Abstract
In higher plants, the Dof transcription factors that harbour a conserved plant-specific DNA-binding domain function in the regulation of diverse biological processes that are unique to plants. Although these factors are present in both higher and lower plants, they have not yet been characterized in lower plants. Here six genes encoding Dof transcription factors in the moss Physcomitrella patens are characterized and two of these genes, PpDof1 and PpDof2, are functionally analysed. The targeted disruption of PpDof1 caused delayed or reduced gametophore formation, accompanied by an effect on development of the caulonema from the chloronema. Furthermore, the ppdof1 disruptants were found to form smaller colonies with a reduced frequency of branching of protonemal filaments, depending on the nutrients in the media. Most of these phenotypes were not apparent in the ppdof2 disruptant, although the ppdof2 disruptants also formed smaller colonies on a particular medium. Transcriptional repressor activity of PpDof1 and PpDof2 and modified expression of a number of genes in the ppdof disruptant lines were also shown. These results thus suggest that the PpDof1 transcriptional repressor has a role in controlling nutrient-dependent filament growth.
Keywords: Dof transcription factor, filament growth, gene targeting, nutrient response, Physcomitrella patens
Introduction
Dof transcription factors are defined by the presence of a Dof DNA-binding domain that includes a C2C2-type zinc finger and is regarded as being plant specific due to its unique amino acid sequence. The Dof DNA-binding domain was initially identified in transcription factors from a monocot, maize (Yanagisawa and Izui, 1993; Yanagisawa, 1995), and then in a number of plant proteins from a variety of higher plants (reviewed in Yanagisawa, 2002, 2004). All known Dof transcription factors have a single copy of the Dof domain, which is located within the N-terminal regions of the respective proteins in general, but very divergent amino acid sequences outside of this domain. As the Dof DNA-binding domain is highly conserved among Dof transcription factors, these proteins bind the AAAG sequence motif similarly in a sequence-specific manner (Yanagisawa and Schmidt, 1999). Furthermore, some Dof domains have been shown to be involved not only in DNA recognition but also in specific protein–protein interactions, namely the interactions with nuclear high-mobility group (HMG) proteins (Yanagisawa, 1997; Krohn et al., 2002) and particular transcription factors including bZIP and MYB proteins (e.g. Zhang et al., 1995; Vicente-Carbajosa et al., 1997; Diaz et al., 2002; Washio, 2003). Hence, the Dof transcription factors show versatility and, regardless of their very similar DNA recognition properties, are thought to be capable of regulating particular gene promoters via specific protein–protein interactions (Singh, 1998; Rubio-Somoza et al., 2006). Indeed, some Dof transcription factors have been shown to act as a transcriptional activator or a repressor and to modulate the activity of particular gene promoters in plant cells (e.g. Mena et al., 1998; Yanagisawa and Sheen, 1998; Diaz et al., 2002; Martínez et al., 2005; Yamamoto et al., 2006).
A number of previous studies have already suggested that angiosperm Dof transcription factors are involved in the control of a variety of biological processes including tissue- or cell type-specific expression in leaves, endosperm, and stomata guard cells, stress responses, light responses or phytochrome signalling, seed maturation and germination, and plant hormone responses (reviewed in Yanagisawa, 2002). Furthermore, Dof transcription factors have also been suggested to be involved in the expression of genes associated with carbon fixation and nitrogen assimilation (Yanagisawa and Sheen, 1998; Yanagisawa, 2000; Rueda-López et al., 2008; Tanaka et al., 2009), photoperiodic flowering (Imaizumi et al., 2005; Iwamoto et al., 2009; Li et al., 2009), secondary metabolism (Skirycz et al., 2006, 2007), lipid metabolism in seeds (Wang et al., 2007), leaf axial patterning (Kim et al., 2010), vascular development (Konishi and Yanagisawa, 2007; Guo et al., 2009; Gardiner et al., 2010), cell cycle regulation (Skirycz et al., 2008), and the control of abscission (Wei et al., 2010). The suggested roles of Dof transcription factors are divergent, but most are tightly associated with plant-specific biological processes, some of which are common in all plant species and others which are unique to vascular or flowering plants.
The Dof transcription factors are present not only in higher plants (vascular plants) but also in lower plants. For instance, Arabidopsis and rice harbour 37 Dof transcription factor genes including one pseudogene (Yanagisawa, 2002) and 30 Dof transcription factor genes (Lijavetzky et al., 2003), respectively. Dof transcription factor genes have also been found in the genome of the moss Physcomitrella patens and the green alga Chlamydomonas reinhardtii. In the P. patens and C. reinhardtii genomes, 19 genes and a single Dof transcription factor gene have been found, respectively (Moreno-Risueno et al., 2007; Shigyo et al., 2007). Because no identifiable Dof transcription factor gene was found in the genomes of a red alga Cyanidioschyzon merolae and a diatom Thalassiosira pseudonana, the origin of these transcription factors has been suggested to pre-date the divergence of the green algae and the ancestors of terrestrial plants (Shigyo et al., 2007). Accordingly, the multiplication of Dof transcription factor genes appears to be linked with the emergence of diverse transcriptional regulation mechanisms during the course of plant evolution, and some Dof transcription factors might play conserved roles in both higher and lower plants. However, further elucidation of this hypothesis awaits the cloning of Dof transcription factor genes from lower plants.
The moss P. patens is used as a model plant as it shares many physiological processes with higher plants (Cove and Knight, 1993; Cove, 2005; Rensing et al., 2008), and gene targeting methods for this organism have been fully developed (Schaefer and Zrÿd, 1997; Reski, 1998). In the life cycle of P. patens, a spore develops into a protonema, which is a filamentous structure composed of two types of cells, chloronema and caulonema. In the protonemal colony, the filaments of the chloronemal cells, which are more photosynthetically active with well-developed chloroplasts, are initially formed. Filaments of caulonemal cells that contain fewer and smaller chloroplasts emerge later. The apical cells of chloronemal filaments extend at a rate of 2–5 μm h−1 and divide every 22–26 h, whereas the apical cells of the caulonemal filaments extend at a rate of 25–40 μm h−1 and divide every 6–8 h (Cove, 2005). The protonemal colonies spread efficiently via the rapid radial growth of caulonemal filaments. Although protonemal filaments generally proliferate through tip growth of their apical cells, they also generate side branches from the subapical cells. Because some side branches of the initial cells of the caulonemal filaments can differentiate into buds and some buds further develop into gametophores (i.e. leafy shoots with sexual organs), the differentiation of the chloronema into the caulonema is the first step of the entry into the reproduction stage (Cove, 2005).
In the present study, the P. patens Dof transcription factor genes are characterized. A previous phylogenetic analysis using the amino acid sequences of the Arabidopsis, rice, P. patens, and C. reinhardtii Dof domains revealed three distinct groups, namely A-type, B-type, and C-type Dof domains (Shigyo et al., 2007). The group A-type Dof domain sequences are unique to C. reinhardtii, as well as to six Dof domains from P. patens and seven Dof domains from Arabidopsis. In the current analyses, the focus is on the P. patens genes that encode the group A-type Dof domain, particularly PpDof1 and PpDof2. The phenotypic analysis reveals that the disruption of PpDof1 induces abnormal vegetative growth, accompanied by reduced filament branching and gametophore formation, and smaller sized colonies. Some of these phenotypes were found to be dependent on carbon and nitrogen nutrients in the media, suggesting involvement of PpDof1 in growth control of protonemal filaments in response to environmental nutrient conditions.
Materials and methods
Growth conditions
Physcomitrella patens (Hedw.) Bruch & Schimp. subsp. patens Tan (Ashton and Cove, 1977) was used. The protonema of wild-type and transformed P. patens was grown on cellophane-covered agar plates at 25 °C under a day/night cycle of 16/8 h with ∼50 μE light. BCD medium (1 mM MgSO4, 1.84 mM KH2PO4, 10 mM KNO3, 45 μM FeSO4, 0.22 μM CuSO4, 10 μM H3BO3, 0.23 μM CoCl2, 0.1 μM Na2MoO4, 0.19 μM ZnSO4, 2 μM MnCl2, 0.17 μM KI, 1 mM CaCl2) was used as standard medium. Media that were essentially BCD medium supplemented with 0.5% glucose and 5 mM ammonium tartrate, or 5 mM ammonium tartrate alone, were also used. Hereafter, these media are referred to as BCD medium supplemented with ammonium and glucose and BCD medium supplemented with ammonium. The regeneration medium was the same as BCD medium supplemented with ammonium, except that the supplementation was with 6% mannitol and 10 mM CaCl2 instead of 1 mM CaCl2.
Determination of the structures of PpDof1–PpDof6
Full-length cDNA clones for PpDof1–PpDof4 and PpDof6 (identification numbers pdp31562, pdp36135, pdp02416, pdp17798, and pdp13251, respectively) were provided by the RIKEN Bioresource center (Tsukuba, Japan). The DNA sequences determined using these clones have been deposited in GenBank (accession numbers: AB626697, AB626698, AB626699, AB626700, and AB626701). The cDNA clones for PpDof5 were isolated separately by PCR amplification of the 5′- and 3′-terminal halves. The 5'-terminal half was amplified using the primers listed in Supplementary Table S4 available at JXB online, whereas the cDNA for the 3'-terminal half was obtained using a 3' RACE (rapid amplification of cDNA ends) system (Invitrogen, Carlsbad, CA, USA) and subsequent PCR amplification. The sequences of the cDNA clones were merged to identify the complete open reading frame (ORF). The GenBank accession numbers of the PpDof5 cDNA clones are AB626702 and AB626703. Alignments of deduced amino acid sequences were performed using the ClustalW program in the Genetyx computer software (Genetyx Corporation, Tokyo, Japan).
RT-PCR
Preparations of total RNA, reverse transcription reactions, and quantitative and semi-quantitative PCR were undertaken as described previously (Konishi and Yanagisawa, 2008, 2010). The primers used are listed in Supplementary Table S3 at JXB online.
Generation of P. patens disruptants
For the isolation of genomic clones for PpDof1 and PpDof2, PCR was performed using the wild-type P. patens genomic DNA and the primers listed in Supplementary Table S4 at JXB online. The 1.4 kb and 1.6 kb PCR products that included a 150 bp sequence encoding the Dof domain were cloned into pGEM-T (Promega, Madison, WI, USA). For the construction of gene targeting vectors, the Dof domain sequences in the PpDof1 and PpDof2 inserts of the resultant plasmids were replaced with the 35S promoter-driven hygromycin resistance gene of pHTS14, and the 35S promoter-driven kanamycin resistance gene of pTN80, respectively, as positive selection markers. The structures of pHTS14 and pTN80 are described at http://www.nibb.ac.jp/evodevo/5-Appendix3.html. Transformation of P. patens with these targeting vectors was performed using the polyethylene glycol method (Schaefer and Zrÿd, 1997). The transformants were then selected on agar plates containing BCD medium supplemented with ammonium and kanamycin or hygromycin B.
Phenotypic analysis
Histological analyses were performed using either a stereomicroscope (MZ16F, Leica Microsystems K.K., Tokyo, Japan) or a light microscope equipped with Nomarski optics (BX51, Olympus Co., Tokyo, Japan). For growth analysis, the protonema was regenerated from single protoplasts on cellophane-covered agar plates containing regeneration medium and grown for 4 weeks, and then transferred onto cellophane-covered plates containing BCD medium, BCD medium supplemented with ammonium, or BCD medium supplemented with ammonium and glucose. The protonema was grown further with a cellophane cover to promote growth in an even direction. The areas occupied by the protonemal colonies were measured using ImageJ software (http://rsb.info.nih.gov/ij). The ratios of chloronema cells to caulonema cells in the protonema that were regenerated from single protoplasts by a 3 d cultivation on regeneration medium plates and subsequent 8 d cultivation on the plates containing BCD medium, BCD medium supplemented with ammonium and glucose, or BCD medium supplemented with ammonium, were measured.
Transient assay using maize protoplasts
A reporter construct was generated by insertion of eight copies of the LexA-binding sites (Yanagisawa et al., 2003) between the 35S enhancer (position –45 to –421) and the 35S minimal promoter truncated at –72 that was fused to the luciferase (LUC) gene. Effector constructs were generated by replacement of the sequence encoding the VP16 transcriptional activation domain in the expression vector of the LexA–VP16 synthetic transcription factor (Yanagisawa et al., 2003) with a MYC tag sequence, the repressor domain of SUPERMAN (Hiratsu et al., 2002), or the entire coding region for PpDof1 or PpDof2. Reporter and effector constructs were co-transfected into maize protoplasts together with an internal control plasmid that contained uidA [β-glucuronidase (GUS) gene] under the control of the maize ubiquitin promoter, as described previously (Yanagisawa, 2000). LUC and GUS activity was measured as described previously (Konishi and Yanagisawa, 2008).
Screening of differentially expressed genes in the double disruptant lines
Probes for use with a 44K microarray were designed using the application software, eArray, of Agilent Technologies (https://earray.chem.agilent.com/earray/) and were based on the data for P. patens cDNAs that are publically provided by the Joint Genome Institute (http://genome.jgi-psf.org/Phypa1_1/Phypa1_1.home.html). As the arrays contain 35 938 cDNAs, one or two probes were designed for each of these sequences. For DNA microarray analysis, total RNA from the protonema of the wild-type P. patens and the ppdof1 ppdof2 double disruptants was used for amplification and labelling with a Low RNA Input Linear Amplification/Labeling kit (Agilent Technologies). Hybridization was conducted according to the protocol recommended by the supplier. The glass slides were scanned using a microarray scanner (G2565BA, Agilent Technologies). More than 3-fold up-regulated and down-regulated genes were selected using four sets of data obtained with independent samples, and significant differences between the values obtained with the wild-type P. patens and the ppdof1 ppdof2 disruptant were evaluated using the t-test (n=4).
Results
Structure and expression of the PpDof1–PpDof6 genes
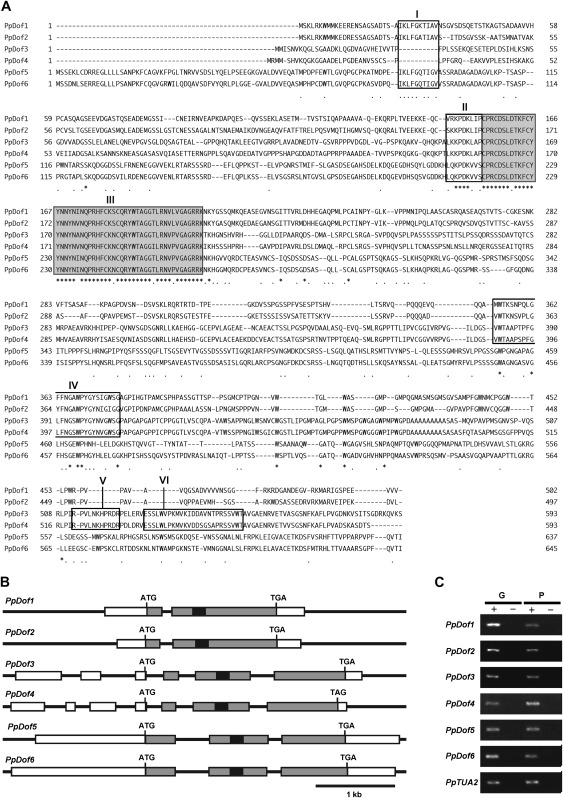
Six DNA sequences encoding the group A-type Dof domain were previously identified in the P. patens genome and denoted PpDof1–PpDof6 (Shigyo et al., 2007). To reveal the primary structures of the products encoded by these putative genes, the corresponding full-length cDNA clones were obtained from a public bioresource centre, except for the PpDof5 cDNA which was isolated by RT-PCR. The DNA sequences of these cDNA clones were determined and the primary structures of the PpDof1–PdDof6 proteins, which are composed of 502, 497, 593, 593, 637, and 645 amino acid residues, respectively, were deduced from the complete ORFs in these cDNA clones (Fig. 1A). An alignment of these amino acid sequences revealed remarkably high similarities outside the Dof domain between PpDof1 and PpDof2, PpDof3 and PpDof4, and PpDof5 and PpDof6, in addition to a high level of conservation within the Dof domains (Fig. 1A). This indicated that the P. patens genes encoding the group A-type Dof domain comprised three pairs of related factors, as suggested previously by phylogenic analysis using amino acid sequences for Dof domains alone (Shigyo et al., 2007). The intron–exon structures, which were determined by comparison of the cDNA sequences with the corresponding genomic DNA sequences, also supported these paired relationships (Fig. 1B). These results were also in good agreement with the conclusions of a recent genome analysis that a large-scale or genome-wide duplication event had occurred in P. patens (Rensing et al., 2008).
Fig. 1.
Physcomitrella patens genes encoding the group A-type Dof domain. (A) Alignment of the deduced amino acid sequences of PpDof1–PpDof6. Amino acid sequence motifs similar to small structural motifs that were previously found in the N-terminal regions of angiosperm group-A Dof factors are indicated by boxes I and II. The Dof domains are indicated by box III. Amino acid sequences that are conserved in PpDof1–PpDof4 are indicated by box IV. Two amino acid sequence motifs similar to small structural motifs that were found in the C-terminal regions of Arabidopsis CDF1–CDF3 are indicated by boxes V and VI. (B) Schematic representation of the intron–exon structures of PpDof1–PpDo6 genes. Exons and introns are indicated by boxes and bars, respectively. The 5′- and 3′-untranslated regions, coding regions, and the sequence encoding the Dof domain are indicated by white, grey, and black boxes, respectively. Translational start codons (ATG) and stop codons (TGA or TAG) are also shown. (C) Expression analysis by RT-PCR. Reverse transcription reactions were performed in the presence (+) or absence (–) of reverse transcriptase using RNA prepared from gametophores (G) and protonema (P). Transcripts of the α-tubulin gene (PpTUA2) (accession number AB096719) were detected as a control.
Although the primary structures, apart from the Dof domain, are largely divergent among the Dof transcription factors in general, small structural motifs that can be hallmarks used for the classification of Dof transcription factors are conserved in some members of this family (Yanagisawa, 2002; Lijavetzky et al., 2003). Two small structural motifs that are specifically present in the N-terminal regions of angiosperm Dof transcription factors with the group A-type Dof domain, which are referred to as group-A Dof factors hereafter, were found in the N-terminal regions of PpDof1–PpDof6 (Fig. 1A, boxes I and II), confirming the close relationship between PpDof1–PpDof6 and the angiosperm group-A Dof factors. Furthermore, additionally conserved amino acid sequence motifs that were previously found in the C-terminal regions of CDF1–CDF3, which are Arabidopsis group-A Dof factors involved in photoperiodic flowering (Imaizumi et al. 2005), but not in other Arabidopsis Dof factors, were found in PpDof3 and PpDof4 (Fig. 1A, boxes V and VI), suggesting that PpDof3 and PpDof4 may be homologues of CDF1–CDF3. Another additional homologous sequence was found in PpDof1– PpDof4 (Fig. 1A, boxes IV), suggesting that PpDof1 and PpDof2 are relatively close to PpDof3 and PpDof4 rather than to PpDof5 and PpDof6.
To clarify whether PpDof1–PpDof6 are functional genes, their expression was examined by RT-PCR analysis. The results indicated the PpDof1–PpDof6 are indeed expressed in both the protonema and the gametophores (Fig. 1C).
Generation of ppdof1 and ppdof2 single disruptant lines and ppdof1 ppdof2 double disruptant lines
As the first step in addressing the biological functions of the Dof transcription factors in P. patens, ppdof1 and ppdof2 disruptants were generated by targeted gene disruption (Supplementary Fig. S1 at JXB online). In addition to two independent lines of both ppdof1 disruptants (ppdof1-#1 and 1-#3 lines) and ppdof2 disruptants (ppdof2-#1 and 2-#2 lines), two independent ppdof1 ppdof2 double disruptant lines were generated by further disruption of the PpDof1 gene in the ppdof2-#1 and ppdof2-#2 lines because of their close relationship (Fig. 1A, B). By RT-PCR analysis in these lines it was confirmed that each gene disruption made the corresponding intact transcript undetectable, and by genomic Southern blot analysis it was confirmed that there was no insertion apart from at the intended locus (Supplementary Fig. S1 at JXB online). However, two independent lines were always analysed in this study to avoid potential interference by somatic mutations or other unexpected events. As consistent results were obtained in every experiment, the data obtained from one line are presented as representative results in this report.
Delayed or reduced gametophore formation in the ppdof1 disruptant
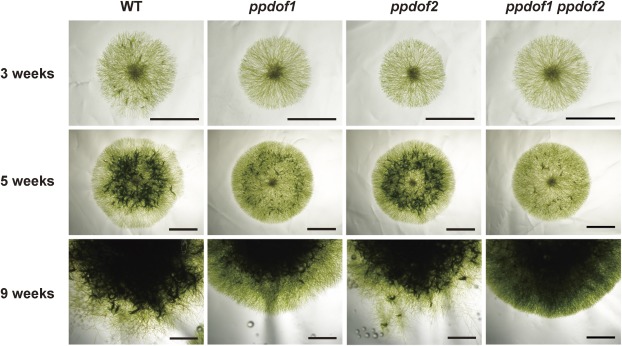
To investigate the impacts of disrupting the PpDof1 and PpDof2 genes, filament growth in the mutant lines was monitored. Single protoplasts of each disruptant line were cultured on regeneration medium for 4 weeks and then further cultured on BCD medium supplemented with ammonium and glucose, so that synchronized filament growth could be induced from a single cell (Fig. 2). A reduced or delayed formation of gametophores, which can be observed as darker green spots, was found in the filamental colonies of the ppdof1 disruptant lines that were cultured for 3 or 5 weeks. Furthermore, a difference was also found in the peripheral parts of 9-week-old filamental colonies. The colonies of the wild-type P. patens showed irregular boundaries probably due to growth of caulonemal filaments, while the ppdof1 disruptants showed a smooth periphery. Gametophores were formed in the peripheral parts of 9-week-old filaments of the wild-type P. patens, but fewer gametophores were formed in those of the ppdof1 disruptant lines. These phenotypes were only very weakly discernible in the ppdof2 disruptant, and the phenotypes were comparable or slightly more severe in the ppdof1 ppdof2 double disruptant lines. These results suggest that the observed phenotypes are more tightly linked with the function of PpDof1 rather than with that of PpDof2.
Fig. 2.
Altered morphology of protonemal colonies of ppdof disruptant lines. Colonies that were formed from a single protoplast of the wild type (WT), ppdof1 or ppdof2 single disruptant line, and the ppdof1 ppdof2 double disruptant line were grown on regeneration medium plates for 4 weeks and then on BCD medium supplemented with ammonium and glucose for the indicated periods, and photographed. Scale bars=3 mm.
Nutrient condition-dependent effects of the disruption of PpDof1
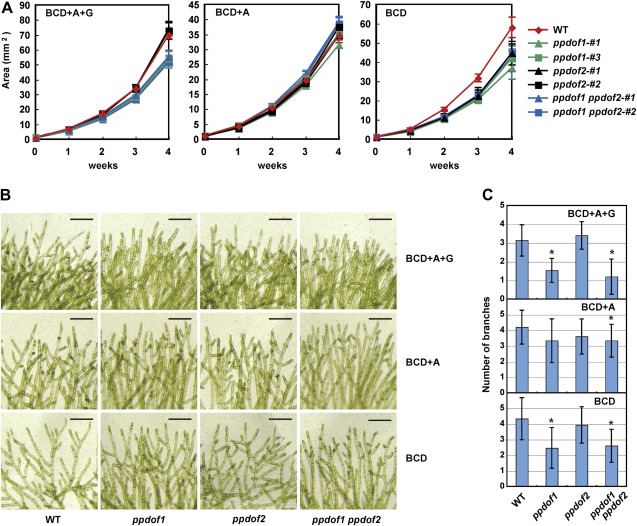
To clarify the effects of the disruption of PpDof1 and PpDof2 on the growth of filamentous colonies, the sizes of the colonies were measured. As it is known that environmental factors such as exogenously supplied sugar and light influence the formation of caulonemal filaments the radial growth of which expanded the protonemal colonies (Olsson et al., 2003; Thelander et al., 2005), three different media were used, namely BCD medium supplemented with ammonium and glucose, BCD medium supplemented with ammonium, and BCD medium. Abnormal filament growth of the disruptant lines was revealed by the smaller sizes of the protonemal colonies on BCD medium supplemented with ammonium and glucose (Fig. 3A). Although this was not evident for the ppdof1 disruptants grown on BCD medium supplemented with ammonium, this phenotype was observed again when using BCD medium. An effect of the disruption of the PpDof2 gene was also found. The sizes of the ppdof2 disruptant colonies were smaller to those of the wild-type colonies when they were grown on BCD medium. The sizes of the ppdof2 disruptant colonies were comparable with those of the ppdof1 disruptant colonies on BCD medium.
Fig. 3.
Nutrient condition-dependent phenotype of the disruptant lines. (A) Areas of protonemal colonies that were initially grown on regeneration medium for 4 weeks and then on BCD medium supplemented with ammonium and glucose (BCD+A+G), BCD medium supplemented with ammonium (BCD+A), or BCD medium for the indicated periods after protoplast formation. Values are the means ±SD of three replicates. (B) Images showing the branching of protonemal filaments of wild-type (WT), ppdof1 or ppdof2 single disruptant lines, and the ppdof1 ppdof2 double disruptant line. Scale bars=200 μm. (C) The numbers of branches within six cells from the filament top. These numbers were measured using protonemal colonies that were initially grown on regeneration medium plates for 4 weeks and then on BCD medium supplemented with ammonium and glucose (BCD+A+G), BCD medium supplemented with ammonium (BCD+A), or BCD medium for 3 weeks. Values are the means ±SD of 15 replicates. Asterisks indicate a statistically significant difference (P < 0.05), compared with the values obtained for wild-type P. patens.
Given the observation of different shaped boundary regions of the protonema (Fig. 2), the effects of the ppdof1 and ppdof2 disruption on the branching frequency were also evaluated by counting the numbers of branches within the first to sixth cells from the tip of protonemal filaments (Fig. 3B, C). It has been shown that the branching frequency of filaments is also affected by both glucose and light (Thelander et al., 2005). Although only slight effects of a difference in the nutrient composition of the medium on the branching frequency could be observed using the wild-type P. patens, it was found that the ppdof1 disruptants showed reduced branching of the filaments, compared with the wild-type P. patens. Furthermore, this effect was also found to be dependent on nutrients in the medium, similar to the effects on colony sizes. The effects of the ppdof2 disruption on the branching frequency were not apparent. However, although the difference between branching frequencies of the ppdof1 disruptants and the wild-type P. patens was not statistically significant when they were grown on BCD medium supplemented with ammonium, the difference between branching frequencies of the ppdof1 ppdof2 double disruptants and the wild-type P. patens was statistically significant (P < 0.05) even on BCD medium supplemented with ammonium. This might imply that PpDof1 and PpDof2 have a redundant role, although PpDof1 appeared to play a more dominant role.
A modified ratio of chloronemal cells to caulonemal cells in the ppdof1 protonema
Gametophores are formed through the development of chloronemal apical cells into caulonemal cells, the differentiation of caulonemal side branch initials into buds, and the development of these buds into gametophores (Cove and Knight, 1993; Cove, 2005). Taken together with the phenotype observed in the peripheral parts of 9-week-old protonemal colonies and colony sizes, it was speculated that a delay in or repression of differentiation of the chloronema into the caulonema might influence gametophore formation in the protonema of the disruptants. Thus, the numbers of chloronemal and caulonemal cells in the protonema that were freshly formed from a single protoplast were counted via a 3 d cultivation on regeneration medium and subsequent 8 d cultivation on BCD medium supplemented with ammonium and glucose (Table 1). The ppdof1 ppdof2 disruptants were used because they appeared to show more severe phenotypes than the ppdof1 single disruptants (Figs 2, 3B, C). The results indicated that the ratio of chloronemal cells to caulonemal cells in the ppdof1 ppdof2 protonema was slightly higher than that in the wild-type P. patens protonema when filaments were grown on BCD medium supplemented with ammonium and glucose (Table 1).
Table 1.
Ratios of chloronema cells to caulonema cells in the protonema
| Growth medium | Plant line | Chloronema (%) | Caulonema (%) | Ratio (chloronema/caulonema) | n |
| BCD+A+G | WT | 65.9 | 34.1 | 1.93 | 211 |
| ppdof1 ppdof2 | 72.1 | 27.9 | 2.58 | 140 | |
| BCD+A | WT | 68.5 | 31.5 | 2.17 | 165 |
| ppdof1 ppdof2 | 69.7 | 30.3 | 2.3 | 165 | |
| BCD | WT | 59.6 | 40.4 | 1.48 | 250 |
| ppdof1 ppdof2 | 60.1 | 39.9 | 1.51 | 248 |
BCD medium supplemented with ammonium and glucose (BCD+A+G), BCD medium supplemented with ammonium (BCD+A), and BCD medium were used for protonemal growth.
PpDof1 and PpDof2 are transcriptional repressors
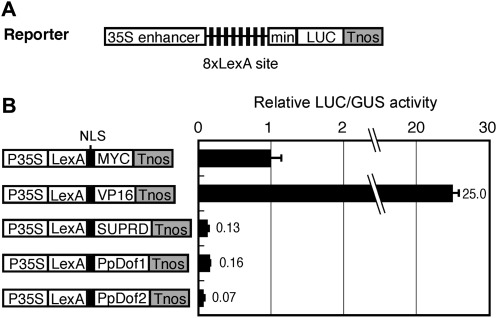
The activity of PpDof1 and PpDof2 as transcription factors was investigated using protoplast transient assays. In the initial experiment where the LUC reporter gene under the control of the LexA-binding sites fused to the 35S minimal promoter was co-transfected into protoplasts together with expression vectors for the bacterial LexA DNA-binding domain fused to PpDof1 or PpDof2, the expression of the fusion proteins slightly repressed transcription (data not shown). This suggested that PpDof1 and PpDof2 might be transcriptional repressors. To test this possibility, a different reporter construct in which eight copies of the LexA-binding sites were inserted between the 35S enhancer and the 35S minimal promoter was used (Fig. 4A). In this co-transfection experiment, the LexA DNA-binding domain fused to a transcriptional activation domain of herpes simplex virus protein VP16 (LexA–VP16) activated transcription, whereas the LexA DNA-binding domain fused to the transcriptional repression domain of SUPERMAN (LexA–SUPRD) repressed it. Both the LexA–PpDof1 and LexA–PpDof2 fusion proteins also repressed transcription to a similar extent to LexA–SUPRD (Fig. 4B), indicating that PpDof1 and PpDof2 can function as transcriptional repressors. Consistent with this transcriptional repressor activity, PpDof2 fused to green fluorescent protein was found to be localized to nuclei, although the subcellular localization of PpDof1 could not be confirmed, probably due to its low expression in protoplasts (Supplementary Fig. S2 at JXB online).
Fig. 4.
Transcriptional repressor activity of PpDof1 and PpDof2. (A) Reporter construct that harbours a LUC gene under the control of eight copies of the LexA-binding site (8×LexA site) located between the 35S enhancer and the 35S minimal promoter. (B) Transrepression assay. The reporter construct was co-transfected into maize protoplasts, together with an internal control plasmid and an effector plasmid for the LexA DNA-binding domain fused to a MYC tag, a VP16 transcriptional activation domain, a repressor domain for SUPERMAN (SUPRD), or full-length PpDof1 or PpDof2. After normalization with the GUS activity derived from the internal control plasmid, the relative LUC activity was calculated as the means ±SD (n=3). The relative LUC activity obtained with the effector plasmid for the expression of LexA–MYC protein was set at 1.
The modified gene expression pattern in the ppdof disruptants
Modulations in the gene expression patterns in the ppdof1 ppdof2 double disruptants were investigated using custom-made DNA microarrays that contained probes for 35 938 independent transcripts and RNA prepared from protonemal colonies grown on BCD medium supplemented with ammonium and glucose. Based on four independent sets of array data, 34 genes and 91 genes were found to be up-regulated and down-regulated >3-fold with statistical significance, respectively (Supplementary Tables S1, S2 at JXB online). Although most of the identified genes encoded proteins of unknown function, some up-regulated or down-regulated genes encoded proteins for which the function has been annotated. However, these annotated functions were revealed to be very divergent, and no strong relationship could be found between the up-regulated and down-regulated genes and any particular physiological process.
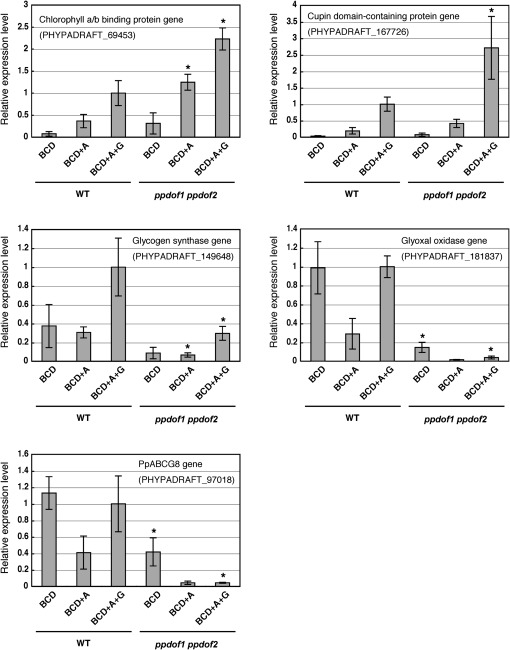
Several of these differentially expressed genes were possibly associated with energy synthesis or cell growth. For instance, strongly up-regulated genes encoded chlorophyll a/b-binding protein and a protein containing the cupin domain, which is a conserved β-barrel structure found in transcriptional regulators and metabolic enzymes (Dunwell et al., 2000). The strongly down-regulated genes included genes for a putative glycogen synthase, a glyoxal oxidase, and an ATP-binding cassette (ABC) transporter subfamily G member. Thus, modified expression levels of these genes in the ppdof1 ppdof2 disruptant were analysed by quantitative RT-PCR using RNA prepared from protonemal colonies (Fig. 5). Consistent with the results of the DNA microarray analysis, the expression levels of all analysed genes were consistently higher or lower in the protonema of the ppdof1 ppdof2 disruptants grown on BCD medium supplemented with ammonium and glucose. Up-regulated and down-regulated expression of these genes was also observed in the protonema grown under the different nutrient conditions.
Fig. 5.
Modified gene expression in the ppdof1 ppdof2 double disruptant line. Quantitative PCR analysis of transcripts of the putative chlorophyll a/b-binding protein, cupin domain-containing protein, glycogen synthase, glyoxal oxidase, and ABC transporter subfamily member (PpABCG8) genes (the locus tags are PHYPADRAFT_69453, PHYPADRAFT_167726, PHYPADRAFT_149648, PHYPADRAFT_181837, and PHYPADRAFT_97018, respectively). RNA was prepared from protonemal colonies that were initially grown in regeneration medium for 3 d after protoplast formation and then with BCD medium supplemented with ammonium and glucose (BCD+A+G), BCD medium supplemented with ammonium (BCD+A), or BCD medium for 10 d. Values are the means ±SD of three replicates relative to the transcript levels of the β-tubulin 1 gene (PpTUB1) (Holm et al., 2010). The expression levels of these genes in the wild-type plants grown on BCD medium supplemented with ammonium and glucose were assigned a reference value of 1 unit. Asterisks indicate statistically significant differences (P < 0.05) compared with the values obtained for the wild-type protonema grown under the same nutrient conditions.
Discussion
In the present report, structural and expression analysis of the PpDof1–PpDof6 genes and functional analysis of PpDof1 and PpDof2 were performed. The results revealed that PpDof1–PpDof6 are members of the group A-type Dof factors and that PpDof1 is a transcriptional repressor involved in the regulation of the vegetative growth of filaments in response to the environmental nutrient conditions. PpDof2 was found to be another transcriptional repressor. Hence, this is the first report that successfully characterizes Dof transcription factors in lower plants. Importantly, the data indicate a novel function for the Dof transcription factors that have not been previously described in higher plants, and the function was suggested to be associated with growth in response to carbon and nitrogen sources in the media. PpDof1 might be a clue to investigate the molecular mechanism underlying plant growth control in response to the carbon/nitrogen balance in the environment.
Group A-type Dof factors in Arabidopsis and P. patens
The structural analysis of P. patens Dof transcription factors established PpDof1–PpDof6 as group A type and suggested that PpDof3 and PpDof4 might be homologues of Arabidopsis CDF1–CDF3, which are direct transcriptional repressors for the promoter of the CONSTANS gene that mediates the functions of the circadian clock and the control of flowering (Imaizumi et al., 2005). Because the homologue of the Arabidopsis CONSTANS gene has already been identified in P. patens (Shimizu et al., 2004), the functional analysis of PpDof3 and PpDof4 may provide new insights into circadian clock-dependent regulation in plants. Furthermore, the finding of amino acid sequence motifs that are conserved between Arabidopsis CDF1–CDF3 and PpDof3 and PpDof4 may offer a future opportunity to reveal the molecular mechanisms underlying the transcriptional control mediated by Arabidopsis CDF1–CDF3 and their homologues.
The relationships among PpDof1 and PpDof2 and PpDof5 and PpDof6 and other Arabidopsis group A-type Dof factors, such as COG1 and three uncharacterized factors (AtDof1.3, AtDof1.10, and AtDof2.3), remain unclear however due to the lack of unique structural features that define their identities. In the Arabidopsis cog1 mutant that was found in an activation tagging pool, the enhanced expression of the COG1 gene has been shown to lead to light-mediated inhibition of hypocotyl elongation, implying a function for COG1 in the negative regulation of phytochrome signalling (Park et al., 2003). Filament growth of the wild type and the ppdof1 ppdof2 disruptant lines was therefore compared under illumination with white, red, and blue light sources. No significant differences in growth under red light conditions were found (data not shown), suggesting that the physiological roles of PpDof1 and PpDof2 and Arabidopsis COG1 may be associated with two distinct regulatory pathways. There are a few previous studies in which P. patens orthologues of Arabidopsis transcription factors were analysed. In the cases of GOLDEN2-LIKE and ABSCISIC ACID INSENSITIVE3 that are involved in chloroplast development and ABA signalling, respectively, P. patens homologues of these factors have been shown to play similar roles in P. patens (Yasumura et al., 2005; Marella et al., 2006). In contrast, the transcription factor LEAFY has been shown to play different roles in angiosperms and P. patens. Arabidopsis and rice LEAFY help to define the floral fates during the reproductive phase, which is a biological process not present in P. patens, whereas LEAFY controls general aspects of the life cycle in P. patens (Maizel et al., 2005; Tanahashi et al., 2005). Hence, further analysis would be necessary to clarify whether some group A-type Dof factors play similar roles in both higher and lower plants.
Phenotypes of the ppdof mutant lines
It is shown that the disruption of the PpDof1 gene resulted in decreased or delayed gametophore formation, the formation of smaller protonemal colonies, and a reduced frequency of branching (Figs 2, 3). These phenotypes can be caused by a delay in the differentiation of chloronemal cells into caulonemal cells. Indeed, a modified ratio of chloronemal cells to caulonemal cells was detected in the filaments of the ppdof1 ppdof2 disruptants (Table 1). However, this effect was not strong and was evident only in the filaments grown on BCD medium supplemented with ammonium and glucose. On the other hand, the formation of smaller protonemal colonies and a reduced branching frequency were observed for the filaments grown either on BCD medium supplemented with ammonium and glucose or on BCD medium (Fig. 3). This discrepancy might be because the effects of the ppdof1 disruption are more evident at the later stages of vegetative filamental growth. The chloronema/caulonema ratio was measured at a very early stage of filament growth (Table 1), whereas other phenotypes were monitored at later stages (Figs 2, 3). Thus, it is speculated that the more the colonies are grown, the more prominent are the effects of the ppdof1 disruption (Figs 2, 3), although the possibility that the effects of the ppdof1 disruption are enhanced by environmental factors during a long period of growth on plates cannot be ruled out.
The nutrient condition-dependent effects of the ppdof disruptions
When the wild-type P. patens was grown on BCD medium supplemented with ammonium, the colony size was smaller than those of the wild-type P. patens grown on BCD medium (Fig. 3A), indicating the repressive effect of ammonium on expansion of colonies. In contrast, the colony size of the wild-type P. patens was larger on BCD medium supplemented with ammonium and glucose, compared with that on BCD medium supplemented with ammonium (Fig. 3A), indicating that sugar functioned antagonistically against ammonia on the expansion of colonies. These observations indicate the regulation of the radial growth in response to carbon and nitrogen sources in the medium. Because the effects of the ppdof1 disruption were observed in the filaments grown on either BCD medium supplemented with ammonium and glucose or BCD medium (Figs 2, 3) but were not evident on BCD medium supplemented with ammonium, a role for PpDof1 might be associated with promotion of radial growth under the high carbon/nitrogen ratio conditions. The carbon/nitrogen balance is also known to be one of the key factors determining biomass and growth in higher plants (reviewed in Ericsson, 1995). Furthermore, a recent study reported identification of a RING-type ubiquitin ligase that functions in the carbon/nitrogen response in Arabidopsis (Sato et al., 2009), suggesting the presence of the molecular mechanism underlying plant growth regulation in response to the carbon/nitrogen ratio in the environment. However, the molecular mechanism remains to be elucidated in both lower and higher plants. Thus, further analyses of PpDof1 and their possible counterparts in Arabidopsis (some group-A Dof factors) might offer an avenue to elucidate the mechanism. The effects of the disruption of ppdof2 were almost unrecognizable, but the ppdof2 disruptants showed smaller sizes of colonies on BCD medium (Fig. 3A). Hence, PpDof2 might have some functional redundancy with PpDof1, particularly under the growth condition where ammonia is absent from the medium. This hypothesis should be evaluated by further analysis.
Physcomitrella patens mutants displaying reduced or delayed formation of the caulonema filaments
The reduced or delayed formation of the caulonema filaments of the ppdof1 mutants is similar to the reported phenotype of two P. patens mutants in which the metabolic enzyme genes, PpHXK1 or PpSiR1, was disrupted. The disruption of PpHXK1, which encodes the major hexokinase, results in the suppression of glucose-inducible caulonema formation, suggesting that high energy growth conditions promote caulonema formation (Olsson et al., 2003; Thelander et al., 2005). Repression of caulonema formation in the presence of glucose in the ppdof1 mutant appears to be similar to that in the hxk1 mutant. However, in the case of the ppdof1 mutants, the phenotypes were not dependent simply on the glucose supply but also on both the carbon and nitrogen sources in the media. It is unlikely, therefore, that PpDof1 is simply involved in regulation of the energy supply, and the effects caused by the ppdof1 disruption should be distinguishable from those caused by the hxk1 disruption. An additional P. patens mutant, ΔSiR1, in which one of three genes encoding sulphite reductase (PpSiR1) was disrupted, also showed reduced formation of sporophytes and caulonemal cells (Wiedemann et al., 2010). Although SiR is the sulphate metabolism enzyme that catalyses the reduction of sulphite to sulphide, the knockout of the PpSiR1 gene exerted minimal effects on sulphur metabolism. Furthermore, this phenotype was not observed in the ΔAPR and ΔAPR-B mutants, additional P. patens mutants possessing a disrupted gene for 5'-phosphosulphate reductase in the sulphate assimilation pathway. The phenotype of ΔSiR1 has been proposed to be independent of the enzymatic activity of SiR1. In the case of the ppdof1 disruptant in the present study, reduced or delayed growth of caulonemal filaments was found to be dependent on the carbon and nitrogen source in the medium (Figs 2, 3). Although this might suggest a connection between the role of PpDof1 and carbon and nitrogen metabolism, no significant modulations were found in the analysed carbon and nitrogen metabolite contents (data not shown). Hence, the modulation of filament growth in response to the environmental nutrient conditions is likely not to be caused not by simple changes of metabolic balance but through a currently unknown mechanism in the ppdof1 disruptants, similar to the case of the ΔSiR1 mutant.
Differentially expressed genes in the ppdof mutants
A number of genes that were up-regulated and down-regulated in the ppdof1 ppdof2 disruptants were identified in the present analysis (Fig. 5; Supplementary Tables S1, S2 at JXB online), including not only genes for metabolic enzymes but also a gene for a putative regulatory protein as well as many genes encoding unknown proteins. Interestingly, the genes whose expression levels were modified by the ppdof1 ppdof2 disruptions were found to show the expression pattern in response to the carbon and nitrogen sources in the medium. Expression levels of the glyoxal oxidase gene and the PpABCG8 gene in wild-type P. patens were lower on BCD medium supplemented with ammonium (the low carbon/nitrogen ratio condition) than on BCD medium and BCD medium supplemented with ammonium and glucose (the high carbon/nitrogen ratio condition). Furthermore, their down-regulated expression levels in the ppdof1 ppdof2 disruptants were comparable with or lower than those in the wild-type P. patens grown on BCD medium supplemented with ammonium. This may imply that expression of these genes is associated with the phenotype of the ppdof1 ppdof2 disruptants and that PpDof1 (and PpDof2) plays a role in promotion of filament growth on high carbon/nitrogen media via modulations of gene expression. However, because it was shown that both PpDof1 and PpDof2 are transcriptional repressors, these down-regulated genes are not the direct targets of PpDof1. Their differential expression is probably a secondary response caused by the modified expression of the direct targets of PpDof1. Furthermore, it was found that the expression levels of PpDof1 and PpDof2 are not affected by the carbon and nitrogen sources in the media (Supplementary Fig. S3 on JXB online). Identifying direct targets of PpDof1 and revealing post-transcriptional regulation of PpDof1 are therefore of importance to reveal how PpDof1 exerts its effects on the growth of filaments in response to environmental nutrient conditions. The preliminary findings of additional experiments implied that the genes for chlorophyll a/b-binding protein and a protein containing the cupin domain were up-regulated in both ppdof1 and ppdof2 single disruptants, while the ppdof1 and ppdof2 disruptions exerted different effects on the expression of other genes analysed (data not shown). This suggests that detailed gene expression analysis using single and double ppdof disruptants might provide clues to clarify redundant and distinct roles for PpDof1 and PpDof2 under different growth conditions.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Targeted disruption of the PpDof1 and PpDof2 genes.
Figure S2. Nuclear localization of PpDof2.
Figure S3. Expression levels of PpDof1 and PpDof2 in protonemal colonies grown under different nutrient conditions.
Table S1. Up-regulated genes in the ppdof1 ppdof2 disruptants.
Table S2. Down-regulated genes in the ppdof ppdof2 disruptants.
Table S3. Primers used for semi-quantitative and quantitative RT-PCR analysis.
Table S4. PCR primers used for the isolation of cDNA and genomic clones and preparation of DNA probes for genomic Southern blot analysis.
Acknowledgments
We thank Dr M. Hasebe (NIBB, Japan), Mr N. Sawaki (our laboratory), Kyowa Hakko Co. (Tokyo, Japan), and the RIKEN Bioresource Center (Tsukuba, Japan) for generously providing pHTS14 and pTN80 plasmids, the LexA–SUPRD construct, driselase, and full-length cDNA clones, respectively. This work was supported by KAKENHI (22380043) and a Grant-in-Aid for Scientific Research on Innovative Areas (21114004) from The Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- Ashton NW, Cove DJ. The isolation and preliminary characterization of auxotrophic and analogue resistant mutants of the moss. Physcomitrella patens. Molecular and General Genetics. 1977;154:87–95. [Google Scholar]
- Cove D. The moss Physcomitrella patens. Annual Review of Genetics. 2005;39:339–358. doi: 10.1146/annurev.genet.39.073003.110214. [DOI] [PubMed] [Google Scholar]
- Cove DJ, Knight CD. The moss Physcomitrella patens, a model system with potential for the study of plant reproduction. The Plant Cell. 1993;5:1483–1488. doi: 10.1105/tpc.5.10.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz I, Vicente-Carbajosa J, Abraham Z, Martínez M, Isabel-La Moneda I, Carbonero P. The GAMYB protein from barley interacts with the DOF transcription factor BPBF and activates endosperm-specific genes during seed development. The Plant Journal. 2002;29:453–464. doi: 10.1046/j.0960-7412.2001.01230.x. [DOI] [PubMed] [Google Scholar]
- Dunwell JM, Khuri S, Gane PJ. Microbial relatives of the seed storage proteins of higher plants: conservation of structure and diversification of function during evolution of the cupin superfamily. Microbiology and Molecular Biology Reviews. 2000;64:153–179. doi: 10.1128/mmbr.64.1.153-179.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericsson T. Growth and shoot:root ratio of seedlings in relation to nutrient availability. Plant and Soil. 1995;168–169:205–214. [Google Scholar]
- Gardiner J, Sherr I, Scarpella E. Expression of DOF genes identifies early stages of vascular development in Arabidopsis leaves. Interntional Journal of Developmental Biology. 2010;54:1389–1396. doi: 10.1387/ijdb.093006jg. [DOI] [PubMed] [Google Scholar]
- Guo Y, Qin G, Gu H, Qu L-J. Dof5.6/HCA2, a Dof transcription factor gene, regulates interfascicular cambium formation and vascular tissue development in Arabidopsis. The Plant Cell. 2009;21:3518–3534. doi: 10.1105/tpc.108.064139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K, Ohta M, Matsui K, Ohme-Takagi M. The SUPERMAN protein is an active repressor whose carboxy-terminal repression domain is required for the development of normal flowers. FEBS Letters. 2002;514:351–354. doi: 10.1016/s0014-5793(02)02435-3. [DOI] [PubMed] [Google Scholar]
- Holm K, Källmann T, Gyllenstrand N, Hedman H, Lagercrantz U. Does the core circadian clock in the moss Physcomitrella patens (Bryophyta) comprise a single loop? BMC Plant Biology. 2010;10:109. doi: 10.1186/1471-2229-10-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science. 2005;309:293–297. doi: 10.1126/science.1110586. [DOI] [PubMed] [Google Scholar]
- Iwamoto M, Higo K, Takano M. Circadian clock- and phytochrome-regulated Dof-like gene, Rdd1, is associated with grain size in rice. Plant, Cell and Environment. 2009;32:592–603. doi: 10.1111/j.1365-3040.2009.01954.x. [DOI] [PubMed] [Google Scholar]
- Kim H-S, Kim SJ, Abbasi N, Bressan RA, Yun D-J, Yoo S-D, Kwon S-Y, Choi S-B. The DOF transcription factor Dof5.1 influences leaf axial patterning by promoting Revoluta transcription in Arabidopsis. The Plant Journal. 2010;64:524–535. doi: 10.1111/j.1365-313X.2010.04346.x. [DOI] [PubMed] [Google Scholar]
- Konishi M, Yanagisawa S. Sequential activation of two Dof transcription factor gene promoters during vascular development in Arabidopsis thaliana. Plant Physiology and Biochemistry. 2007;45:623–629. doi: 10.1016/j.plaphy.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Konishi M, Yanagisawa S. Ethylene signaling in Arabidopsis involves feedback regulation via the elaborate control of EBF2 expression by EIN3. The Plant Journal. 2008;55:821–831. doi: 10.1111/j.1365-313X.2008.03551.x. [DOI] [PubMed] [Google Scholar]
- Konishi M, Yanagisawa S. Identification of the nitrate-responsive cis-element in the Arabidopsis NIR1 promoter defines the presence of multiple cis-regulatory elements for nitrogen response. The Plant Journal. 2010;63:269–282. doi: 10.1111/j.1365-313X.2010.04239.x. [DOI] [PubMed] [Google Scholar]
- Krohn NM, Yanagisawa S, Grasser KD. Specificity of the stimulatory interaction between chromosomal HMGB proteins and the transcription factor Dof2 and its negative regulation by protein kinase CK2-mediated phosphorylation. Journal of Biological Chemistry. 2002;277:32438–32444. doi: 10.1074/jbc.M203814200. [DOI] [PubMed] [Google Scholar]
- Li D, Yang C, Li X, Gan Q, Zhao X, Zhu L. Functional characterization of rice OsDof12. Planta. 2009;229:1159–1169. doi: 10.1007/s00425-009-0893-7. [DOI] [PubMed] [Google Scholar]
- Lijavetzky D, Carbonero P, Vicente-Carbajosa J. Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evolutionay Biology. 2003;3:17. doi: 10.1186/1471-2148-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizel A, Busch MA, Tanahashi T, Perkovic J, Kato M, Hasebe M, Weigel D. The floral regulator LEAFY evolves by substitutions in the DNA binding domain. Science. 2005;308:260–263. doi: 10.1126/science.1108229. [DOI] [PubMed] [Google Scholar]
- Marella HH, Sakata Y, Quatrano RS. Characterization and functional analysis of ABSCISIC ACID INSENSITIVE3-like genes from Physcomitrella patens. The Plant Journal. 2006;46:1032–1044. doi: 10.1111/j.1365-313X.2006.02764.x. [DOI] [PubMed] [Google Scholar]
- Martínez M, Rubio-Somoza I, Fuentes R, Lara P, Carbonero P, Díaz I. The barley cystatin gene (Icy) is regulated by DOF transcription factors in aleurone cells upon germination. Journal of Experimental Botany. 2005;56:547–556. doi: 10.1093/jxb/eri033. [DOI] [PubMed] [Google Scholar]
- Mena M, Vicente-Carbajosa J, Schmidt RJ, Carbonero P. An endosperm-specific DOF protein from barley, highly conserved in wheat, binds to and activates transcription from the prolamin-box of a native B-hordein promoter in barley endosperm. The Plant Journal. 1998;16:53–62. doi: 10.1046/j.1365-313x.1998.00275.x. [DOI] [PubMed] [Google Scholar]
- Moreno-Risueno MA, Martínez M, Vicente-Carbajosa J, Carbonero P. The family of DOF transcription factors: from green unicellular algae to vascular plants. Molecular Genetics and Genomics. 2007;277:379–390. doi: 10.1007/s00438-006-0186-9. [DOI] [PubMed] [Google Scholar]
- Olsson T, Thelander M, Ronne H. A novel type of chloroplast stromal hexokinase is the major glucose phosphorylating enzyme in the moss Physcomitrella patens. Journal of Biological Chemistry. 2003;278:44439–44447. doi: 10.1074/jbc.M306265200. [DOI] [PubMed] [Google Scholar]
- Park DH, Lim PO, Kim JS, Cho DS, Hong SH, Nam HG. The Arabidopsis COG1 gene encodes a Dof domain transcription factor and negatively regulates phytochrome signaling. The Plant Journal. 2003;34:161–171. doi: 10.1046/j.1365-313x.2003.01710.x. [DOI] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- Reski R. Physcomitrella and Arabidopsis: the David and Goliath of reverse genetics. Trends in Plant Science. 1998;3:209–210. [Google Scholar]
- Rubio-Somoza I, Martinez M, Abraham Z, I. Diaz I, Carbonero P. Ternary complex formation between HvMYBS3 and other factors involved in transcriptional control in barley seeds. The Plant Journal. 2006;47:269–281. doi: 10.1111/j.1365-313X.2006.02777.x. [DOI] [PubMed] [Google Scholar]
- Rueda-López M, Crespillo R, Cánovas FM, Avila C. Differential regulation of two glutamine synthetase genes by a single Dof transcription factor. The Plant Journal. 2008;56:73–85. doi: 10.1111/j.1365-313X.2008.03573.x. [DOI] [PubMed] [Google Scholar]
- Sato T, Maekawa S, Yasuda S, et al. CNI1/ATL31, a RING type ubiquitin ligase that functions in the carbon/nitrogen response for growth phase transition in Arabidopsis seedlings. The Plant Journal. 2009;60:852–864. doi: 10.1111/j.1365-313X.2009.04006.x. [DOI] [PubMed] [Google Scholar]
- Schaefer DG, Zrÿd J-P. Efficient gene targeting in the moss Physcomitrella patens. The Plant Journal. 1997;11:1195–1206. doi: 10.1046/j.1365-313x.1997.11061195.x. [DOI] [PubMed] [Google Scholar]
- Skirycz A, Jozefczuk S, Stobiecki M, Muth D, Zanor MI, Witt I, Mueller-Roeber B. Transcription factor AtDOF4;2 affects phenylpropanoid metabolism in Arabidopsis thaliana. New Phytologist. 2007;175:425–438. doi: 10.1111/j.1469-8137.2007.02129.x. [DOI] [PubMed] [Google Scholar]
- Skirycz A, Radziejwoski A, Busch W, et al. The DOF transcription factor OBP1 is involved in cell cycle regulation in Arabidopsis thaliana. The Plant Journal. 2008;56:779–792. doi: 10.1111/j.1365-313X.2008.03641.x. [DOI] [PubMed] [Google Scholar]
- Skirycz A, Reichelt M, Burow M, et al. DOF transcription factor AtDof1.1 (OBP2) is part of a regulatory network controlling glucosinolate biosynthesis in Arabidopsis. The Plant Journal. 2006;47:10–24. doi: 10.1111/j.1365-313X.2006.02767.x. [DOI] [PubMed] [Google Scholar]
- Shimizu M, Ichikawa K, Aoki S. Photoperiod-regulated expression of the PpCOL1 gene encoding a homolog of CO/COL proteins in the moss Physcomitrella patens. Biochemical and Biophysical Research Communications. 2004;324:1296–1301. doi: 10.1016/j.bbrc.2004.09.194. [DOI] [PubMed] [Google Scholar]
- Singh KB. Transcriptional regulation in plants: the importance of combinatorial control. Plant Physiology. 1998;118:1111–1120. doi: 10.1104/pp.118.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigyo M, Tabei N, Yoneyama K, Yanagisawa S. Evolutionary processes during the formation of the plant-specific Dof transcription factor family. Plant and Cell Physiology. 2007;48:179–185. doi: 10.1093/pcp/pcl044. [DOI] [PubMed] [Google Scholar]
- Tanahashi T, Sumikawa N, Kato M, Hasebe M. Diversification of gene function: homologs of the floral regulator FLO/LFY control the first zygotic cell division in the moss Physcomitrella patens. Development. 2005;132:1727–1736. doi: 10.1242/dev.01709. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Takahata Y, Nakayama H, Nakatani M, Tahara M. Altered carbohydrate metabolism in the storage roots of sweet potato plants overexpressing the SRF1 gene, which encodes a Dof zinc finger transcription factor. Planta. 2009;230:737–746. doi: 10.1007/s00425-009-0979-2. [DOI] [PubMed] [Google Scholar]
- Thelander M, Olsson T, Ronne H. Effect of the energy supply on filamentous growth and development in Physcomitrella patens. Journal of Experimental Botany. 2005;56:653–662. doi: 10.1093/jxb/eri040. [DOI] [PubMed] [Google Scholar]
- Vicente-Carbajosa J, Moose SP, Parsons RL, Schmidt RJ. A maize zinc-finger protein binds the prolamin box in zein gene promtoters and interacts with the basic leucine zipper transcriptional activator Opaque2. Proceedings of the National Academy of Sciences, USA. 1997;94:7685–7690. doi: 10.1073/pnas.94.14.7685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H-W, Zhang B, Hao Y-J, Huang J, Tian A-G, Liao Y, Zhang J-S, Chen S-Y. The soybean Dof-type transcription factor genes, GmDof4 and GmDof11, enhance lipid content in the seeds of transgenic Arabidopsis plants. The Plant Journal. 2007;52:716–729. doi: 10.1111/j.1365-313X.2007.03268.x. [DOI] [PubMed] [Google Scholar]
- Washio K. Functional dissections between GAMYB and Dof transcription factors suggest a role for protein–protein associations in the gibberellin-mediated expression of the RAmy1A gene in the rice aleurone. Plant Physiology. 2003;133:850–863. doi: 10.1104/pp.103.027334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P-C, Tan F, Gao X-Q, Zhang X-Q, Wang G-Q, Xu H, Li L-J, Chen J, Wang X-C. Overexpression of AtDOF4.7, an Arabidopsis DOF family transcription factor, induces floral organ abscission deficiency in Arabidopsis. Plant Physiology. 2010;153:1031–1045. doi: 10.1104/pp.110.153247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann G, Hermsen C, Melzer M, Büttner-Mainik A, Rennenberg H, Reski R, Kopriva S. Targeted knock-out of a gene encoding sulfite reductase in the moss Physcomitrella patens affects gametophytic and sporophytic development. FEBS Letters. 2010;584:2271–2278. doi: 10.1016/j.febslet.2010.03.034. [DOI] [PubMed] [Google Scholar]
- Yamamoto MP, Onodera Y, Touno SM, Takaiwa F. Synergism between RPBF Dof and RISBZ1 bZIP activators in the regulation of rice seed expression genes. Plant Physiology. 2006;141:1694–1707. doi: 10.1104/pp.106.082826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S. A novel DNA binding domain that may form a single zinc finger motif. Nucleic Acids Research. 1995;23:3403–3410. doi: 10.1093/nar/23.17.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S. Dof DNA-binding domains of plant transcription factors contribute to multiple protein–protein interactions. European Journal of Biochemistry. 1997;250:403–410. doi: 10.1111/j.1432-1033.1997.0403a.x. [DOI] [PubMed] [Google Scholar]
- Yanagisawa S. Dof1 and Dof2 transcription factors are associated with expression of multiple genes involved in carbon metabolism in maize. The Plant Journal. 2000;21:281–288. doi: 10.1046/j.1365-313x.2000.00685.x. [DOI] [PubMed] [Google Scholar]
- Yanagisawa S. The Dof family of plant transcription factors. Trends in Plant Science. 2002;7:555–560. doi: 10.1016/s1360-1385(02)02362-2. [DOI] [PubMed] [Google Scholar]
- Yanagisawa S. Dof domain proteins: plant-specific transcription factors associated with diverse phenomena unique to plants. Plant and Cell Physiology. 2004;45:386–391. doi: 10.1093/pcp/pch055. [DOI] [PubMed] [Google Scholar]
- Yanagisawa S, Izui K. Molecular cloning of two DNA binding proteins of maize that are structurally different but interact with the same sequence motif. Journal of Biological Chemistry. 1993;268:16028–16036. [PubMed] [Google Scholar]
- Yanagisawa S, Schmidt RJ. Diversity and similarity among recognition sequences of Dof transcription factors. The Plant Journal. 1999;17:209–214. doi: 10.1046/j.1365-313x.1999.00363.x. [DOI] [PubMed] [Google Scholar]
- Yanagisawa S, Sheen J. Involvement of maize Dof zinc finger proteins in tissue-specific and light-regulated gene expression. The Plant Cell. 1998;10:75–89. doi: 10.1105/tpc.10.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S, Yoo S-D, Sheen J. Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature. 2003;425:521–525. doi: 10.1038/nature01984. [DOI] [PubMed] [Google Scholar]
- Yasumura Y, Moylan EC, Langdale JA. A conserved transcription factor mediates nuclear control of organelle biogenesis in anciently diverged land plants. The Plant Cell. 2005;17:1894–1907. doi: 10.1105/tpc.105.033191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Chen W, Foley RC, Büttner M, Singh KB. Interactions between distinct types of DNA binding proteins enhance binding to ocs element promoter sequences. The Plant Cell. 1995;7:2241–2252. doi: 10.1105/tpc.7.12.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.