Abstract
α-Linolenic acid (ALA) deficiency and a skewed of ω6:ω3 fatty acid ratio in the diet are a major explanation for the prevalence of cardiovascular diseases and inflammatory/autoimmune diseases. There is a need to enhance the ALA content and to reduce the ratio of linoleic acid (LA) to ALA. Six ω-3 (Δ-15) fatty acid desaturase (FAD) genes were cloned from rice and soybean. The subcellular localizations of the proteins were identified. The FAD genes were introduced into rice under the control of an endosperm-specific promoter, GluC, or a Ubi-1 promoter to evaluate their potential in increasing the ALA content in seeds. The ALA contents in the seeds of endoplasmic reticulum (ER)-localized GmFAD3-1 and OsFAD3 overexpression lines increased from 0.36 mg g−1 to 8.57 mg g−1and 10.06 mg g−1, respectively, which was 23.8- and 27.9-fold higher than that of non-transformants. The trait of high ALA content was stably inheritable over three generations. Homologous OsFAD3 is more active than GmFAD3-1 in catalysing LA conversion to ALA in rice seeds. Overexpression of ER-localized GmFAD3-2/3 and chloroplast-localized OsFAD7/8 had less effect on increasing the ALA content in rice seeds. The GluC promoter is advantageous compared with Ubi-1 in this experimental system. The enhanced ALA was preferentially located at the sn-2 position in triacylglycerols. A meal-size portion of high ALA rice would meet >80% of the daily adult ALA requirement. The ALA-rich rice could be expected to ameliorate much of the global dietary ALA deficiency.
Keywords: Endosperm-specific expression, ω-3 fatty acid desaturase, hyperfortification, α-linolenic acid, transgenic rice
Introduction
Long-chain polyunsaturated fatty acids (LC-PUFAs) are important for human health in maintaining the cellular membrane by regulating cholesterol synthesis and transportation (Simopoulos, 1991) and eicosanoid synthesis (Kankaanpaa et al., 1999). The recommended dietary ω-6:ω-3 fatty acid ratio is 5:1; however, the ratio in the modern diet is ∼15:1 to 20:1 (Simopoulos, 2001). The skewed ω-6:ω-3 fatty acid ratio is thought to be a major explanation for the prevalence of cardiovascular diseases and inflammatory/autoimmune diseases (Stark et al., 2008).
α-Linolenic acid (ALA) is the precursor of the most important LC-ω-3-PUFAs, eicosapentaenoic acid (EPA), docosapentaenoic acid (DPA), and docosahexaenoic acid (DHA), which can be synthesized in the body. Humans cannot convert linoleic acid (LA) into ALA because they lack ω-3 (Δ-15) fatty acid desaturase (FAD), which catalyses the conversion of LA into ALA in plants (Yadav et al., 1993); therefore, humans must obtain ALA from the diet. Presently, the main dietary sources of ALA are only some deep sea fish and some specific oilseed plants (e.g. flax, soybean, rape, walnut, and perilla). Because of the decline in the world fish supply and heavy metal pollution in the sea, sources of ALA from deep sea fish are limited. Also, ALA-rich oilseed plants are few. One of the goals of oilseed breeding is to develop new varieties with decreased ALA content (Vrinten et al., 2005; Clemente and Cahoon, 2009) because a high ALA content reduces the oxidative stability of oil, thus leading to rancidity and ‘off’ flavours in food products (Feussner and Wasternakc, 2002). Therefore, there is a need to develop non-oilseed sources of ALA.
In plants, the desaturation of LA to ALA occurs both in the plastids and in the endoplasmic reticulum (ER). However, ALA synthesis in seeds is mainly catalysed by microsomal ω-3 FAD (Vrinten et al., 2005). In Arabidopsis FAD3 mutant lines, the ALA content is reduced to 1–2% of total seed fatty acids from ∼20% in wild-type plants (James and Dooner, 1990; Lemieux et al., 1990). In flax LuFAD3A and LuFAD3B double mutant lines, the seed ALA content is <2% as compared with ∼52% in the wild-type parent (Vrinten et al., 2005). Puttick et al. (2009) reported that the ALA content of seed increased from 19% to nearly 40% of total fatty acids in Arabidopsis by seed-specific overexpression of FAD3. Ectopic expression of FAD3 in soybean seed also greatly increased both the ALA content and the ω-3:ω-6 ratio (Damude et al., 2006; Eckert et al., 2006).
Rice is one of the most important staple foods for a large part of the world’s population. Storage components, including fatty acids, accumulated in seed are highly stable. However, rice seed has a very low ALA content (<0.4 mg g−1) and a high ratio of LA to ALA (∼24:1); therefore, varieties containing high amounts of ALA could ameliorate much of the global dietary ALA deficiency. To date, some efforts have employed heterologous FAD3 expression strategies to enhance the ALA content in rice seeds. Shimada et al. (2000) expressed tobacco FAD3 in rice under the control of a Cauliflower mosaic virus (CaMV) 35S promoter and obtained stable transgenic rice with ALA content increased up to 2.5-fold compared with wild-type seed. Anai et al. (2003) obtained rice seed with 13-fold the ALA content by introducing soybean FAD3 under the control of the maize ubiquitin-1 (Ubi-1) promoter. CaMV 35S and Ubi-1 are constitutive expression promoters that are not strong enough in rice seed (Qu and Takaiwa, 2004). The use of a strong endosperm-specific promoter might help to increase the accumulation of ALA in rice endosperm.
The rice genome contains at least three FAD3 genes, which are expressed abundantly in root but at a very low level in leaf and seed. Kodama et al. (1997) cloned a microsomal FAD3 gene from rice (OsFAD3), and demonstrated that the gene was related to chilling tolerance. The subcellular localizations of the rice ω-3 FAD genes and their functions in increasing ALA content in rice seeds remained unclear. In this study, three ω-3 FAD genes were cloned from rice and soybean, respectively, and their subcellular localization was identified. The ω-3 FAD genes were introduced into rice under the control of an endosperm-specific expression promoter, GluC (Qu et al., 2008), or a constitutive expression promoter, Ubi-1, to evaluate their potential in increasing the ALA content in rice seeds. The ALA contents in seeds of transgenic lines in the T1, T2, and T3 generation and the distribution of fatty acid in triacylglycerols (TAGs) were also analysed.
Materials and methods
Isolation of OsFAD3/7/8 and GmFAD3-1/2/3
Total RNA from soybean (Glycine max) and rice (Oryza sativa) seeds at 12 days after flowering (DAF) was isolated using Trizol reagent (Takara, Japan) according to the manufacturer’s instructions. First-strand cDNA was generated as described (Qu et al., 2002). The generated first-strand cDNA was used as a template for PCR. Primer pairs for three rice ω-3 FADs (OsFAD3/7/8, GenBank accession nos AK071185, AB232382, and AB232383) and three soybean FAD3s (GmFAD3-1/2/3, GenBank accession nos AY204710, AY204711, and AY204712) are listed in Table 1. The PCR products were cloned into a pMD18-T vector (Takara), and then the inserts were sequenced.
Table 1.
Primers used for cloning of FAD genes
| Primer name | Primer sequence (with the restriction site underlined) |
| OsFAD3F | 5′-AACCCGGGATGGCGGCGTCGGCGACCCAG-3′ (SmaI) |
| OsFAD3R | 5′-AAGAGCTCTCACTTGTGCTTAGCATCTTC-3′ (SacI) |
| OsFAD7F | 5′-AAGGATCCATGGCACGGCTCGTCCTCTCCG-3′(BamHI) |
| OsFAD7R | 5′-AAGAGCTCTCAATCCGAGCTTTGTGCAGAA-3′ (SacI) |
| OsFAD8F | 5′-AAGGATCCATGGCCCGGCTGCTGCTCTCCG-3′(BamHI) |
| OsFAD8R | 5′-AAGAGCTCTTAGTTAGCAGGGTCGGTCTGG-3′ (SacI) |
| GmFAD3-1F | 5′-AACCCGGGATGGTTAAAGACACAAAG-3′ (SmaI) |
| GmFAD3-1R | 5′-AAGAGCTCTCAGTCTCGTTGCGAGTGGA-3′ (SacI) |
| GmFAD3-2F | 5′-AAGCGGCCGCCATGGTTAAAGACACAAAGCCTT-3′ (NotI) |
| GmFAD3-2R | 5′-AAGCGGCCGCTCAGTCTCGTTGCGAGTGGAGG-3′ (NotI) |
| GmFAD3-3F | 5′-AAGCGGCCGCCATGGTTCAAGCACAGCCTCTAC-3′ (NotI) |
| GmFAD3-3R | 5′-AAGCGGCCGCTTAGTTGGACTGGGTCCAAGAA-3′ (NotI) |
GFP and RFP fusion constructs for transient expression in rice protoplasts
OsFAD3 and GmFAD3 were fused to green fluorescent protein (GFP) in the N-terminus (GmFAD3-1, GmFAD3-2, GmFAD3-3, and OsFAD3) or in the C-terminus (OsFAD7 and OsFAD8), respectively. Monomeric red fluorescent protein (mRFP) was fused to the ER retention signal (mRFP–KDEL) or transit peptide (1–73 amino acids) of the small subunit of ribulose bisphosphate carboxylase (Rubisco–mRFP), respectively. The chimeric genes were subcloned into pBI221, replacing β-glucuronidase (GUS) to obtain the transient expression vectors. The transient expression vectors were co-transformed into the rice protoplast as described previously (Liu et al., 2010). The transformed cells were cultured on K3 medium for 12 h and observed under a confocal microscope (Zeiss, Germany).
Construction and transformation of the chimeric gene
The OsFAD3 and GmFAD3-1 fragments were inserted into the binary vector pGPTV-GluC-GUS-35S-HPT (Qu et al., 2008) containing the GluC promoter by replacing the GUS gene. The binary vector was introduced into rice (O. sativa cv. Kitaake) by Agrobacterium tumefaciens-mediated transformation as described (Qu et al., 2005). Transformants were grown in a greenhouse as described previously (Qu et al., 2008). The successful transformation was verified by PCR and self-pollinated for three generations. The T1–T3 plants were used for further studies.
Southern blot analysis
Rice genomic DNA was extracted from leaves by the cetyltrimethyl ammonium bromide (CTAB) method. A 20 μg aliquot of genomic DNA was digested with EcoRV. DNA digests were fractioned on a 0.8% agarose gel, transferred onto a nylon membrane (Hybond N+), and hybridized with a specific probe to detect the hpt (hygromycin phosphotransferase) gene, prepared by PCR amplification with a primer set (5′-GCAAGGAATCGGTCAATACA-3′ and 5′-TTCTACACAGCCATCGGTC-3′). After pre-hybridization of ∼5 h, the 32P-labelled probe was added and then hybridized for another 20 h at 60 °C. The membrane was washed twice with 2× SSC plus 0.1% SDS at 65 °C and once with 1× SSC plus 0.1% SDS at 65 °C. The membrane was exposed to X-ray film (Kodak, Rochester, NY, USA) at –80 °C for ≥2 d.
Northern blot analysis
RNA was prepared from developing rice seeds 10–14 DAF. RNA extraction, probe labelling, hybridization, and signal detection were carried as described previously (Qu et al. 2002). The full-length GmFAD3-1 or OsFAD3 cDNAs were used as probes.
Western blot analysis
The expression patterns and level of GmFAD3-1 and OsFAD3 in mature seeds were examined by in situ western analysis and western blot analysis as described previously (Qu et al., 2005). Antisera against OsFAD3 and GmFAD3-1 were raised in rabbits that had been immunized with amino acid residues 153–248 and 149–244 of OsFAD3 and GmFAD3-1 proteins, respectively. The intensities of visualized bands were measured by use of Quantity one (Bio-Rad).
Lipid analysis
Fatty acid and individual lipid contents in endosperm were analysed according to Wu et al. (2005). The conditions of digestion and the isolation procedure for positional analysis of TAGs were described previously (Mattson and Beck, 1954). An ALA standard sample (Sigma-Aldrich, St Louis, MO, USA) was used to confirm ALA in the non-transformed (NT) and transgenic lines. Heptadecanoic acid (Sigma-Aldrich) was used as the internal standard for ALA content. Results are presented as the means ±SDs.
Statistical analysis
Mean comparisons were calculated by Student’s test, with P-values indicated in the Results.
Results
Subcellular localization of FAD3 proteins
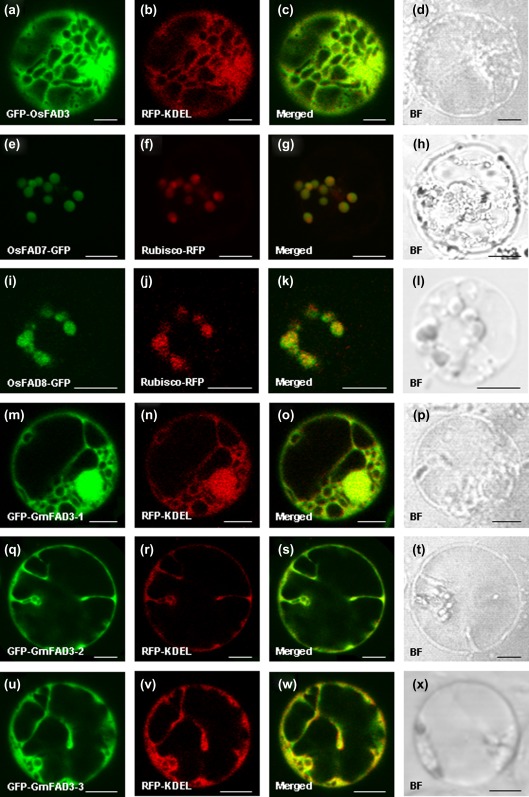
ω-3 FAD catalyses LA desaturation to ALA in both the ER and the chloroplast. Three ω-3 FAD genes were cloned from rice and soybean, respectively, by reverse transcription-PCR (RT-PCR). To test whether the ω-3 FAD proteins are localized in the ER or the chloroplast surface, a translational fusion was made between ω-3 FAD and GFP, RFP tagged with KDEL (a marker for the ER), and the Rubisco small subunit (a marker for the chloroplast) and RFP, and the fusion proteins were transiently expressed in rice protoplast. Confocal images of RFP–KDEL marker line protoplasts that were transformed with GFP–OsFAD3, GFP–GmFAD3-1, GFP–GmFAD3-2, and GFP–GmFAD3-3 showed almost 100% overlap of RFP and GFP fluorescence (Fig. 1c, o, s, and w). Transient expression of GFP–OsFAD3, GFP–GmFAD3-1, GFP–GmFAD3-2, and GFP–GmFAD3-3 in Rubisco–RFP protoplasts leads to distinct fluorescence patterns of GFP and RFP (Supplementary Fig. S1c, o, s, and w available at JXB online). In contrast, OsFAD7–GFP and OsFAD8–GFP were expressed in the same compartment with Rubisco–RFP (Fig. 1g, k) but distinct from RFP–KDEL (Supplementary Fig. S1g, k). These results indicated that OsFAD3, GmFAD3-1, GmFAD3-2, and GmFAD3-3 proteins localized in the ER, whereas OsFAD7 and OsFAD8 localized in the chloroplast.
Fig. 1.
Subcellular localization of the OsFAD3/7/8 and GmFAD3-1/2/3 proteins. The green GFP and red RFP signals obtained by confocal microscopy indicate fusion proteins OsFAD7/8–GFP (e and i) or GFP–FAD3 (a, m, q, and u) and Rubisco–RFP (chloroplast marker protein; f and j) or RFP–KDEL (endoplasmic reticulum marker protein; b, n, r, and v) which were transiently expressed in rice protoplasts. The overlap of green and red fluorescent signals is indicated in merged images (c, g, k, o, s, and w). Bright fields are showed in d, h, l, p, t, and x. Bars, 5 μm.
Characterization of transgenic rice plants
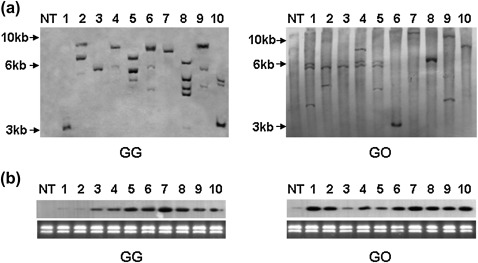
The ER-localized OsFAD3 and GmFAD3-1 were introduced into rice under control of the rice endosperm-specific GluC promoter. Successful transformation was determined by PCR analysis with genomic DNA extracted from leaves of T0 plants. Transformants expressing GluC/GmFAD3-1 and GluC/OsFAD3 were named GG and GO lines, respectively. Twenty-one GG and 23 GO lines were obtained. Southern hybridization with genomic DNA from T0 transgenic rice digested with EcoRV (there is no EcoRV site within GmFAD3-1 and OsFAD3) with hpt cDNA used as a probe showed 1–5 bands at different positions in GG and GO lines, which confirmed that the GmFAD3-1 or OsFAD3 gene was successfully integrated into the rice genome, and independent transgenic rice lines were obtained (Fig. 2a). Northern hybridization with total RNA extracted from T1 seeds (10–14 DAF) gave a single band of 1.1 kb, which corresponded to the expected size of GmFAD3-1 and OsFAD3, indicating that the gene is expressed in seeds of each line (Fig. 2b). The transcript levels of OsFAD3 in GO lines were 2.5- to 18.2-fold higher than those of the NT lines.
Fig. 2.
Southern and northern analysis of rice lines. (a) A 20 μg aliquot of genomic DNA was digested by EcoRV, fractioned on a 0.8% agarose gel, transferred onto a nylon membrane, and hybridized with a fragment of hpt. Size markers are shown on the left in kilobases. (b) A 10 μg aliquot of total RNA was fractioned on a 1.2% agarose–formaldehyde gel, transferred onto a nylon membrane, and hybridized with a fragment encoding GmFAD3-1 or OsFAD3. NT, Kitaake, non-transformants; GG, transformant with the GmFAD3-1 gene directed by the GluC promoter; GO, transformants with the OsFAD3 gene directed by the GluC promoter.
Accumulation and distribution of GmFAD3-1 or OsFAD3 in transgenic rice seeds
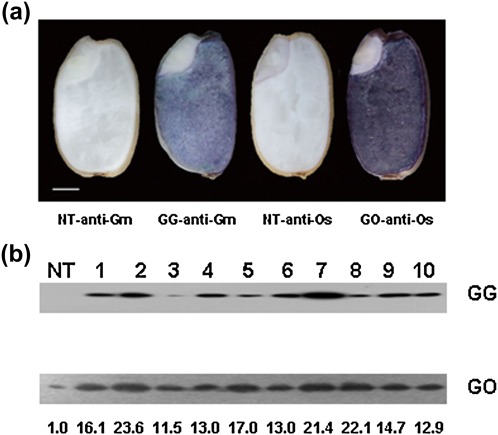
The protein expression pattern of GmFAD3-1 or OsFAD3 directed by the GluC promoter was determined by in situ western hybridization (Fig. 3a). GmFAD3-1 protein expression was restricted to the endosperm of the GG lines. In GO lines, OsFAD3 was expressed strongly in the endosperm and very weakly in the embryo, which reflects the expression of homologous protein. Of note, GmFAD3-1 or OsFAD3 were expressed almost equally throughout the endosperm of the transgenic plants. The expression patterns of GmFAD3-1 and OsFAD3 in the GG and GO lines were consistent with previously reported tissue-specific expression of the GluC promoter (Qu et al., 2008). Western blot was used to analyse the total protein extracted from mature seed to investigate the protein expression of GmFAD3-1 and OsFAD3 in the GG and GO lines (Fig. 3b). Polyclonal antibody directed against GmFAD3-1 bound to a 44 kDa band from GG plants, with no band observed in the NT lines. In GO lines, with anti-OsFAD3, a unique band of ∼44 kDa was found in both transgenic and non-transgenic plants, with levels in transformants being 11.5- to 23.6-fold higher than those in the NT plants. Of note, GmFAD3-1 was not expressed in any tissues other than the endosperm in any GG lines, whereas the expression of OsFAD3 was weak in vegetative tissues of GO lines and did not differ from that of the NT lines (data not shown). In addition, the transgene expression level was not related to copy number.
Fig. 3.
In situ western hybridization and western blot analysis of FAD3 in transgenic rice seeds. (a) In situ western hybridization of FAD3 transgenic rice seeds. Scale bar=0.1 mm. (b) Western blot analysis of FAD3 in transgenic rice seeds. Total proteins extracted from each transgenic and non-transgenic seed were fractioned by SDS–PAGE and immunoblotted with the GmFAD3-1 or OsFAD3 rabbit polyclonal antibodies. The numbers above each lane indicate the relative multiples of FAD3 levels. NT, GG, and GO are as described in Fig. 2.
ALA content in seed of transgenic lines
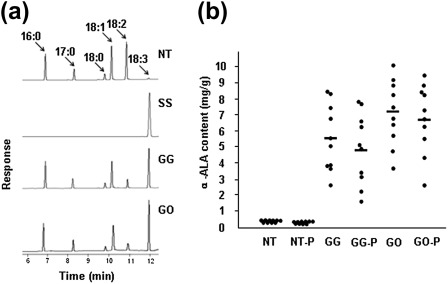
The ALA content in the GG and GO lines was analysed by gas chromatography (GC). The ALA standard sample confirmed that the fatty acid whose content was increased in the GG and GO lines was indeed ALA (Fig. 4a). In the GG and GO lines, mean ALA content was 5.64±0.16 mg g−1 (range from 2.66±0.11 mg g−1 to 8.57±0.20 mg g−1) and 7.19±0.22 mg g−1 (3.74±0.19 mg g−1 to 10.06±0.23 mg g−1), respectively (Fig. 4b), but was only 0.36±0.02 mg g−1 for NT lines. The mean ALA content in GG and GO lines was 15.7-fold (range from 7.4- to 23.8-fold, P < 0.01) and 20.0-fold (10.4- to 27.9-fold, P < 0.01), respectively, higher than that of NT plants. The mean ALA content was 27.2% (up to 41.3%) and 33.18% (46.4%) of the total fatty acids in the GG and GO lines, respectively, as compared with 1.68% in the NT lines (Table 2). The LA contents decreased from 40.5% in the NT lines to as low as 15.7% and 15.0% in the GG and GO lines, respectively (Table 2). The mean ratio of LA to ALA was 1.22 (0.38–2.72) and 0.79 (0.32–1.95) for the GG and GO lines, respectively, and 24.1 for the NT lines (Table 2). The ratio of LA to ALA in GG and GO lines was 30.7-fold (8.9- to 63.3-fold, P < 0.01) and 40.6-fold (12.3- to 75.2-fold, P < 0.01) lower than that of the NT lines, respectively. Thus, the ALA content was greatly enhanced in rice seed by ectopic expression of GmFAD3-1 or overexpression of OsFAD3 under control of the GluC promoter. Also, homologous OsFAD3 was more efficient than heterologous GmFAD3-1 in converting LA to ALA in rice. The total lipid contents in the seeds of NT, GG, and GO lines were 2.1±0.2, 2.1±0.1, and 2.2±0.1%, respectively. The total seed lipid content of the transgenic lines showed no obvious differences from that of the NT lines (P > 0.05).
Fig. 4.
ALA content in transgenic rice seeds. (a) GC analysis of seed fatty acid methyl esters. SS, ALA standard sample; 16:0, palmitic acid; 17:0, heptadecanoic acid (which served as the internal standard); 18:0, stearic acid; 18:1, oleic acid; 18:2, linoleic acid; 18:3, α-linolenic acid. (b) ALA content in transgenic rice seeds. NT, GG, and GO are as described in Fig. 2. NT-P, GG-P, and GO-P are polished seeds corresponding to NT, GG, and GO. A dot represents an average value of each experimental line. A horizontal bar represents the mean of each construction.
Table 2.
Seed fatty acid composition (weight%) of the NT and transgenic plants
| 16:0 | 18:0 | 18:1 | 18:2 | 18:3 | 18:2/18:3 | |
| NT | 21.62±0.53 | 2.20±0.15 | 34.04±0.76 | 40.47±1.04 | 1.68±0.05 | 24.09 |
| GG lines | ||||||
| 7 | 20.88±0.35 | 2.19±0.15 | 19.90±0.61 | 15.68±0.35 | 41.34±0.36 | 0.38 |
| 2 | 21.40±0.66 | 2.19±0.20 | 20.42±0.53 | 15.95±0.75 | 40.60±0.30 | 0.39 |
| 9 | 21.48±0.57 | 2.41±0.16 | 21.10±0.68 | 18.13±0.31 | 36.01±0.28 | 0.50 |
| 1 | 21.49±0.14 | 2.36±0.07 | 23.45±0.67 | 19.82±0.84 | 33.48±1.75 | 0.59 |
| 4 | 21.54±0.38 | 2.35±0.15 | 24.99±1.23 | 27.13±1.17 | 26.97±0.76 | 1.01 |
| 10 | 21.45±0.94 | 2.32±0.19 | 25.18±0.86 | 30.66±1.72 | 24.31±0.22 | 1.26 |
| 6 | 21.54±0.09 | 2.16±0.09 | 27.73±0.76 | 32.69±1.74 | 19.25±0.90 | 1.70 |
| 5 | 21.52±0.91 | 2.49±0.06 | 29.60±0.17 | 33.01±1.53 | 19.03±0.18 | 1.73 |
| 8 | 21.38±0.16 | 2.56±0.16 | 29.92±1.39 | 34.11±1.53 | 18.15±0.51 | 1.88 |
| 3 | 21.51±0.47 | 2.25±0.10 | 30.17±1.90 | 34.94±0.50 | 12.83±0.35 | 2.72 |
| GO lines | ||||||
| 2 | 19.27±0.39 | 2.44±0.15 | 16.84±1.06 | 15.04±0.24 | 46.40±0.42 | 0.32 |
| 8 | 19.32±0.33 | 2.20±0.21 | 17.55±0.40 | 16.40±0.59 | 42.37±2.00 | 0.39 |
| 7 | 19.34±0.63 | 2.14±0.18 | 18.96±1.01 | 17.68±0.63 | 41.25±1.66 | 0.43 |
| 5 | 19.58±0.47 | 2.20±0.28 | 19.38±0.92 | 19.58±0.77 | 39.12±0.93 | 0.50 |
| 1 | 20.12±0.21 | 2.29±0.15 | 20.04±1.04 | 20.23±0.92 | 34.73±0.68 | 0.58 |
| 9 | 20.57±0.42 | 2.19±0.26 | 22.38±1.29 | 21.54±0.94 | 32.01±1.60 | 0.67 |
| 10 | 20.51±0.29 | 2.28±0.12 | 23.20±0.57 | 22.20±1.24 | 29.79±0.30 | 0.75 |
| 4 | 20.63±0.37 | 2.14±0.10 | 23.53±0.93 | 26.04±1.44 | 26.68±1.14 | 0.98 |
| 6 | 20.54±0.40 | 2.08±0.16 | 24.11±0.38 | 29.11±0.81 | 22.14±1.06 | 1.31 |
| 3 | 20.61±0.50 | 2.20±0.26 | 27.88±0.94 | 33.59±1.65 | 17.26±0.98 | 1.95 |
The mean ALA content in the polished seeds of GG and GO lines was 4.94±0.15 mg g−1 (1.64±0.05 mg g−1 to 7.93±0.11 mg g−1) and 6.83±0.16 mg g−1 (2.67±0.05 mg g−1 to 9.50±0.20 mg g−1), respectively, whereas that of the NT lines was 0.27±0.01 mg g−1 (Fig. 4b). The ALA content in the polished seeds of GG and GO lines was up to 29.4-fold (P < 0.01) (18.3-fold on average) and 35.2-fold (P < 0.01) (25.3-fold on average) higher than those of NT seeds. The ALA accounted for a mean of 30.61% (up to 48.7%) and 40.1% (up to 52.81%) of the total fatty acids in the polished seeds of the GO and GG lines, respectively, as compared with 1.71% in the NT lines.
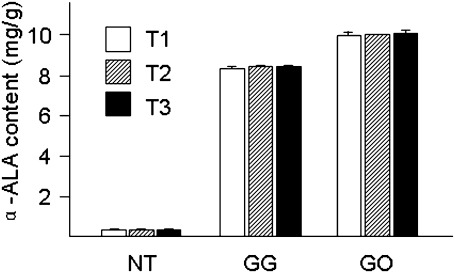
ALA content in the seed of transgenic lines was constant over three generations
To evaluate the stability of ALA accumulation, the ALA contents in the seeds of GG-7 and GO-2, the highest ALA lines (Table 2), were measured through three generations from T1 to T3. The ALA contents in T1, T2, and T3 seeds of GG-7 were 8.49±0.14, 8.58±0.12, and 8.54±0.03 mg g−1 (8.54±0.05 mg g−1 on average), respectively. The ALA contents in T1, T2, and T3 seeds of GO-2 were 9.99±0.16, 10.01±0.06, and 10.08±0.13 mg g−1 (10.03±0.04 mg g−1 on average), respectively. In contrast, those of the NT lines were 0.36±0.01, 0.36±0.02, and 0.36±0.02 mg g−1, respectively (Fig. 5). The ALA contents showed almost no difference through three generations in both GG-7 and GO-2 lines, indicating that the trait of high ALA content was stably inheritable in the transgenic lines. The results also showed that ALA content in GO-2 seed was always higher than that in GG-7 in all generations.
Fig. 5.
Seed ALA concentrations from T1 to T3 generations. NT, GG, and GO are as described in Fig. 2. Data are the mean ±SD of three independent biological replicates.
Fatty acid composition and proportion of individual lipids
To illustrate the effect of overexpressing FAD3 genes on the proportion of individual lipids and the fatty acid profile, total lipids were extracted from the endosperm. Phosphatidylinositol (PI), sulphoquinovosyldiacylglycerol (SQDG), phosphatidylcholine (PC), phosphatidylglycerol (PG), digalactosyldiacylglycerol (DGDG), and phosphatidylethanolamine (PE) were separated and their contents were measured. PI, SQDG, PC, PG, DGDG, and PE accounted for 28.5, 27.8, 18.7, 12.9, 8.1, and 4.0% of total lipids, respectively, in the NT rice endosperm (Table 3). In the endosperm of GO and GG lines, the proportion of the six classes of lipid was almost the same as that of the NT lines (P > 0.05); however, the ALA contents in PC, PE, SQDG, PI, PG, and DGDG were 19.1-, 18.2-, 17.1-, 15.7-, 13.5-, and 1.6-fold and 22.9-, 22.5-, 20.6-, 18.9-, 16.5-, and 1.9-fold higher than those of the NT plant (P < 0.01, except for DGDG, P < 0.05 ) (Table 3). These results indicated that ALA enhancement did not significantly affect the synthesis of each lipid class.
Table 3.
Fatty acid composition and proportion of individual lipids from the NT and transgenic rice endosperm
| Lipid | LA (%) | ALA (%) | % total lipids | |
| NT | 40.8±1.9 | 1.9±0.3 | 28.5±0.8 | |
| PI | GG | 20.5±2.7 | 29.8±1.8 | 28.8±1.7 |
| GO | 17.1±1.7 | 35.9±1.7 | 29.0±1.6 | |
| NT | 43.8±1.5 | 1.8±0.2 | 27.8±2.1 | |
| SQDG | GG | 19.6±1.8 | 30.8±2.1 | 26.9±1.7 |
| GO | 16.6±1.1 | 37.0±1.4 | 26.4±1.1 | |
| NT | 45.7±0.7 | 1.6±0.1 | 18.7±0.3 | |
| PC | GG | 17.2±1.0 | 30.6±2.0 | 19.3±1.6 |
| GO | 14.1±1.0 | 36.6±1.3 | 20.1±2.1 | |
| NT | 46.3±0.9 | 1.7±0.1 | 12.9±0.9 | |
| PG | GG | 24.1±1.4 | 23.0±0.4 | 12.2±1.6 |
| GO | 20.1±2.0 | 28.0±0.7 | 11.6±1.1 | |
| NT | 24.1±1.3 | 1.6±0.1 | 8.1±0.8 | |
| DGDG | GG | 25.7±0.7 | 2.5±0.3 | 8.1±0.7 |
| GO | 21.1±1.2 | 3.1±0.3 | 7.8±0.4 | |
| NT | 25.5±1.3 | 2.0±0.1 | 4.0±0.6 | |
| PE | GG | 2.1±0.2 | 36.4±1.0 | 4.2±0.3 |
| GO | 1.7±0.2 | 44.9±1.1 | 4.3±0.3 |
Each value represents the mean ±SD of three independent biological replicates. NT, GG, and GO are as described in Fig. 2. PC, phosphatidylcholine; PE, phosphatidylethanolamine; SQDG, sulphoquinovosyldiacylglycerol; PI, phosphatidylinositol; PG, phosphatidylglycerol; DGDG, digalactosyldiacylglycerol.
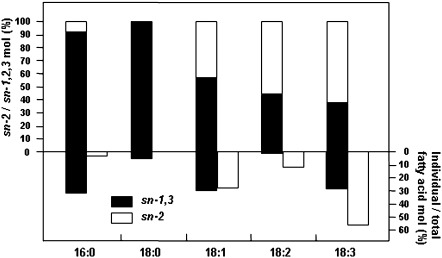
The distribution of fatty acids in TAGs
Fatty acids bound at the sn-2 position in TAGs are digested, absorbed, and metabolized more effectively than those in the sn-1 and sn-3 positions (Carnielli et al., 1995). Positional analysis of fatty acids in the GO line showed that ALA and LA at the sn-2 position accounted for 61.3±3.6% and 56.6±3.8% of the total ALA and LA bound to TAGs, respectively (Fig. 6). Palmitic acid (PA, 16:0) and stearic acid (SA, 18:0) were almost exclusively found at sn-1 and sn-3 positions. Oleic acid (OA, 18:1) was distributed almost equally at sn-2 and sn-1 and sn-3 positions. Both GG and GO lines showed a similar distribution of fatty acids in TAGs (data not shown). Thus, the high content ALA in transgenic rice is bioavailable in humans.
Fig. 6.
Distribution of fatty acids in triacylglycerols (TAGs). The proportion of individual fatty acids in the sn-2 or sn-1,3 positions (upper panel) and proportion of individual to total fatty acids in the sn-2 or sn-1,3 positions (lower panel). TAGs from mature seed endosperm were digested with pancreatic lipase, and products were separated by thin-layer chromatography, transmethylated, and analysed by GC. 16:0, 18:0, 18:1, 18:2, and 18:3 are as described in Fig. 4. Data are the mean of three independent biological replicates.
Discussion
ω-3 Fatty acid deficiency is one of the most prevalent nutritional problems in the world. Because of a declining supply of deep sea fish and a reduced content of ω-3 fatty acids in land-based oil crops, developing a ω-3 fatty acid-enriched staple plant food through traditional plant breeding or molecular biological strategies would be beneficial. Cereal seeds contain a low level of ALA, which might be due to the low level of FAD3, the major LA desaturase in seeds (Vrinten et al., 2005). It was reported that ALA synthesis in seeds was mainly catalysed by microsomal FAD (FAD3) (Vrinten et al., 2005). Six FAD3 genes were cloned from rice and soybean, and their subcellular localizations were identified. GmFAD3-1/2/3 and OsFAD3 were localized in the ER, while OsFAD7 and OsFAD8 were localized in the chloroplast (Fig. 1). The ER-localized OsFAD3 and GmFAD3-1 were overexpressed in rice under the control of the strong endosperm-specific promoter GluC. The OsFAD3 mRNA and protein level in GO seeds was increased 18.2- and 23.6-fold, respectively, as compared with the NT seeds (Figs 2b, 3b). GmFAD3-1 was also expressed at a high level in GG lines (Figs 2b, 3b). High expression of FAD3 proteins greatly increased the content of ALA and decreased that of LA in transgenic rice endosperm, with no detrimental effects on growth, development, seed weight, or other yield traits (data not shown). The ALA content in seeds of GG and GO lines was increased 23.8- and 27.9-fold, respectively, compared with that of the NT lines (Fig. 4). However, endosperm-specific expression of GmFAD3-2 and GmFAD3-3 slightly increased the ALA level in transgenic rice seeds (Supplementary Table S1 at JXB online). These results reflect the functional differences of ER-localized FAD isoforms. The chloroplast-localized FADs can also convert LA to ALA, but the overexpression of the two chloroplast-localized ω-3 FAD genes OsFAD7 and OsFAD8 under the control of the GluC promoter resulted in only a slight increase in the ALA level in rice seeds (Supplementary Table S1).
Flax and soybean seeds contain more ALA than cereal crops, suggesting that their FAD3 might be more active. Anai et al. (2003) introduced a soybean FAD3 gene into rice under the control of the Ubi-1 promoter, which resulted in up to a 13-fold increase in ALA content in transformed seeds. Rice has its own FAD3 genes, but its ALA content is low. The mean ALA content in the transgenic rice endosperm of GO lines was 38.26% (P < 0.01) higher than that of GG lines. Similarly, the ALA content in the endosperm of transgenic lines of Ubi-1-OsFAD3 was 18.31% (P < 0.01) higher than that of Ubi-1-GmFAD3-1 transgenic lines (data not shown). These results indicated that homologous OsFAD3 is more active than heterologous GmFAD3-1 in catalysing LA to ALA conversion in rice seeds, although the amino acid sequences showed 64% identity between OsFAD3 and GmFAD3-1 genes. Plant homologous genes might work better than heterologous genes in improving natural components via transgenic strategies.
It has been reported that the ALA content in transgenic rice seeds was increased up to 2.5- and 13-fold by introducing heterologous microsomal FAD3 genes driven by the CaMV 35S and Ubi-1 promoters, respectively (Shimada et al., 2000; Anai et al., 2003). These results raised the possibility that a higher ALA content in planta might be achieved by increasing the expression level of FAD3 in transgenic rice seeds, because, in general, the Ubi-1 promoter is stronger than the CaMV 35S promoter in cereal. Based on this hypothesis, the rice glutelin GluC promoter, which is much stronger than the Ubi-1 promoter in rice endosperm (Qu et al., 2008), was selected to express the soybean and rice FAD3 cDNA in the transformation. The ALA contents in endosperm of the GG and GO lines were ∼39.15% (P < 0.01) and 62.62% (P < 0.01) higher than those driven by the Ubi-1 promoter (Ubi-1-GmFAD3-1 and Ubi-1-OsFAD3), respectively (data not shown), confirming the advantage of the GluC promoter over Ubi-1 in this experimental system.
In addition to the chain length and degree of saturation, the distribution of fatty acids in TAGs has an important role in determining lipid nutrition by affecting the digestibility, absorbability, and metabolic ability of fatty acids (Carnielli et al., 1995). Fatty acids at the sn-2 position in TAGs were absorbed more effectively than those at the sn-1 and sn-3 positions. In GG and GO lines, ALA was mainly located at the sn-2 position (61.7±3.2% and 61.3±3.6%, respectively, Fig. 6) in TAGs. Thus, the high content of ALA in the transgenic rice endosperm can be sufficiently utilized by the human body, which could balance the low ratio of ω-3 to ω-6 from other foods.
Rice is usually consumed after polishing the outer layers and embryo. Because the FAD3 gene directed by the Ubi-1 promoter is mainly expressed in the subaleurone layer (Qu and Takaiwa, 2004) (data not shown), polishing will cause considerable loss of ALA. The GluC promoter led to specific expression and accumulation of protein within the whole endosperm rather than the outer layer of that tissue as with most of the other endosperm-specific promoters (Qu and Takaiwa, 2004; Qu et al., 2008). The expression patterns of OsFAD3 and GmFAD3-1 in whole endosperm of GO and GG lines (Fig. 3a) agreed well with previous studies of GUS used as the reporter gene (Qu et al., 2008). The ALA content in the polished rice seeds of the transgenic line accumulated up to 35-fold more ALA (9.50±0.20 mg g−1) than normal seeds (0.27±0.01 mg g−1), and the trait is stably heritable (Figs 4, 5). The ALA content in a meal-size portion of ‘ω-3 rice’ (∼200 g) would meet >86% of the daily adult ALA requirement (∼2.2 g of ALA) (Simopoulos, 2001). This achievement suggests that ‘ω-3 rice’ might contribute to the alleviation of human health problems caused by ALA deficiency.
It is reported that a high ALA content reduces the oxidative stability of oil (Feussner and Wasternakc, 2002); however, the ALA stored in seed is considered to be highly stable. Kolodziejczyk and Fedec (1995) stated that ALA in the intact seed of flax (with an ALA content of 17.5–26.5%) was remarkably resistant to oxidation and rancidity. The performances of high ALA-content rice seeds in storage, germination, and cooking will be investigated in future studies.
ALA is the precursor of LC-ω3-PUFAs, such as EPA and DHA. Although higher plants do not possess the enzymes to follow the elongation and desaturation steps to convert C18-PUFAs into very long-chain (VLC)-PUFAs, recent advances in plant metabolic engineering have made it possible to produce EPA and DHA in higher plants. Qi et al. (2004) reported the production of EPA in transgenic Arabidopsis leaves, accounting for ∼3% of total fatty acids. Wu et al. (2005) demonstrated the feasibility of making VLC-PUFAs in transgenic Brassica juncea, which resulted in the accumulation of EPA (8%) and DHA (0.2%). It is reasonable to expect to be able to produce EPA- and DHA-containing rice by synchronously overexpressing Δ6 and C20 elongases, and Δ6, Δ5, and Δ4 desaturases using these high ALA rice as hosts.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Subcellular localization of the OsFAD3/7/8 and GmFAD3-1/2/3 proteins.
Table S1. ALA content of transgenic rice seeds.
Acknowledgments
This work was supported by the National Program of Transgenic Variety Development of China (2011ZX08001-006), the National High Technology Research and Development Program (863) (2011AA100604), and the Natural Science Foundation of China (no. 30971562).
References
- Anai T, Koga M, Tanaka H, Kinoshita T, Rahman SM, Takagi Y. Improvement of rice (Oryza sativa L.) seed oil quality through introduction of a soybean microsomal ω-3 fatty acid desaturase gene. Plant Cell Reports. 2003;21:988–992. doi: 10.1007/s00299-003-0609-6. [DOI] [PubMed] [Google Scholar]
- Carnielli VP, Luijendijk IH, van Beek RH, Boerma GJ, Degenhart HJ, Sauer PJ. Effect of dietary triacylglycerol fatty acid positional distribution on plasma lipid classes and their fatty acid composition in preterm infants. American Journal of Clinical Nutrition. 1995;62:776–781. doi: 10.1093/ajcn/62.4.776. [DOI] [PubMed] [Google Scholar]
- Clemente TE, Cahoon EB. Soybean oil: genetic approaches for modification of functionality and total content. Plant Physiology. 2009;151:1030–1040. doi: 10.1104/pp.109.146282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damude HG, Zhang H, Farrall L, Ripp KG, Tomb JF, Hollerbach D, Yadav NS. Identification of bifunctional Δ12/ω-3 fatty acid desaturases for improving the ratio of ω-3 to ω-6 fatty acid in microbes and plants. Proceedings of the National Academy of Sciences, USA. 2006;103:9446–9451. doi: 10.1073/pnas.0511079103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert H, LaVallee B, Schweiger BJ, Kinney AJ, Cahoon EB, Clemente T. Co-expression of the borage Δ6 desaturase and the Arabidopsis Δ15 desaturase results in high accumulation of stearidonic acid in the seeds of transgenic soybean. Planta. 2006;224:1050–1057. doi: 10.1007/s00425-006-0291-3. [DOI] [PubMed] [Google Scholar]
- Feussner I, Wasternakc C. The lipoxygenase pathway. Annual Review of Plant Biology. 2002;53:275–297. doi: 10.1146/annurev.arplant.53.100301.135248. [DOI] [PubMed] [Google Scholar]
- James DW, Dooner HK. Isolation of EMS-induced mutants in Arabidopsis altered in seed fatty acid composition. Theoretical and Applied Genetics. 1990;80:241–245. doi: 10.1007/BF00224393. [DOI] [PubMed] [Google Scholar]
- Kankaanpaa P, Sutas Y, Salminen S, Lichtenstein A, Isolauri E. Dietary fatty acids and allergy. Annals of Medicine. 1999;31:282–287. doi: 10.3109/07853899908995891. [DOI] [PubMed] [Google Scholar]
- Kodama H, Akagi H, Kusumi K, Fujimura T, Iba K. Structure, chromosomal location and expression of a rice gene encoding the microsome ω-3 fatty acid desaturase. Plant Molecular Biology. 1997;33:493–502. doi: 10.1023/a:1005726210977. [DOI] [PubMed] [Google Scholar]
- Kolodziejczyk P, Fedec P. In: Processing flaxseed for human consumption. Flaxseed in human nutrition. Cunnane SC, Thompson LU, editors. Champaign, IL: AOCS Press; 1995. pp. 261–280. [Google Scholar]
- Lemieux B, Miquel M, Somerville C, Browse J. Mutants of Arabidopsis with alterations in seed lipid fatty acid composition. Theoretical and Applied Genetics. 1990;80:234–240. doi: 10.1007/BF00224392. [DOI] [PubMed] [Google Scholar]
- Liu WX, Liu HL, Chai ZJ, Xu XP, Song YR, Qu LQ. Evaluation of seed storage protein gene 5'-UTR in enhancing gene expression in transgenic rice seed. Theoretical and Applied Genetics. 2010;121:1267–1274. doi: 10.1007/s00122-010-1386-6. [DOI] [PubMed] [Google Scholar]
- Mattson FH, Beck LW. The digestion in vitro of triglycerides by pancreatic lipase. Journal of Biological Chemistry. 1954;214:115–125. [PubMed] [Google Scholar]
- Puttick D, Dauk M, Lozinsky S, Smith MA. Overexpression of a FAD3 desaturase increases synthesis of a polymethylene-interrupted dienoic fatty acid in seeds of Arabidopsis thaliana L. Lipids. 2009;44:753–757. doi: 10.1007/s11745-009-3315-5. [DOI] [PubMed] [Google Scholar]
- Qi BX, Fraser T, Mugford S, Dobson G, Sayanova O, Butler J, Napier JA, Stobart AK, Lazarus CM. Production of very long chain polyunsaturated omega-3 and omega-6 fatty acids in plants. Nature Biotechnology. 2004;22:739–745. doi: 10.1038/nbt972. [DOI] [PubMed] [Google Scholar]
- Qu LQ, Takaiwa F. Evaluation of tissue specificity and expression strength of rice seed component gene promoters in transgenic rice. Plant Biotechnology Journal. 2004;2:113–125. doi: 10.1111/j.1467-7652.2004.00055.x. [DOI] [PubMed] [Google Scholar]
- Qu LQ, Wei XL, Satoh H, Kumamaru T, Ogawa M, Takaiwa F. Inheritance of alleles for glutelin α-2 subunit genes in rice and identification of their corresponding cDNA clone. Theoretical and Applied Genetics. 2002;105:1099–1108. doi: 10.1007/s00122-002-1046-6. [DOI] [PubMed] [Google Scholar]
- Qu LQ, Xing YP, Liu WX, Xu XP, Song YR. Expression pattern and activity of six glutelin gene promoters in transgenic rice. Journal of Experimental Botany. 2008;59:2417–2424. doi: 10.1093/jxb/ern110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu LQ, Yoshihara T, Ooyama A, Goto F, Takaiwa F. Iron accumulation does not parallel the high expression level of ferritin in transgenic rice seeds. Planta. 2005;222:225–233. doi: 10.1007/s00425-005-1530-8. [DOI] [PubMed] [Google Scholar]
- Shimada T, Wakita Y, Otani M, Iba K. Modification of fatty acid composition in rice plants by transformation with a tobacco microsomal ω-3 fatty acid desaturase gene (NtFAD3) Plant Biotechnology. 2000;17:43–48. [Google Scholar]
- Simopoulos AP. Omega-3 fatty acids in health and disease and in growth and development. American Journal of Clinical Nutrition. 1991;54:438–463. doi: 10.1093/ajcn/54.3.438. [DOI] [PubMed] [Google Scholar]
- Simopoulos AP. N-3 fatty acids and human health, defining strategies for public policy. Lipids. 2001 doi: 10.1007/s11745-001-0687-7. 36 Suppl, S83–89. [DOI] [PubMed] [Google Scholar]
- Stark AH, Crawford MA, Reifen R. Update on α-linolenic acid. Nutrition Reviews. 2008;66:326–332. doi: 10.1111/j.1753-4887.2008.00040.x. [DOI] [PubMed] [Google Scholar]
- Vrinten P, Hu Z, Munchinsky MA, Rowland G, Qiu X. Two FAD3 desaturase genes control the level of linolenic acid in flax seed. Plant Physiology. 2005;139:79–87. doi: 10.1104/pp.105.064451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Truksa M, Datla N, Vrinten P, Bauer J, Zank T, Cirpus P, Heinz E, Qiu X. Stepwise engineering to produce high yields of very long-chain polyunsaturated fatty acids in plants. Nature Biotechnology. 2005;23:1013–1017. doi: 10.1038/nbt1107. [DOI] [PubMed] [Google Scholar]
- Yadav NS, Wierzbicki A, Aegerter M, et al. Cloning of higher plant ω-3 fatty acid desaturases. Plant Physiology. 1993;103:467–476. doi: 10.1104/pp.103.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.