Abstract
Plant programmed cell death (PCD) is a genetically controlled process that plays an important role in development and stress responses. Reactive oxygen species (ROS) are key inducers of PCD. The addition of 50 mM H2O2 to tobacco Bright Yellow-2 (TBY-2) cell cultures induces PCD. A comparative proteomic analysis of TBY-2 cells treated with 50 mM H2O2 for 30 min and 3 h was performed. The results showed early down-regulation of several elements in the cellular redox hub and inhibition of the protein repair–degradation system. The expression patterns of proteins involved in the homeostatic response, in particular those associated with metabolism, were consistently altered. The changes in abundance of several cytoskeleton proteins confirmed the active role of the cytoskeleton in PCD signalling. Cells undergoing H2O2-induced PCD fail to cope with oxidative stress. The antioxidant defence system and the anti-PCD signalling cascades are inhibited. This promotes a genetically programmed cell suicide pathway. Fifteen differentially expressed proteins showed an expression pattern similar to that previously observed in TBY-2 cells undergoing heat shock-induced PCD. The possibility that these proteins are part of a core complex required for PCD induction is discussed.
Keywords: H2O2, PCD, proteome, redox homeostasis, TBY-2 cells
Introduction
Programmed cell death (PCD) is a genetically regulated process vitally important in cellular differentiation, organ development/abortion, cellular senescence, and in response to biotic and abiotic stresses (Gunawardena et al., 2004; Williams and Dickman, 2008). PCD hallmarks described in plants or plant cultured cells include condensed cell morphology, nuclear shrinkage, DNA laddering, mitochondrial release of cythocrome c, and organelle swelling. Changes in H2O2 homeostasis trigger genetic programmes that promote stress acclimation or induce PCD (Gechev and Hille, 2005; de Pinto et al., 2012). The overexpression of the H2O2-detoxifying enzyme ascorbate peroxidase (APX) suppresses H2O2-induced PCD (Murgia et al., 2004), while decreasing catalase activity causes perturbations of H2O2 homeostasis inducing PCD (Dat et al., 2003; Palma and Kermode, 2003). H2O2 increases early in the PCD process (Locato et al., 2008), and different concentrations of exogenous H2O2 induce cell death (Houot et al., 2001; Gechev et al., 2006). The direct addition of 50 mM H2O2 to tobacco Bright Yellow-2 (TBY-2) cell suspension is able to induce PCD (de Pinto et al., 2006).
To identify new elements involved in the early phases of H2O2-induced PCD, changes in protein expression were analysed in TBY-2 cells treated with 50 mM H2O2 for 30 min and 3 h, when cell viability was 95% and 85%, respectively. Using two-dimensional electrophoresis (2-DE) in combination with tandem mass spectrometry (MS/MS) analysis, 150 H2O2-responsive proteins were identified. Their mapping to various cellular processes gave a global view of the changes elicited in TBY-2 cells by an oxidative stress inducing PCD. To improve our understanding of the characteristics common to PCD processes induced by different activators, the proteomic data of H2O2-induced PCD were compared with those previously reported for the same cell line undergoing PCD induced by heat shock (HS; Marsoni et al., 2010). In both cases cell death occurred through non-autolytic PCD, the most common class of PCD occurring in plants (van Doorn, 2011).
Materials and methods
Cell culture and growth conditions
The suspension of TBY-2 cells was cultured at 27 °C as previously described (de Pinto et al., 2002). For H2O2 treatment, a stationary culture was diluted 4:100 (v/v; 100 ml), cultured for 4 d, and treated with 50 mM H2O2. At the times indicated, the cells were collected for analyses and stored at –80 °C until use. Where indicated, 2 mM spermidine was added to the cell suspension 15 min before the H2O2 addition. For HS-induced PCD, cells were treated as previously described (Marsoni et al., 2010). Cell viability was measured in three biological replicates by trypan blue staining as previously described (de Pinto et al., 2002).
Ascorbate and glutathione contents
Cells were collected by filtration on Whatman 3MM paper, weighed, and homogenized with 2 vols of cold 5% (w/v) metaphosphoric acid at 4 °C in a porcelain mortar. The homogenate was centrifuged at 20 000 g for 15 min at 4 °C, and the supernatant was collected. Analyses of ascorbate (ASC) and glutathione (GSH) contents were performed as described by de Pinto et al. (2000).
Proteasome activity
TBY-2 cells were ground in ice-cold homogenization buffer containing 20 mM TRIS-HCl (pH 7.2), 0.1 mM EDTA, 1 mM 2-mercaptoethanol, 5 mM ATP, 20% glycerol, 0.04% NP-40. After centrifugation, 40 μg of cell lysates were incubated at 37 °C with 50 μM of the fluorescent substrate succinyl-Leu-Leu-Val-Tyr-7-amido-4-methylcoumarin (Suc-LLVY-MCA) in 150 μl of 50 mM HEPES–TRIS (pH 8.0), 5 mM EGTA, for 20 min. The reaction was stopped by adding 1350 μl of 1% SDS. The proteasome activity was monitored by measuring the hydrolysis of the substrate using a RF-1501 Shimadzu spectrofluorimeter (380 nm excitation and 460 nm emission).
Protein extraction and 2-DE
The total proteins were extracted by phenol as previously described (Marsoni et al., 2008). Three independent protein extractions were performed from each sample. A 800 μg aliquot of total proteins was loaded onto 18 cm and pH 4–7 linear gradient IPG strips (GE Healthcare, Uppsala, Sweden). The separation of proteins in the first and second dimension was carried out as reported in Marsoni et al. (2010). Gels were visualized by the modified Colloidal Coomassie Brilliant Blue (CCBB) staining method (Candiano et al., 2004). Each separation was repeated three times for each biological replicate.
Stained gels were analysed by using the Image Master 2D Platinum software version 5.0 (Amersham Biosciences) as described in Marsoni et al. (2010). Statistical analysis (Student’s t-test at a level of 95%) identified proteins that significantly increased or decreased (at least 1.5-fold in relative abundance) after the different treatments with respect to the control. These spots were selected for MS/MS analysis.
In-gel digestion and mass spectrometry analysis
Selected spots were manually excised from the 2-D gels and digested as described in Marsoni et al. (2010). The tryptic fragments were analysed by MS/MS after reverse phase separation of peptides [liquid chromatography–electrospray ionization tandem mass spectrometry (LC-ESI-MS/MS); Marsoni et al., 2008]. Protein identification was performed by searching in the National Center for Biotechnology Information (NCBI) viridiplantae and/or EST-viridiplantae protein database using the MASCOT program (http://www.matrixscience.com). The following parameters were adopted for database searches: complete carbamidomethylation of cysteines, partial oxidation of methionines, peptide mass tolerance 1.2 Da, fragment mass tolerance 0.8 Da, and missed cleavage 1. For positive identification, the score of the result of [–10×log(P)] had to be over the significance threshold level (P < 0.05). Unsuccessful protein identifications were submitted to de novo analysis by PepNovo software using default parameters (http://proteomics.ucsd.edu/Software/PepNovo.html). Only those PepNovo results were accepted that received a mean probability score of at least 0.5. Peptide sequence candidates were edited according to MS BLAST rules, and an MS BLAST search was performed against the NCBI non-redundant database at http://www.dove.embl-heidelberg.de/Blast2/msblast.html. Statistical significance of hits was evaluated according to the MS BLAST scoring scheme. Other than the Mowse and MS BLAST scoring system to assign correct identification, a minimum of two matched peptides was necessary.
For the subcellular localization, the CELLO v.2.5: subCELlular LOcalization predictor was used (Yu et al., 2004).
Protein expression clustering
Significant differences in protein expression were analysed through the two-way hierarchical clustering methodology using the PermutMatrix software (http://www.lirmm.fr/∼caraux/PermutMatrix/; Caraux and Pinloche, 2005; Meunier et al., 2007). The row-by-row normalization of data was performed using the classical zero-mean and unit-standard deviation technique. Pearson’s distance and Ward’s algorithm were used for the analysis. PermutMatrix allows for clustering result visualization with a dendrogram of the samples and a dendrogram of the protein spots.
Western blotting
For monodimensional western blots, total soluble proteins were extracted as described above and resuspended in Laemmli sample buffer. A 60 μg aliquot of proteins was loaded onto a 14% SDS–polyacrylamide gel and transferred to polyvinylidene fluoride (PVDF) membranes (Westran CS, 0.45 μm, Whatman). Membranes were probed with 1:2000 AtSUMO antibody (AbCam, Cambridge, UK), using the Supersignal West Dura Extended Duration Chemiluminescent Substrate for HRP (horseradish peroxidase) system (Pierce). Protein loading was verified by Ponceau staining of the membrane.
For 2-D western blots, the samples (400 μg) were separated in the first and second dimension as previously described and transferred to PVDF membranes. Membranes were probed with APX monoclonal antibody, kindly supplied by Dr Akihiro Kubo (Environmental Biology Division National Institute for Environmental Studies, Onogawa, Japan). Densitometric analysis of antibody responses was performed with Phoretrix 2D v2004 (Amersham, Bioscences). The mean levels of three proteins not differentially expressed (based on proteomic analysis) were used to normalize western blot signals of APX in treated cells in comparison with the control. The analyses were carried out in triplicate.
Statistical analysis
For all analyses, at least three replicates were performed for each treatment and the values represent the means (±SD). Statistical analysis was done using a two-tailed Student’s t-test at a significance level of 95% or 99%.
Results and Discussion
Morphological and physiological responses induced by 50 mM H2O2 in TBY-2 cultured cells
TBY-2 cell cultures exposed to 50 mM H2O2 were evaluated for cell viability. Cell viability 30 min and 3 h after H2O2 treatment decreased by 5% and 15%, respectively. As previously reported (de Pinto et al., 2006), a further decrease was detected when cells were evaluated at greater intervals after the treatment. Three hours after treatment, nearly 90% of the trypan blue-dyed cells exhibited cytoplasmic shrinkage, a PCD hallmark widely reported in cultured cells (de Pinto et al., 2012). The occurrence of PCD was confirmed by the appearance in the following hours of formation of micronuclei and DNA laddering (data not shown), consistent with data obtained previously using the same experimental conditions (Vacca et al., 2004; de Pinto et al., 2006).
2-D separation and identification of differentially accumulated proteins of control and H2O2-treated TBY-2 cells
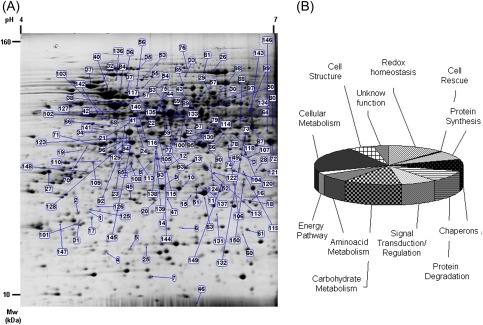
To identify proteins differentially expressed in the early phases of H2O2-induced PCD in TBY-2 cells after 30 min and 3 h, protein profiles were examined by 2-DE. Following CCBB staining, ∼1300 reproducible protein spots were detected from each sample (Fig. 1A). When comparing samples from control and H2O2-treated cells, 210 protein spots exhibited a significant (t-test; P ≤ 0.05) differences in relative abundance (±1.5 fold). Of these, 150 spots were successfully identified by LC-MS/MS analysis (Table 1). Of the differentially expressed proteins, 41 and 32 were specifically expressed at 30 min and 3 h following H2O2 treatment, respectively. A total of 77 proteins showed changes common to both treatments. Some of the identified proteins displayed discrepancies in their theoretical Mw or pI, a common phenomenon in 2-D gels. Several reasons may explain these discrepancies, including protein modifications during the extraction or the separation procedures, various isoforms of the same gene product, proteolytic cleavage, and post-translational modifications. Twenty-one unique proteins were present as different identities (isoforms) and exhibited opposite expression pattern within each set of isoforms. These isoforms may exhibit different activities or have different roles in modulating the H2O2 response.
Fig. 1.
(A) Image of a representative 2-DE gel: differentially expressed spots are indicated by their relative numbers. (B) The functional category distribution of all identified proteins in response to H2O2 stress. (This figure is available in colour at JXB online.)
Table 1.
List of the differentially H2O2-responsive proteins in TBY-2 cells
| Spot | Protein name and plant species | NCBI accession number | EST NCBI accession number | Fold variation |
Statistical significancea |
Mascot score/peptide number | Mol. wt (kDa)/pI |
|||
| 30 min | 3 h | 30 min | 3 h | Theoretical | Expected | |||||
| Redox homeostasis | ||||||||||
| 1 | Thioredoxin peroxidase, Nicotiana tabacum | gi|21912927 | –6 | <0.01 | 0.0001 | – | 225/5 | 30.0/8.2 | 24.0/5.0 | |
| 2 | Thioredoxin peroxidase, Nicotiana tabacum | gi|21912927 | –2.4 | –1.7 | 0.005 | 0.007 | 187/5 | 30.0/8.2 | 23.0/4.7 | |
| 3 | Thioredoxin peroxidase, Nicotiana tabacum | gi|21912927 | 1 | –1.9 | – | 0.01 | 314/7 | 30.0/8.2 | 32.0/6.6 | |
| 4 | Peroxiredoxin-2E, chloroplastic, Arabidopsis thaliana | gi|143360522 | –1.5 | –1.7 | 0.05 | 0.04 | 101/2 | 22.5/7.6 | 17.0/5.1 | |
| 5 | Peroxiredoxin, putativeb, Ricinus communis | gi|255575353 | gi|225317455 | <0.01 | 1 | – | – | 189/5 | 23.7/7.6 | 16.5/5.4 |
| 6 | Peroxiredoxinb, Ipomoea batatas | gi|37783267 | gi|52834488 | 3.9 | 8 | 0.002 | 0.003 | 275/6 | 20.7/8.8 | 20.0/5.9 |
| 7 | Thioredoxin H-type 1, Nicotiana tabacum | gi|267124 | –1.9 | 1 | 0.030 | – | 178/4 | 14.1/5.6 | 13.7/5.5 | |
| 8 | Disulphide oxidoreductase, putativeb, Ricinus communis | gi|255575237 | gi|761563 | –1.6 | 1 | 0.04 | – | 321/6 | 39.3/8.2 | 35.0/5.6 |
| 9 | Ascorbate peroxidase, Nicotiana tabacum | gi|559005 | 2.3 | 3.6 | 0.03 | 0.03 | 125/3 | 27.3/5.4 | 27.0/5.4 | |
| 10 | Ascorbate peroxidase, Nicotiana tabacum | gi|559005 | 1 | –1.5 | – | 0.05 | 507/11 | 27.3/5.4 | 27.0/5.7 | |
| 11 | Ascorbate peroxidase, Nicotiana tabacum | gi|76869309 | 1 | –4 | – | 0.03 | 441/9 | 27.0/5.4 | 27.0/6.1 | |
| 12 | Thylakoid-bound ascorbate peroxidase, Nicotiana tabacum | gi|4996604 | –2.4 | –4 | 0.03 | 0.02 | 891/15 | 42.0/7.6 | 32.0/5.7 | |
| 13 | Thylakoid-bound ascorbate peroxidase, Nicotiana tabacum | gi|4996604 | –1.7 | –2.4 | 0.03 | 0.04 | 150/5 | 42.0/7.6 | 32.0/6.0 | |
| 14 | Superoxide dismutase [Fe]chloroplastic, Nicotiana plumbaginifolia | gi|134642 | <0.01 | <0.01 | – | – | 300/6 | 23.0/5.5 | 23.0/5.7 | |
| 15 | Probable glutathione S-transferaseb, Capsicum annuum | gi|60459397 | gi|76871463 | 4.7 | 6 | 0.01 | 0.002 | 135/4 | 25.4/5.5 | 26.0/5.9 |
| 16 | Glutathione S-transferaseb, Solanum commersonii | gi|148616162 | gi|83420783 | –1.9 | 1 | 0.03 | – | 202/5 | 23.8/5.9 | 25.0/6.8 |
| 17 | Glyoxalase Ia, Solanum lycopersicum | gi|2494844 | gi|190145651 | –1.6 | 1 | 0.02 | – | 268/6 | 20.7/5.3 | 21.0/4.9 |
| 18 | Flavodoxin-like quinone reductase 1b, Arabidopsis thaliana | gi|15239652 | gi|83420109 | –1.8 | 1 | 0.006 | – | 357/7 | 21.7/5.9 | 25.0/6.7 |
| Cell rescue | ||||||||||
| 19 | Chilling-responsive protein, Nicotiana tabacum | gi|153793260 | –2 | 2.3 | 0.05 | 0.03 | 114/4 | 36.0/4.9 | 39.0/4.9 | |
| Protein synthesis | ||||||||||
| 20 | Eukaryotic translation initiation factor 3 subunit, putativeb, Ricinus communis | gi|255550315 | gi|76867840 | –1.8 | 1.7 | 0.05 | 0.03 | 459/10 | 26.9/5.8 | 25.6/5.6 |
| 21 | Eukaryotic initiation factor 4A-11, Nicotiana tabacum | gi|2500518 | 1.6 | 3.2 | 0.05 | 0.007 | 735/17 | 47.2/5.4 | 47.0/5.3 | |
| 22 | Eukaryotic initiation factor 4A-9b, Nicotiana tabacum | gi|2500517 | gi|190874337 | 1 | –1.8 | – | 0.001 | 580/11 | 47.0/5.5 | 45.0/5.4 |
| 23 | Eukaryotic initiation factor 4A-9, Nicotiana tabacum | gi|2500517 | 1.7 | 2.5 | 0.04 | 0.008 | 691/15 | 47.0/5.5 | 46.0/5.3 | |
| 24 | Eukaryotic initiation factor 4A-15, Nicotiana tabacum | gi|2500521 | 1 | 2 | – | 0.008 | 1214/23 | 46.9/5.4 | 45.0//5.4 | |
| 25 | Eukaryotic translation initiation factor 5A-2c, Nicotiana plumbaginifolia | gi|124226 | 1 | <0.01 | – | – | 60/1 | 17.6/5.6 | 17.3/5.5 | |
| 26 | Eukaryotic translation elongation factor, putativeb, Ricinus communis | gi|255544686 | gi|92027234 | 2 | 1 | 0.001 | – | 441/15 | 94.0/5.9 | 105.0/6.3 |
| 27 | Elongation factor 1-beta/EF-1-betav, Arabidopsis thaliana | gi|145324076 | gi|76867651 | 1 | –1.7 | – | 0.006 | 493/11 | 28.7/4.6 | 31.4/4.5 |
| 28 | Ribosomal protein L2-like, Solanum tuberosum | gi|81074776 | 7.3 | 1 | 0.03 | – | 100/3 | 28.7/10.6 | 31.0/6.7 | |
| 29 | Glycyl-tRNA synthetase, putativeb, Ricinus communis | gi|255543218 | gi|190806778 | 2.6 | 1 | 0.07 | – | 223/6 | 77.3/6.6 | 66.5/6.2 |
| 30 | Glycyl-tRNA synthetase, putativea, Ricinus communis | gi|255543218 | gi|190878443 | <0.01 | 1 | – | – | 75/2 | 77.3/6.6 | 69.0/6.3 |
| Chaperones | ||||||||||
| 31 | Mitochondrial small heat shock proteinc, Solanum lycopersicum | gi|3492854 | gi|92010828 | 1 | 1.9 | – | 0.04 | 78/1 | 23.8/6.5 | 21.3/4.8 |
| 32 | Heat shock protein, putative, Ricinus communis | gi|255581792 | 1 | –1.5 | – | 0.04 | 498/8 | 90.0/5.2 | 92.0/5.1 | |
| 33 | Heat shock protein, Solanum lycopersicum | gi|68989120 | <0.01 | –2.9 | – | 0.009 | 276/6 | 110/6.2 | 102.0/6.0 | |
| 34 | Hsp70-binding protein, putative, Ricinus communis | gi|255581500 | gi|224704536 | 1 | –1.6 | – | 0.009 | 169/4 | 39.2/5.2 | 41.4/5.0 |
| 35 | Heat shock 70 kDa protein, putativeb, Ricinus communis | gi|255574576 | gi|92037393 | 1 | –1.6 | – | 0.009 | 227/4 | 94.0/5.2 | 110.0/5.6 |
| 36 | Heat shock 70 kDa protein, putativeb, Ricinus communis | gi|255574576 | gi|92027886 | 1 | –1.8 | – | 0.02 | 262/2 | 94.0/5.2 | 110.0/5.5 |
| 37 | Molecular chaperone Hsp90-1, Nicotiana benthamiana | gi|38154482 | 1 | –2.2 | – | 0.02 | 404/9 | 80.0/4.9 | 83.0/4.9 | |
| 38 | Heat shock protein, putative, Ricinus communis | gi|255554571 | –5.2 | 1 | 0.03 | – | 527/10 | 71.0/6.1 | 60.2/5.3 | |
| 39 | Hypothetical protein cpn60, Vitis vinifera | gi|225442531 | –2 | –1.7 | 0.02 | 0.04 | 589/11 | 65.3/5.6 | 60.2/5.3 | |
| 40 | Putative luminal binding proteina, Corylus avellana | gi|10944737 | gi|190753562 | <0.01 | –2 | – | 0.02 | 618/23 | 73.0/4.9 | 74.0/5.1 |
| 41 | Chaperonin containing t-complex protein 1, epsilon subunit, tcpe, putativeb, Ricinus communis | gi|255547962 | gi|92040148 | 2.3 | 1.6 | 0.03 | 0.04 | 488/11 | 59.2/5.5 | 60.0/5.6 |
| 42 | Chaperonin containing t-complex protein 1, epsilon subunit, tcpe, putativeb, Ricinus communis | gi|255547962 | gi|92040148 | <0.01 | <0.01 | – | – | 173/4 | 59.2/5.5 | 55.4/5.2 |
| 43 | Chaperonin containing t-omplex protein 1, gamma subunit, tcpe, putativeb, Ricinus communis | gi|255577568 | gi|76867096 | 1 | 1.7 | – | 0.003 | 604/11 | 60.0/5.9 | 63.0/5.9 |
| 44 | Chaperonin T-complex protein 1 subunit epsilon, Zea mays | gi|226506102 | 1.5 | 2.5 | 0.05 | 0.002 | 204/5 | 59.6/5.7 | 59.0/5.6 | |
| Protein degradation | ||||||||||
| 45 | Chaperonin 21 precursorb, Solanum lycopersicum | gi|7331143 | gi|52839170 | –1.9 | 1 | 0.04 | 1 | 212/4 | 26.5/6.8 | 25.2/5.4 |
| 46 | Ubiquitin, Nicotiana benthamiana | gi|213868277 | 1 | 2.7 | – | 0.03 | 222/4 | 7.5/5.7 | 7.5/6.1 | |
| 47 | 20S proteasome subunit beta-6b, Petunia hybrida | gi|17380185 | gi|92037305 | 1 | 1.7 | – | 0.003 | 367/5 | 24.6/6.3 | 26.7/5.7 |
| 48 | 20S proteasome alpha 6 subunit, Nicotiana benthamiana | gi|22947842 | <0.01 | <0.01 | – | – | 153/4 | 29.8/5.0 | 33.0/5.2 | |
| 49 | 26S proteasome non-ATPase regulatory subunit, putative, Ricinus communis | gi|255538376 | 1 | <0.01 | – | – | 97/2 | 27.2/6.1 | 32.0/6.5 | |
| 50 | 26S protease regulatory subunit, putativeb, Ricinus communis | gi|255570523 | gi|94328305 | 1.6 | –1.7 | 0.05 | 0.03 | 610/11 | 49.5/5.9 | 58.0/6.2 |
| 51 | Putative alpha7 proteasome subunit, Nicotiana tabacum | gi|14594925 | –1.6 | 1 | 0.03 | – | 116/4 | 27.2/6.1 | 27.0/6.1 | |
| 52 | Cysteine proteinase aleuran typeb, Nicotiana benthamiana | gi|71482942 | gi|76866797 | –4.7 | <0.01 | 0.008 | – | 158/3 | 39.2/6.9 | 29.5/6.2 |
| 53 | ATP-dependent Clp protease ATP–binding subunit clpA homologue CD4B, chloroplastic, Solanum lycopersicum | gi|399213 | –2.4 | –2 | 0.04 | 0.02 | 692/15 | 102.5/5.7 | 96.0/5.5 | |
| 54 | Oligopeptidase Ab, Ricinus communis | gi|255572579 | gi|190876913 | –8 | 1 | 0.03 | – | 247/4 | 88.0/5.2 | 76.0/5.6 |
| 55 | Oligopeptidase A, putativeb, Ricinus communis | gi|255572579 | gi|190876913 | –5.6 | 1 | 0.03 | – | 87/2 | 88.0/5.2 | 78. /5.5 |
| 56 | Oligopeptidase A, putative, Ricinus communis | gi|255572579 | gi|76868752 | 1.9 | –1.8 | 0.05 | 0.01 | 524/11 | 88.0/5.2 | 77.0/5.4 |
| 57 | Mitochondrial processing peptidaseb, Solanum tuberosum | gi|587566 | gi|190874528 | 1 | –1.8 | – | 0.003 | 459/8 | 59.9/6.2 | 60.0/6.0 |
| 58 | Mitochondrial processing peptidase, Solanum tuberosum | gi|587564 | –2.6 | 1 | 0.04 | – | 281/8 | 59.4/6.2 | 60.0/5.6 | |
| 59 | Cytochrome c reductase-processing, peptidase subunit IIb, Solanum tuberosum | gi|410634 | gi|224697460 | 1 | –2 | – | 0.013 | 59.3/6.2 | 59.0/5.7 | |
| Signal transduction/regulation | ||||||||||
| 60 | BTF3, Nicotiana benthamiana | gi|90823167 | <0.01 | <0.01 | – | – | 100/3 | 17.3/6.3 | 19.0/6.5 | |
| 61 | BTF3, Nicotiana benthamiana | gi|90823167 | 2.4 | 2 | 0.05 | 0.0009 | 195/4 | 17.3/6.3 | 19.4/6.6 | |
| 62 | Nucleic acid-binding protein, putativeb, Ricinus communis | gi|255558037 | gi|190847542 | <0.01 | –1.7 | – | 0.03 | 454/7 | 28.6/4.9 | 32.0/5.4 |
| 63 | DNA-binding protein GBP16, Oryza sativa Japonica Group | gi|2511541 | <0.01 | <0.01 | – | – | 74/2 | 43.4/6.5 | 26.0/6.1 | |
| 64 | EBP1, Solanum tuberosum | gi|116292768 | gi|94324881 | 1 | –2.3 | – | 0.02 | 288/7 | 42.8/6.3 | 49.0/6.4 |
| 65 | SGT1-like protein, Nicotiana tabacum | gi|29468339 | <0.1 | <0.1 | – | – | 138/3 | 41.4/5.2 | 33.0/5.4 | |
| 66 | SGT1-like protein, Nicotiana tabacum | gi|29468339 | 2.4 | 1.6 | 0.015 | 0.03 | 167/3 | 41.4/5.2 | 46.0/5.1 | |
| 67 | Hypothetical protein RNA bindingb, Vitis vinifera | gi|296081884 | gi|190733368 | –1.7 | 1 | 0.02 | – | 253/5 | 43.7/5.8 | 62.5/5.4 |
| 68 | Hypothetical protein DNA helicase, putativeb, Vitis vinifera | gi|147858961 | gi|190806396 | 2.5 | 1 | 0.002 | – | 219/5 | 51.4/5.3 | 53.0/5.6 |
| 69 | DNA helicase, putative, Ricinus communis | gi|255565715 | 2.1 | 1 | 0.034 | – | 476/8 | 50.2/5.8 | 54.0/5.8 | |
| 70 | Adenosine kinase isoform 1T, Nicotiana tabacum | gi|51949796 | 1.5 | 1.6 | 0.05 | 0.02 | 125/2 | 37.8/5.1 | 39.0/4.8 | |
| 71 | Adenosine kinase isoform 1T, Nicotiana tabacum | gi|51949796 | 1 | –1.5 | – | 0.007 | 608/11 | 37.8/5.1 | 40.0/4.9 | |
| Carbohydrate metabolism | ||||||||||
| 72 | GAPDH, Nicotiana tabacum | gi|120676 | –1.8 | –2.2 | 0.05 | 0.0005 | 135/4 | 35.5/6.1 | 36.0/6.8 | |
| 73 | GAPDH, Nicotiana tabacum | gi|120676 | 4.8 | 6.4 | 0.001 | 0.002 | 750/17 | 35.5/6.1 | 36.5/6.6 | |
| 74 | GAPDH, Nicotiana tabacum | gi|120676 | >100 | 1 | – | – | 453/10 | 35.5/6.1 | 36.5/6.3 | |
| 75 | Enolase, Nicotiana tabacum | gi|119354 | 2.2 | 2 | 0.008 | 0.02 | 223/4 | 48.0/5.6 | 50.0/5.9 | |
| 76 | Phosphoenolpyruvate carboxylase, Glycine max | gi|399182 | 1 | –2.7 | – | 0.0001 | 74/2 | 110.6/5.7 | 110/5.8 | |
| 77 | Phosphoglycerate kinase, chloroplastic pecursor, Nicotiana tabacum | gi|2499497 | 1 | –1.7 | – | 0.04 | 563/10 | 50.3/8.5 | 40.0/5.8 | |
| 78 | Alcohol dehydrogenase, Nicotiana tabacum | gi|551257 | –1.8 | –2.7 | 0.007 | 0.01 | 661/14 | 41.9/6.6 | 42.0/6.4 | |
| 79 | Alcohol dehydrogenase, Nicotiana tabacum | gi|551257 | 2.1 | 6 | 0.01 | 0.05 | 442/10 | 41.9/6.6 | 42.0/6.1 | |
| 80 | Alcohol dehydrogenase class IIIb, Solanum lycopersicum | gi|283825505 | gi|285193921 | 2.2 | 8 | 0.05 | 0.001 | 100/4 | 40.7/6.3 | 43.0/6.9 |
| 81 | Cytosolic aconitase, Nicotiana tabacum | gi|11066033 | 1 | 5 | – | 0.03 | 134/3 | 98.7/5.8 | 108/6.1 | |
| 82 | Putative aconitaseb, Capsicum chinense | gi|171854675 | gi|92028044 | 1 | <0.01 | – | 0.041 | 131/3 | 108/7.0 | 48.0/6.0 |
| 83 | Pyruvate decarboxylase isozyme 1, Nicotiana tabacum | gi|1706327 | 2.3 | 1 | 0.003 | – | 407/7 | 45.7/6.6 | 60.0/5.7 | |
| 84 | Succinate dehydrogenase, putativeb, Ricinus communis | gi|255579273 | gi|190782074 | <0.1 | <0.1 | – | – | 181/3 | 68.5/6.2 | 73.0/5.2 |
| 85 | NADH-ubiquinone oxidoreductase, putative, Ricinus communis | gi|255582280 | 1 | –1.7 | – | 0.001 | 360/4 | 80.8/5.9 | 85.0/6.1 | |
| 86 | NAD-dependent isocitrate dehydrogenase, Nicotiana tabacum | gi|3790188 | 1 | 7 | – | 0.03 | 334/7 | 40.6/7.2 | 40.6/ 6.2 | |
| 87 | ADH-like UDP-glucose dehydrogenase, Nicotiana tabacum | gi|48093455 | –1.9 | –2.2 | 0.03 | 0.05 | 796/17 | 42.0/6.2 | 42.0/6.7 | |
| 88 | ADH-like UDP-glucose dehydrogenase, Nicotiana tabacum | gi|48093455 | 3.7 | 2.2 | 0.04 | 0.03 | 819/17 | 42.0/6.2 | 42.0/6.5 | |
| 89 | UTP-glucose-1-phosphate uridylyltransferase, Solanum tuberosum | gi|17402533 | 2:00 | 1 | 0.03 | – | 494/9 | 52.0/5.4 | 54.0/5.85 | |
| 90 | UDP-glucose:protein transglucosylase-likeb, Solanum tuberosum | gi|77416931 | gi|123218663 | 1.7 | 1 | 0.001 | – | 362/8 | 41.1/5.6 | 38.0/6.1 |
| Amino acid metabolism | ||||||||||
| 91 | Diaminopimelate epimerase, putativeb, Ricinus communis | gi|255584553 | gi|83422108 | 1 | –2 | – | 0.0005 | 377/9 | 40.1/6.0 | 35.0/5.2 |
| 92 | Aspartate semialdehyde dehydrogenase family proteinvb, Arabidopsis thaliana | gi|15223910 | gi|39863720 | 6.1 | 9 | 0.001 | 0.008 | 266/6 | 40.7/6.5 | 39.0/5.5 |
| 93 | Aspartate semialdehyde dehydrogenase, putativeb, Nicotiana sylvestris | gi|255584961 | gi|190769575 | –1.8 | –1.7 | 0.04 | 0.03 | 306/7 | 41.2/8.2 | 37.2/5.6 |
| 94 | Putative 3-isopropylmalate dehydrogenase large subunit, Capsicum annuum | gi|193290700 | <0.01 | 1 | – | – | 170/4 | 43.7/5.9 | 40.0/5.3 | |
| 95 | Isovaleryl-CoA dehydrogenase 2, mitochondrialb, Solanum tuberosum | gi|25453061 | gi|83422448 | 1.8 | 1 | 0.01 | – | 166/63 | 43.9/6.1 | 40.0/6.1 |
| 96 | Glutamine synthetase, Nicotiana plumbaginifolia | gi|121373 | 1.8 | 2.3 | 0.05 | 0.04 | 117/4 | 39.0/5.5 | 38.0/5.2 | |
| 97 | D−3−phosphoglyceratedehydrogenase,putative, Ricinus communis | gi|255555301 | gi|39856030 | –5.6 | 1 | 0.007 | – | 575/9 | 63.0/7.7 | 62.7/6.3 |
| 98 | EDA9 (embryo sac development arrest 9), ATP binding, Arabidopsis thaliana | gi|15235282 | >100 | 1 | – | – | 97/2 | 63.5/6.2 | 64.0/6.3 | |
| 99 | Chain A, structure of threonine synthase, Arabidopsis thaliana | gi|15825882 | <0.01 | 1 | – | – | 92/2 | 53.5/5.6 | 54.7/6.6 | |
| 100 | S-Adenosylmethionine synthase 2, Solanum lycopersicum | gi|1170938 | <0.01 | 1 | – | – | 112/3 | 43.0/5.4 | 45.0/5.9 | |
| Energy pathway | ||||||||||
| 101 | ATP synthase subunit delta', mitochondrialc, Ipomoea batatas | gi|2493046 | –1.8 | 2.2 | 0.007 | 0.01 | 55/1 | 21.3/5.9 | 20.0/4.7 | |
| 102 | Mitochondrial ATPase beta subunit, Nicotiana sylvestris | gi|11228579 | <0.01 | <0.01 | – | – | 622/15 | 59.6/5.2 | 48.0/5.1 | |
| 103 | Vacuolar H+-ATPase B subunit, Nicotiana tabacum | gi|6715512 | 1.6 | 1.5 | 0.004 | 0.04 | 351/8 | 53.8/5.1 | 55.0/5.1 | |
| 104 | Electron carrier/oxidoreductaseb, Arabidopsis thaliana | gi|15232542 | gi|92032774 | –1.8 | –2.3 | 0.05 | 0.001 | 218/4 | 37.2/5.7 | 36.0/6.2 |
| Cellular metabolism | ||||||||||
| 105 | Putative carbamoyl phosphate synthase small subunit, Nicotiana tabacum | gi|21535793 | –2.7 | 1 | 0.04 | – | 100/4 | 47.7/6.0 | 42.0/5.9 | |
| 106 | N-Carbamoylputrescine amidaseb, Solanum tuberosum | gi|118572820 | gi|92015448 | 1.5 | 2.2 | 0.05 | 0.007 | 220/6 | 33.4/5.9 | 33.0/6.4 |
| 107 | N-Carbamoylputrescine amidaseb, Solanum tuberosum | gi|118572820 | gi|92015448 | –5.4 | <0.01 | 0.03 | – | 199/5 | 33.4/5.9 | 33.4/6.6 |
| 108 | Spermidine synthase, Nicotiana sylvestris | gi|6094336 | 1.7 | 1 | 0.01 | – | 460/8 | 34.5/5.2 | 31.4/5.3 | |
| 109 | Spermidine synthase, Nicotiana sylvestris | gi|6094336 | 2.4 | 1 | 0.02 | – | 347/8 | 34.5/5.2 | 31.8/4.9 | |
| 110 | Spermidine synthase, Nicotiana sylvestris | gi|6094336 | <0.01 | 1 | – | – | 198/4 | 34.5/5.2 | 32.0/5.1 | |
| 111 | Putative pyridoxine biosynthesis protein isoform A, Nicotiana tabacum | gi|46399269 | 2.1 | 1.7 | 0.03 | 0.0002 | 709/12 | 33.0/5.9 | 31.0/5.8 | |
| 112 | Putative pyridoxine biosynthesis protein isoform A, Nicotiana tabacum | gi|46399269 | –1.9 | 1 | – | 347/5 | 33.0/5.9 | 31.0/6.1 | ||
| 113 | 5'-Aminoimidazole ribonucleotide synthetaseb, Solanum tuberosum | gi|37983566 | gi|190730691 | –1.9 | –1.8 | 0.03 | 0.004 | 109/2 | 42.9/5.2 | 33.0/5.3 |
| 114 | Putative 4-methyl-5 (β-hydroxyethyl)-thiazol monophosphate biosynthesis enzymec, Capsicum chinense | gi|171854671 | –1.5 | <0.01 | 0.04 | – | 77/1 | 41.7/5.4 | 40.0/5.9 | |
| 115 | Putative 4-methyl-5 (β-hydroxyethyl)-thiazol monophosphate biosynthesis enzymeb, Capsicum chinense | gi|171854671 | gi|190775218 | <0.01 | <0.01 | – | – | 225/5 | 41.7/5.4 | 37.0/5.6 |
| 116 | Putative 4-methyl-5 (β-hydroxyethyl)-thiazol monophosphate biosynthesis enzymeb, Capsicum chinense | gi|171854671 | gi|47004010 | >100 | >100 | – | – | 232/3 | 41.7/5.4 | 40.0/5.5 |
| 117 | Betaine-aldehyde dehydrogenaseb, Nicotiana tabacum | gi|92037527 | 1 | –1.6 | – | 0.006 | 187/3 | 59.7/5.4 | 57.3/5.6 | |
| 118 | Putative cinnamyl alcohol dehydrogenaseb, Nicotiana tabacum | gi|156763848 | gi|190749158 | 1 | 1.8 | – | 0.02 | 82/2 | 38.9/6.6 | 39.8/6.5 |
| 119 | NAD-dependent epimerase/dehydratase, putativeb, Ricinus communis | gi|255537241 | gi|190751083 | >100 | >100 | – | – | 261/5 | 34.0/6.2 | 31.0/6.6 |
| 120 | NAD dependent epimerase/dehydratase, putativeb, Ricinus communis | gi|255537241 | gi|92028282 | 3.5 | 2.9 | 0.007 | 0.004 | 440/10 | 34.0/6.2 | 30.0/6.5 |
| 121 | NAD-dependent epimerase/dehydratase, putativeb, Ricinus communis | gi|255537241 | gi|92028282 | –8.1 | –7 | 0.04 | 0.004 | 476/10 | 34.0/6.2 | 30.0/6.8 |
| 122 | Type 2 proly 4–hydroxylaseb, Nicotiana tabacum | gi|215490181 | gi|190760427 | >100 | >100 | – | – | 219/3 | 32.7/6.3 | 29.0/6.2 |
| 123 | Gibberellin 20 oxidase, putative, Ricinus communis | gi|255556243 | –2.5 | <0.01 | 0.02 | – | 126/2 | 42.0/5.7 | 39.0/5.3 | |
| 124 | 1-Aminocyclopropane-1-carboxylate oxidaseb, Solanum lycopersicum | gi|50830975 | gi|39853392 | 1.7 | –1.6 | 0.001 | 0.03 | 114/2 | 34.4/6.1 | 32.0/6.2 |
| 125 | Acireductone dioxygenaseb, Solanum tuberosum | gi|158325159 | gi|76867763 | <0.01 | <0.01 | – | – | 237/4 | 23.3/4.8 | 24.0/4.8 |
| 126 | Acireductone dioxygenaseb, Solanum tuberosum | gi|158325159 | gi|76866491 | 1 | <0.01 | – | – | 149/4 | 23.3/4.8 | 24.6/4.9 |
| 127 | Rubisco subunit binding-protein alpha subunitb, Ricinus communis | gi|255587664 | gi|190794529 | –1.5 | –1.5 | 0.05 | 0.04 | 895/9 | 53.2/5.2 | 61.0/4.8 |
| 128 | SAL1; phosphatidylinositol phosphataseb, Arabidopsis thaliana | gi|145359623 | gi|190143393 | 1.5 | 1.6 | 0.05 | 0.02 | 353/6 | 43.4/6.0 | 40.0/5.3 |
| 129 | Patatin homologue, Nicotiana tabacum | gi|1546817 | –1.9 | –1.8 | 0.05 | 0.02 | 363/6 | 42.5/5.1 | 41.0/5.2 | |
| 130 | Acyl-[acyl-carrier-protein] desaturase, chloroplasticb, Solanum commersoni | gi|94730426 | gi|92012186 | <0.01 | –1.9 | – | 0.03 | 376/8 | 44.8/6.3 | 38.6/5.9 |
| 131 | DH putative beta-hydroxyacyl-ACP dehydrataseb, Capsicum annuum | gi|193290688 | gi|52834057 | >100 | >100 | – | – | 146/2 | 23.9/9.4 | 23.6/6.2 |
| 132 | Putative pyruvate dehydrogenase E1 alpha subunit, Capsicum annuum | gi|193290722 | <0.01 | <0.01 | – | – | 159/5 | 48.0/6.3 | 40.0/6.6 | |
| 133 | Glutamate–cysteine ligase, chloroplastic, Nicotiana tabacum | gi|122194121 | –2.5 | –1.9 | 0.05 | 0.05 | 304/6 | 59.4/6.2 | 46.3/5.7 | |
| 134 | L-Galactono-gamma-lactone dehydrogenase, Nicotiana tabacum | gi|6519872 | <0.01 | –2 | – | 0.04 | 350/9 | 67.1/7.7 | 54.0/6.6 | |
| 135 | Putative ketol-acid reductoisomerase, Capsicum annuum | gi|193290660 | –2.6 | 1 | 0.04 | – | 281/8 | 63.7/6.5 | 60.0/5.7 | |
| 136 | Prolyl endopeptidase, putativeb, Ricinus communis | gi|255539116 | gi|190876295 | –2.3 | –1.9 | 0.04 | 0.008 | 301/5 | 80.0/5.3 | 95.0/5.2 |
| 137 | Short chain dehydrogenaseb, Solanum tuberosum | gi|77403673 | gi|254640456 | <0.01 | 1 | – | – | 139/3 | 27.2/6.2 | 26.8/6.1 |
| Cell structure | ||||||||||
| 138 | Actin isoform Bb, Mimosa pudica | gi|6683504 | gi|39877476 | <0.01 | <0.01 | – | – | 153/4 | 41.7/5.3 | 36.0/5.7 |
| 139 | Actin, Nicotiana tabacum | gi|197322805 | <0.01 | <0.01 | – | – | 72/1 | 41.7/5.3 | 35.0/5.7 | |
| 140 | Actin-binding protein ABP29b, Lilium longiflorum | gi|117553552 | gi|190829672 | –2.7 | –5.5 | 352/6 | 29.4/6.0 | 38.0/5.5 | ||
| 141 | Actin, Nicotiana tabacum | gi|50058115 | gi|39805809 | 2.5 | 2.6 | 0.05 | 0.03 | 147/5 | 41.8/5.3 | 43.0/5.2 |
| 142 | alpha-Tubulin, Nicotiana tabacum | gi|11967906 | 1 | –2 | 1 | 0.002 | 124/2 | 50.4/4.9 | 49.0/6.5 | |
| 143 | Tubulin beta-2 chainb, Anemia phyllitidis | gi|297826947 | gi| 56691710 | <0.01 | 1 | – | – | 114/4 | 46.8/4.9 | 55/6.8 |
| 144 | Predicted proteinb,. Populus trichocarpa | gi|224125262 | gi|39875797 | 1.5 | 1.5 | 0.04 | 0.02 | 322/7 | 26.0/5.1 | 30.0/5.7 |
| 145 | Villin 3 fragmentc, Arabidopsis thaliana | gi|6735320 | <0.01 | 1 | – | – | 63/1 | 64.8/5.5 | 38.0/5.4 | |
| 146 | Hypothetical protein stomatin-like proteinb, Vitis vinifera | gi|225442194 | gi|92027149 | 1.7 | 1 | 0.03 | – | 553/8 | 45.6/9.0 | 39.0/6.8 |
| 147 | Hypothetical protein isoform 2b, Vitis vinifera | gi|225454579 | gi|190805443 | 3.2 | 1 | 0.0003 | – | 230/5 | 17.0/4.7 | 21.0/4.6 |
| 148 | Hypothetical protein coatomer delta subunitb, Vitis vinifera | gi|270239956 | gi|190738155 | <0.01 | <0.01 | – | – | 226/5 | 61.5/5.6 | 30.0/4.9 |
| 149 | Stem-specific protein TSJT1, putativeb, Ricinus communis | gi|255552037 | gi|92026938 | 1 | –2.4 | – | 0.002 | 380/7 | 27.0/6.0 | 27.8/6.3 |
| 150 | Stem-specific protein TSJT1, putativeb, Ricinus communis | gi|255552037 | gi|92026938 | 3.8 | 1 | 0.02 | – | 448/10 | 27.0/6.0 | 28.0/6.6 |
A value >100 (<0.01) means that the protein spot is detected (is not present) under the condition reporting this value.
Tests of statistical significance of changes in spot volumes utilized a Student’s t-test, and levels of P ≤ 0.05 were considered significant.
Protein found in the EST NCBI database. The name, the accession number, the molecular weight, and the isoelectric point were annotated by BLAST search
Protein confirmed by de novo analysis.
According to their putative physiological functions, the identified proteins were classified into different functional categories (Fig. 1B): redox homeostasis (18), cell rescue (1), protein synthesis (11), chaperones (15), protein degradation (14), signal transduction/regulation (12), carbohydrate metabolism (19), amino acid metabolism (10), energy pathways (four), cellular metabolism (33), cell structure (8), and unknown function (5). The involvement of different processes in the early phase of PCD is in agreement with the fact that this cell suicide is an active process which is metabolically regulated.
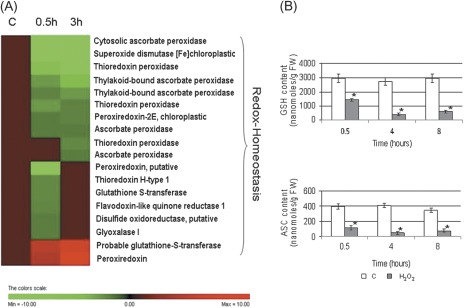
Cellular redox homeostasis
Of the 18 differentially expressed proteins implicated in redox homeostasis, 16 were down-regulated (Fig. 2A). Six protein spots (spots 1–6) were members of the peroxiredoxin (PRX) family. All of these protein spots displayed more acidic experimental pI values compared with the theoretical values and four of the identified PRXs showed smaller Mw values (Table 1). In redox-stressed HeLa cells, the acidic shift of the PRX spot position is due to irreversible protein oxidation, which leads to PRX inactivation (Wagner et al., 2002). Moreover, site-specific protein oxidation has been reported to signal ubiquitination, thus triggering protein degradation (Iwai et al., 1998). Therefore, the observed decrease in the PRX quantity may be explained by the acceleration of PRX turnover under conditions inducing a high level of oxidative damage to proteins. Consistently, the analysis showed that the level of a thioredoxin (spot 7), which regenerates the active form of PRX (Dietz, 2003), was decreased.
Fig. 2.
(A) Hierarchical clustering of proteins implicated in cellular redox homeostasis after treatment with H2O2. (B) Contents of GSH and ASC after H2O2 treatment. * indicates values that are significantly different from those of control cells with P < 0.01 (Student’s t-test).
PRXs can function as peroxidases, redox sensors, and molecular chaperones (Neuman et al., 2003; Kim et al., 2009). Furthermore, they are involved in a variety of cellular functions including apoptosis. In mammalian cells, PRX5 overexpression prevents p53-dependent ROS generation and apoptosis (Zhou et al., 2000). Overexpression of PRX1 and PRX2 leads to the elimination of H2O2, thereby protecting cells from apoptosis (Kim et al., 2000). Furthermore, the depletion of PRX3 by RNA interference in HeLa cells or the suppression of 1-Cys PRX in rat lung epithelial cells leads to increased susceptibility to peroxide-induced apoptosis (Chang et al., 2001; Pak et al., 2002). During H2O2-induced PCD, it is plausible that the observed decrease in PRXs promotes the oxidative cellular environment and deregulates protein stability due to an inhibition of chaperone function (see the following paragraph).
Two cytosolic APXs (c-APXs, spots 10 and 11) and thylakoidal APXs (t-APXs, spots 12 and 13) were down-regulated in cells undergoing H2O2-induced PCD, while spot 9, also corresponding to another isoform of c-APX, was clearly increased. The behaviour of APX isoenzymes is different from that previously observed during HS-induced PCD, where only c-APX levels decreased (Marsoni et al., 2010).
H2O2-treated cells showed a decreased level of glutamate-cysteine ligase (γ-ECS, spot 133), the rate-limiting enzyme involved in the synthesis of GSH and of L-galactono-γ-lactone dehydrogenase (GLDH, spot 134), the last enzyme of the ASC biosynthetic pathway. It is known that ASC and GSH are critical for the removal of reactive oxygen species (ROS) in plants and the control of redox homeostasis (Mittler, 2002; Shigeoka et al., 2002; Foyer and Noctor, 2011). Previous studies using Arabidopsis mutants deficient in ASC demonstrated that low ASC levels trigger PCD (Pavet et al., 2005). Alteration in glutathione pool is also part of the signalling cascade leading to PCD (Kranner et al., 2006). The rapid and drastic decrease of GSH and ASC in cells undergoing H2O2-induced PCD seems to be correlated with the decrease of the enzymes mentioned above. (Fig. 2B, C). When PCD is triggered by HS, there is no change in the amount of the two enzymes (Marsoni et al., 2010). Consistently, in HS-induced PCD, the decrease of the two redox metabolites occurred after a more prolonged time from the PCD induction (Locato et al., 2008).
GLDH is an integral part of the plant mitochondrial Complex I. It has been reported that the cellular redox state affects GLDH catalysis (Millar et al., 2003). The down-regulation of the NADH-ubiquinone oxidoreductase of Complex I (spot 85) observed in TBY-2 cells treated with H2O2 for 3 h suggests a correlation between the GLDH level and Complex I inhibition. These data are consistent with the identified role of GLDH in the assembly and accumulation of Complex I in plant mitochondria (Pineau et al., 2008).
Impairment of GSH metabolism in H2O2-treated cells may also correlate with the decrease in glyoxalase I (spot 17). Indeed, glyoxalase I converts toxic 2-oxoaldehydes into less active 2-hydroxyacids using GSH as a cofactor. In mammalian tumour cells, the inhibition of glyoxalase I is able to induce apoptosis (Thornalley et al., 1996).
Two other enzymes involved in ROS detoxification were lowered by 50 mM H2O2 treatment: superoxide dismutase (SOD, spot 14) and flavodoxin-like quinone reductase 1 (FQR, spot 18). Laskowski et al. (2002) hypothesized that FQR1 may protect cells against oxidative stress by preventing the formation of semiquinones that may contribute to ROS accumulation.
It is interesting to note that by 30 min after PCD induction, most of these ROS-scavenging enzymes were already strongly reduced and showed a further decrease when examined at longer time intervals (Table 1).
The data presented above, together with the decrease in ASC and GSH levels, confirm that antioxidant systems play a role as regulators of PCD pathways activated under oxidative stress. They also further substantiate that the lowering of antioxidant defences favours the oxidative cellular environment that characterizes PCD activated by different elicitors (de Pinto et al., 2012, and references therein).
Protein synthesis and degradation
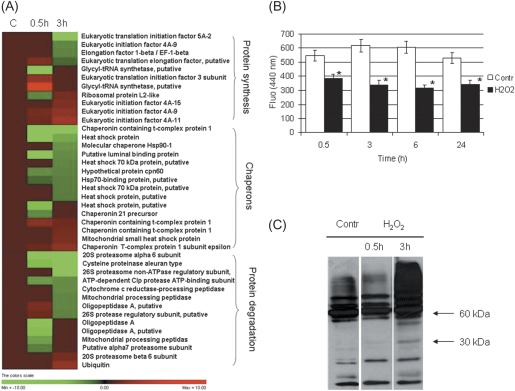
A total of 40 proteins whose levels changed in response to H2O2 treatment were involved in protein metabolism (protein synthesis, chaperons, protein degradation; Table 1, Fig. 3A). This indicates that the active control of protein biosynthesis, folding, and degradation is important in cells undergoing H2O2-dependent PCD.
Fig. 3.
(A) Hierarchical clustering of proteins implicated in protein synthesis and degradation upon treatment with H2O2. (B) Proteasome activity in H2O2- stressed cells. * indicates values that are significantly different from control cells with P < 0.01 (Student’s t-test). (C) Western blot analysis with SUMO-1 antibody in control and H2O2-treated samples.
One protein group consisted of several eukaryotic initiation and elongation factors of translation (spots 20–27), two glycyl-tRNA synthetases (spots 29, 30), and a ribosomal protein (spot 28). The 50 mM H2O2 treatment was observed to increase the levels of some proteins, while decreasing the levels of others. These data do not allow a conclusion to be drawn on whether H2O2 treatment has a positive or negative effect on protein synthesis. However, after phenolic extraction, the amount of total extracted protein did not differ significantly between the control and H2O2-treated cells during the experimental conditions tested (data not shown).
A second group consisted of 15 proteins involved in proper protein folding. Eleven of these proteins were decreased. In particular, seven heat shock proteins (HSPs, spots 32–38) exhibited reduced levels. This suggests that these plant proteins, as observed in animal systems, may possess an anti-PCD function that is not necessarily related to their chaperone role (Beere, 2005; Didelot et al., 2006).
Interestingly, in cells treated with H2O2, a decrease in a luminal binding protein (BiP, spot 40) was observed after 30 min of treatment. This protein shares high homology with the well-studied human protein GRP78, a member of the HSP family required for endoplasmic reticulum (ER) integrity. Its role in promoting cell growth and antagonizing apoptosis has been demonstrated in several tumour cell lines (Zhao et al., 2010).
It has recently been reported that BiP prevents stress-induced cell death in plants (Reis et al., 2011; Ye et al., 2011). Down-regulation of BiP at early stages of H2O2-induced PCD confirms that the ER stress/unfolded protein response (UPR) may be involved in the control of cell death.
The third protein group consisted of 14 proteins involved in protein degradation. Twelve of these were down-regulated (Fig. 3A), including three isoforms of oligopeptidase A (spots 54–56), two mitochondrial processing peptidases (spot 57 and 58), a cysteine proteinase (spot 52), a Clp protease (spot 53), and five proteasome subunits (spots 47–51). Intriguingly, the α6 and α7 proteasome subunit genes are down-co-expressed in Arabidopsis during PCD induced by the hypersensitive response (http://atted.jp, Obayashi et al., 2009). Changes in proteasome subunit composition reveal potential modifications of its substrate specificity. Based on proteasome subunit alterations revealed by proteomic analysis, H2O2-treated cells were examined to determine whether and how proteasome activity changed. The proteasome activity decreased up to 50% with respect to the control from 30 min up to 24 h of treatment (Fig. 3B). In animal cells, proteasome inhibitors induce apoptosis (Wojcik, 1999). In tobacco plants, virus-induced gene silencing of the α6 subunit of the 20S proteasome reduces proteasome activity, leading to the accumulation of polyubiquitinated proteins and activation of PCD (Kim et al., 2003). Interestingly, the proteomic data showed a drastic decrease of the same α6 subunit (spot 48) in cells undergoing H2O2-induced PCD.
HSPs and the ubiquitin–proteasome (UPM) machinery do not represent mutually exclusive pathways. Instead, both are directly linked to the ER quality control system that deals with unfolded protein accumulation. In animal cells, HSP/UPM controls apoptotic cell death (Garrido and Solary, 2003). The present results and the decrease in the BiP content support the proposed role of ER in the cell death signalling cascade also in plants (Urade, 2007; Cacas, 2010; Liu and Howell, 2010). Additionally, protein sumoylation influences ubiquitination and protein stability and is involved in apoptosis (Hatake et al., 2009). In mammalian cells, high H2O2 concentrations inhibit sumo-deconjugation from target proteins, resulting in increased sumoylation levels (Veal et al., 2007). Thus, the sumoylation level of the H2O2-treated cells was evaluated, although proteome analysis did not detect any variation in the free SUMO content between control and H2O2-treated cells. An initial slight decrease and a subsequent strong accumulation in the level of SUMO-conjugates was found during H2O2-induced PCD (Fig. 3C). These results could be explained by the differential sensitivity of the conjugation and de-conjugation machinery to ROS-induced inactivation, as suggested by Bossis and Melchior (2006). It is tempting to speculate that 30 min after H2O2 addition, only the conjugation is affected, leading to a slight decrease of SUMO-conjugates, whereas after 3 h de-conjugation is strongly impaired, which results in an increased sumoylation level.
Metabolism
Proteomic data confirmed that oxidative stress inducing PCD affects central metabolic pathways including glycolysis, the tricarboxylic acid cycle (TCA), fermentation, and amino acid metabolism. Treatment of TBY-2 cells with H2O2 resulted in a decrease in the abundance of some key mitochondrial proteins. In particular, two enzymes involved in the electron transport chain (ETC) were down-regulated, including succinate dehydrogenase (spot 84), involved in the ETC and TCA cycle, and NADH-ubiquinone oxidoreductase (75 kDa subunit, spot 85), which is a core component of the mitochondrial membrane respiratory chain NADH dehydrogenase (Complex I). Enzymes of the glycolytic and fermentation pathways were differentially regulated in cells undergoing H2O2-induced PCD compared with control cells. These enzymes include glyceraldehyde 3-phosphate dehydrogenase (GAPDH, spots 72–74), phosphoglycerate kinase (spot 77), enolase (spots 75), alcohol dehydrogenase (spots 78–80), and pyruvate decarboxylase (spot 76). The up-regulation of some enzymes involved in ethanol fermentation suggests that fermentation may compensate for mitochondrial energy dysfunction. Similar results were obtained under abiotic stress (Kürsteiner et al., 2003).
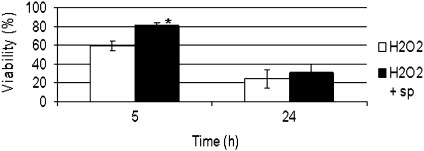
A number of enzymes involved in polyamine (spots 106–110) and glycine betaine biosynthesis (spot 117) were significantly altered in the H2O2-treated cells. Thus, the effects of exogenously added spermidine on PCD were analysed. Based on results presented in Fig 4, it can be concluded that spermidine delays H2O2-induced PCD. This finding confirmed previous results suggesting that exogenous spermidine application could modify oxidative stress intensity by altering the expression or activity of some scavenging enzymes and the cellular ROS levels (He et al., 2008; Marsoni et al., 2010).
Fig. 4.
Effect of spermidine pre-treatment on cell viability of H2O2-stressed cells. The values are the means of three different experiments. * indicates values that are significantly different from those of control cells with P < 0.01 (Student’s t-test).
The abundance of carbamoyl phosphate synthetase II (CPSII, spot 105), a cytosolic enzyme that catalyses the first step in pyrimidine biosynthesis, was decreased by H2O2 treatment. CPSII is a target for caspase-dependent regulation during apoptosis (Huang et al., 2002). Alterations in pyrimidine nucleotide synthesis have been shown to be strictly associated with the early phase of H2O2-induced PCD in TBY-2 cells (Stasolla et al., 2004).
In cells undergoing H2O2-induced PCD, proteomic analysis revealed the differential expression of two proteins involved in purine metabolism: 5′-aminoimidazole ribonucleotide synthetase (AIR, spot 113), an enzyme involved in purine biosynthesis; and two isoforms of adenosine kinase1T (ADK1T, spots 70 and 71), an enzyme involved in both the phosphorylation of adenosine and the purine salvage pathway. The decrease in AIR may be related to intracellular energy depletion, consistent with the observation made during PCD senescence in tulip petals (Azad et al., 2008). The intracellular phosphorylation of isopentenyladenosine (iPA) is necessary for the activation of caspase-like proteases and for the induction of PCD in TBY-2 cells (Mlejnek and Prochazka, 2002; Mlejnek et al., 2003). Among the four tobacco ADK isoforms altered in H2O2-induced PCD, ADK1T exhibits a 10-fold higher affinity for iPA when compared with other substrates (Kwade et al., 2005). These data, together with the transient increase of ADK1T in cells undergoing H2O2-induced PCD, suggest that this enzyme is a component of the PCD machinery acting at a very early step of the process.
To regulate PCD, H2O2 participates in complex interactions with plant hormones. An initial increase and subsequent decrease in the amount of ACC oxidase (spot 124), the last enzyme in ethylene biosynthesis, was found during H2O2-induced PCD. Ethylene is a positive regulator of several types of H2O2-induced PCD, including lysigenic aerenchyma in roots and the camptothecin-induced PCD in tomato cells (He et al., 1996; de Jong et al., 2002). Interestingly, ASC is a cofactor of ACC oxidase. Therefore, the down-regulation of this enzyme 3 h after PCD induction could also be correlated with the strong depletion of ASC (∼90%) occurring at this time. Two isoforms of acireductone dioxygenase (ARD, spots 125 and 126), an enzyme involved in methionine salvage, were also decreased and potentially related to alterations in ethylene synthesis and signalling. Similar results were obtained for ARD in response to biotic and abiotic stresses in wheat (Xu et al., 2010).
Cell structure
The levels of three enzymes involved in the biosynthesis of cell wall polysaccharides and lignin were altered in cells experiencing H2O2-induced PCD: UTP-glucose-1-phosphate uridyltransferase (spot 89), UDP-glucose dehydrogenase (spots 87 and 88), and cinnamyl-alcohol dehydrogenase (spot 118). Moreover a prolyl 4-hydroxylase (spot 122) accumulated following H2O2 treatment. This enzyme hydroxylates proline-rich structural glycoproteins of the cell walls. The plant cell wall is rich in hydroxyproline-rich glycoproteins, which are developmentally regulated and correlated with changes in cellular morphology. Specific hydroxyproline-rich glycoproteins bridge the cell wall and cytoskeleton (Knox, 1995). It is noteworthy that cytoskeleton proteins (spots 138–145) were also altered.
In the PCD induced during tracheary element differentiation, the cell wall undergoes reinforcement and thickening (Gadjev et al., 2008). Alterations in cell wall thickness were not evident during H2O2-induced PCD (data not shown). Further studies will be necessary to characterize whether this compartment is structurally affected during a type of PCD that does not lead to vessel formation.
Proteins common to H2O2- and HS-induced PCD: towards the ‘core complex’ of the PCD process
PCD activators can be very different: chemical or physical agents, phytopathogens, endogenous metabolic signals, and others. When a specific agent activates PCD it could produce other kinds of effects in the cell which are specific to that particular agent and not necessary involved in PCD signalling. Therefore, these effects must be distinguished from those directly involved in PCD. Indeed, the nature and activities of core complex regulators of PCD are poorly understood, being masked by homeostatic alteration induced by the specific stressor. In an attempt to identify proteins common to a minimum of two PCD pathways, proteomic data from cells experiencing H2O2-induced PCD were compared with those previously obtained from cells undergoing HS-induced PCD (Marsoni et al., 2010). In both cases, 3 h after the induction of PCD, cell mortality was ∼15%. This indicates that cells collected 3 h after treatment in each PCD system can be considered comparable. However, the PCD process proceeds more rapidly following H2O2 treatment when compared with HS treatment. Cell death reaches values of 70% and 50% after 24 h in H2O2-induced PCD and in HS-induced PCD, respectively (de Pinto et al., 2006; Locato et al., 2008). Therefore, the results obtained 6 h after HS treatments were also analysed in an effort to identify the ‘core complex proteins’ involved in PCD.
The comparison of the proteomic profiles of H2O2-induced PCD and HS-induced PCD (Table 1; Marsoni et al., 2010) demonstrates that there are many differences between the two PCD pathways. However, 15 protein spots exhibited similar trends in both types of PCD (Table 2). The intensity of 12 of these spots was decreased and the intensity of 3 spots increased in both PCD pathways.
Table 2.
List of the putative core complex proteins required for PCD induction in TBY-2 cells
| Spot | Protein name | NCBI accession number | Fold variation |
||
| H2O2, 3 h | HS, 3 h | HS, 6 h | |||
| Redox homeostasis | |||||
| 1 | Thioredoxin peroxidase, Nicotiana tabacum | gi|21912927 | <0.01 | 1 | –4.3 |
| 10 | Ascorbate peroxidase, Nicotiana tabacum | gi|76869309 | –1.5 | –1.5 | –1.5 |
| Chaperones | |||||
| 35 | Heat shock 70 kDa protein, putative, Ricinus communis | gi|255574576 | –1.6 | –1.6 | –2 |
| 36 | Heat shock 70 kDa protein, putative. Ricinus communis | gi|255574576 | –1.8 | 1 | –2 |
| Protein degradation | |||||
| 48 | 20S proteasome alpha 6 subunit, Nicotiana benthamiana | gi|22947842 | <0.01 | –1.8 | –1.6 |
| Signal transduction | |||||
| 65 | SGT1-like protein, Nicotiana tabacum | gi|29468339 | <0.01 | –3.2 | 1 |
| Metabolism | |||||
| 72 | GAPDH, Nicotiana tabacum | gi|120676 | –2.2 | –1.5 | 1 |
| 75 | Enolase, Nicotiana tabacum | gi|119354 | 2 | 2 | 2.7 |
| 76 | Phosphoenolpyruvate carboxylase, Glycine max | gi|399182 | –2.7 | –1.7 | 1 |
| 103 | Vacuolar H+-ATPase B subunit, Nicotiana tabacum | gi|6715512 | 1.5 | 1 | 1.9 |
| 117 | Betaine-aldehyde dehydrogenase, Nicotiana tabacum | gi|92037527 | –1.6 | 1 | –2.2 |
| Cell structure | |||||
| 138 | Actin isoform B, Mimosa pudica | gi|6683504 | <0.01 | –2 | –2.4 |
| 139 | Actin, Nicotiana tabacum | gi|197322805 | <0.01 | –2.8 | –3.2 |
| 141 | Actin, Nicotiana tabacum | gi|50058115 | 2.6 | 2 | 1 |
| 142 | Alpha tubulin, Nicotiana tabacum | gi|11967906 | –2 | –2.6 | 1 |
Among the core complex proteins, five proteins were related to the cytoskeleton. Swidzinski et al. (2002) reported that after PCD-inducing treatments in cell cultures of Arabidopsis thaliana, the expression of the alpha tubulin and actin2 genes was inhibited. Studies using drugs that affect actin turnover suggest that either actin stabilization or depolymerization can induce PCD in yeast and mammalian cells (Gourlay and Ayscough, 2005; Thomas et al., 2006). Consistently, cytoskeleton reorganization has been very recently suggested to be an active and common player in the initiation and regulation of plant PCD (Smertenko and Franklin-Tong, 2011). Alteration in the cytoskeleton as a precocious step in PCD signalling could be relevant for cytoplasmic shrinkage, a PCD hallmark occurring in TBY-2 cells undergoing both H2O2- and HS-induced PCD (Vacca et al., 2004; de Pinto et al., 2006).
For some of the core proteins listed in Table 2, it is difficult to understand if their alteration is part of the signalling leading to PCD or a homeostatic response aimed at maintaining cellular metabolism, in particular for those proteins with enzymatic activity, the amount of which increase during PCD. The possibility remains that during plant PCD altered proteins may play other novel roles in addition to the already known functions. For example, Hsp70 proteins play an important role in the maintenance and survival of mammalian cells by acting as anti-apoptosis proteins, a function that appears to be independent of their chaperone activity (Beere et al., 2000). The fact that two Hsp70 isoforms were down-regulated both in H2O2- and HS-induced PCD support an anti-PCD role for this protein also in plants.
As in animals (Chuang et al., 2005), plant GAPDH might have multiple functions, one of which may be regulation of H2O2 signalling in the cell (Hancock et al., 2005; Baek et al., 2008). As a consequence, the decrease in the amount of this enzyme might be aimed at impairing ROS defence responses during H2O2-induced PCD, even if further studies are required to support this hypothesis. In relation to the proteins typically involved in redox homeostasis, it was quite surprising to identify only two proteins (thioredoxin peroxidase and APX) the alterations of which were similarly induced in both HS- and H2O2-induced PCD, although a strong redox impairment occurs under the two kinds of PCD (de Pinto et al., 2006; Locato et al., 2008) and several proteins involved in redox regulation were altered in both H2O2- and HS-induced PCD (Table 1; Marsoni et al., 2010). This result is in agreement with the view that redox regulation/homeostasis involves a complex network of metabolites and enzymes. This makes redox regulation extremely flexible and particularly useful as a nodal regulatory point of developmental processes as well as of defence responses (Foyer and Noctor, 2011).
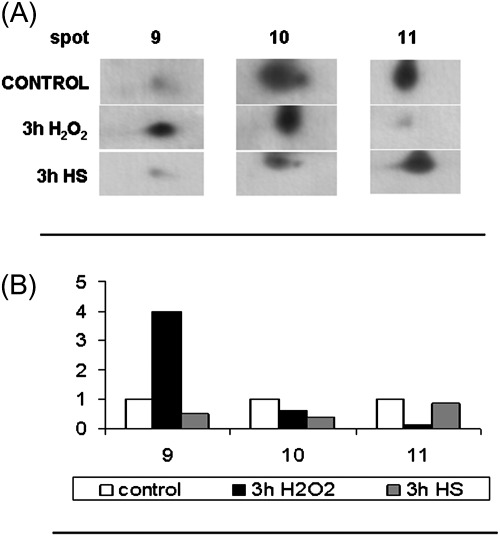
Among the two peroxidases down-regulated in the cells leading to PCD, a more thorough analysis of the c-APX isoenzymes was performed in TBY-2 cells undergoing H2O2- and HS-induced PCD. Figure 5 shows the western blot of APX separated by 2-DE and identified by a monoclonal antibody that specifically recognizes the cytosolic isoenzymes of APXs. The three c-APX spots (9, 10, and 11) were recognized by the antibody in H2O2-treated cells, consistent with the data in Table 1. Comparing the intensity of these spots with those obtained from cells treated with HS for 3 h only, spot 10 significantly decreases in both types of PCD with respect to the control. In spite of the fact that c-APX activity strongly decreases in both HS- and H2O2-induced PCD (Locato et al., 2008), under H2O2 treatment spot 9 of c-APX strongly increased. Such an increase is probably due to protein oxidation by H2O2, induced by the treatment used for inducing PCD.
Fig. 5.
2-DE changes in the level of ascorbate peroxidase (APX) protein in cells exposed to heat shock and H2O2 treatments. (A) Immunoblotting analysis was performed using a specific APX antibody (see Materials and methods) in control cells, in cells treated at 55 °C for 10 min and recovered for 3 h (3 h HS), and in cells exposed to H2O2 for 3 h. (B) Densitometric analysis of antibody responses. The relative optical density is expressed in arbitrary units. The levels of three protein not differentially expressed (based on proteomic analysis) were used to normalize western blot signals. The image represents one of three independent replicas.
The identification of c-APX among the core proteins involved in PCD underlines the relevance of this enzymes in contributing to create the redox environment necessary for PCD progression.
Conclusion
Exogenous application of 50 mM H2O2 induces a strong oxidative stress in TBY-2 cells (de Pinto et al., 2006). The data presented here indicate that, under these conditions, the cell fails to cope with oxidative stress. The antioxidant defence system and the anti-PCD signalling cascades are inhibited. This promotes a genetically programmed cell suicide pathway. Proteomic and physiological data indicate the following:
(i) The inhibition of several players in the cellular redox hub as a key point for PCD induction. The occurrence of a redox impairment during PCD is also supported by the protective effect of spermidine, a potent scavenger of hydroxyl radicals, occurring in PCD triggered by two different stimuli. The involvement of ASC and GSH, non-specific ROS scavengers, and spermidine suggests that other forms of ROS are important as positive regulators of PCD induction. Similar conclusions have been reported by Doyle and McCabe (2010).
(ii) The inhibition of the protein repair–degradation system and of several chaperonins results in the accumulation of abnormal, oxidized, and misfolded proteins that triggers fine-tuned signalling mechanims devoted to the alleviation of the stress. If stress cannot be resolved, cells commit suicide.
(iii) A consistent part of the proteins altered in H2O2-treated cells are involved in metabolism and the cytoskeleton. The changes of the former group could be a homeostatic response to the metabolic changes induced during PCD, in particular to the strong impairment in energy metabolisms. The variation of the latter group could be correlated with PCD cytological markers.
References
- Azad AK, Ishikawa T, Sawa Y, Shibata H. Intracellular energy depletion triggers programmed cell death during petal senescence in tulip. Journal of Experimental Botany. 2008;59:2085–2095. doi: 10.1093/jxb/ern066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D, Jin Y, Jeong JC, et al. Suppression of reactive oxygen species by glyceraldehyde-3-phosphate dehydrogenase. Phytochemistry. 2008;69:333–338. doi: 10.1016/j.phytochem.2007.07.027. [DOI] [PubMed] [Google Scholar]
- Beere HM. Death versus survival: functional interaction between the apoptotic and stress-inducible heat shock protein pathways. Journal of Clinical Investigation. 2005;115:2633–2639. doi: 10.1172/JCI26471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beere HM, Wolf BB, Cain K, Mosser DD, Mahboubi A, Kuwana T, Tailor P, Morimoto RI, Cohen GM, Green DR. Heat-shock protein 70 inhibits apoptosis by preventing recruitment of procaspase-9 to the Apaf-1 apoptosome. Nature Cell Biology. 2000;2:469–475. doi: 10.1038/35019501. [DOI] [PubMed] [Google Scholar]
- Bossis G, Melchior F. Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Molecular Cell. 2006;21:349–357. doi: 10.1016/j.molcel.2005.12.019. [DOI] [PubMed] [Google Scholar]
- Candiano G, Bruschi M, Musante L, Santucci L, Ghiggeri GM, Crnemolla B, Orechhia P, Zardi L, Righetti PG. Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis. 2004;25:1327–1333. doi: 10.1002/elps.200305844. [DOI] [PubMed] [Google Scholar]
- Cacas JL. Devil inside: does plant programmed cell death involve the endomembrane system? Plant. Cell and Environment. 2010;33:1453–1473. doi: 10.1111/j.1365-3040.2010.02117.x. [DOI] [PubMed] [Google Scholar]
- Caraux G, Pinloche S. PermutMatrix: a graphical environment to arrange gene expression profiles in optimal linear order. Bioinformatics. 2005;21:1280–1281. doi: 10.1093/bioinformatics/bti141. [DOI] [PubMed] [Google Scholar]
- Chang JW, Jeon HB, Lee JH, Yoo JS, Chun JS, Kim JH, Yoo YJ. Augmented expression of peroxiredoxin I in lung cancer. Biochemical and Biophysical Research Communications. 2001;289:507–512. doi: 10.1006/bbrc.2001.5989. [DOI] [PubMed] [Google Scholar]
- Chuang D, Hough C, Senatorov V. Glyceraldehyde-3-phosphate dehydrogenase, apoptosis, and neurodegenerative diseases. Annual Review of Pharmacology and Toxicology. 2005;45:269–290. doi: 10.1146/annurev.pharmtox.45.120403.095902. [DOI] [PubMed] [Google Scholar]
- Dat JF, Pellinen R, Beeckman T, Van De Cotte B, Langebartels C, Kangasjärvi J, Inzè D, Van Breusegem F. Changes in hydrogen peroxide homeostasis trigger an active cell death process in tobacco. The Plant Journal. 2003;33:621–632. doi: 10.1046/j.1365-313x.2003.01655.x. [DOI] [PubMed] [Google Scholar]
- Didelot C, Schmitt E, Brunet M, Maingret L, Parcellier A, Garrido C. Heat shock proteins: endogenous modulators of apoptotic cell death. Handbook of Experimental Pharmacology. 2006;5:171–198. doi: 10.1007/3-540-29717-0_8. [DOI] [PubMed] [Google Scholar]
- Dietz KJ. Plant peroxiredoxins. Annual Review of Plant Biology. 2003;54:93–107. doi: 10.1146/annurev.arplant.54.031902.134934. [DOI] [PubMed] [Google Scholar]
- Doyle SM, McCabe PF. Type and cellular location of reactive oxygen species determine activation or suppression of programmed cell death in Arabidopsis suspension cultures. Plant Signaling and Behavior. 2010;5:467–468. doi: 10.4161/psb.5.4.11399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer C, Noctor G. Ascorbate and glutathione: the heart of the redox hub. Plant Physiology. 2011;155:2–18. doi: 10.1104/pp.110.167569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadjev IJ, Stone M, Gechev TS. Programmed cell death in plants: new insights into redox regulation and the role of hydrogen peroxide. International Review of Cell and Molecular Biology. 2008;270:87–144. doi: 10.1016/S1937-6448(08)01403-2. [DOI] [PubMed] [Google Scholar]
- Garrido C, Solary E. A role of HSPs in apoptosis through ‘protein triage’? Cell Death and Differentiation. 2003;10:619–620. doi: 10.1038/sj.cdd.4401229. [DOI] [PubMed] [Google Scholar]
- Gechev TS, Hille J. Hydrogen peroxide as a signal controlling plant programmed cell death. Journal of Cell Biology. 2005;168:17–20. doi: 10.1083/jcb.200409170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gechev TS, Van Breusegem F, Stone JM, Denev I, Laloi C. Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. Bioessays. 2006;28:1091–1101. doi: 10.1002/bies.20493. [DOI] [PubMed] [Google Scholar]
- Gourlay CW, Ayscough KR. The actin cytoskeleton in ageing and apoptosis. FEMS Yeast Research. 2005;5:1193–1198. doi: 10.1016/j.femsyr.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Gunawardena A, Greenwood JS, Dengler NG. Programmed cell death remodels lace plant leaf shape during development. The Plant Cell. 2004;16:60–73. doi: 10.1105/tpc.016188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatake K, Kuniyoshi R, Mishima Y, Terui Y. Sumoylation and apoptosis. In: Wilson V, editor. SUMO regulation of cellular processes. Berlin: Springer; 2009. pp. 217–230. [Google Scholar]
- Hancock JT, Henson D, Nyirenda M, Desikan R, Harrison J, Lewis M, Hughes J, Neill SJ. Proteomic identification of glyceraldehyde 3-phosphate dehydrogenase as an inhibitory target of hydrogen peroxide in Arabidopsis. Plant Physiology and Biochemistry. 2005;43:828–835. doi: 10.1016/j.plaphy.2005.07.012. [DOI] [PubMed] [Google Scholar]
- He C, Morgan P, Drew M. Transduction of an ethylene signal is required for cell death and lysis in the root cortex of maize during aerenchyma formation induced by hypoxia. Plant Physiology. 1996;112:463–472. doi: 10.1104/pp.112.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Ban Y, Inoue H, Matsuda N, Liu J, Moriguchi T. Enhancement of spermidine content and antioxidant capacity in transgenic pear shoots overexpressing apple spermidine synthase in response to salinity and hyperosmosis. Phytochemistry. 2008;69:2133–2141. doi: 10.1016/j.phytochem.2008.05.015. [DOI] [PubMed] [Google Scholar]
- Houot V, Etienne P, Petitot A-S, Barbier S, Blein J-P, Suty L. Hydrogen peroxide induces programmed cell death features in cultured tobacco BY-2 cells, in a dose dependent manner. Journal of Experimental Botany. 2001;52:1721–1730. [PubMed] [Google Scholar]
- Huang M, Kozlowski P, Collins M, Wang Y, Haystead TA, Graves LM. Caspase-dependent cleavage of carbamoyl phosphate synthetase II during apoptosis. Molecular Pharmacology. 2002;61:569–577. doi: 10.1124/mol.61.3.569. [DOI] [PubMed] [Google Scholar]
- Iwai K, Drake SK, Wehr NB, Weissman AM, LaVaute T, Minato N, Klausner RD, Levine RL, Rouault TA. Iron-dependent oxidation, ubiquitination, and degradation of iron regulatory protein-2: implications for degradation of oxidized proteins. Proceedings of the National Academy of Sciences, USA. 1998;95:4924–4928. doi: 10.1073/pnas.95.9.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong AJ, Yakimova ET, Kachina VM, Woltering EJ. A critical role for ethylene in hydrogen peroxide release during programmed cell death in tomato suspension cells. Planta. 2002;214:537–545. doi: 10.1007/s004250100654. [DOI] [PubMed] [Google Scholar]
- de Pinto MC, Locato V, De Gara L. Redox regulation in plant programmed cell death. Plant, Cell and Environment. 2012;35:234–244. doi: 10.1111/j.1365-3040.2011.02387.x. [DOI] [PubMed] [Google Scholar]
- de Pinto MC, Paradiso A, Leonetti P, De Gara L. Hydrogen peroxide, nitric oxide and cytosolic ascorbate peroxidase at the crossroad between defence and cell death. The Plant Journal. 2006;48:784–795. doi: 10.1111/j.1365-313X.2006.02919.x. [DOI] [PubMed] [Google Scholar]
- de Pinto MC, Tommasi F, De Gara L. Changes in the antioxidant systems as part of the signalling pathway responsible for the programmed cell death activated by nitric oxide and reactive oxygen species in tobacco BY-2 cells. Plant Physiology. 2002;130:698–708. doi: 10.1104/pp.005629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pinto MC, Tommasi F, De Gara L. Enzymes of the ascorbate biosynthesis and ascorbate–glutathione cycle in cultured cells of tobacco bright yellow 2. Plant Physiology and Biochemistry. 2000;38:541–550. [Google Scholar]
- Kim H, Lee T H, Park ES, Suh JM, Park SJ, Chung HK, Kwon OY, Kim YK, Ro HK, Shong M. Role of peroxiredoxins in regulating intracellular hydrogen peroxide and hydrogen peroxide-induced apoptosis in thyroid cells. Journal of Biological Chemistry. 2000;275:18266–18270. doi: 10.1074/jbc.275.24.18266. [DOI] [PubMed] [Google Scholar]
- Kim M, Ahn JW, Jin UH, Chai D, Poek KH, Pai HS. Activation of the programmed cell death pathway by inhibition of proteasome function in plant. Journal of Biological Chemistry. 2003;278:19406–19415. doi: 10.1074/jbc.M210539200. [DOI] [PubMed] [Google Scholar]
- Kim SY, Jang HH, Lee JR, et al. Oligomerization and chaperone activity of a plant 2-Cys peroxiredoxin in response to oxidative stress. Plant Science. 2009;177:227–232. [Google Scholar]
- Knox JP. The extracellular matrix in higher plants. 4. Developmentally regulated proteoglycans and glycoproteins of the plant cell surface. FASEB Journal. 1995;9:1004–1012. doi: 10.1096/fasebj.9.11.7544308. [DOI] [PubMed] [Google Scholar]
- Kranner I, Birtic S, Anderson K, Pritchard HW. Glutathione half-cell reduction potential: a universal stress marker and modulator of programmed cell death? Free Radical Biology and Medicine. 2006;40:2155–2165. doi: 10.1016/j.freeradbiomed.2006.02.013. [DOI] [PubMed] [Google Scholar]
- Kürsteiner O, Dupuis I, Kuhlemeier C. The pyruvate decarboxylase1 gene of Arabidopsis is required during anoxia but not other environmental stresses. Plant Physiology. 2003;132:968–978. doi: 10.1104/pp.102.016907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwade Z, Swiatek A, Azmi A, Goossens A, Inzé D, Van Onckelen H, Roef L. Identification of four adenosine kinase isoforms in tobacco BY-2 cells and their putative role in the cell cycle-regulated cytokinin metabolism. Journal of Biological Chemistry. 2005;280:17512–17519. doi: 10.1074/jbc.M411428200. [DOI] [PubMed] [Google Scholar]
- Laskowski MJ, Dreher KA, Gehring MA, Abel S, Gensler AL, Sussex IM. FQR1, a novel primary auxin-response gene, encodes a flavin mononucleotide-binding quinine reductase. Plant Physiology. 2002;128:578–590. doi: 10.1104/pp.010581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Howell SH. Endoplasmic reticulum protein quality control and its relationship to environmental stress responses in plants. The Plant Cell. 2010;22:2930–2942. doi: 10.1105/tpc.110.078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locato V, Gadaleta C, De Gara L, de Pinto MC. Production of reactive species and modulation of antioxidant network in response to heat shock: a critical balance for cell fate. Plant, Cell and Environment. 2008;31:1606–1619. doi: 10.1111/j.1365-3040.2008.01867.x. [DOI] [PubMed] [Google Scholar]
- Marsoni M, Bracale M, Espen L, Prinsi B, Negri AS, Vannini C. Proteomic analysis of somatic embryogenesis in Vitis vinifera. Plant Cell Reports. 2008;27:347–356. doi: 10.1007/s00299-007-0438-0. [DOI] [PubMed] [Google Scholar]
- Marsoni M, Cantara C, de Pinto C, Gadaleta C, De Gara L, Bracale M, Vannini C. Exploring the soluble proteome of Tobacco Bright Yellow-2 cells at the switch towards different cell fates in response to heat shocks. Plant, Cell and Environment. 2010;33:1161–1175. doi: 10.1111/j.1365-3040.2010.02137.x. [DOI] [PubMed] [Google Scholar]
- Meunier B, Dumas E, Piec I, Béchet D, Hébraud M, Hocquette JF. Assessment of hierarchical clustering methodologies for proteomic data mining. Journal of Proteome Research. 2007;6:358–366. doi: 10.1021/pr060343h. [DOI] [PubMed] [Google Scholar]
- Millar AH, Mittova V, Kiddle G, Heazlewoood JL, Bartoli CG, Theodoulou FL, Foyer CH. Control of ascorbate synthesis by respiration and its implications for stress responses. Plant Physiology. 2003;133:443–447. doi: 10.1104/pp.103.028399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R. Oxidative stress, antioxidants and stress tolerance. Trends in Plant Science. 2002;7:405–410. doi: 10.1016/s1360-1385(02)02312-9. [DOI] [PubMed] [Google Scholar]
- Mlejnek P, Dolezel P, Procházka S. Intracellular phosphorylation of benzyladenosine is related to apoptosis induction in tobacco BY-2 cells. Plant, Cell and Environment. 2003;26:1723–1735. [Google Scholar]
- Mlejnek P, Procházka S. Activation of caspase-like proteases and induction of apoptosis by isopentenyladenosine in tobacco BY-2 cells. Planta. 2002;215:158–166. doi: 10.1007/s00425-002-0733-5. [DOI] [PubMed] [Google Scholar]
- Murgia ID, Tarantino D, Vannini C, Bracale M, Caravieri S, Soave C. Arabidopsis thaliana plants overexpressing thylakoidal ascorbate peroxidase show increased resistance to paraquat-induced photooxidative stress and to nitric oxide-induced cell death. The Plant Journal. 2004;38:940–953. doi: 10.1111/j.1365-313X.2004.02092.x. [DOI] [PubMed] [Google Scholar]
- Neumann CA, Krause DS, Carman CV, Das S, Dubey DP, Abraham JL, Bronson RT, Fujiwara Y, Orkin SH, van Etten RA. Essential role for the peroxiredoxin Prdx1 in erythrocyte antioxidant defence and tumour suppression. Nature. 2003;424:561–565. doi: 10.1038/nature01819. [DOI] [PubMed] [Google Scholar]
- Obayashi T, Hayashi S, Saeki M, Ohta H, Kinoshita K. ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Research. 2009;37:987–991. doi: 10.1093/nar/gkn807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak JH, Manevich Y, Kim HS, Feinstein SI, Fisher AB. An antisense oligonucleotide to 1-cys peroxiredoxin causes lipid peroxidation and apoptosis in lung epithelial cells. Journal of Biological Chemistry. 2002;277:49927–49934. doi: 10.1074/jbc.M204222200. [DOI] [PubMed] [Google Scholar]
- Palma K, Kermode AR. Metabolism of hydrogen peroxide during reserve mobilization and programmed cell death of barley (Hordeum vulgare L.) aleurone layer cells. Free Radical Biology and Medicine. 2003;35:1261–1270. doi: 10.1016/s0891-5849(03)00511-2. [DOI] [PubMed] [Google Scholar]
- Pavet V, Olmos E, Kiddle G, Mowla S, Kumar S, Antoniw J. Ascorbic acid deficiency activates cell death and disease resistance responses in Arabidopsis. Plant Physiology. 2005;139:1291–1303. doi: 10.1104/pp.105.067686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineau B, Layoune O, Danon A, De Paepe R. l-Galactono-1,4-lactone dehydrogenase is required for the accumulation of plant respiratory complex I. Journal of Biological Chemistry. 2008;283:32500–32505. doi: 10.1074/jbc.M805320200. [DOI] [PubMed] [Google Scholar]
- Reis PA, Rosado GL, Silva LA, Oliveira LC, Oliveira LB, Costa MD, Alvim FC, Fontes EP. The binding protein BiP attenuates stress-induced cell death in soybean via modulation of the N-rich protein-mediated signaling pathway. Plant Physiology. 2011;157:1853–1865. doi: 10.1104/pp.111.179697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeoka S, Ishikawa T, Tamoi M, Miyagawa Y, Takeda T, Yabuta Y, Yoshimura K. Regulation and function of ascorbate peroxidase isoenzymes. Journal of Experimental Botany. 2002;53:1305–1319. [PubMed] [Google Scholar]
- Smertenko A, Franklin-Tong VE. Organisation and regulation of the cytoskeleton in plant programmed cell death. Cell Death and Differentiation. 2011;18:1263–1270. doi: 10.1038/cdd.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasolla C, Loukanina N, Yeung EC, Thorpe TA. Alterations in pyrimidine metabolism as an early signal during the execution of programmed cell death in tobacco BY-2 cells. Journal of Experimental Botany. 2004;55:2513–2522. doi: 10.1093/jxb/erh259. [DOI] [PubMed] [Google Scholar]
- Swidzinski JA, Sweetlove LJ, Leaver CJ. A custom microarray analysis of gene expression during programmed cell death in Arabidopsis thaliana. The Plant Journal. 2002;30:431–446. doi: 10.1046/j.1365-313x.2002.01301.x. [DOI] [PubMed] [Google Scholar]
- Thornalley J, Edwards G, Kang Y, Wyatt C, Davies N, Ladan MJ, Double J. Antitumour activity of S-p-bromobenzylglutathione cyclopentyl diester in vitro and. in vivo. Biochemical Pharmacology. 1996;51:1365–1372. doi: 10.1016/0006-2952(96)00059-7. [DOI] [PubMed] [Google Scholar]
- Thomas SG, Huang S, Li S, Staiger CJ, Franklin-Tong VE. Actin depolymerization is sufficient to induce programmed cell death in selfincompatible pollen. Journal of Cell Biology. 2006;174:221–229. doi: 10.1083/jcb.200604011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urade R. Cellular response to unfolded proteins in the endoplasmic reticulum of plants. FEBS Journal. 2007;274:1152–1171. doi: 10.1111/j.1742-4658.2007.05664.x. [DOI] [PubMed] [Google Scholar]
- Vacca RA, de Pinto MC, Valenti D, Passerella S, Marra E, De Gara L. Reactive oxygen species production, impairment of glucose oxidation and cytosolic ascorbate peroxidase are early events in heat-shock induced programmed cell death in tobacco BY-2 cells. Plant Physiology. 2004;134:1100–1112. doi: 10.1104/pp.103.035956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn W. Classes of programmed cell death in plants, compared to those in animals. Journal of Experimental Botany. 2011;62:4749–4761. doi: 10.1093/jxb/err196. [DOI] [PubMed] [Google Scholar]
- Veal EA, Day AM, Morgan BA. Hydrogen peroxide sensing and signaling. Molecular Cell. 2007;26:1–14. doi: 10.1016/j.molcel.2007.03.016. [DOI] [PubMed] [Google Scholar]
- Wagner E, Luche S, Penna L, Chevallet L, Van Dorsselaer A, Leize-Wagner E, Rabilloud T. A method for detection of overoxidation of cysteines: peroxiredoxins are oxidized in vivo at the active-site cysteine during oxidative stress. Biochemical Journal. 2002;366:777–785. doi: 10.1042/BJ20020525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B, Dickman M. Plant programmed cell death: can’t live with it; can’t live without it. Molecular Plant Pathology. 2008;9:531–544. doi: 10.1111/j.1364-3703.2008.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik C. Proteasomes in apoptosis: villains or guardians? Cellular and Molecular Life Science. 1999;56:908–917. doi: 10.1007/s000180050483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Jia J, Lv J, Liang X, Han D, Huang L, Kang Z. Characterization of the expression profile of a wheat aci-reductone-dioxygenase-like gene in response to stripe rust pathogen infection and abiotic stresses. Plant Physiology and Biochemistry. 2010;48:461–468. doi: 10.1016/j.plaphy.2010.03.002. [DOI] [PubMed] [Google Scholar]
- Ye C, Dickman MB, Whitham SA, Payton M, Verchot J. The unfolded protein response is triggered by a plant viral movement protein. Plant Physiology. 2011;156:741–755. doi: 10.1104/pp.111.174110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CS, Chen YC, Lu CH, Hwang JK. Predicting subcellular localization. Proteins: Structure, Function and Bioinformatics. 2004;64:643–651. [Google Scholar]
- Zhao C, Zhang W, Tian X, Fang C, Lu H, Yuan Z, Yang P, Wen Y. Proteomic analysis of cell lines expressing small hepatitis b surface antigen revealed decreased glucose-regulated protein 78kDa expression in association with higher susceptibility to apoptosis. Journal of Medical Virology. 2010;82:14–22. doi: 10.1002/jmv.21654. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Kok KH, Chun AC, Wong CM, Wu H, Lin MC, Fung PC, Kung H, Jin DY. Mouse peroxiredoxin V is a thioredoxin peroxidase that inhibits p53-induced apoptosis. Biochemical and Biophysical Research Communications. 2000;268:921–927. doi: 10.1006/bbrc.2000.2231. [DOI] [PubMed] [Google Scholar]