and the signalling pathway of ethylene.
In conclusion, our data for the first time showed that ABA potentially regulated the degree of pigmentation and carotenoid composition during tomato fruit ripening. In addition, ABA could control, at least in part, the production and effects of ethylene in climacteric tomato fruit. These findings contribute new information to the field of hormonal control of climacteric fruit ripening.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Fig. S1. Hairpin construct of the SlNCED1 gene for double-stranded RNAi under the control of the tomato E8 promoter.
Supplementary Fig. S2. Genetic transformation of SlNCED1-RNAi tomato.
Supplementary Fig. S3. Histochemical GUS assay for transgenic tomato plants.
Supplementary Fig. S4. Detection of transformed tomato plants.
Supplementary Fig. S5. Wavelength scanning of standards for lycopene and carotenoids.
Supplementary Fig. S6. HPLC chromatogram of standards for lutein and zeaxanthin.
Supplementary Fig. S7. HPLC chromatogram of the four carotenoid standards.
Acknowledgments
The authors thank Drs Akitsugu Inaba, Ryohei Nakano, Koichiro Ushijima, and Yasutaka Kubo (Faculty of Agriculture, Okayama University, Japan) for their assistance in the study. This work was supported by the Beijing Municipal Science and Technology Commission (D0706002000091).
References
- Adams-Phillips L, Barry C, Giovannoni J. Signal transduction systems regulating fruit ripening. Trends in Plant Science. 2004;9:331–338. doi: 10.1016/j.tplants.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Bovy A, Vosa R, Kempera M, et al. High-flavonol tomatoes resulting from heterologous expression of the maize transcription factor genes LC and CI. The Plant Cell. 2002;14:2509–2526. doi: 10.1105/tpc.004218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd J, Gai Y, Nelson KM, et al. Sesquiterpene-like inhibitors of a 9-cis-epoxycarotenoid dioxygenase regulating abscisic acid biosynthesis in higher plants. Bioorganic and Medicinal Chemistry. 2009;17:2902–2912. doi: 10.1016/j.bmc.2009.01.076. [DOI] [PubMed] [Google Scholar]
- Bramley P, Teulieres C, Blain I, Bird C, Schuch W. Biochemical characterization of transgenic tomato plants in which carotenoid synthesis has been inhibited through the expression of antisense RNA to pTOM5. The Plant Journal. 1992;2:343–349. [Google Scholar]
- Buesa C, Dominguez M, Vendrell M. Abscisic acid effects on ethylene production and respiration rate in detached apple fruits at different stages of development. Revista Espanola de Cienciay Tecnologia de Alimentos. 1994;34:495–506. [Google Scholar]
- Burbidge A, Grieve T, Terry C, Corlett J, Thompson A, Taylor I. Structure and expression of a cDNA encoding zeaxanthin epoxidase, isolated from a wilt-related tomato (Lycopersicon esculentum Mill.) library. Journal of Experimental Botany. 1997;48:1749–1750. [Google Scholar]
- Butelli E, Titta L, Giorgio M, et al. Enrichment of tomato fruit with health-promoting antochyanins by expression of select transcription factors. Nature Biotechnology. 2008;26:1301–1308. doi: 10.1038/nbt.1506. [DOI] [PubMed] [Google Scholar]
- Corona V, Aracri B, Kosturkova G, Bartley GE, Pitto L, Giorgetti L, Scolnik PA, Giuliano G. Regulation of a carotenoid biosynthesis gene promoter during plant development. The Plant Journal. 1996;9:505–512. doi: 10.1046/j.1365-313x.1996.09040505.x. [DOI] [PubMed] [Google Scholar]
- Crozier A, Michael EJL, McDonald MS, Black C. Quantitative analysis of the flavonoid content of commercial tomatoes, onions, lettuce, and celery. Journal of Agricultural and Food Chemistry. 1997;45:590–595. [Google Scholar]
- Cunningham FX, Gantt E. Genes and enzymes of carotenoid biosynthesis in plants. Annual Review of Plant Physiology and Plant Molecular Biology. 1998;49:557–583. doi: 10.1146/annurev.arplant.49.1.557. [DOI] [PubMed] [Google Scholar]
- Davidovich-Rikanati R, Sitrit Y, Tadmor Y, et al. Enrichment of tomato flavor by diversion of the early plastidial terpenoid pathway. Nature Biotechnology. 2007;25:899–901. doi: 10.1038/nbt1312. [DOI] [PubMed] [Google Scholar]
- Davuluri GR, Tuinen A, Fraser PD, et al. Fruit-specific RNAi-mediated suppression of DET1 enhances carotenoid and flavonoid content in tomatoes. Nature Biotechnologyl. 2005;23:890–895. doi: 10.1038/nbt1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deikman J, Randy K, Robert LF. Organization of ripening and ethylene regulatory regions in a fruit-specific promoter from tomato (Lycopersicon esculentum) Plant Physiology. 1992;100:2013–2017. doi: 10.1104/pp.100.4.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DellaPenna D, Pogson BJ. Vitamin synthesis in plants: tocopherols and carotenoids. Annual Reviews of Plant Biology. 2006;57:711–738. doi: 10.1146/annurev.arplant.56.032604.144301. [DOI] [PubMed] [Google Scholar]
- Exposito-Rodriguez M, Borges AA, Borges-Perez A, Perez JA. Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biology. 2008;8:131–143. doi: 10.1186/1471-2229-8-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser PD, Bramley PM. The biosynthesis and nutritional uses of carotenoids. Progress in Lipid Research. 2004;43:228–265. doi: 10.1016/j.plipres.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Fraser PD, Kiano JW, Truesdale MR, Schuch W, Bramley PM. Phytoene synthase-2 enzyme activity in tomato does not contribute to carotenoid biosynthesis in ripening fruit. Plant Molecular Biology. 1999;40:687–698. doi: 10.1023/a:1006256302570. [DOI] [PubMed] [Google Scholar]
- Fraser PD, Romer S, Shipton CA, Mills PB, Kiano JW, Misawa N, Drake RG, Schuch W, Bramley PM. Evaluation of transgenic tomato plants expressing an additional phytoene synthase in a fruit-specific manner. Proceedings of the National Academy of Sciences, USA. 2002;99:1092–1097. doi: 10.1073/pnas.241374598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galpaz N, Wang Q, Menda N, Zamir D, Hirschberg J. Abscisic acid deficiency in the tomato mutant high-pigment 3 leading to increased plastid number and higher fruit lycopene content. The Plant Journal. 2008;53:717–730. doi: 10.1111/j.1365-313X.2007.03362.x. [DOI] [PubMed] [Google Scholar]
- Giovannoni JJ. Genetic regulation of fruit development and ripening. The Plant Cell. 2004;16:S170–S180. doi: 10.1105/tpc.019158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano G, Bartley GE, Scolnik PA. Regulation of carotenoind biosynthesis during tomato development. The Plant Cell. 1993;5:379–387. doi: 10.1105/tpc.5.4.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. A species of small antisense RNA in post-transcriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Hart JD, Scott KJ. Development and evaluation of an HPLC method for the analysis of carotenoids in foods and the measurement of the carotenoid content of vegetables and fruits commonly consumed in the UK. Food Chemistry. 1995;54:101–111. [Google Scholar]
- Hirschberg J. Carotenoid biosynthesis in flowering plants. Current Opinion in Plant Biology. 2001;4:210–218. doi: 10.1016/s1369-5266(00)00163-1. [DOI] [PubMed] [Google Scholar]
- Inaba Y, Zhong WQ, Zhang XH, Jack MW. Specificity of expression of the GUS reporter gene (uidA) driven by the tobacco ASA2 promoter in soybean plants and tissue cultures. Journal of Plant Physiology. 2007;164:824–834. doi: 10.1016/j.jplph.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Leng P, Zhang GL, Li XX, Wang LH, Zheng ZM. Cloning of 9-cis-epoxycarotenoid dioxygenase (NCED) gene encoding a key enzyme during abscisic acid (ABA) biosynthesis and ABA-regulated ethylene production in detached young persimmon calyx. Chinese Science Bulletinl. 2009;54:2830–2838. [Google Scholar]
- Marin E, Nussaume L, Quesada A, Gonneau M, Sotta B, Hugueney P, Frey A, Marion-Poll A. Molecular identification of zeaxanthin epoxidase of Nicotiana plumbaginifolia, a gene involved in abscisic acid biosynthesis and corresponding to the ABA locus of Arabidopsis thaliana. EMBO Journal. 1996;15:2331–2342. [PMC free article] [PubMed] [Google Scholar]
- Mehta RA, Cassol T, Li N, Ali N, Handa AK, Mattoo AK. Engineered polyamine accumulation in tomato enhances phytonutrient content, juice quality, and vine life. Nature Biotechnology. 2002;20:613–618. doi: 10.1038/nbt0602-613. [DOI] [PubMed] [Google Scholar]
- Niggeweg R, Michael AJ, Martin C. Engineering plants with increased levels of the antioxidant chlorogenic acid. Nature Biotechnology. 2004;22:746–754. doi: 10.1038/nbt966. [DOI] [PubMed] [Google Scholar]
- Pech JC, Bouzayen M, Latche A. Climacteric fruit ripening: ethylene-dependent and independent regulation of ripening pathways in melon fruit. Plant Science. 2008;175:114–120. [Google Scholar]
- Prasad K, Zhang XW, Tobon E, Ambrose BA. The Arabidopsis B-sister MADS-box protein, GORDITA, represses fruit growth and contributes to integument development. The Plant Journal. 2010;62:203–214. doi: 10.1111/j.1365-313X.2010.04139.x. [DOI] [PubMed] [Google Scholar]
- Ren J, Sun L, Wang C, Zhao S, Leng P. Expression analysis of the cDNA for magnesium chelatase H subunit (CHLH) during sweet cherry fruit ripening and under stress conditions. Plant Growth Regulation. 2011;63:301–307. [Google Scholar]
- Ren J, Sun L, Wu J, Zhao S, Wang C, Wang Y, Ji K, Leng P. Cloning and expression analysis of cDNAs for ABA 8’-hydroxylase during sweet cherry fruit maturation and under stress conditions. Journal of Plant Physiology. 2010;167:1486–1493. doi: 10.1016/j.jplph.2010.05.027. [DOI] [PubMed] [Google Scholar]
- Rodriguez JAM, Morcillo RL, Vierheilig H, Ocampo JA, Ludwig-Muller J, Garrido JMG. Mycorrhization of the notabilis and sitiens tomato mutants in relation to abscisic acid and ethylene contents. Journal of Plant Physiology. 2010;167:606–613. doi: 10.1016/j.jplph.2009.11.014. [DOI] [PubMed] [Google Scholar]
- Ronen G, Carmel-Goren L, Zamir D, Hirschberg J. An alternative pathway to β-carotene formation in plant chromoplasts discovered by map-based cloning of Beta and old-gold color mutations in tomato. Proceedings of the National Academy of Sciences, USA. 2000;97:11102–11107. doi: 10.1073/pnas.190177497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romer S, Fraser PD. Recent advances in carotenoid biosynthesis, regulation and manipulation. Planta. 2005;221:305–308. doi: 10.1007/s00425-005-1533-5. [DOI] [PubMed] [Google Scholar]
- Romer S, Fraser PD, Kiano JW, Shipton CA, Misawa N, Schuch W, Bramley PM. Elevation of the provitamin A content of transgenic tomato plants. Nature Biotechnology. 2000;18:666–669. doi: 10.1038/76523. [DOI] [PubMed] [Google Scholar]
- Spanu P, Grosskopf DG, Felix G, Boller T. The apparent turnover of 1-aminocyclopropane-1-carboxylate synthase in tomato cells is regulated by protein phosphorylation and dephosphorylation. Plant Physiology. 1994;106:529–535. doi: 10.1104/pp.106.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Wang YP, Chen P, Ren J, Ji K, Li Q, Li P, Dai SJ, Leng P. Transcriptional regulation of SlPYL, SlPP2C and SlSnRK2 gene families encoding ABA signal core components during tomato fruit development and drought stress. Journal of Experimental Botany. 2011;62:5659–5669. doi: 10.1093/jxb/err252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Zhang M, Ren J, Qi JX, Zhang GJ, Leng P. Reciprocity between abscisic acid and ethylene at the onset of berry ripening and after harvest. BMC Plant Biology. 2010;10:257–268. doi: 10.1186/1471-2229-10-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AJ, Tor M, Barry CS, Vrebalov J, Orfila C, Jarvis MC, Giovannoni JJ, Grierson D, Seymour GB. Molecular and genetic characterisation of a novel pleiotropic tomato-ripening mutant. Plant Physiology. 1999;120:383–389. doi: 10.1104/pp.120.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AJ, Thorne ET, Burbidge A, Jackson AC, Sharp RE, Taylor IB. Complementation of notabilis, an abscisic acid-deficient mutant of tomato: importance of sequence context and utility of partial complementation. Plant, Cell and Environment. 2004;27:459–471. [Google Scholar]
- Tieman DM, Ciardi JA, Taylor MG, Klee HJ. Members of the tomato LeEIL (EIN3-like) gene family are functionally redundant and regulate ethylene responses throughout plant development. The Plant Journal. 2001;26:47–58. doi: 10.1046/j.1365-313x.2001.01006.x. [DOI] [PubMed] [Google Scholar]
- Tung SA, Smeeton R, White CA, Black CR, Taylor IB, Hilton HW, Thompson AJ. Over-expression of LeNCED1 in tomato (Solanum lycopersicum L.) with the rbcS3C promoter allows recovery of lines that accumulate very high levels of abscisic acid and exhibit severe phenotypes. Plant, Cell and Environment. 2008;31:968–981. doi: 10.1111/j.1365-3040.2008.01812.x. [DOI] [PubMed] [Google Scholar]
- Wan CY, Wilkins TA. A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.) Analytical Biochemistry. 1994;223:7–12. doi: 10.1006/abio.1994.1538. [DOI] [PubMed] [Google Scholar]
- Zhang M, Leng P, Zhang G, Li X. Cloning and functional analysis of 9-cis-epoxycarotenoid dioxygenase (NCED) genes encoding a key enzyme during abscisic acid biosynthesis from peach and grape fruits. Journal of Plant Physiology. 2009a;166:1241–1252. doi: 10.1016/j.jplph.2009.01.013. [DOI] [PubMed] [Google Scholar]
- Zhang M, Yuan B, Leng P. The role of ABA in triggering ethylene biosynthesis and ripening of tomato fruit. Journal of Experimental Botany. 2009b;60:1579–1588. doi: 10.1093/jxb/erp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.