Abstract
The relationship between asparagine metabolism and protein concentration was investigated in soybean seed. Phenotyping of a population of recombinant inbred lines adapted to Illinois confirmed a positive correlation between free asparagine levels in developing seeds and protein concentration at maturity. Analysis of a second population of recombinant inbred lines adapted to Ontario associated the elevated free asparagine trait with two of four quantitative trait loci determining population variation for protein concentration, including a major one on chromosome 20 (linkage group I) which has been reported in multiple populations. In the seed coat, levels of asparagine synthetase were high at 50 mg and progressively declined until 150 mg seed weight, suggesting that nitrogenous assimilates are pre-conditioned at early developmental stages to enable a high concentration of asparagine in the embryo. The levels of asparaginase B1 showed an opposite pattern, being low at 50 mg and progressively increased until 150 mg, coinciding with an active phase of storage reserve accumulation. In a pair of genetically related cultivars, ∼2-fold higher levels of asparaginase B1 protein and activity in seed coat, were associated with high protein concentration, reflecting enhanced flux of nitrogen. Transcript expression analyses attributed this difference to a specific asparaginase gene, ASPGB1a. These results contribute to our understanding of the processes determining protein concentration in soybean seed.
Keywords: Asparaginase, asparagine, asparagine synthetase, quantitative trait locus, seed protein concentration, soybean
Introduction
In soybean [Glycine max (L.) Merr.], seed protein concentration is an important trait for end use and therefore relevant to crop value. In general, there are well-documented negative relationships between the concentration of protein and oil and between protein concentration and yield (Brim and Burton, 1979), although the latter is not observed in all breeding populations (Cober and Voldeng, 2000). Several studies have mapped quantitative trait loci (QTLs) determining protein concentration. A major QTL has been mapped to chromosome 20 [linkage group (LG) I] from various sources including wild soybean, and has been detected in multiple different populations (Diers et al., 1992; Brummer et al., 1997; Sebolt et al., 2000; Chung et al., 2003; Tajuddin et al., 2003; Nichols et al., 2006; Zhao-ming et al., 2011). Introgression of this QTL may be associated with pleiotropic effects, including increased stature, reduced yield and oil concentration, reduced seed weight, and earlier maturity. Recently, this QTL has been mapped to an 8.4 Mbp region, and candidate genes have been identified within this interval through transcript profiling in a pair of near-isogenic lines (Bolon et al., 2010). High protein concentration is associated with the accumulation of specific subunits of the 11S globulin glycinin (Krishnan et al., 2007; Krishnan and Nelson, 2011).
Understanding the biological mechanisms governing the accumulation of protein and oil is a pre-requisite to developing improved, high-yielding soybean varieties for future generations (Ainsworth et al., 2012). A major physiological question has been whether seed protein concentration is controlled by the maternal plant, through the provision and delivery of nitrogenous assimilates, or by the developing embryo, through its intrinsic capacity for the uptake of nitrogenous assimilates and synthesis of storage proteins. The developing embryo receives carbon assimilates as sucrose, and nitrogenous assimilates mainly as glutamine and asparagine. Metabolism and interconversion of the amides into other amino acids is initiated in the seed coat, a maternal tissue, before release into the apoplast (Rainbird et al., 1984). Uptake of assimilates by the embryo is mediated by a process of active transport (Tegeder and Rentsch, 2010). Asparagine is dominant in the developing cotyledon, representing up to 50% of total free amino acids (Hernández-Sebastià et al., 2005). Ammonia derived from asparagine breakdown by asparaginase (ASPG) is reassimilated through the glutamine synthetase/glutamate synthase cycle (Gomes and Sodek, 1984; Haga and Sodek, 1987). Assimilation of nitrogen and interconversion of amino acids require organic acids mostly derived from sucrose as carbon skeletons. In addition to sucrose, re-fixation of CO2 by ribulose bisphosphate carboxylase can make a significant contribution to the provision of organic acids via phosphoglycerate and phosphoenolpyruvate (Ruuska et al., 2004; Schwender et al., 2004). α-Ketoglutarate derived from glutamate represents a major point of entry into the tricarboxylic acid cycle which can generate malate and oxaloacetate for the biosynthesis of aspartate-derived amino acids, and malic enzyme can synthesize part of the pyruvate required for the biosynthesis of alanine, valine, leucine, and isoleucine (Schwender, 2008; Allen et al., 2009).
The results of metabolic engineering experiments emphasize the importance of several processes in the control of seed protein accumulation. The first one is the biosynthesis of nitrogenous assimilates in source leaves. Constitutive overexpression of asparagine synthetase (AS) 1 (ASN1) in Arabidopsis thaliana led to higher levels of free asparagine in developing siliques and increased seed protein concentration (Lam et al., 2003). According to results from similar experiments in Brassica napus, seed protein concentration may be increased only under nitrogen-sufficient conditions (Seiffert et al., 2004). In these studies, the use of a constitutive promoter did not allow the effects in source and sink tissues to be distinguished. In soybean, high AS1 transcript levels in source leaves are positively correlated with seed protein concentration (Wan et al., 2006), and, in roots, are associated with an elevated ratio of asparagine versus aspartate in xylem sap transported to the shoot (Antunes et al., 2008). A second mechanism exerting control is phloem loading of transported amino acids (Zhang et al., 2010). Inactivation of the phloem-localized amino acid transporter AAP2 in A. thaliana led to increased seed yield and oil concentration, consistent with the above noted negative relationship with protein concentration. Results obtained with the aap2 mutant were consistent with the observation of an increased rate of nitrogen translocation to the phloem in rape cultivars with high protein concentration (Lohaus and Moellers, 2000). Thirdly, uptake of amino acids by the embryo mediated by amino acid transporters has been demonstrated as a key step controlling seed protein accumulation (Rolletschek et al., 2005; Gotz et al., 2007; Weigelt et al., 2008; Sanders et al., 2009; Tan et al., 2010). Lastly, enhanced provision of organic acids for nitrogen assimilation and interconversion of amino acids may stimulate protein accumulation. This was observed after seed-specific transgenic expression of a bacterial phosphoenolpyruvate carboxylase (Rolletschek et al., 2004; Radchuk et al., 2007). Transgenic expression of a barley sucrose transporter in endosperm led to increased sucrose uptake, and enhanced nitrogen and prolamine concentration in wheat grain (Weichert et al., 2010).
A previous study identified an association between high levels of free asparagine in developing embryo and protein concentration at maturity in soybean (Hernández-Sebastià et al., 2005). In a pair of genetically related lines, higher levels of free asparagine and alanine were consistently detected in the high protein genotype during both the light and dark period. In the dark, the low protein line overaccumulated glutamate-derived amino acids, whereas the high protein line maintained higher levels of the pyruvate- and phosphoenolpyruvate-derived amino acids, indicating a more efficient process of nitrogen assimilation and amino acid interconversion. Similar observations have been made in kernels of maize lines differing in nitrogen use efficiency in relation to yield (Cañas et al., 2011). The association between high levels of free asparagine and protein concentration was reported in barley and maize grain (Dembinski and Bany, 1991; Dembinski et al., 1991; Lohaus et al., 1998). Higher levels of asparagine were present in leaf, phloem, and xylem of a high protein maize strain (Lohaus et al., 1998). Positive correlations between free asparagine, total free amino acids, and grain protein concentration were observed in mature kernels of transgenic maize segregating for reduced zein levels (Huang et al., 2006).
The results cited above suggested that free asparagine in the soybean embryo at mid-maturation may represent a physiological marker, and possibly a metabolite signal regulating protein concentration. Enhanced nitrogen flux to storage protein does not necessarily imply a higher steady-state concentration of free asparagine and other free amino acids. Therefore, asparagine levels are likely to be tightly regulated in the developing embryo. In the present study, experiments were conducted to investigate further the relationship between asparagine metabolism and protein concentration in soybean seed. Two populations of recombinant inbred lines (RILs) segregating for protein concentration, adapted to Illinois (IL, USA) and Ontario (ON, Canada) were analysed. The first population was used to confirm and evaluate the relationships between protein and free asparagine, carbon, and mineral assimilates. In the second population, RILs were characterized to investigate associations between the free asparagine trait and individual QTLs determining the population variation for protein concentration. Expression of the asparagine metabolic genes and enzymes, AS and ASPG, was compared in seed coat and embryo between genetically related cultivars contrasted in protein concentration, to better understand how steady-state levels of asparagine are regulated in the developing embryo.
Materials and methods
Plant materials
The RIL population of soybean adapted to IL was generated through a cross between the oilseed type cultivar Williams 82 (Bernard and Cremeens, 1988) and the high protein breeding line LG00-13365. LG00-13365 is an F8 line selected for high protein from the cross of Williams 82×PI 437088A. PI 437088A was introduced into the USA from Russia in 1980 (http://www.ars-grin.gov/npgs/acc/acc_queries.html/; accessed 16 November 2011). This population was evaluated in 2004 and 2005 for protein, oil, and phytate concentration in mature seed, and the concentration of free amino acids, simple sugars, including glucose, fructose, sucrose, and maltose, and inorganic and organic anions, chloride, phosphate, sulphate, malate, and oxaloacetate in developing seeds as described below. The RIL population adapted to ON, named X4050, was developed from a cross between X3145-B-B-3-15 (abbreviated X3145), a high protein breeding line, and AC Brant, an early maturing oilseed type cultivar (Voldeng et al., 1996a). The F2 population was grown in 1995, and single pods were picked to initiate single seed descent, which resulted in the development of 201 F4:5 lines. The RIL population was phenotyped in the field from 1998 to 2000 at eight station-years in two replicate trials. Near-infrared analysis of seed from each plot was carried out to determine seed protein concentration. The first 96 RILs, a random subset of the whole population, were genotyped with ∼130 simple sequence repeat (SSR or microsatellite) markers using standard techniques for PCR, acrylamide gel electrophoresis, silver staining of gels, and scoring of allelic bands as previously described (Molnar et al., 2003). SSR markers developed by Cregan et al. (1999) were selected from the USDA consensus map to achieve broad genome coverage. A recombination map was produced using the computer program MapMaker (Lander et al., 1987). There was strong co-linearity of markers with the consensus map. QTL analysis was performed using the computer program MQTL (Tinker and Mather, 1995) using the mean of 3 years of field data for protein concentration. Four QTLs were detected (SJM and ERC, unpublished). A large QTL at SSR marker Satt496 on chromosome 20 (LG I) explained 56% of the population variation in protein concentration. Three smaller QTLs at Satt575 (chromosome 15, LG E), Satt077 (chromosome 1, LG D1a), and Satt520 (chromosome 6, LG C2) explained 18, 15, and 13% of the population variation, respectively. The total variance explained by these four QTLs was 68%.
Lines were selected as follows to analyse the relationship between individual QTLs and free asparagine levels in developing seeds. Given that at each of the four protein concentration QTLs, a RIL can have either the maternal (A) or paternal (B) allele, 16 different genotypes are possible. However, to study the effect of a single A allele individually at each of the four loci, a choice was made to focus on the four genotypes composed of one A plus three B alleles in comparison with the all B parent, and similarly to study the effect of a single B allele by focusing on the four genotypes composed of one B plus three A alleles, in comparison with the all A parent. As a test of reproducibility, two additional RILs were included such that for one of the genotypes, three independently derived RILs were studied. In total, 10 RILs and the two parents were included. Specific RILs matching the desired genotypes were selected for entrance into this study based on their genotype at the SSR locus most closely linked to each of the four protein concentration QTLs. If several RILs had a suitable genotype, the RIL with protein concentration (mean of 8 station-years) closest to the mean protein concentration for that genotypic class was chosen to represent the class. These lines were grown in the field during summer 2008 in London, ON.
Maple Arrow, AC Hercule (93.75% Maple Arrow genetic background) (Voldeng et al., 1997), and AC Proteus (87.5% Maple Arrow background) (Voldeng et al., 1996b) are three genetically related cultivars adapted to ON which vary in protein concentration. Their protein concentration was measured in the field as the average of two successive seasons, 2004 and 2005, in Ottawa, ON. They were grown in the field during summer 2008, in London, ON to quantify free amino acids in developing seeds. Maple Arrow and AC Proteus were also grown in growth cabinets (Conviron E8H, Controlled Environments, Winnipeg, Manitoba, Canada). Seeds were sown in small pots (8×12 cm) containing Pro-Mix PGX (Premier Horticulture, Rivière-du-Loup, Québec, Canada). Seedlings were then transplanted to larger pots (17×20 cm) containing Pro-Mix BX (Premier Horticulture). Plants were given 16 h light (300–400 μmol photons m−2 s−1) and 8 h dark, with a temperature cycling between 18 °C and 24 °C (Supplementary Table S1 available at JXB online). When plants started their reproductive cycle, short day conditions were created by reducing the photoperiod to 12 h, by switching off lights at 18:00 h (Supplementary Table S1) in order to enhance flowering (Raper et al., 1984). Plants were fertilized with 3 g l−1 20:20:20 [Plant-Prod, Laval, Quebec (QE), Canada] once weekly.
Biochemical analyses from developing seeds
For soybeans grown in IL, seeds were harvested in the field and frozen in liquid nitrogen. Seeds, powdered in liquid nitrogen, were extracted twice with 80% ethanol and once with 50% ethanol at room temperature. The supernatants were combined, and dried under vacuum prior to being resuspended in water. For sugar analysis (glucose, fructose, and sucrose), reconstituted supernatants were cleared by centrifugation at 10 000 g for 20 min, and pre-treated with a mixed-bed ion exchanger (100 mg resin/100 ml extract; Serdolit MB-1, Boehringer, Ingelheim, Germany) in order to remove charged molecules such as amino acids. Sugars were analysed by isocratic anion exchange-high-performance liquid chromatography (HPLC) using a CarboPac PA1 guard column (4×50 mm) and separation column (4×250 mm), and pulsed amperometric detection (4500i; Dionex, Sunnyvale, CA, USA). Inorganic (chloride, phosphate, and sulphate) and organic anions (malate and oxalate) were separated by anion chromatography with continuously suppressed conductivity detection (Biotronic, Maintal, Germany). Free amino acids were measured in the same extracts used for sugar analysis (but without ion exchange pre-treatment), following deproteinization, with an amino acid analyser Biochrom 20 Plus (Biochrom Ltd, Cambridge, UK). In all procedures, standards were either subjected to the same procedures as samples or were added as internal standards, where possible. Recovery was usually ∼95%. For materials grown in ON, free amino acids were analysed by HPLC after derivatization with phenylisothiocyanate as previously described (Taylor et al., 2008). Approximately 1.5 g of seed tissue was pooled from different plants and ground in liquid nitrogen, and 100 mg was used to extract and quantify free amino acids.
Protein extraction and immunoblot analysis
Frozen tissue was ground in liquid nitrogen and soluble protein extracted by homogenizing with 300–400 μl of 50 mM TRIS-HCl pH 8.0, 50 mM KCl, 1 mM CaCl2, 10% (v/v) glycerol, 1 mM phenylmethylsulphonyl fluoride (PMSF), 2 mM dithiothreitol (DTT), and Complete Mini EDTA-free protease inhibitor cocktail tablet (Roche Applied Science, Laval, QE, Canada) per 100 mg of tissue. Samples were centrifuged at 17 530 g for 20 min at 4 °C. Supernatant was collected into a fresh tube and used for protein quantification with the Bio-Rad Protein Assay solution (Mississauga, ON, Canada) and bovine serum albumin (BSA) as standard. The expression of AS and ASPG protein was analysed by western blot using rabbit polyclonal ant-peptide antibodies. An antibody was raised against the following peptide in the α-subunit of mature ASPGB1a and -b: NH2-ASIMDGPKRRCGAVSC-COOH, by Genemed Synthesis (San Antonio, TX, USA). The antibody was affinity purified against the peptide antigen. The peptide used for affinity purification was synthesized by Bio-Synthesis (Lewisville, TX, USA). The peptide was coupled to Affi-Gel 10 (Bio-Rad Laboratories), serum was purified, and the antibody was eluted with 3 M sodium isothiocyanate, and dialysed into phosphate-buffered saline (PBS)/azide according to Sun and Arlinghaus (2004). An anti-AS antibody was raised against the following peptide: NH2-CEHPYLPKHILYRQKE-COOH, which was coupled to keyhole limpet haemocyanin as carrier using the N-terminal cysteine residue. The antibodies were produced and affinity purified by Bethyl Laboratories (Montgomery, TX, USA).
Protein extracts (15 μg) were separated by SDS–PAGE. Proteins were transferred to a polyvinylidene fluoride (PVDF) membrane (9×6 cm) using a semi-dry transfer apparatus (Bio-Rad Laboratories) at 15 V during 20 min for ASPG, and 50 min for AS. The membrane was washed in PBS–Tween (136 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, 1.76 mM KH2PO4, 0.1% (v/v) Tween-20, pH 7.4]. For ASPG, blocking was performed in 5% (w/v) non-fat dry milk in PBS–Tween; and for AS, in 8% (w/v) non-fat dry milk in PBS–Tween at room temperature for 1 h. The membrane was incubated with 1:1000 purified anti-ASPG antibody in PBS–Tween supplemented with 5% (w/v) non-fat dry milk, or in 1:10 000 purified anti-AS antibody in PBS–Tween supplemented with 8% (w/v) non-fat dry milk, for 2 h at room temperature. The membrane was washed and incubated with 1:10 000 horseradish peroxidase-conjugated goat anti-rabbit IgG (KPL, Gaithersburg, MD, USA) for 1 h at room temperature. The membrane was developed using enhanced chemiluminescence (ECL) plus western blotting reagents (GE Healthcare Life Sciences, Baie d’Urfé, QE, Canada). Signals were quantified by scanning densitometry using the Quantity One software (Bio-Rad Laboratories).
ASPG extraction and assay
Seeds of ∼150 mg were harvested, flash-frozen in liquid nitrogen, and stored at −80 °C. Ten seeds per replicate per cultivar were used in the experiment. Seeds were homogenized in 2 ml of buffer A [20 mM TRIS-HCl pH 7.5, 50 mM KCl, 1 mM CaCl2, 10% (v/v) glycerol, 1 mM DTT, and plant protease inhibitor (Sigma-Aldrich, Mississauga, ON, Canada)] and centrifuged for 30 min at 20 000 g at 4 °C. Ammonium sulphate was added to the supernatant to 25% saturation, and the sample was vortexed and centrifuged for 15 min at 20 000 g at 4°C. The pellet was resuspended in 300 μl of buffer B [20 mM sodium phosphate pH 8.0, 50 mM KCl, 1 mM CaCl2, 10% (v/v) glycerol, 1 mM DTT, and plant protease inhibitor] and desalted on a PD-10 column (GE Healthcare Life Sciences) pre-equilibrated with buffer B. To 50 μg of protein extract, 50 mM KCl and 50 μM asparagine were added and incubated at room temperature for 35 min. Asparagine was omitted from control assays. Free amino acids were separated from globular molecules using Microcon centrifugal filter devices (Millipore, Billerica, MA, USA), and quantified by HPLC after derivatization with o-phthalaldehyde and 3-mercaptopropionic acid as previously described (Ivanov et al., 2011).
RNA isolation and gene expression analysis
RNA was extracted as previously described (Bruneau et al., 2006). RNA was quantified by spectrophotometry on a NanoDrop 1000 (Thermo Scientific, Wilmington, DE, USA) and its quality evaluated from the A260/A280 ratio and agarose gel electrophoresis. Total RNA was treated with amplification-grade DNase I (Invitrogen, Burlington, ON, Canada). First-strand cDNA was synthesized from 1.5 μg of total RNA using qScript cDNA SuperMix (Quanta BioSciences, Gaithersburg, MD, USA) in a volume of 30 μl. Gene-specific primers were designed using Primer-BLAST (Rozen and Skaletsky, 2000). The transcript of the soybean ubiquitin-3 (SUBI-3) gene does not vary among seed tissues and was used as a reference to calculate relative mRNA levels (Yi et al., 2010). Primers were: for SUBI-3, qUbi-F, 5′-GTGTAATGTTGGATGTGTTCCC-3′ and qUbi-R, 5′-ACACAATTGAGTTCAACACAAACCG-3′; for AS1a, qAS1_11g27480F, 5′-AACTAGTAGTTGCCCTAGAACCAA-3′ and qAS1_11g27480R, 5′-GTGGATGGCACAAAGACGACT-3′; for AS1b, qAS1_11g27720F, 5′-TAGTAGTTGCCCTAGAACCAAA-3′ and qAS1_11g27720R, 5′-ATGGAACAACACTTAAGACAGCA-3′; for AS1c, qAS1_18g06840F, 5′-TGAGCCATAGCAAGGACTAGTA-3′ and qAS1_18g06840R, 5′-GGCACAACACTTAAGACAGCT-3′; for AS2b, qAS2_14g37440F, 5′-TGGCTCGGGATGCTATTGGGGT-3′ and qAS2_14g37440R, 5'-TGTACCATCTGCGGAACCCTCT-3′ (the latter may amplify AS2a since qAS2_14g37440F has a perfect match to this template and qAS2_14g37440R a single mismatch); for AS3a, qAS3_11g38130F, 5′-AGTGCCTGGAGGTCCTAGTG-3′ and qAS3_11g38130R, 5′-TGCATGAACACCAAGCGCTG-3′; for AS3b, qAS3_18g02060F, 5′-AGTGCCAGGAGGTCCTAGTG-3′ and qAS3_18g02060R, 5′-AGCATCATGAATACCAAGTGCGG-3′; for ASPGB1a, ASPG1_14g09510F, 5′-GCGCGGCAGCAACATCCACG-3′ and ASPG1_14g09510R, 5′-CTTCACCCGTGCACGATACGCC-3′; for ASPGB1b, ASPG1_17g35650F, 5′-CGGCAGCGACATCCACAGGG-3′ and ASPG1_17g35650R, 5′-CTCGCCCGTGCACGATACACC-3′; for ASPGB2a, ASPG2_04g04470F, 5′-GGGCTGATAGCGGTGTCGAATA-3′ and ASPG2_04g04470R, 5′-CAAGAGAGAGTACGTGACCACTGAC-3′; and for ASPGB2b, ASPG2_06g04590F, 5′-TGAGGTGGCAGCGGTGATGG-3′ and ASPG2_06g04590R, 5′-ACCAGAACTTGACACGGCGATC-3′. Primers were optimized for use at an annealing temperature of 61.5 °C. Quantitative PCR was performed using SsoFast EvaGreen Supermix with a CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories). Reactions contained primers at a concentration of 0.5 μM. They were carried out in Hard-Shell 96-well clear PCR plates (Bio-Rad Laboratories) in a final volume of 10 μl. In each plate, controls without template were performed in duplicate. The PCR program consisted of an initial step of 3 min at 95 °C followed by 40 cycles of 15 s at 95 °C and 30 s at 61.5 °C. Data were expressed as the cycle number necessary to reach a threshold fluorescence value (Cq). The reported values are the means of three biological replicates consisting of independent RNA extracts, with each biological replicate the average of three technical replicates. Data were normalized to the mean Cq of the reference gene SUBI-3, for which the variation between cultivars was ≤0.1 for seed coat, and 0.3 for embryo. The specificity of primer pairs was confirmed by melt curve analysis in comparison with controls without template and by agarose gel electrophoresis of PCR products. PCR efficiency was calculated from a standard curve of Cq versus the logarithm of starting template quantity. Each assay was optimized so that the efficiency ranged between 95% and 105%, with a coefficient of determination (R2) >0.98.
Statistical analysis
Unpaired, two-tailed t-test assuming homogeneity of the variances was performed with Excel software (Dytham, 1999). Analysis of variance (ANOVA) was performed using the SuperANOVA statistical program (Abacus Concepts, Berkeley, CA, USA) or SAS version 9.2 (Toronto, ON, Canada). Homogeneity of the variances was inspected by residual graphic analysis.
Accession numbers
Sequence data can be found under accession numbers (GenBank ID) D28123 for SUBI-3; (Phytozome ID) Glyma11g27480 for AS1a; (Phytozome ID) Glyma11g27720 for AS1b; (Phytozome ID) Glyma18g06840 for AS1c; (Phytozome ID) Glyma02g39320 for AS2a; (Phytozome ID) Glyma14g37440 for AS2b; (Phytozome ID) Glyma11g38130 for AS3a; (Phytozome ID) Glyma18g02060 for AS3b; (Phytozome ID) Glyma14g09510 for ASPGB1a; (Phytozome ID) Glyma17g35650 for ASPGB1b; (Phytozome ID) Glyma04g04470 for ASPGB2a; and (Phytozome ID) Glyma06g04590 for ASPGB2b.
Results
Relationship between seed protein concentration at maturity and the concentration of assimilates in developing seeds
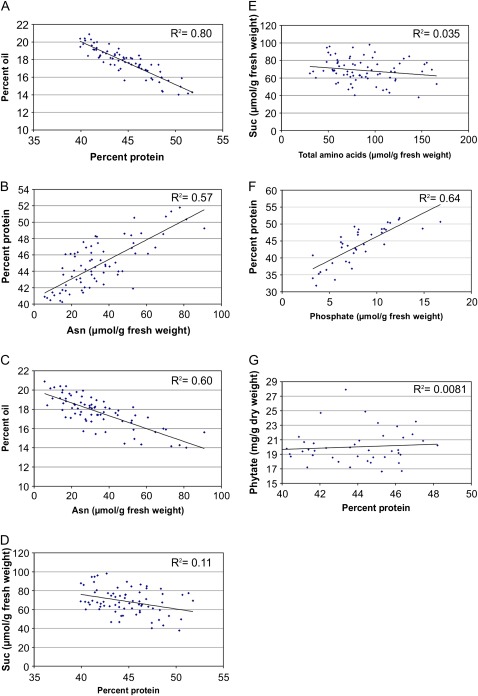
A population of 73 RILs derived from Williams 82 and LG00-13365 was grown over two successive seasons and phenotyped. As expected, a strong negative correlation between protein and oil concentration was observed (Pearson correlation coefficient, R2= –0.80) (Fig. 1A). The free asparagine concentration at mid-maturation was positively correlated with protein concentration (R2=0.57), consistent with previous observations, and negatively correlated with oil concentration (R2= –0.60) (Fig. 1B, C). Similar correlations were observed between total free amino acids and protein and oil concentration, with R2 values of 0.53 and –0.58, respectively. Levels of the nitrogen-rich amino acid, arginine, were also negatively correlated with oil concentration (R2= –0.57). The values of correlation coefficients were lower for the relationships between other central intermediates of amino acid metabolism, aspartate, glutamate, and glutamine, and protein or oil concentration. The relationships observed with free asparagine and total free amino acids did not extend to carbon assimilates. No significant relationships were observed between sucrose concentration and total free amino acids in developing seed, or protein concentration at maturity (Fig. 1D, E). Among organic and inorganic anions, the levels of free phosphate in developing seeds were positively correlated with protein concentration (Fig. 1F). No similar correlation was observed between protein concentration and the levels of soluble phytic acid in mature seed (Fig. 1G).
Fig. 1.
Relationships between biochemical parameters in seed in a population of 73 recombinant inbred lines adapted to Illinois, derived from Williams 82 and LG00-13365: (A) protein and oil concentration at maturity; (B) asparagine concentration in developing seed and protein concentration at maturity; (C) asparagine concentration in developing seed and oil concentration at maturity; (D) protein concentration at maturity and sucrose concentration in developing seed; (E) total free amino acids and sucrose concentration in developing seed; (F) phosphate concentration in developing seed and protein concentration in mature seed; (G) protein and phytate concentration in mature seed.
Association between the free asparagine trait and QTLs determining protein concentration
To investigate further the genetic factors underlying free asparagine levels in developing seed in relation to protein concentration, a second inbred population was selected for study. Population X4050 had been developed through a cross between AC Brant and X3145, and a molecular marker recombination map developed using a subset of 96 RILs. Four QTLs influencing protein concentration had been detected located on chromosomes 20, 15, 1, and 6 (LGs I, E, D1a, and C2, respectively) (SJM and ERC, unpublished). The chromosome 20 (LG I) QTL had a major effect, determining 56% of the population variation for the trait (Table 1). This result is consistent with the description of a major QTL determining protein concentration at the same location in several other populations (Zhao-ming et al., 2011). Protein concentration QTLs located on chromosomes 15 (LG E) near marker Satt575 and 1 (LG D1) near Satt077 have been reported by Diers et al. (1992) and Csanadi et al. (2001), respectively. To the authors’ knowledge, a protein concentration QTL has not been previously reported on chromosome 6 (LG C2) near Satt520. To evaluate the effect of each QTL on free asparagine levels in developing seeds, a subset of two parents and 10 RILs representing eight genotypes were selected to study the effect of the maternal A allele at each of the four QTLs in combination with the paternal B allele at the other three QTLs, and also the reciprocal four allelic combinations. Relative to the oilseed type parent AC Brant, which has the A allele at all four QTLs, lines with the B allele from X3145 at loci on LGs I and E had a significantly increased free asparagine concentration and total free amino acids. This was not the case for QTLs on LGs D1a and C2. Consistent with this, relative to X3145, which has the B allele at all four QTLs, lines with the A allele at the QTL on the LGs I and E had significantly lower free asparagine and total free amino acids. Line 46 with the A allele at the LG D1a QTL also had significantly lower free asparagine levels and total free amino acids relative to X3145, but line 47 with the A allele at the LG C2 QTL was not significantly different from X3145 for the two parameters. This analysis associated the protein concentration QTLs on LGs I and E with the high free asparagine trait in developing seed.
Table 1.
Free asparagine and total free amino acids in developing seeds at the 100 mg stage in two parental and 10 RILs from the X4050 population with differing genotypes at four QTLs for protein concentration
| Linkage group | I | E | D1a | C2 | Lines | Seed protein concentration (%) | Asparagine | Total free amino acids |
| 56%a | 18%a | 18%a | 18%a | |||||
| Marker | Satt496 | Satt575 | Satt077 | Satt520 | ||||
| Genotypic classes | ||||||||
| Parent | A | A | A | A | AC Brant | 40.9 | 36.9 | 81.0 |
| RILs (1B+3A) alleles | B | A | A | A | 1 | 46.0 | 46.2 | 101.4 |
| A | B | A | A | 103 | 45.9 | 53.2 | 101.1 | |
| A | A | B | A | 14 | 44.5 | 38.6 | 82.5 | |
| A | A | A | B | 59 | 44.9 | 30.0 | 63.9 | |
| RILs (1A+3B) alleles | B | B | B | A | 47 | 48.0 | 53.7 | 100.8 |
| B | B | A | B | 46 | 48.6 | 42.6 | 93.0 | |
| B | A | B | B | 31 | 45.9 | 40.0 | 82.7 | |
| A | B | B | B | 75 | 46.4 | 45.1 | 86.0 | |
| A | B | B | B | 60 | 45.1 | 31.5 | 78.6 | |
| A | B | B | B | 62 | 47.3 | 36.4 | 85.6 | |
| Parent | B | B | B | B | X3145 | 49.6 | 53.4 | 103.4 |
| LSD | 3.9 | 8.6 | ||||||
| ANOVA P-value | 0.0001 | 0.0001 | ||||||
Parents AC Brant (genotype AAAA) and X3145 (BBBB) have 40.9% and 49.6% protein concentration, respectively. QTLs are identified by linkage group and linked microsatellite marker. Free asparagine and total free amino acids are expressed in nmol mg−1 seed weight. n=4; LSD, Fisher’s protected least significant difference at P ≤ 0.05; ANOVA, analysis of variance.
Percentage contributed by each QTL to the population variation for seed protein concentration.
Expression of AS and ASPG in seed of genetically related cultivars differing in protein concentration
Suitable genetically related cultivars were selected in order to investigate the regulation of free asparagine levels in developing seeds. Maple Arrow, AC Hercule, and AC Proteus have a progressive increase in protein concentration (Table 2). Seeds were harvested at mid-maturation from field-grown plants, and free amino acids were extracted and quantified. This analysis confirmed the correlation between free asparagine levels in developing seeds and protein concentration. In this case, the levels of all free amino acids were highly correlated and progressively increased with protein concentration, similar to what was previously reported by Wan et al. (2006). The two cultivars exhibiting the highest contrast in protein concentration, Maple Arrow and AC Proteus, were selected for subsequent analyses.
Table 2.
Profiles of free amino acids in developing seeds at the 100 mg stage of genetically related soybean cultivars differing in protein concentration
| Cultivara | Aspartate | Glutamate | Asparagine | Serine | Gluatmine | Glycine | Histidine | Citrulline |
| Maple Arrow | 0.22±0.02 | 0.78±0.07 | 2.2±0.2 | 0.20±0.01 | 1.0±0.1 | 0.047±0.005 | 0.55±0.07 | 0.098±0.012 |
| AC Hercule | 0.40±0.16 | 1.54±0.58 | 4.9±1.8 | 0.46±0.21 | 3.7±1.5 | 0.102±0.045 | 1.10±0.56 | 0.253±0.112 |
| AC Proteus | 1.07±0.24 | 4.03±1.04 | 15.8±3.2 | 1.19±0.21 | 9.2±1.3 | 0.276±0.057 | 3.26±0.64 | 0.772±0.177 |
| LSD | 0.29 | 1.24 | 2.7 | 0.29 | 2.0 | 0.087 | 0.86 | 0.215 |
| ANOVA P-value | 0.002 | 0.005 | 0.001 | 0.0001 | 0.0001 | 0.0003 | 0.0001 | 0.0001 |
| Cultivar | Arginine | GABA | Threonine | Alanine | Proline | ANBA | Tyrosine | Valine |
| Maple Arrow | 0.36±0.04 | 0.16±0.04 | 0.055±0.004 | 0.42±0.03 | 0.034±0.002 | 0.003±0.005 | 0.030±0.002 | 0.069±0.005 |
| AC Hercule | 0.95±0.37 | 0.31±0.15 | 0.111±0.050 | 0.91±0.46 | 0.076±0.036 | 0.024±0.018 | 0.091±0.041 | 0.166±0.072 |
| AC Proteus | 2.95±0.81 | 0.72±0.15 | 0.285±0.061 | 2.09±0.84 | 0.189±0.033 | 0.056±0.011 | 0.204±0.028 | 0.447±0.062 |
| LSD | 0.93 | 0.215 | 0.079 | 0.99 | 0.048 | 0.020 | 0.048 | 0.092 |
| ANOVA P-value | 0.0003 | 0.0005 | 0.0002 | 0.009 | 0.0001 | 0.0006 | 0.0001 | 0.0001 |
| Cultivar | Methionine | Isoleucine | Leucine | Phenylalanine | Ornithine | Lysine | Total free amino acids |
| Maple Arrow | 0.078±0.010 | 0.043±0.003 | 0.043±0.004 | 0.033±0.004 | 0.008±0.003 | 0.023±0.004 | 6.5±0.5 |
| AC Hercule | 0.211±0.126 | 0.107±0.044 | 0.132±0.051 | 0.074±0.034 | 0.013±0.007 | 0.047±0.025 | 15.7±6.5 |
| AC Proteus | 0.597±0.145 | 0.265±0.027 | 0.365±0.049 | 0.226±0.041 | 0.062±0.010 | 0.144±0.038 | 44.3±7.6 |
| LSD | 0.192 | 0.048 | 0.069 | 0.053 | 0.013 | 0.047 | 10.0 |
| ANOVA P-value | 0.0004 | 0.0001 | 0.0001 | 0.0001 | 0.0001 | 0.0005 | 0.0001 |
Values are expressed in nmol mg−1 seed weight. Mean ±SD; n=4.
Protein concentration was equal to 40.9% for Maple Arrow, 45.7% for AC Hercule, and 48.0% for AC Proteus.
ANBA, amino-N-butyric acid; GABA, γ-aminobutyric acid.
To understand how the levels of free asparagine are regulated in developing seed, expression of the asparagine metabolic enzymes AS and ASPG was evaluated at the protein and transcript level in seed coat and embryo. Three AS and two ASPG genes had been originally isolated from soybean (Hughes et al., 1997; Bruneau et al., 2006; Wan et al., 2006; Cho et al., 2007; Antunes et al., 2008). Soybean predominantly expresses K+-dependent ASPG genes which are part of the ASPGB subfamily, according to a nomenclature developed for A. thaliana (Ivanov et al., 2011). Each of the originally defined AS and ASPGB genes has two paralogues in the soybean genome, except for AS1 which has three (http://www.phytozome.org) (Schmutz et al., 2010). They are designated here with the suffix -a, -b, or -c.
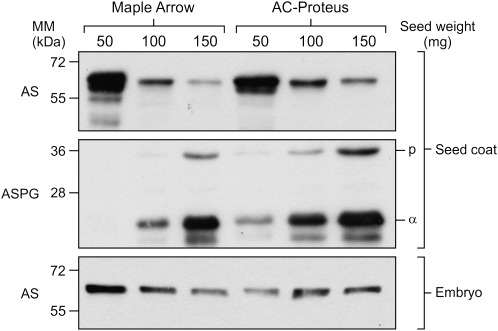
The levels of AS and ASPG were evaluated by western blot in seed coat and embryo extracts at three seed development stages (Fig. 2). Anti-AS antibodies detected a protein band migrating at an apparent molecular mass of 68.5 kDa, close to the predicted molecular mass of 65.3 kDa for AS1 (Hughes et al., 1997). The anti-ASPGB1 antibody detected a major protein band of 24.4 kDa, corresponding to the α-subunit, having a predicted molecular mass of 20.8 kDa, and a minor protein band of 37.7 kDa, corresponding to the uncleaved polypeptide precursor, with a predicted molecular mass of 34.5 kDa (Cho et al., 2007). AS and ASPGB1 isoforms were developmentally regulated in seed coat. AS levels were highest at 50 mg and progressively declined until 150 mg seed weight. The opposite was observed for ASPGB1. Levels were low or undetectable at 50 mg and progressively increased until 150 mg. In the developing embryo, the levels of AS were relatively stable, while ASPGB1 could not be detected. The most noticeable difference between cultivars was a higher level of ASPGB1 in the seed coat of AC Proteus as compared with Maple Arrow at all three developmental stages. This difference was equal to 2.4-fold at 100 mg seed weight, as determined by scanning densitometry. To validate this observation, ASPG activity was quantified in seed coat extracts. The specific activity measured in AC Proteus was equal to 1.55±0.13 pkatal mg−1 as compared with 0.91±0.03 pkatal mg−1 in Maple Arrow, a 1.7-fold difference (average ±SD; n=3 independent extracts; t-test P ≤ 0.001).
Fig. 2.
Western blots of asparagine synthetase (AS) and asparaginase (ASPG) in seed coat and embryo of developing seeds from Maple Arrow (low protein) and AC Proteus (high protein). The molecular mass (MM) of markers is indicated on the left; p, polypeptide precursor; α, α-subunit.
To investigate further the regulation of AS and ASPG, transcripts were quantified in seed coat and cotyledon by reverse transcription-PCR (RT-PCR) relative to the levels of the reference gene ubiquitin. Since a similar amount of cDNA template was used in assays, the average Cq value provides information about absolute transcript levels in each tissue. In seed coat, AS1c was predominantly expressed among AS genes (Table 3). Transcripts of AS1a and -c were more abundant in AC Proteus than in Maple Arrow at 100 mg, by 1.9- and 1.7-fold [cultivar (C), P ≤ 0.04 and 0.02], respectively, in parallel with the 2.4-fold higher levels of AS detected by western blot at 150 mg (Fig. 2). Among ASPG genes, ASPGB1a was predominantly expressed. Its transcript levels were higher in AC Proteus than in Maple Arrow at 100 mg seed weight, by 2.2-fold [C, P ≤ 0.0001; C×stage (S), P ≤ 0.03]. These results strongly suggest that the differences observed in ASPG protein and activity levels are due to the differential expression of this gene.
Table 3.
Relative transcript expression of AS and ASPG genes in seed coat of Maple Arrow (low protein) and AC Proteus (high protein) determined by quantitative reverse transcription-PCR
| Cultivar | Stage | AS1a (27) | AS1b (29) | AS1c (24) | AS2b (26) | AS3a (26) | AS3b (26) | ASPGB1a (22) | ASPGB1b (25) | ASPGB2a (28) | ASPGB2b (26) |
| Maple Arrow | 50 mg | 0.41 | 0.43 | 0.91 | 1.05 | 1.07 | 1.13 | 0.75 | 0.70 | 0.35 | 0.44 |
| 100 mg | 0.27 | 0.39 | 0.39 | 0.30 | 0.52 | 0.81 | 0.54 | 0.92 | 0.29 | 0.34 | |
| AC Proteus | 50 mg | 0.44 | 0.35 | 0.98 | 0.97 | 0.99 | 1.05 | 1.15 | 1.22 | 0.24 | 0.36 |
| 100 mg | 0.51 | 0.47 | 0.67 | 0.58 | 0.75 | 0.91 | 1.20 | 1.07 | 0.76 | 0.79 | |
| Source of variation | df | ANOVA P-values | |||||||||
| Cultivar (C) | 1 | 0.04 | NS | 0.02 | NS | NS | NS | 0.0001 | 0.009 | NS | NS |
| Stage (S) | 1 | NS | NS | 0.0001 | 0.0001 | 0.0002 | 0.008 | NS | NS | NS | NS |
| C×S | 1 | NS | NS | NS | 0.05 | 0.03 | NS | 0.03 | NS | 0.06 | 0.06 |
| Error | 8 | ||||||||||
Data were normalized to the mean Cq of the reference gene, ubiquitin. Values are the means of three biological replicates, with each biological replicate the average of three technical replicates. Statistically significant ANOVA P-values are shown in bold. Mean Cq is indicated in parentheses. The same volume of template cDNA was used in all assays. The mean Cq value for ubiquitin was equal to 19. df, degrees of freedom; NS, non-significant (P > 0.05).
In cotyledon, AS3a and -b were predominantly expressed among AS genes, and ASPGB2a among ASPG genes (Table 4). Transcript levels of ASPGB2a were higher in Maple Arrow than in AC Proteus, by 2.1-fold at 50 mg (C, P ≤ 0.0005). A similar observation was made for ASPGB2b (C, P ≤ 0.0001). Transcript levels of ASPGB1a were higher in AC Proteus than in Maple Arrow by 2.3- to 3.8-fold at 50 mg and 100 mg, respectively (C, P ≤ 0.0005). The lower transcript levels of this gene in cotyledon as compared with seed coat, as determined by Cq values, are consistent with the lack of detection of ASPGB1 isoforms by western blot in the former tissue.
Table 4.
Relative transcript expression of AS and ASPG genes in cotyledon of Maple Arrow (low protein) and AC Proteus (high protein) determined by quantitative reverse transcription-PCR
| Cultivar | Stage | AS1a (30) | AS1b (33) | AS1c (31) | AS2b (32) | AS3a (24) | AS3b (24) | ASPGB1a (25) | ASPGB1b (29) | ASPGB2a (22) | ASPGB2b (24) |
| Maple Arrow | 50 mg | 1.08 | 0.77 | 1.04 | 0.74 | 1.19 | 1.07 | 0.48 | 1.29 | 1.21 | 1.30 |
| 100 mg | 0.94 | 0.71 | 0.83 | 0.55 | 0.96 | 1.00 | 0.26 | 1.05 | 0.95 | 1.16 | |
| AC Proteus | 50 mg | 0.90 | 0.71 | 0.76 | 0.75 | 0.86 | 0.95 | 1.10 | 0.81 | 0.58 | 0.71 |
| 100 mg | 0.76 | 0.52 | 0.62 | 0.50 | 0.72 | 0.89 | 0.98 | 0.86 | 0.64 | 0.68 | |
| Source of variation | df | ANOVA P-values | |||||||||
| Cultivar (C) | 1 | 0.05 | NS | 0.04 | NS | 0.003 | 0.05 | 0.0001 | 0.0004 | 0.0005 | 0.0001 |
| Stage (S) | 1 | NS | NS | NS | NS | 0.03 | NS. | 0.006 | NS | NS | 0.02 |
| C×S | 1 | NS | NS | NS | NS | NS | NS | NS | 0.04 | NS | NS |
| Error | 8 | ||||||||||
Data were normalized to the mean Cq of the reference gene, ubiquitin. Values are the means of three biological replicates, with each biological replicate the average of three technical replicates. Statistically significant ANOVA P-values are shown in bold. Mean Cq is indicated in parentheses. The same volume of template cDNA was used in all assays. The mean Cq value for ubiquitin was equal to 20. df, degrees of freedom; NS, non-significant (P > 0.05).
Discussion
Understanding the mechanisms determining seed protein concentration is important for crop improvement. A generic negative relationship between protein concentration and yield in major crops has meant that yield increases have been associated with a decreased nutritional value (Triboi and Triboi-Blondel, 2009). The present study further explored a correlation previously established between free asparagine levels in developing embryo at mid-maturation and protein concentration at maturity in soybean seed (Hernández-Sebastià et al., 2005). The high free asparagine trait may represent a physiological marker associated with high seed protein concentration. Free asparagine may also act as a metabolite signal in the developing embryo and influence seed protein accumulation. Asparagine levels are probably tightly regulated through a control of its metabolism.
Phenotyping the RIL population adapted to IL confirmed the relationship between the free asparagine concentration at mid-maturation and seed protein concentration. This relationship extends to total free amino acids and may include other free amino acids beside asparagine, depending on the genotypes investigated. A positive correlation was observed between free phosphate levels in developing seed and protein concentration. This echoes similar findings in wheat grain where phosphorus is part of a group of mineral elements whose levels are positively correlated with protein nitrogen (Triboi and Triboi-Blondel, 2009). Intriguingly, protein concentration was not correlated with the concentration of soluble phytic acid, even though it constitutes the most abundant reserve of phosphate in mature seed (Lott et al., 2000).
Analysis of the X4050 RIL population adapted to ON conclusively associated two out of four QTLs determining population variation for protein concentration with the high free asparagine trait, including the major QTL on chromosome 20 (LG I). The results suggest that the contribution of the QTL on chromosome 6 (LG C2) to protein concentration is independent of an effect on free asparagine levels in developing seeds. Results were ambiguous for the QTL on chromosome 1 (LG D1a). When the gene underlying the major QTL on chromosome 20 is identified, it will be interesting to investigate its function relative to the high free asparagine trait.
Results on AS and ASPG provide insight into the high concentration of free asparagine present in the developing soybean embryo. High levels of AS were present in the seed coat early during seed development. ASPGB1 isoforms accumulated later, coinciding with the onset of storage reserve accumulation. In the embryo, AS is likely to contribute to the high levels of free asparagine, whereas ASPGB1 isoforms were undetectable, indicating that they were expressed at low levels or rapidly turned over. The high levels of AS detected in the seed coat suggest that at early developmental stages, the supply of nitrogenous assimilates is pre-conditioned via the biosynthesis of asparagine, prior to uptake by the developing embryo. This has been suggested in maize where a high ratio of asparagine to glutamine in the cob was proposed to condition kernel development (Seebauer et al., 2004).
The higher ASPGB1a transcript, enzyme, and activity levels in seed coat represent a major difference between cultivars associated with high protein concentration. This difference is likely to reflect enhanced flux of nitrogen to the protein pool in the high protein genotype. Despite the higher ASPG activity in the seed coat, the high protein cultivar maintained higher steady-state levels of free asparagine in the embryo. The lower transcript levels of ASPGB2 paralogues may partly explain the higher free asparagine levels. In summary, the results presented here highlighted several differences between high and low protein soybean genotypes related to the metabolism of asparagine in developing seed. In future, these data may help to interpret the function of genes determining protein concentration, a key trait for soybean genetic improvement.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Temperature cycle used for growth in growth cabinets.
Acknowledgments
This research was supported in part by the United Soybean Board. We are grateful to Rey Interior, at the Advanced Protein Technology Centre, Hospital for Sick Children, for amino acid analyses. We are indebted to Alex Molnar, at the Southern Crop Protection and Food Research Center, for preparation of figures. We thank Denis Maxwell, at the University of Western Ontario, for serving as SP’s co-supervisor during his MSc programme.
Glossary
Abbreviations
- AS
asparaginase synthetase
- ASPG
asparaginase
- LG
linkage group
- QTL
quantitative trait locus
- RIL
recombinant inbred line
- SSR
simple sequence repeat
References
- Ainsworth EA, Yendrek CR, Skoneczka JA, Long SP. Accelerating yield potential in soybean: potential targets for biotechnological improvement. Plant, Cell and Environment. 2012;35:38–52. doi: 10.1111/j.1365-3040.2011.02378.x. [DOI] [PubMed] [Google Scholar]
- Allen DK, Ohlrogge JB, Shachar-Hill Y. The role of light in soybean seed filling metabolism. The Plant Journal. 2009;58:220–234. doi: 10.1111/j.1365-313X.2008.03771.x. [DOI] [PubMed] [Google Scholar]
- Antunes F, Aguilar M, Pineda M, Sodek L. Nitrogen stress and the expression of asparagine synthetase in roots and nodules of soybean (Glycine max) Physiologia Plantarum. 2008;133:736–743. doi: 10.1111/j.1399-3054.2008.01092.x. [DOI] [PubMed] [Google Scholar]
- Bernard RL, Cremeens CR. Registration of ‘Williams 82’ soybean. Crop Science. 1988;28:1027–1028. [Google Scholar]
- Bolon YT, Joseph B, Cannon SB, et al. Complementary genetic and genomic approaches help characterize the linkage group I seed protein QTL in soybean. BMC Plant Biology. 2010;10:41. doi: 10.1186/1471-2229-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brim CA, Burton JW. Recurrent selection in soybeans. II. Selection for increased percent protein in seeds. Crop Science. 1979;19:494–498. [Google Scholar]
- Brummer EC, Graef GL, Orf J, Wilcox JR, Shoemaker RC. Mapping QTL for seed protein and oil content in eight soybean populations. Crop Science. 1997;37:370–378. [Google Scholar]
- Bruneau L, Chapman R, Marsolais F. Co-occurrence of both L-asparaginase subtypes in Arabidopsis: At3g16150 encodes a K+-dependent L-asparaginase. Planta. 2006;224:668–679. doi: 10.1007/s00425-006-0245-9. [DOI] [PubMed] [Google Scholar]
- Cañas RA, Amiour N, Quilleré I, Hirel B. An integrated statistical analysis of the genetic variability of nitrogen metabolism in the ear of three maize inbred lines (Zea mays L.) Journal of Experimental Botany. 2011;62:2309–2318. doi: 10.1093/jxb/erq373. [DOI] [PubMed] [Google Scholar]
- Cho CW, Lee HJ, Chung E, et al. Molecular characterization of the soybean L-asparaginase gene induced by low temperature stress. Molecules and Cells. 2007;23:280–286. [PubMed] [Google Scholar]
- Chung J, Babka HL, Graef GL, Staswick PE, Lee DJ, Cregan PB, Shoemaker RC, Specht JE. The seed protein, oil, and yield QTL on soybean linkage group I. Crop Science. 2003;43:1053–1067. [Google Scholar]
- Cober ER, Voldeng HD. Developing high-protein, high-yield soybean populations and lines. Crop Science. 2000;40:39–42. [Google Scholar]
- Cregan PB, Jarvik T, Bush AL, et al. An integrated genetic linkage map of the soybean genome. Crop Science. 1999;39:1464–1490. [Google Scholar]
- Csanadi G, Vollmann J, Stift G, Lelley T. Seed quality QTLs identified in a molecular map of early maturing soybean. Theoretical and Applied Genetics. 2001;103:912–919. [Google Scholar]
- Dembinski E, Bany S. The amino acid pool of high and low protein rye inbred lines (Secale cereale L.) Journal of Plant Physiology. 1991;138:494–496. [Google Scholar]
- Dembinski E, Rafalski A, Wisniewska I. Effect of long-term selection for high and low protein content on the metabolism of amino acids and carbohydrates in maize kernel. Plant Physiology and Biochemistry. 1991;29:549–558. [Google Scholar]
- Diers BW, Keim P, Fehr WR, Shoemaker RC. RFLP analysis of soybean seed protein and oil content. Theoretical and Applied Genetics. 1992;83:608–612. doi: 10.1007/BF00226905. [DOI] [PubMed] [Google Scholar]
- Dytham C. Choosing and using statistics: a biologist’s guide. Oxford: Blackwell Science Ltd; 1999. [Google Scholar]
- Gomes MAF, Sodek L. Allantoinase and asparaginase activities in maturing fruits of nodulated and non-nodulated soybeans. Physiologia Plantarum. 1984;62:105–109. [Google Scholar]
- Gotz K-P, Staroske N, Radchuk R, Emery RJN, Wutzke K-D, Herzog H, Weber H. Uptake and allocation of carbon and nitrogen in Vicia narbonensis plants with increased seed sink strength achieved by seed-specific expression of an amino acid permease. Journal of Experimental Botany. 2007;58:3183–3195. doi: 10.1093/jxb/erm164. [DOI] [PubMed] [Google Scholar]
- Haga KI, Sodek L. Utilization of nitrogen sources by immature soybean cotyledons in culture. Annals of Botany. 1987;59:597–602. [Google Scholar]
- Hernández-Sebastià C, Marsolais F, Saravitz C, Israel D, Dewey RE, Huber SC. Free amino acid profiles suggest a possible role for asparagine in the control of storage-product accumulation in developing seeds of low- and high-protein soybean lines. Journal of Experimental Botany. 2005;56:1951–1963. doi: 10.1093/jxb/eri191. [DOI] [PubMed] [Google Scholar]
- Huang S, Frizzi A, Florida CA, Kruger DE, Luethy MH. High lysine and high tryptophan transgenic maize resulting from the reduction of both 19- and 22-kD alpha-zeins. Plant Molecular Biology. 2006;61:525–535. doi: 10.1007/s11103-006-0027-6. [DOI] [PubMed] [Google Scholar]
- Hughes CA, Beard HS, Matthews BF. Molecular cloning and expression of two cDNAs encoding asparagine synthetase in soybean. Plant Molecular Biology. 1997;33:301–311. doi: 10.1023/a:1005784202450. [DOI] [PubMed] [Google Scholar]
- Ivanov A, Kameka A, Pajak A, Bruneau L, Beyaert R, Hernandez-Sebastia C, Marsolais F. Arabidopsis mutants lacking asparaginases develop normally but exhibit enhanced root inhibition by exogenous asparagine. Amino Acids. 2011 doi: 10.1007/s00726-011-0973-4. (in press) [DOI] [PubMed] [Google Scholar]
- Krishnan HB, Natarajan SS, Mahmoud AA, Nelson RL. Identification of glycinin and β-conglycinin subunits that contribute to the increased protein content of high-protein soybean lines. Journal of Agricultural and Food Chemistry. 2007;55:1839–1845. doi: 10.1021/jf062497n. [DOI] [PubMed] [Google Scholar]
- Krishnan HB, Nelson RL. Proteomic analysis of high protein soybean (Glycine max) accessions demonstrates the contribution of novel glycinin subunits. Journal of Agricultural and Food Chemistry. 2011;59:2432–2439. doi: 10.1021/jf104330n. [DOI] [PubMed] [Google Scholar]
- Lam HM, Wong P, Chan HK, Yam KM, Chen L, Chow CM, Coruzzi GM. Overexpression of the ASN1 gene enhances nitrogen status in seeds of Arabidopsis. Plant Physiology. 2003;132:926–935. doi: 10.1104/pp.103.020123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newberg LA. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Lohaus G, Buker M, Hussmann M, Soave C, Heldt HW. Transport of amino acids with special emphasis on the synthesis and transport of asparagine in the Illinois Low Protein and Illinois High Protein strains of maize. Planta. 1998;205:181–188. [Google Scholar]
- Lohaus G, Moellers C. Phloem transport of amino acids in two Brassica napus L. genotypes and one B. carinata genotype in relation to their seed protein content. Planta. 2000;211:833–840. doi: 10.1007/s004250000349. [DOI] [PubMed] [Google Scholar]
- Lott JNA, Ockenden I, Raboy V, Batten GD. Phytic acid and phosphorus in crop seeds and fruits: a global estimate. Seed Science Research. 2000;10:11–33. [Google Scholar]
- Molnar SJ, Rai S, Charette M, Cober ER. Simple sequence repeat (SSR) markers linked to E1, E3, E4, and E7 maturity genes in soybean. Genome. 2003;46:1024–1036. doi: 10.1139/g03-079. [DOI] [PubMed] [Google Scholar]
- Nichols DM, Glover KD, Carlson SR, Specht JE, Diers BW. Fine mapping of a seed protein QTL on soybean linkage group I and its correlated effects on agronomic traits. Crop Science. 2006;46:834–839. [Google Scholar]
- Radchuk R, Radchuk V, Gotz K-P, Weichert H, Richter A, Emery RJN, Weschke W, Weber H. Ectopic expression of phosphoenolpyruvate carboxylase in Vicia narbonensis seeds: effects of improved nutrient status on seed maturation and transcriptional regulatory networks. The Plant Journal. 2007;51:819–839. doi: 10.1111/j.1365-313X.2007.03196.x. [DOI] [PubMed] [Google Scholar]
- Rainbird RM, Thorne JH, Hardy RWF. Role of amides, amino acids, and ureides in the nutrition of developing soybean seeds. Plant Physiology. 1984;74:329–334. doi: 10.1104/pp.74.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raper CD, Jr, Patterson RP, List ML, Obendorf RL, Downs RJ. Photoperiod effects on growth rate of in vitro cultured soybean embryos. Botanical Gazette. 1984;145:157–162. doi: 10.1086/337441. [DOI] [PubMed] [Google Scholar]
- Rolletschek H, Borisjuk L, Radchuk R, Miranda M, Heim U, Wobus U, Weber H. Seed-specific expression of a bacterial phosphoenolpyruvate carboxylase in Vicia narbonensis increases protein content and improves carbon economy. Plant Biotechnology Journal. 2004;2:211–219. doi: 10.1111/j.1467-7652.2004.00064.x. [DOI] [PubMed] [Google Scholar]
- Rolletschek H, Hosein F, Miranda M, Heim U, Gotz KP, Schlereth A, Borisjuk L, Saalbach I, Wobus U, Weber H. Ectopic expression of an amino acid transporter (Vfaap1) in seeds of Vicia narbonensis and pea increases storage proteins. Plant Physiology. 2005;137:1236–1249. doi: 10.1104/pp.104.056523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods in Molecular Biology. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Ruuska SA, Schwender J, Ohlrogge JB. The capacity of green oilseeds to utilize photosynthesis to drive biosynthetic processes. Plant Physiology. 2004;136:2700–2709. doi: 10.1104/pp.104.047977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders A, Collier R, Trethewy A, Gould G, Sieker R, Tegeder M. AAP1 regulates import of amino acids into developing Arabidopsis embryos. The Plant Journal. 2009;59:540–552. doi: 10.1111/j.1365-313X.2009.03890.x. [DOI] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- Schwender J. Metabolic flux analysis as a tool in metabolic engineering of plants. Current Opinion in Biotechnology. 2008;19:131–137. doi: 10.1016/j.copbio.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Schwender J, Goffman F, Ohlrogge JB, Shachar-Hill Y. Rubisco without the Calvin cycle improves the carbon efficiency of developing green seeds. Nature. 2004;432:779–782. doi: 10.1038/nature03145. [DOI] [PubMed] [Google Scholar]
- Sebolt AM, Shoemaker RC, Diers BW. Analysis of a quantitative trait locus allele from wild soybean that increases seed protein concentration in soybean. Crop Science. 2000;40:1438–1444. [Google Scholar]
- Seebauer JR, Moose SP, Fabbri BJ, Crossland LD, Below FE. Amino acid metabolism in maize earshoots. Implications for assimilate preconditioning and nitrogen signaling. Plant Physiology. 2004;136:4326–4334. doi: 10.1104/pp.104.043778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiffert B, Zhou ZW, Wallbraun M, Lohaus G, Mollers C. Expression of a bacterial asparagine synthetase gene in oilseed rape (Brassica napus) and its effect on traits related to nitrogen efficiency. Physiologia Plantarum. 2004;121:656–665. [Google Scholar]
- Sun T, Arlinghaus RB. Preparation and application of polyclonal and monoclonal sequence-specific anti-phosphoamino acid antibodies. Current Protocols in Protein Science. 2004 doi: 10.1002/0471140864.ps1306s34. 13.16.11–13.16.27. [DOI] [PubMed] [Google Scholar]
- Tajuddin T, Watanabe S, Yamanaka N, Harada K. Analysis of quantitative trait loci for protein and lipid contents in soybean seeds using recombinant inbred lines. Breeding Science, 2003;53:133–140. [Google Scholar]
- Tan Q, Zhang L, Grant J, Cooper P, Tegeder M. Increased phloem transport of S-methylmethionine positively affects sulfur and nitrogen metabolism and seed development in pea plants. Plant Physiology. 2010;154:1886–1896. doi: 10.1104/pp.110.166389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M, Chapman R, Beyaert R, Hernández-Sebastià C, Marsolais F. Seed storage protein deficiency improves sulfur amino acid content in common bean (Phaseolus vulgaris L.): redirection of sulfur from gamma-glutamyl-S-methyl-cysteine. Journal of Agricultural and Food Chemistry. 2008;56:5647–5654. doi: 10.1021/jf800787y. [DOI] [PubMed] [Google Scholar]
- Tegeder M, Rentsch D. Uptake and partitioning of amino acids and peptides. Molecular Plant. 2010;3:997–1011. doi: 10.1093/mp/ssq047. [DOI] [PubMed] [Google Scholar]
- Tinker NA, Mather DE. MQTL: software for simplified composite interval mapping of QTL in multiple environments. Journal of Quantitative Trait Loci. 1995;1:1. [Google Scholar]
- Triboi E, Triboi-Blondel A-M. Productivity and seed composition. In: Krishnan H, editor. Modification of seed composition to promote health and nutrition. Madison, WI: American Society of Agronomy, Crop Science Society of America, Soil Science Society of America; 2009. pp. 1–38. [Google Scholar]
- Voldeng HD, Guillemette RJD, Leonard DA, Cober ER. AC Brant soybean. Canadian Journal of Plant Science. 1996a;76:149–150. [Google Scholar]
- Voldeng HD, Guillemette RJD, Leonard DA, Cober ER. AC Proteus soybean. Canadian Journal of Plant Science. 1996b;76:153–154. [Google Scholar]
- Voldeng HD, Guillemette RJD, Leonard DA, Cober ER. AC Hercule soybean. Canadian Journal of Plant Science. 1997;77:257–258. [Google Scholar]
- Wan TF, Shao GH, Shan XC, Zeng NY, Lam HM. Correlation between AS1 gene expression and seed protein contents in different soybean (Glycine max [L.] Merr.) cultivars. Plant Biology. 2006;8:271–275. doi: 10.1055/s-2006-923876. [DOI] [PubMed] [Google Scholar]
- Weichert N, Saalbach I, Weichert H, et al. Increasing sucrose uptake capacity of wheat grains stimulates storage protein synthesis. Plant Physiology. 2010;152:698–710. doi: 10.1104/pp.109.150854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt K, Kuster H, Radchuk R, Muller M, Weichert H, Fait A, Fernie AR, Saalbach I, Weber H. Increasing amino acid supply in pea embryos reveals specific interactions of N and C metabolism, and highlights the importance of mitochondrial metabolism. The Plant Journal. 2008;55:909–926. doi: 10.1111/j.1365-313X.2008.03560.x. [DOI] [PubMed] [Google Scholar]
- Yi J, Derynck MR, Chen L, Dhaubhadel S. Differential expression of CHS7 and CHS8 genes in soybean. Planta. 2010;231:741–753. doi: 10.1007/s00425-009-1079-z. [DOI] [PubMed] [Google Scholar]
- Zhang L, Tan Q, Lee R, Trethewy A, Lee YH, Tegeder M. Altered xylem–phloem transfer of amino acids affects metabolism and leads to increased seed yield and oil content in Arabidopsis. The Plant Cell. 2010;22:3603–3620. doi: 10.1105/tpc.110.073833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao-ming Q, Ya-nan S, Qiong W, Chun-yan L, Guo-hua H, Qing-shan C. A meta-analysis of seed protein concentration QTL in soybean. Canadian Journal of Plant Science. 2011;91:221–230. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.