Abstract
Salinity poses a major threat for agriculture worldwide. Rice is one of the major crops where most of the high-yielding cultivars are highly sensitive to salinity. Several studies on the genetic variability across rice cultivars suggest that the activity and composition of root plasma membrane transporters could underlie the observed cultivar-specific salinity tolerance in rice. In the current study, it was found that the salt-tolerant cultivar Pokkali maintains a higher K+/Na+ ratio compared with the salt-sensitive IR20 in roots as well as in shoots. Using Na+ reporter dyes, IR20 root protoplasts showed a much faster Na+ accumulation than Pokkali protoplasts. Membrane potential measurements showed that root cells exposed to Na+ in IR20 depolarized considerably further than those of Pokkali. These results suggest that IR20 has a larger plasma membrane Na+ conductance. To assess whether this could be due to different ion channel properties, root protoplasts from both Pokkali and IR20 rice cultivars were patch-clamped. Voltage-dependent K+ inward rectifiers, K+ outward rectifiers, and voltage-independent, non-selective channels with unitary conductances of around 35, 40, and 10 pS, respectively, were identified. Only the non-selective channel showed significant Na+ permeability. Intriguingly, in both cultivars, the activity of the K+ inward rectifier was drastically down-regulated after plant growth in salt but gating, conductance, and activity of all channel types were very similar for the two cultivars.
Keywords: Non-selective channel, patch clamp, salinity tolerance, rice, root protoplasts
Introduction
Soil salinity negatively impacts on agricultural production. Rice, one of the most important crops globally, is particularly affected since it is relatively salt-sensitive amongst cereals. As is the case with other glycophytes, rice responds to salt stress using a number of strategies that include minimizing influx, maintaining efflux, and translocation and compartmentation of potentially toxic ions such as Na+ and Cl- (Yeo and Flowers, 1982; Tester and Davenport, 2003; Anil et al., 2005, 2007; Kader and Lindberg, 2005; Kader et al., 2006; Hauser and Horie, 2010). To improve these faculties, and therefore rice salt tolerance, extensive breeding programmes are being carried out using the vast genetic variability that exists across rice cultivars. This genetic variation can also be employed to identify potential processes and molecular components that underlie cultivar (cv.)-specific responses to salt tolerance. For example, comparative studies using genome-wide microarrays show that, in both shoots (Walia et al., 2005) and in roots (Senadheera et al., 2009), various cv.-specific transcripts are induced in response to salt stress. At the whole plant level it appears that tolerant cvs limit Na+ uptake at the root soil boundary, retain more K+ throughout their tissues, and translocate less Na+ to photosynthesizing organs. Cellular approaches reveal that tolerant varieties maintain lower Na+ influx, accumulate less Na+ in their cytoplasm and possess a greater capacity for Na+ extrusion from the cytosol (Kader and Lindberg, 2005; Anil et al., 2007). Inhibitor profiles of Na+ uptake may also differ between cvs. For example, Kader and Lindberg (2005) suggested that Na+ influx in sensitive varieties may be mediated by a different set of ion channels and/or carriers; Na+ influx in root protoplasts from sensitive ecotypes was reduced in the presence of pharmaceuticals that block K+ channels and non-selective channels whereas influx in protoplasts from more tolerant rice was predominantly responsive to inhibitors of non-selective ion channels. In addition, and in agreement with whole plant studies, work with suspension cells points to a generally lower Na+ conductance in the plasma membranes of cells derived from tolerant cvs such as Pokkali (Anil et al., 2007).
In all, these data imply that the activity and composition of plasma membrane transporters in root cells is a key factor in determining overall Na+ tolerance but details on the exact mechanisms are extremely scarce, particularly in rice. Thus the identification of these systems is therefore of great use in understanding intercultivar differences and as a source for molecular handles to improve rice salt tolerance via molecular breeding and/or engineering. However, in spite of the enormous agricultural value of rice and its prominent status as a cereal model system, electrophysiological characterization of rice membranes is virtually absent.
In this study, Na+ reporter dyes and conventional electrophysiology were used and it was found that the Na+ permeability in root cell plasma membranes of the salt-tolerant Pokkali is lower than that of the more sensitive IR20. To establish if ion channels, and if so which ion channel type, could underlie this difference, the patch-clamp technique was applied to root protoplasts of Pokkali and IR20. Our extensive survey shows that root cells of both cvs contain a similar set of cation channels. Only one type shows significant Na+ conductance and in both cvs the activity of the K+ inward rectifier decreases after growth in the presence of NaCl.
Materials and methods
Plant growth
Seeds of two indica rice cultivars, Pokkali and IR20, were obtained from the University of Agricultural Sciences, Bangalore. Seeds were germinated and seedlings were transferred to hydroponic medium (1.25 mM KNO3, 0.5 mM Ca(NO3)2.4H2O, 0.5 mM MgSO4.7H2O, 42.5 μM FeNaEDTA, 0.625 mM KH2PO4, and 1.0×10−2 μM Cu2+, Zn2+, Mn2+, B3+, Mo2+, and Co2+ ) 10 d after sowing (DAS) and grown in controlled conditions at 22/19 °C day and night temperatures, 100 μmol m−2 s−1 of irradiance for 16 h d−1, and 40% relative humidity. For growth measurements, tissue ion content, and xylem sap composition, seedlings were grown in control conditions or exposed to salinity stress by adding 50 mM NaCl to the hydroponic solution at 15 DAS. Hydroponic solution was renewed every 4 d. Plants were harvested at different time intervals for analyses. To measure the relative growth rates (RGRs) of plants, a minimum of three plants from three independent replicates were randomly selected from the two treatments (control, 50 mM NaCl) at the beginning and end of the 10 d treatment. For monovalent cation analysis, tissue was collected at the end of treatment, washed with cold 20 mM LaCl3 solution for 2–5 min, dried at 80 °C for 48 h, and extracted in 5 ml of 20 mM LaCl3 for 24 h. Measurements were recorded using a flame photometer (Sherwood flame photometer-410 Cambridge, UK).
Xylem sap analysis
For xylem sap analyses, 4-week-old plants grown in the presence of 50 mM NaCl were used. Plants were cut 20 mm above the root:shoot junction and cut roots were mounted in a pressure chamber (EL540-300, Wagtech, Berkshire, UK). Pressure was applied which just exceeded the osmotic pressure of the external solution and exuding xylem sap was collected for 30 min from three plants and pooled. Na+ and K+ concentrations in xylem sap were measured by flame photometry. Three biological replicates were carried (using a total of nine plants for each cultivar).
Membrane potential recordings
Plants were grown as described above. Intact (4–5 cm) roots from 3–4-week-old seedlings were fixed on a stage as described previously (Carden et al., 2003). Roots were immersed in growth medium and individual cells impaled with 0.2 M KCl filled glass pipettes. Cortical cells were selected as those impaled after passing though the outer cell layer. During impalement, the bath solution was continuously refreshed and increasing salt was applied by changing the same solution with added NaCl to a stepwise increasing concentrations of 25, 50, 75 or 100 mM. Larger concentration changes of NaCl always resulted in lost electrode impalements (data not shown).
Quantitative real-time PCR
Total RNA was extracted from the roots of control-grown plants or plants exposed to 100 mM NaCl for 2 d and reverse transcribed using MMLV Reverse Transcriptase (Invitrogen Inc.) following the manufacturer’s instructions. Real-time PCR was carried out with a KAPA SYBR FAST qPCR Kit (KAPA Biosystems, Woburn, MA, USA) using Rotor-Gene 3000 (Corbett Life science). The primers used were: 5′-TCCATTGCTGACCTTGAAGA-3′ (forward) and 5′-ACACCAAAAACCACCCAAAA-3′ (reverse) for OsAKT1 and 5′-TTGGACTCTGGTGATGGTGT-3′ (forward) and 5′-GCCGTTGTGGTGAATGAG-3′ (reverse) for Actin-1. Data analysis was done using rotor gene 6.1 software.
Protoplast isolation
Protoplasts were isolated from the roots of 5–7-d-old plants grown in control conditions or exposed to 100 mM NaCl for 2 d. The enzyme solution contained 1.5% (w/v) cellulase (Onozuka R-10), 0.5% macerozyme (Onozuka R-10), 0.5% hemicellulase (Sigma), 0.1% bovine serum albumin, 0.05% polyvinylpyrrolidone, 1% (v/v) pectinase (Sigma), 10 mM CaCl2, and 10 mM MES/TRIS, pH 5.7. The osmolarity of the enzyme solution was adjusted to 300 mosmol kg−1 using D-sorbitol. Roots were incubated in enzyme solution at 30 °C for 1 h in a shaking water bath. The released protoplasts were filtered through 50 μm nylon mesh and washed in 2 mM CaCl2 buffer (osmolarity, 400 mosmol kg−1, pH 5.6) by centrifugation (10 min at 500 rpm at room temperature). The protoplasts were finally suspended in holding buffer which contained (in mM): 5 KCl, 2 CaCl2, 1 MgCl2, 10 sucrose, 10 glucose, and 10 MES/TRIS, pH 5.7. The osmolarity of the holding buffer was adjusted to 400 mosmol kg−1 using D-sorbitol. The protoplast suspension was stored on ice and aliquots used for patch-clamp measurement.
Patch-clamp protocols and data analysis
Measurements were performed in cell-attached and excised patch mode using a D6100 patch-clamp amplifier (List-Medical-Electronic, Darmstadt, Germany). Data were low-pass-filtered with an eight pole Bessel filter with a cut-off frequency of 1 kHz and sampled at 2.5 times the filter frequency. Data acquisition and analyses were done using Clampex and Clampfit 9.0.2.03 software (Axon instruments Inc. Union city, CA). Patch pipettes were prepared from Kimax-51 glass capillaries (Kimble products, Vineland, NY, USA). Pipette solutions contained (in mM): 100 KCl, 1 MgCl2, 10 MES/TRIS (pH 5.6). The standard external solutions contained (in mM): 10 KCl, 10 CaCl2, 1 MgCl2, and 10 MES/TRIS (pH 5.6). The osmolarity of the solutions was adjusted to 300 mosmol kg−1 using D-sorbitol.
Na+ dye recordings
Acetoxymethyl (AM) ester of CoroNa Green (Molecular Probes, Inc., Eugene, OR) at 10 μM was loaded into rice root protoplasts suspended in dye loading buffer that contained (in mM): 4 KCl, 1 MgCl2, 0.1 CaCl2, 10 MES/TRIS (pH 5.6), osmolarity adjusted to 400 mosmol kg−1 with D-sorbitol. Dye loading was carried out in the dark at 37 °C for 30 min in the presence of 600 μM Eserine. Unincorporated dye was washed off with wash buffer that contained (in mM): 4 KCl, 1 MgCl2, 1 CaCl2, 10 MES/TRIS (pH 5.6), and the osmolarity was adjusted to 300 mosmol kg−1 with D-sorbitol. The dye loaded protoplasts were re-suspended in holding buffer and were made to settle on coverslips coated with poly-D-lysine for microscopy.
Confocal microscopy was carried out on an LSM 510 meta system (Carl Zeiss, Jena, Germany) using the argon laser at 488 nm wavelength. The laser beam was focused on to the sample with a ×40, 1.4 numerical aperture (NA) oil immersion objective using a primary dichroic beam splitter (HFT 488), and the emitted fluorescence was collected through a secondary dichroic LP490 and 525±25 nm bandpass filter. Corona green intensities were determined before (control fluorescence level) and after 2 min exposure to various NaCl concentration in individual protoplasts using ImageJ 1.43m (National Institute of Health, USA).
Results and discussion
Pokkali rice is more salt tolerant than IR20
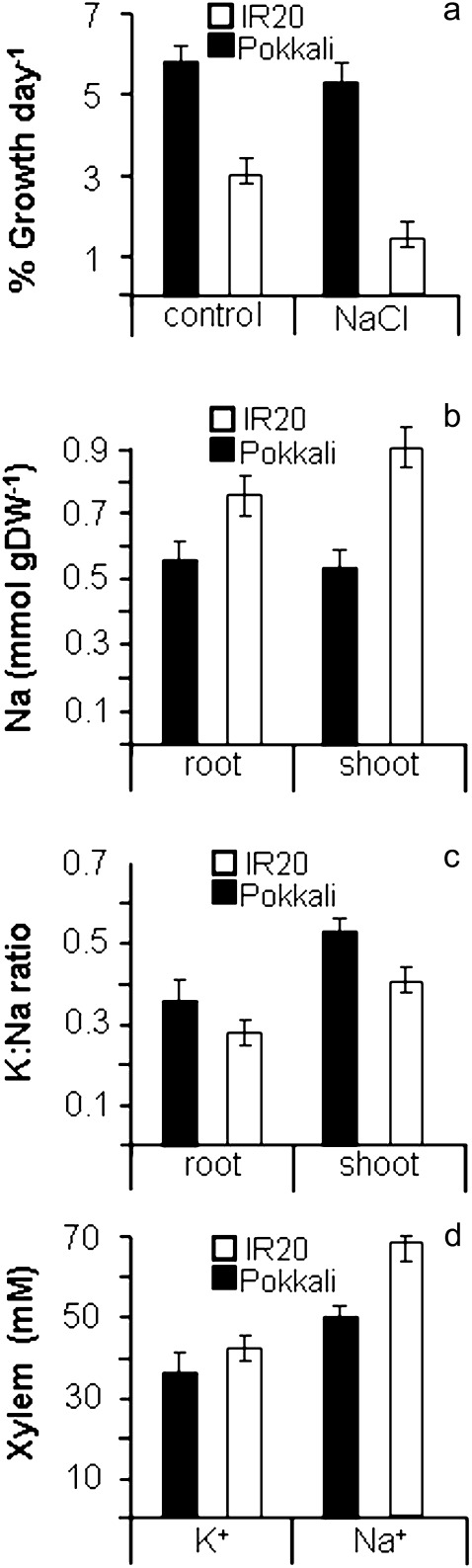
Figure 1a shows relative growth rates (RGRs) for Pokkali and IR20 rice in the absence and presence of 50 mM NaCl in 3–6-week-old plants. Pokkali growth in general is more vigorous and its RGR is hardly reduced in the presence of 50 mM NaCl. By contrast, the RGR of IR20 approximately halves when plants are exposed to salt, demonstrating a significant degree of sensitivity toward salt compared with Pokkali. Rice salt sensitivity greatly depends on growth stage (Walia et al., 2005) but superior Pokkali growth in the presence of NaCl was also evident at other growth stages and also using different NaCl concentrations (data not shown).
Fig. 1.
Relative growth rates and ion concentrations. (a) Relative growth rates (RGRs) for Pokkali and IR20 in the presence and absence of 50 mM NaCl. (b) Na+ contents in root and shoot tissue of Pokkali and IR20 grown in the presence of 50 mM NaCl for 4 d. (c) K+:Na+ ratios for Pokkali and IR20 root and shoot tissue. (d) K+ and Na+ concentrations in the xylem sap of salt-grown Pokkali and IR 20.
Pokkali maintains a higher K+:Na+ ratio than IR20
Many reports have alluded to the beneficial effects of high K+:Na+ ratios with regard to salt tolerance (Maathuis and Amtmann, 1999; Asch et al., 2000; Shabala et al., 2010). For rice, comparative studies have shown that tissue levels of K+ and Na+ show distinctly different patterns in salt-tolerant and -sensitive cvs with tolerant rice cvs such as FL487 maintaining K+:Na+ ratios that are around twice those found in IR29, a variety that is very similar to IR20 (Walia et al., 2005; Senadheera et al., 2009). Root and shoot Na+ levels were considerably higher in IR20 than Pokkali, particularly in shoot tissue (Fig. 1b) whereas K+ levels were comparable (data not shown). In roots, Pokkali thus shows a higher K+:Na+ ratio although this is only marginally so (Fig. 1c). However, in leaves, the Pokkali K+:Na+ ratio is considerably higher than in IR20 (Fig. 1c). To investigate the potential cause of the lower Na+ level in Pokkali shoot tissue, the xylem sap was analysed for K+ and Na+ concentrations. Figure 1d shows that, during salinization, xylem K+ levels are not significantly different between the cvs. However, as previously reported by Krishnamurthy et al. (2011), it was found that the Na+ concentration in IR20 xylem sap is around 40% higher than that in Pokkali when plants are grown in saline conditions. In rice, some Na+ enters the plant apoplastically via the bypass flow (Gong et al., 2006) and it has been shown that part of the intercultivar variation in tolerance may be due to differences in bypass conductance (Krishnamurthy et al., 2011). However, at higher NaCl concentrations, the majority of Na+ (70–90%; Faiyue et al., 2010) enters the rice root symplast and the higher level of Na+ in IR20 xylem sap implies that the net Na+ uptake flux of IR20 at the root soil boundary is considerably larger than that in Pokkali.
Root protoplast from IR20 show rapid Na+ accumulation
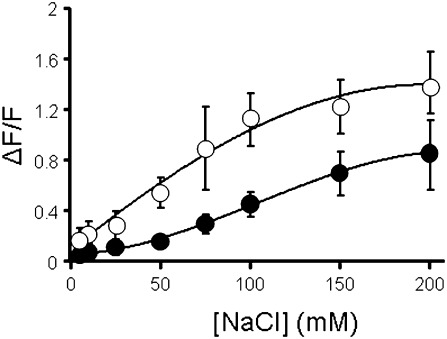
To establish whether the above-described differences in tolerance might originate in membrane transport properties at the cellular level, the Na+ reporter dye CoroNa green (Meier et al., 2006) was used. After dye loading, protoplasts were exposed to a range of [NaCl] and changes in fluorescence relative to the zero NaCl condition were recorded after 2 min. This short time span ensures that the Na+ flux is essentially unidirectional. Figure 2 shows that the change in fluorescence in IR20 protoplasts was consistently greater than that observed in Pokkali protoplasts. Although these data are comparative only, they clearly show that cellular Na+ concentrations in IR20 protoplasts are considerably higher than in Pokkali protoplasts, when monitored after 2 min exposure to NaCl. In turn, this points to a considerably higher Na+ conductance of the plasma membrane of IR20 root cells relative to its Pokkali counterpart.
Fig. 2.
CoroNa Green recordings in root protoplasts. Root protoplasts isolated from control grown rice seedlings from IR20 and Pokkali were loaded with the Na-reporter dye CoroNa Green. CoroNa Green fluorescence was recorded in individual protoplasts at 0 NaCl and after a 2 min exposure to increasing concentration of NaCl. Data shown are average and SD for 5–7 protoplasts of each cultivar.
Root cells of IR20 depolarize more in response to Na+
Membrane potentials of root cells were measured by electrode impalement with complete growth medium solutions bathing the roots. After reaching stable values, roots were exposed to stepwise increasing concentrations of NaCl (25–100 mM) to record changes in membrane potential (see Supplementary Fig. S1 at JXB online). The membrane potential (Em) of Pokkali root cells was considerably more negative than that of IR20 (Table 1). This means that the driving force for uptake of cations such as Na+ and K+ is considerably larger in the Pokkali cultivar. In both cultivars, the steady-state Em in the presence of increased medium NaCl (up to 100 mM;) became more positive but more so in IR20 (Table 1). The magnitude of the initial depolarization, which reflects the Na+ membrane permeability (Maathuis et al., 1996; Walker et al., 1996) was much smaller in Pokkali. Thus, despite a more negative resting potential in Pokkali which increases the driving force for Na+ root influx, there is less Na+ accumulation in this cultivar.
Table 1.
Membrane potentials in root cells of Pokkali and IR20
| Pokkali | IR20 | |
| Growth medium | –117.5±13.3 (n=11) | –53.3±5.2 (n=20) |
| ΔmV (25 NaCl) | 8.0±1.4 (n=6) | 18.4±2.1 (n=5) |
Rice plants were grown in control medium and membrane potentials and NaCl-induced depolarizations were measured in cortical cells of intact seedlings with their roots immersed in full growth medium or medium supplemented with 25 mM. Data are in mV from 3–5 plants for each cultivar and given as means across 5–20 roots ±SD.
Patch-clamp studies show similar cation channels in root cells from Pokkali and IR20
The previous data, derived using different methods, show that Na+ accumulation and uptake in Pokkali are reduced compared with IR20 and that the plasma membrane Na+ conductance of Pokkali root cells is lower than that of IR20 cells. Many different types of transporter are likely to contribute to the plasma membrane Na+ conductance, but the molecular identity of the predominant Na+ uptake pathways in plants is still unclear. Nevertheless, several studies have provided evidence that non-selective ion channels are a main conduit for Na+ uptake in plants (see Demidchik and Maathuis, 2007, for a review). In addition, HKT proteins have been implicated as contributing significantly to Na+ uptake (Uozumi et al., 2000; Horie et al., 2007; Hauser and Horie, 2010). To establish whether the observed lower conductance in Pokkali roots is due to the presence of different ion channels that allow Na+ movement across the membranes, the patch-clamp technique was applied to root cells from Pokkali and IR20 grown in control conditions or exposed to 100 mM NaCl. In these assays, the cell attached mode was primarily used which, unlike excised patch or whole cell recordings, retains the intact cellular machinery and thus potential regulatory factors that impact on channel activity. The cell attached configuration is, therefore, more physiologically relevant and more appropriate to compare channel properties between cultivars.
Figure 3 shows typical traces obtained in the cell attached configuration for Pokkali and IR20. An inward rectifying channel was observed that displays steep voltage dependence in both Pokkali and IR20 (Fig. 3a). This channel has a unitary conductance of around 35 pS and 33 pS in Pokkali and IR20, respectively (Table 2) in the presence of 100 mM KCl in the pipette and bath. The voltage dependence and single channel conductance are similar to the OsAKT1-mediated currents described by Fuchs et al. (2005) and, therefore, henceforth are referred to as OsAKT1-like channels. A voltage dependent conductance that was outward rectifying was also recorded in cell-attached patches (Fig. 3b) with a single channel conductance of around 33 pS in Pokkali and 44 pS in IR20 (Table 2). A third conductance (Fig. 3c) showed far more noisy current traces and no or very little voltage dependence. Amplitude histograms (see Supplementary Fig. S2 at JXB online) suggest that inward and outward unitary conductance of this instantaneously activating channel is around 9 pS in Pokkali and 8 pS in IR20 with 100 KCl symmetrical conditions (Table 2).
Fig. 3.
Single channel recordings. Single-channel activity in cell-attached patches from Pokkali (left hand panels) and IR20 (right hand panels) root protoplasts of (a) potassium inward rectifier; (b) potassium outward rectifier; (c) non-selective cation channel. Step potentials are indicated to the right of the recorded current traces. Closed levels are indicated by the arrows. Patch pipette contained 100 mM KCl.
Table 2.
Unitary conductance of cation channels in cell attached rice protoplasts from Pokkali and IR20
| Inward conductance (pS) | Outward conductance (pS) | Voltage-independent conductance (K+/Na+) (pS) | |
| Pokkali(con) | 35.4±3.8 (n=5) | 33.2±9.6 (n=4) | 8.9±0.6 (n=5)/10.3±1.5 (n=6) |
| Pokkali(NaCl) | – | n.d. | 9.8±0.7 (n=4)/10.9±1.2 (n=5) |
| IR(con) | 33.2±5.3 (n=3) | 43.9 (n=3) | 9.7±2.0 (n=5)/10.2±2.0 (n=6) |
| IR(NaCl) | – | n.d. | 12.2±0.5 (n=5)/10.2±1.8 (n=5) |
Unitary conductances were derived from current voltage relationships for various conductances recorded in cell-attached patches from control (con) and salt-grown (NaCl) rice plants. n.d., Not determined.
To establish whether any of these channels is likely to contribute to Na+ uptake, their Na+ conductance was tested, reversal potentials were determined (Table 3), and the number of times activity was present was scored (Table 4). Using a large number of recordings where KCl was replaced with NaCl in the pipette, OsAKT1-like currents were never observed in either cv. in 38 independent cell-attached recordings (Table 2). This strongly suggests that this channel conducts K+ but not Na+ as has previously been described for other inward-rectifying channels in plant protoplasts (Schroeder et al., 1987; Bertl et al., 1995). Similarly, the exchange of KCl for NaCl in the bath after obtaining inside out excised patches always resulted in a complete cessation of outward single channel currents. This indicates that either gating of the outward rectifying channel is inhibited by cytoplasmic Na+ or that this channel has a negligible Na+ conductance. Evidence for the latter also stems from the reversal potentials of this channel measured in the presence of Na+ and K+ (see Supplementary Table S1 at JXB online). By contrast, the voltage-independent channel occurred as frequently with NaCl in the pipette as when KCl was used and showed an inward unitary conductance that was very similar to that recorded with KCl in the pipette (Table 2). Reversal potentials (Table 3) also show that Na+ and K+ permeability are similar and together these data suggest the voltage-independent channel does not discriminate between K+ and Na+. This conductance thus displays the hallmarks of voltage-independent non-selective cation channels (Demidchik and Maathuis, 2007) that have been characterized in other species such as Arabidopsis (Pei et al., 1998), wheat (Tyerman et al., 1997), and barley (Amtmann et al., 1997) and proposed to form a major conduit for Na+ uptake (Amtmann and Sanders, 1999; Demidchik and Tester, 2002).
Table 3.
Reversal potentials of voltage-independent ion channels in protoplasts from Pokkali and IR20
| 100/100 K/K | 100/10 K/K | 100/100 Na/K | 100/10 Na/K | |
| IR20 | –2.6±6.0 | 44.5±7.8 | 2.5±2.8 | 44.0±3.6 |
| Pokkali | 1.3±2.5 | 42.4±8.2 | 1.00±3.7 | 43.0±2.6 |
Reversal potentials were calculated from current–voltage relationships in excised inside out patches from Pokkali and IR20 root protoplasts. Pipette and bath ionic conditions are given as millimolar K and Na. Data show averages ±SD for 3–4 independent experiments.
Table 4.
Frequency of occurrence for different channel types in cell-attached patches of Pokkali and IR20
| Inward conductance | Outward conductance | Voltage-independent conductance | Total number of patches | |
| Pokkali(con) | 16% | 11% | 76% | 38 |
| Pokkali(NaCl) | 0% | n.d. | 75% | 16 |
| IR20(con) | 12% | 5% | 78% | 39 |
| IR20(NaCl) | 0% | n.d. | 77% | 26 |
The occurrence of inward, outward, and voltage-independent conductances with unitary conductances listed in Table 2 was determined for each cell-attached patch from control (con) and salt-grown (NaCl) plants. n.d., Not determined.
Salinity depresses inward K+ channel activity in both Pokkali and IR20
The above results show the presence of very similar cation channels in both cvs. To acquire an estimate of how often the respective conductances appear, and therefore how much they would contribute to K+ and Na+ fluxes, the frequency of occurrence in cell-attached recordings was calculated. Table 2 shows that, in control-grown plants, the non-selective conductance is observed most often whereas the inward current is only evident in about one out of six or seven protoplasts. It also suggests that the distribution of cation channels is very similar in the two cvs. However, the composition and activity of ion channels can be altered by growth conditions (Murata et al., 1994; Su et al., 2001; Fuchs et al., 2005; Demidchik and Maathuis, 2007) and therefore cell-attached recordings were also analysed in root protoplasts derived from salt-grown plants. Out of 16 recordings no OsAKT1-like current was observed in root protoplasts from salt-grown Pokkali (Table 2). In salt-grown IR20 root protoplasts, an OsAKT1-like current was also not observed (n=26 independent cells). These frequencies are significantly lower (P <0.05) than those in control plants and show that the activity of OsAKT1-like channels is drastically reduced in plants exposed to salt.
One earlier study showed that OsAKT1 was transcriptionally down-regulated in response to salt stress in the tolerant cvs Pokkali and BK but not in the sensitive cv. IR29 (Golldack et al., 2003). Therefore, quantitative RT-PCR analyses were carried out on root tissue of control and salt-grown Pokkali and IR20 (Fig. 4). Our data corroborate earlier ones, showing about a 7-fold reduction in OsAKT1 transcript levels in Pokkali. However, in contrast to earlier work, a similar reduction in OsAKT1 transcript level was observed in roots derived from the sensitive cultivar IR20. The reduction in observed transcript levels could limit the activity of the AKT1-like channel. Patch-clamp recordings by Fuchs et al. (2005) also showed reduced inward current in protoplasts from the salt-grown Nihonmasari japonica cultivar which is relatively salt tolerant. Our data support those of Fuchs et al. (2005) but suggest that the reduced activity of an OsAKT1-like current is not specific for tolerant varieties or japonica subspecies, but is equally pronounced in the indica salt-sensitive IR20.
Fig. 4.
Expression analysis of OsAKT1. (a) OsAKT1 transcript levels in roots of control (con) and 100 mM NaCl (NaCl) treated plants for Pokkali and IR20 rice cultivars. Relative transcript levels were normalized with respect to actin and show averages and SD for three biological replicates.
Conclusions
The relative salt tolerance of Pokkali is probably based on a number of phenomena which includes a lower plasma membrane Na+ conductance of root cells. Our patch-clamp recordings show the presence of three cation conductances of which only one has Na+ permeability. These non-selective channels have comparable unitary conductance and frequency of occurrence in the two cvs, irrespective of the growth conditions. Patch clamp recordings were made on protoplasts derived from various tissues. Therefore, it cannot be ruled out that the tissue distribution of various ion channels in Pokkali is different from that in IR20 and that this could impact on overall Na+ uptake. However, the simplest interpretation of our findings is that the observed lower overall Na+ conductance in Pokkali roots is not caused by reduced activity of non-selective ion channels. Indeed, it appears that the differences in tolerance and Na+ uptake between Pokkali and IR20 do not originate in distinct ion channel properties. This is in contrast to comparative studies on Arabidopsis thaliana and Thellungiella halophila which showed a higher K+/Na+ selectivity ratio of non-selective ion channels in T. halophila (Volkov and Amtmann, 2006) and it was argued that this property could form the basis for the higher level of tolerance in T. halophila.
Our approach does not allow us to assess what the contribution of non-selective channels is to the overall Na+ influx. However, with similar properties in Pokkali and IR20, the non-selective channels must conduct a larger Na+ influx in Pokkali because its root cells are more hyperpolarized. The observation that the total Na+ influx in Pokkali is smaller indicates that pathways other than non-selective channels are dominant. Thus, variation in the activity of other rice plasma membrane transporters that are known to mediate Na+ uptake may therefore be more relevant as an explanation for the difference in salt sensitivity between Pokkali and IR20. In this respect, members of the HKT family particularly spring to mind since these have been shown to contribute to japonica rice Na+ uptake (Horie et al., 2007) and transcriptomics studies suggest they are differentially regulated in sensitive and tolerant cvs during salt stress (Kader et al., 2006).
Our study also provides direct evidence that it is extremely unlikely that K+ selective channels contribute significantly to Na+ uptake, as suggested on the basis of inhibitor profiles of Na+ influx for Suaeda maritima (Kader and Lindberg, 2005; Wang et al., 2007) and rice (Kader and Lindberg, 2005; Wang et al., 2007). Indeed, the role of K+ selective channels in K+ uptake during salinity is unclear and it is tempting to assign a considerable proportion of root K+ uptake to OsAKT1, as has been shown for AKT1 homologues in other species (Hirsch et al., 1998). An intriguing outcome of our experiments and those of others (Fuchs et al., 2005) is the drastic reduction in OsAKT1-like activity after salt exposure. Salinity often compromises adequate K+ nutrition and a reduction in uptake capacity in these conditions would therefore appear to be counterproductive. However other functions, such as maintaining membrane polarization or greater kinetic control, could require the down-regulation of OsAKT1-like channels in saline conditions.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Fig. 1. Example of a membrane potential recording of a Pokkali root cell (top) and an IR20 root cell (bottom).
Supplementary Fig. 2. Representative amplitude histogram for Pokkali (a) and IR20 (b) of instantaneous activating channels recorded in excised, inside out patches.
Acknowledgments
We acknowledge the financial support from the British Council (RXP award 2008–2009) and the Royal Society, UK (ref JP0871438).
References
- Amtmann A, Laurie S, Leigh R, Sanders D. Multiple inward channels provide flexibility in Na+/K+ discrimination at the plasma membrane of barley suspension culture cells. Journal of Experimental Botany. 1997;48:481–497. doi: 10.1093/jxb/48.Special_Issue.481. [DOI] [PubMed] [Google Scholar]
- Amtmann A, Sanders D. Mechanisms of Na+ uptake by plant cell. Advances in Botanical Research. 1999;29:75–112. [Google Scholar]
- Anil VS, Krishnamurthy H, Mathew MK. Limiting cytosolic Na+ confers salt tolerance to rice cells in culture: a two-photon microscopy study of SBFI-loaded cells. Physiologia Plantarum. 2007;129:607–621. [Google Scholar]
- Anil VS, Krishnamurthy P, Kuruvilla S, Sucharitha K, Thomas G, Mathew MK. Regulation of the uptake and distribution of Na+ in shoots of rice (Oryza sativa) variety Pokkali: role of Ca2+ in salt tolerance response. Physiologia Plantarum. 2005;124:451–464. [Google Scholar]
- Asch F, Dingkuhn M, Dörffling K, Miezan K. Leaf K/Na ratio predicts salinity induced yield loss in irrigated rice. Euphytica. 2000;113:109–118. [Google Scholar]
- Bertl A, Anderson JA, Slayman CL, Gaber RF. Use of Saccharomyces cerevisiae for patch-clamp analysis of heterologous membrane proteins: characterization of Kat1, an inward-rectifying K+ channel from Arabidopsis thaliana, and comparison with endogenous yeast channels and carriers. Proceedings of the National Academy of Sciences, USA. 1995;92:2701–2705. doi: 10.1073/pnas.92.7.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carden DE, Walker DJ, Flowers TJ, Miller AJ. Single-cell measurements of the contributions of cytosolic Na+ and K+ to salt tolerance. Plant Physiology. 2003;131:676–683. doi: 10.1104/pp.011445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidchik V, Maathuis FJ. Physiological roles of nonselective cation channels in plants: from salt stress to signalling and development. New Phytologist. 2007;175:387–404. doi: 10.1111/j.1469-8137.2007.02128.x. [DOI] [PubMed] [Google Scholar]
- Demidchik V, Tester M. Sodium fluxes through nonselective cation channels in the plasma membrane of protoplasts from Arabidopsis roots. Plant Physiology. 2002;128:379–387. doi: 10.1104/pp.010524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faiyue B, Vijayalakshmi C, Nawaz S, Nagato Y, Taketa S, Ichii M, Al-Azzawi M, Flowers TJ. Studies on sodium bypass flow in lateral rootless mutants lrt1 and lrt2, and crown rootless mutant crl1 of rice (Oryza sativa L.) Plant, Cell and Environment. 2010;33:687–701. doi: 10.1111/j.1365-3040.2009.02077.x. [DOI] [PubMed] [Google Scholar]
- Fuchs I, Stolzle S, Ivashikina N, Hedrich R. Rice K+ uptake channel OsAKT1 is sensitive to salt stress. Planta. 2005;221:212–221. doi: 10.1007/s00425-004-1437-9. [DOI] [PubMed] [Google Scholar]
- Golldack D, Quigley F, Michalowski CB, Kamasani UR, Bohnert HJ. Salinity stress-tolerant and -sensitive rice (Oryza sativa L.) regulate AKT1-type potassium channel transcripts differently. Plant Molecular Biology. 2003;51:71–81. doi: 10.1023/a:1020763218045. [DOI] [PubMed] [Google Scholar]
- Gong HJ, Randall DP, Flowers TJ. Silicon deposition in the root reduces sodium uptake in rice (Oryza sativa L.) seedlings by reducing bypass flow. Plant, Cell and Environment. 2006;29:1970–1979. doi: 10.1111/j.1365-3040.2006.01572.x. [DOI] [PubMed] [Google Scholar]
- Hauser F, Horie T. A conserved primary salt tolerance mechanism mediated by HKT transporters: a mechanism for sodium exclusion and maintenance of high K+/Na+ ratio in leaves during salinity stress. Plant, Cell and Environment. 2010;33:552–565. doi: 10.1111/j.1365-3040.2009.02056.x. [DOI] [PubMed] [Google Scholar]
- Hirsch RE, Lewis BD, Spalding EP, Sussman MR. A role for the AKT1 potassium channel in plant nutrition. Science. 1998;280:918–921. doi: 10.1126/science.280.5365.918. [DOI] [PubMed] [Google Scholar]
- Horie T, Costa A, Kim TH, Han MJ, Horie R, Leung HY, Miyao A, Hirochika H, An G, Schroeder JI. Rice OsHKT2;1 transporter mediates large Na+ influx component into K+-starved roots for growth. EMBO Journal. 2007;26:3003–3014. doi: 10.1038/sj.emboj.7601732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kader MA, Lindberg S. Uptake of sodium in protoplasts of salt-sensitive and salt-tolerant cultivars of rice, Oryza sativa L. determined by the fluorescent dye SBFI. Journal of Experimental Botany. 2005;56:3149–3158. doi: 10.1093/jxb/eri312. [DOI] [PubMed] [Google Scholar]
- Kader MA, Seidel T, Golldack D, Lindberg S. Expressions of OsHKT1, OsHKT2, and OsVHA are differentially regulated under NaCl stress in salt-sensitive and salt-tolerant rice (Oryza sativa L.) cultivars. Journal of Experimental Botany. 2006;57:4257–4268. doi: 10.1093/jxb/erl199. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy P, Ranathunge R, Nayak S, Schreiber L, Mathew MK. Root barriers block Na+ traffic to shoots in rice (Oryza sativa L.) Journal of Experimental Botany. 2011;62:4215–4228. doi: 10.1093/jxb/err135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maathuis F, Verlin D, Smith FA, Sanders D, Fernandez JA, Walker NA. The physiological relevance of Na+-coupled K+-transport. Plant Physiology. 1996;112:1609–1616. doi: 10.1104/pp.112.4.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maathuis FJM, Amtmann A. K+ nutrition and Na+ toxicity: the basis of cellular K+/Na+ ratios. Annals of Botany. 1999;84:123–133. [Google Scholar]
- Meier SD, Kovalchuk Y, Christine R, Rose CR. Properties of the new fluorescent Na+ indicator CoroNa Green: comparison with SBFI and confocal Na+ imaging. Journal of Neuroscience Methods. 2006;155:251–259. doi: 10.1016/j.jneumeth.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Murata Y, Obi I, Yoshihashi M, Noguchi M, Kakutani T. Reduced permeability to K+ and Na+ ions of K+ channels in the plasma membrane of tobacco cells in suspension after adaptation to 50 mM NaCl. Plant and Cell Physiology. 1994;35:87–92. [Google Scholar]
- Pei ZM, Schroeder JI, Schwarz M. Background ion channel activities in Arabidopsis guard cells and review of ion channel regulation by protein phosphorylation events. Journal of Experimental Botany. 1998;49:319–328. [Google Scholar]
- Schroeder JI, Raschke K, Neher E. Voltage dependence of K channels in guard-cell protoplasts. Proceedings of the National Academy of Sciences, USA. 1987;84:4108–4112. doi: 10.1073/pnas.84.12.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senadheera P, Singh RK, Maathuis FJ. Differentially expressed membrane transporters in rice roots may contribute to cultivar dependent salt tolerance. Journal of Experimental Botany. 2009;60:2553–2563. doi: 10.1093/jxb/erp099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabala S, Cuin TA, Pang J, Percey W, Chen Z, Conn S, Eing C, Wegner LH. Xylem ionic relations and salinity tolerance in barley. The Plant Journal. 2010;61:839–853. doi: 10.1111/j.1365-313X.2009.04110.x. [DOI] [PubMed] [Google Scholar]
- Su H, Golldack D, Katsuhara M, Zhao C, Bohnert HJ. Expression and stress-dependent induction of potassium channel transcripts in the common ice plant. Plant Physiology. 2001;125:604–614. doi: 10.1104/pp.125.2.604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tester M, Davenport R. Na+ tolerance and Na+ transport in higher plants. Annals of Botany. 2003;91:503–527. doi: 10.1093/aob/mcg058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyerman SD, Skerrett M, Garill A, Findlay GP, Leigh R. Pathways for the permeation of Na+ and Cl– into protoplasts derived from the cortex of wheat roots. Journal of Experimental Botany. 1997;48:459–480. doi: 10.1093/jxb/48.Special_Issue.459. [DOI] [PubMed] [Google Scholar]
- Uozumi N, Kim EJ, Rubio F, Yamaguchi T, Muto S, Tsuboi A, Bakker EP, Nakamura T, Schroeder JI. The Arabidopsis HKT1 gene homolog mediates inward Na+ currents in Xenopus laevis oocytes and Na+ uptake in Saccharomyces cerevisiae. Plant Physiology. 2000;122:1249–1259. doi: 10.1104/pp.122.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkov V, Amtmann A. Thellungiella halophila, a salt-tolerant relative of Arabidopsis thaliana, has specific root ion-channel features supporting K+/Na+ homeostasis under salinity stress. The Plant Journal. 2006;48:342–353. doi: 10.1111/j.1365-313X.2006.02876.x. [DOI] [PubMed] [Google Scholar]
- Walia H, Wilson C, Condamine P, Liu X, Ismail AM, Zeng L, Wanamaker SI, Mandal J, Xu J, Cui X, Close TJ. Comparative transcriptional profiling of two contrasting rice genotypes under salinity stress during the vegetative growth stage. Plant Physiology. 2005;139:822–835. doi: 10.1104/pp.105.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker NA, Sanders D, Maathuis FJM. High affinity potassium uptake by plants. Science. 1996;373:977–978. doi: 10.1126/science.273.5277.977. [DOI] [PubMed] [Google Scholar]
- Wang SM, Zhang JL, Flowers TJ. Low-affinity Na+ uptake in the halophyte Suaeda maritima. Plant Physiology. 2007;145:559–571. doi: 10.1104/pp.107.104315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo AR, Flowers TJ. Accumulation and localization of sodium ions within the shoots of rice (Oryza sativa L.) varieties differing in salinity resistance. Physiologia Plantarum. 1982;56:343–348. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.