Abstract
HD2 proteins are plant specific histone deacetylases. Four HD2 proteins, HD2A, HD2B, HD2C, and HD2D, have been identified in Arabidopsis. It was found that the expression of HD2A, HD2B, HD2C, and HD2D was repressed by ABA and NaCl. To investigate the function of HD2 proteins further, two HD2C T-DNA insertion lines of Arabidopsis, hd2c-1 and hd2c-3 were identified. Compared with wild-type plants, hd2c-1 and hd2c-3 plants displayed increased sensitivity to ABA and NaCl during germination and decreased tolerance to salt stress. These observations support a role of HD2C in the ABA and salt-stress response in Arabidopsis. Moreover, it was demonstrated that HD2C interacted physically with a RPD3-type histone deacetylase, HDA6, and bound to histone H3. The expression of ABA-responsive genes, ABI1 and ABI2, was increased in hda6, hd2c, and hda6/hd2c-1 double mutant plants, which was associated with increased histone H3K9K14 acetylation and decreased histone H3K9 dimethylation. Taken together, our results suggested that HD2C functionally associates with HDA6 and regulates gene expression through histone modifications.
Keywords: ABA, Arabidopsis, HD2C, HDA6, histone deacetylases
Introduction
In eukaryotes, genomic DNA is tightly compacted into a complex structure known as chromatin. To facilitate cellular activities, the accessibility of chromatin is dynamically regulated during growth and development (Berger, 2002). Two types of chromatin modification that correlate with either positive or negative transcriptional states are DNA methylation and histone post-translational modifications. The amino-terminal tails of core histones protruding from the nucleosomes interact with DNA and thereby facilitate the chromatin assembly via post-translational modifications including acetylation, phosphorylation, methylation, ubiquitination, and sumoylation. Histone acetylation and deacetylation are catalysed by histone acetyltransferases and histone deacetylases (HDAs), respectively. The acetylation of conserved lysine residues neutralizes the positive charge of the histone tails and decreases their affinity for negatively charged DNA; thereby promoting the accessibility of chromatin to transcriptional regulators. Conversely, removing the acetyl group by histone deacetylation can result in a reduction of gene expression. The level of histone acetylation can, therefore, be used as a marker to understand the activity of target genes (Earley et al., 2006).
Four types of HDAs have been identified in various species of plants (Lusser et al., 2001; Pandey et al., 2002; Alinsug et al., 2009). Three of them are homologous to yeast RPD3 (reduced potassium dependency protein 3), HDA1 (histone deacetylase 1 protein), and SIR2 (silent information regulator protein 2) proteins. The fourth type of HDAs, the HD2-like proteins, appears to be plant-specific HDAs (Lusser et al., 1997). HD2-type histone deacetylase was first discovered in maize as an acidic nucleolar phosphoprotein in a high molecular weight complex (Brosch et al., 1996; Lusser et al., 1997). It was found that the maize HD2 accepted all core histones as substrate in vitro, and it was highly sensitive to deacetylase inhibitors, Trichostain and cyclic tetrapeptides. Four HD2 proteins, HD2A, HD2B, HD2C, and HD2D, have been identified in Arabidopsis (Wu et al., 2000; Dangl et al., 2001; Zhou et al., 2004). HD2A, HD2B, and HD2C can mediate repression of a reporter gene (Wu et al., 2003). Furthermore, HD2 proteins may be involved in fertilization (Lagace et al., 2003), seed development (Wu et al., 2000; Zhou et al., 2004), ABA response (Sridha and Wu, 2006), and leaf development (Ueno et al., 2007). However, the molecular mechanism of how HD2 proteins are involved in these processes is not clear. HD2 proteins do not show sequence homologies with other known HDA proteins, but they share some sequence similarities with nuclear FK506-binding protein family (FKBP) members (Aravind, 1998). The findings that some FKBPs are required for the reduction of gene expression and can interact with RPD3-type HDAs raise the possibility that HD2 proteins may also interact with other known HDAs (Yang et al., 2001; Kuzuhara and Horikoshi, 2004; Earley et al., 2006).
Recent studies indicated the involvement of histone acetylation and deacetylation in the plant abiotic stress response. Both tobacco and Arabidopsis cells show a nucleosomal response to high salinity and cold stress, manifested by transient up-regulation of H3 phosphoacetylation and histone H4 acetylation (Sokol et al., 2007). Furthermore, it was found that histone acetylation on the histone H3 was altered with gene activation on the coding regions of drought stress-responsive genes under drought stress conditions (Kim et al., 2008). Both abscisic acid (ABA) and salt stress can induce histone H3K9K14 acetylation and H3K4 trimethylation but can decrease the H3K9 dimethylation of some ABA and abiotic stress-responsive genes, suggesting that functionally related gene groups are regulated co-ordinately through histone modifications in response to abiotic stress in plant cells (Chen et al., 2010).
In this study, the function of a HD2-type HDA, HD2C, was investigated in ABA- and abiotic-stress responses as well as its interaction with a RPD3-type HDA, HDA6. Bimolecular fluorescence complementation (BiFC), in vitro pull-down assays, and co-immunoprecipitation (Co-IP) indicated that HD2C interacts physically with HDA6. Moreover, HD2C can bind to histone H3 and affect the levels of histone H3K9K14 acetylation, H3K4 trimethylation, and H3K9 dimethylation. In addition, the HD2C T-DNA insertion plants, hd2c-1 and hd2c-3, are more sensitive to ABA and NaCl. The expression and histone modification of ABA-related genes, ABI1, ABI2, and AtERF4, were affected in hda6, hd2c-1, and hda6/hd2c-1 double mutant plants. Our studies provided evidence indicating that HD2C is involved in the ABA and salt-stress response by interacting with HDA6 and modulating stress-responsive genes.
Materials and methods
Plant materials
Plants were germinated and grown at 23 °C under a long day condition (16/8 h light/dark cycle). The T-DNA insertion mutants, hd2c-1 (HD2CT99, Salk_129799.19.60N) and hd2c-3 (HD2CT84, Salk_039784.52.90x) were obtained from the Arabidopsis Resource Centre (http://www.arabidopsis.org/). An hda6 mutant line, axe1-5, is a splice site mutant that has a base change at an intron splice site resulting in two HDA6 transcripts with altered lengths (Murfett et al., 2001; Yu et al., 2011).
Measurement of germination rates
Seeds were treated with 50% bleach solution for 15 min and then rinsed three times with distilled water. The surface-sterilized seeds were incubated at 4 °C for 3 d to synchronize germination and then planted in Petri dishes containing different growth media. The media contained half-strength Murashige and Skoog salts, 1% sucrose, and 0.8% agar, and were supplemented with or without ABA and NaCl. All plates were transferred to a growth chamber and incubated at 23 °C under long-day conditions. For germination-rate tests, seeds were germinated on media with or without ABA (0.5 μM, 1 μM, and 2 μM) or NaCl (100 mM, 125 mM, and 150 mM) in a growth chamber under long-day conditions, and seed germination rates were analysed after 2 d.
For the per cent survival of seedlings under high salinity treatment, 5-d-old seedlings growing in Perti dishes were transferred to a medium containing 150 mM NaCl, and the per cent survival of seedlings was measured after 5 d.
Semi-quantitative RT-PCR analysis
To isolate total RNA, 0.1–0.2 g of Arabidopsis thaliana leaves were ground with liquid nitrogen in a mortar and pestle and mixed with 1 ml TRIZOL Reagent (Invitrogen, Carlsbad, CA, USA) to isolate total RNA. One microgram of total RNA was used for the first-strand cDNA synthesis after incubation at 65 °C for 10 min. cDNA was synthesized in a volume of 20 μl that contained the MoMLV reverse transcriptase buffer (Promega, Madison, Wisconsin, USA), 10 mM dithiothreitol, 1.5 μM poly(dT) primer, 0.5 mM dNTPs, and 2 U of MoMLV reverse transcriptase at 37 °C for 1 h. All PCR reactions were performed with 0.5 U of Taq polymerase, the buffer provided by the supplier, 0.2 mM dNTPs, and a pair of primers (0.1 μM each) in a final volume of 20 μl. The thermocycling conditions were 94 °C for 4 min, followed by 22–35 cycles of 94 °C for 30 s, 50–65 °C for 1 min, and 72 °C for 1 min, with a final polymerization step at 72 °C for 7 min.
Quantitative real-time PCR (qPCR)
cDNAs (diluted ×100) obtained from RT-PCR were used as a template to run real-time PCR. The following components were added to a reaction tube: 9 μl of iQ™ SYBR Green Supermix solution (Bio-Rad; Catalogue no. 170-8882), 1 μl of 5 μM specific primers, and 8 μl of the diluted template. UBIQUITIN was used as an internal control in real-time quantitative RT-PCR. The thermocycling conditions were 95 °C for 3 min, followed by 40 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 20 s, with a melting curve detected at 95 °C for 1 min, 55 °C for 1 min, and the denature time detected from 55 °C to 95 °C. The gene-specific primer pairs are listed in Supplementary Table S1 at JXB online. Each experiment was repeated with three biological and three technical replicates.
Chromatin immunoprecipitation assay
The chromatin immunoprecipitation assay was performed as described by (Gendrel et al. (2005). The chromatin extract was prepared from 18-d-old leaves. Antibodies specific for histone H3K9K14Ac and H3K9Me2 (Millipore) were used in this study. The primers used for real-time PCR analysis in ChIP assays are listed in Supplementary Table S2 at JXB online. Each of the immunoprecipitations was replicated three times, and each sample was quantified at least in triplicate.
Bimolecular fluorescence complementation assay
To generate the construct for BiFC assay, full-length coding sequences of HD2A, HD2B, HD2C, HD2D, and HDA6 were PCR-amplified. The PCR products were subcloned into the pENTR/SD/D-TOPO or pCR8/GW/TOPO vector, and then recombined into the pEarleyGate201-YN and pEarleyGate202-YC vectors (Lu et al., 2010). The resulting constructs were used for transient assays by PEG transfection of Arabidopsis protoplasts (Yoo et al., 2007). Transfected cells were imaged using an Olympus BX51 fluorescence microscope, or a Leica SP5 confocal microscope.
Protein expression and purification
The full-length HDA6, HD2C, and AtFKBP53 cDNA were subcloned into pET25b+ and pGEX-4T-3 expression vectors to generate HDA6-His, GST-HD2C, and GST-AtFKBP53 fusion protein constructs and expressed in E. coli strain BL21 (DE3). The His- and GST-fusion proteins expressed in bacteria were induced by 0.1 mM isopropylthio-β-galactoside at 20 °C for 18 h. For protein extraction, cells were collected by centrifugation and then sonicated in a lysis buffer (50 mM phosphate buffer, pH 8.0, 300 mM NaCl, 20 mM β-mercaptoethanol, 0.1% Triton X-100 and 10 mM imidazole for the His-fusion protein, 4.3 mM Na2HPO4,1.47 mM KH2PO4, 137 mM NaCl, and 2.7 mM KCl, pH 7.3 for the GST-fusion protein). The HDA6-His, GST-HD2C, and GST-AtFKBP53 recombinant fusion proteins were purified by Ni-NTA resin and GST Bind Resin, respectively.
In vitro pull-down assay
The pull-down assay was performed as previously described by Yang et al. (2008). GST-HD2C and HDA6-His fusion proteins or HeLa Core Histones (Active Motif) were incubated in a binding buffer (50 mM TRIS-Cl, pH 7.5, 100 mM NaCl, 0.25% Triton X-100, and 35 mM β-mercaptoethanol) for 2 h at 4 °C, then the binding reaction was performed by mixing 30 μl of GST binding resin for an additional 2 h. After extensive washing (at least six times), the pulled-down proteins were eluted by boiling , separated by 10% SDS-PAGE, and detected by Western blotting using an anti-His or an anti-H3 antibody.
Co-immunoprecipitation assay
The co-immunoprecipitation assay was carried out using the tobacco transient expression system as described previously by Yu et al. (2011). Tobacco leaves were infiltrated with Agrobacterium tumefaciens carring 35S-HDA6-Myc and 35S:HD2C-HA, and the leaf extracts were used to perform co-immunoprecipitation. The co-immunoprecipitated proteins were analysed by Western blotting using anti-HA and anti-Myc antibodies (Sigma).
Determination of histone H3 modifications
Total histone proteins were prepared according to the method of Lu et al. (2011). Proteins were separated by 12% SDS-PAGE and transferred onto a PVDF membrane (Millipore). Antibodies specific for histone H3K9K14Ac, H3K4Me3, and H3K9Me2 (Millipore), were used as a primary antibody. Signals were detected by using the Millipore Immobilon Western chemiluminescent horseradish peroxidase substrate (Millipore).
Results
Expression of HD2A, HD2B, HD2C, and HD2D was repressed by ABA and NaCl
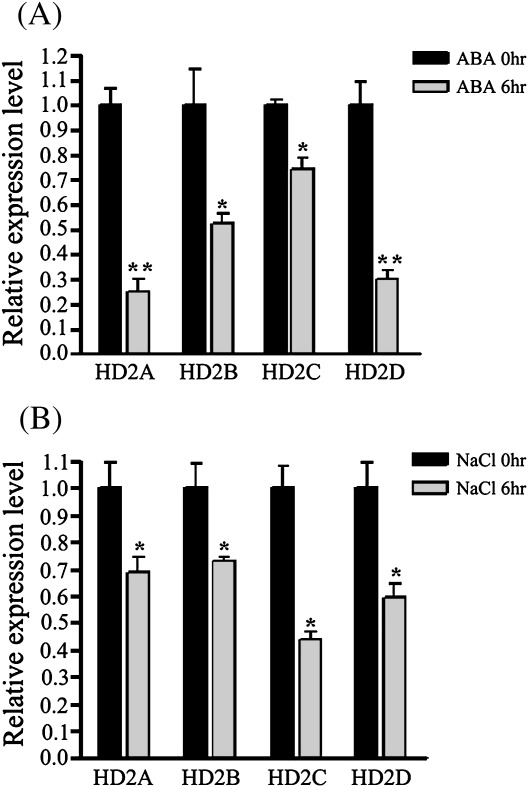
The mRNA accumulation patterns of HD2A, HD2B, HD2C, and HD2D in Arabidopsis plants treated with ABA and NaCl were examined by quantitative PCR. Under 100 μM ABA treatment, the expression of HD2A, HD2B, HD2C, and HD2D was reduced after 6 h (Fig. 1A). For salt treatment, plants were watered with 250 mM NaCl. As shown in Fig. 1B, the expression of HD2A, HD2B, HD2C, and HD2D was repressed when treated with NaCl at 6 h.
Fig. 1.
Expression of HD2A, HD2B, HD2C, and HD2D in response to ABA and NaCl. 2-week-old plants were treated with 100 μM of ABA (A) or 250 mM NaCl (B). Relative mRNA levels were determined by quantitative RT-PCR analysis. Two biological replicates were carried out and gave similar results. Asterisks mark values that are significantly different from the wild type (t test, *P <0.05, **P <0.01).
HD2C T-DNA insertion mutants were hypersensitive to ABA and NaCl
Two Arabidopsis lines, hd2c-1 (Salk_129799.19.60N) and hd2c-3 (Salk_039784.52.90), that contain a T-DNA insertion in the 5th intron and 7th exon of HD2C, respectively (Fig. 2A) were identified. It was not possible to obtain the knock-out mutants for other HD2 genes, so this study was therefore focused on HD2C. The T-DNA insertions were confirmed by PCR screening and sequencing. As shown in Fig. 2B, homozygous hd2c-1 and hd2c-3 T-DNA insertion lines were identified using LBa1/HD2C pr4 and HD2C-RT1/HD2C pr4 primer pairs. Moreover, our sequencing analysis showed that the sites of T-DNA insertion are at 1253 bp and 1732 bp in the 5th intron and 7th exon of HD2C in hd2c-1 and hd2c-3 T-DNA mutants, respectively. In addition, RT-PCR analysis indicated that the HD2C transcript was absent in hd2c-1 and hd2c-3 homozygous plants (Fig. 2C).
Fig. 2.
Identification of T-DNA insertion mutants of HD2C. (A) Schematic structure of HD2C and T-DNA insertion sites of hd2c-1 and hd2c-3. The relative locations of PCR primers were indicated by the arrows. (a) LBa1 (5′-GTTCACGTAGTGGGCCATCG-3′), (b) HD2C-RT1 (5′-TGACGCTGACGGTAGTGAAG-3′), and (c) HD2C pr4 (5′-AATTAGATCTGCACTGTGTTTGGCCTTTG-3′). (B) The HD2C wild-type allele was detected in the wild type using HD2C-RT1 and HD2C pr4 primers, whereas T-DNA alleles were detected in hd2c-1 and hd2c-3 using LBa1 and HD2C pr4 primers. (C) RT-PCR analysis indicated that the HD2C transcript was detected in wild-type plants, but absent in hd2c-1 and hd2c-3.
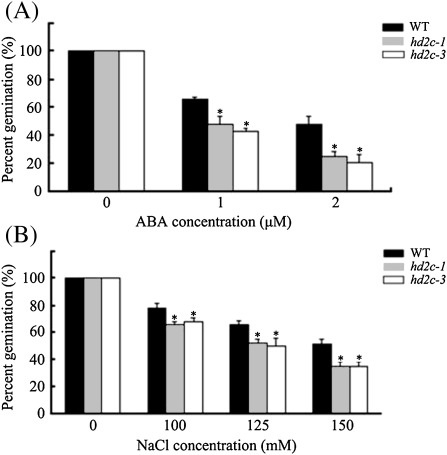
To analyse the involvement of HD2C in the ABA and abiotic stress response, hd2c-1 and hd2c-3 seeds were tested for their response to ABA and NaCl during seed germination. The germination rates were measured after 2 d planting on media containing different concentrations of ABA or NaCl. Without ABA and NaCl treatment, no difference was found in the germination rates among the wild-type and mutant lines. Compared with the wild type, hd2c-1 and hd2c-3 mutants displayed decreased germination rates under ABA and salt treatment conditions (Fig 3). At 2 μM ABA, the germination rate of the wild type was 48%, whereas those of hd2c-1 and hd2c-3 were 25% and 21%, respectively (Fig. 3A). At 150 mM NaCl, the germination rate of the wild type was 52%, whereas those of hd2c-1 and hd2c-3 were 25%, respectively (Fig. 3B).
Fig. 3.
Seed germination rates of hd2c-1 and hd2c-3 plants treated with ABA and NaCl. (A) Germination rates of 2-d-old wild-type (WT), hd2c-1, and hd2c-3 seedlings treated with ABA. (B) Germination rates of 2-d-old wild-type (WT), hd2c-1, and hd2c-3 seedlings treated with NaCl. Three biological replicas were performed with three technical replicates for each treatment (n ≥100), Asterisks mark values that are significantly different from the wild type (t test, *P <0.05, **P <0.01).
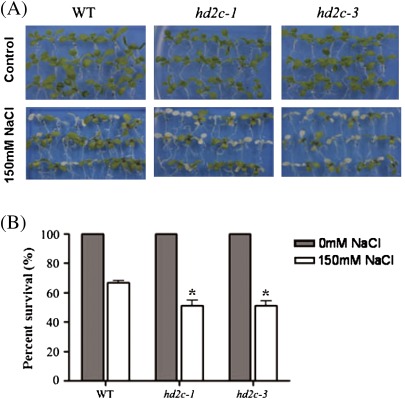
The response of HD2C T-DNA insertion plants to high salinity stress was also investigated. 5-d-old Arabidopsis seedlings were transferred to a medium containing 150 mM NaCl for 2 d. The leaf survival rates were measured as the percentages of green leaves left after treatment. As shown in Fig. 4, hd2c-1 and hd2c-3 plants were more sensitive to salt stress and displayed a lower survival rate. The survival rate of wild-type plants was 67%, whereas the survival rates of both hd2c-1 and hd2c-3 plants were 51% only (Fig- 4B).
Fig. 4.
Phenotype comparison of hd2c-1 and hd2c-3 plants in response to salt stress. (A) 5-d-old seedlings of the wild type (WT), hd2c-1, and hd2c-3 were transferred to a medium containing 150 mM NaCl for 5 d. (B) 5-d-old seedlings were transferred to a medium containing 150 mM NaCl, and the percentage survival of seedlings was measured after 5 d. Three biological replicas were performed with three technical replicates for each treatment (n ≥100). Asterisks mark values that are significantly different from the wild type (t test, *P <0.05).
HD2C interacted with HDA6 in vitro and in vivo
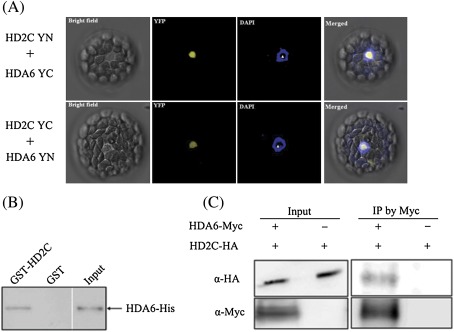
HD2 proteins share some sequence similarities with nuclear FK506-binding protein family members that interact with RPD3-type histone deacetylases, HDAC1 and HDAC2, in human (Aravind, 1998; Yang et al., 2001). To determine whether HD2C could interact with HDA6 in Arabidopsis, BiFC was used to determine their in vivo protein interactions. HD2C was fused to the N-terminal 174 amino acid portion of YFP in the pEarleyGate201 vector (pEarleyGate201-YN) (Lu et al., 2010), whereas HDA6 was fused to the C-terminal 66 amino acid portion of YFP in the pEarleyGate202 vector (pEarleyGate202-YC). The corresponding constructs were co-delivered into protoplasts of Arabidopsis, and fluorescence was observed using a confocal microscope. As shown in Fig. 5A, HD2C interacted with HDA6 in nucleoli, which was in accordance with the localization of HD2C and HDA6 (Pontes et al., 2007; Earley et al., 2010). Similar results were also obtained when the HD2C was fused to YC and HDA6 was fused to YN (Fig. 5A). As the negative controls, no YFP signals were detected when HD2C fused with YN/YC or HDA6 fused with YN/YC was co-transfected with empty vectors (YN and YC) (see Supplementary Fig. S1 at JXB online).
Fig. 5.
HD2C interacted with HDA6. (A) BiFC assays. HD2C and HDA6 fused with N-terminal (YN) or C-terminal (YC) of YFP were co-transfected into protoplasts, and visualized using confocal microscope. The nucleolus is indicated by white triangle. (B) In vitro pull-down assays. Purified His-HDA6 recombinant protein was incubated with GST or GST-HD2C protein. After extensive washing, the pulled-down proteins were eluted and the retention of HDA6-His proteins by GST-HD2C was detected by Western blotting using an anti-His antibody. (C) Co-immunoprecipitation analysis of HD2C interaction with HDA6 in N. benthamiana transient expression system. Agrobacterium cultures carrying 35S-HDA6-Myc and 35S:HD2C-HA were co-infiltrated into tobacco leaves. Crude extracts (input) were immunopreciped (IP) with an anti-Myc antibody and analysed by Western blotting.
An in vitro GST pull-down assay was also performed to examine the interaction between HD2C and HDA6. Purified His-HDA6 recombinant protein was incubated with GST-HD2C protein. As shown in Fig. 5B, HDA6-His was pulled down by GST-HD2C, indicating that HD2C can directly associate with HDA6. Moreover, the interaction between HD2C and HDA6 was further confirmed by the co-immunoprecipitation (co-IP) assay. As shown in Fig. 5C, the HD2C-HA was co-immunoprecipitated by HDA6-Myc. Taken together, these results indicate that HD2C can physically interact with HDA6.
hda6/hd2c-1 double mutants displayed hypersensitivity to ABA and NaCl
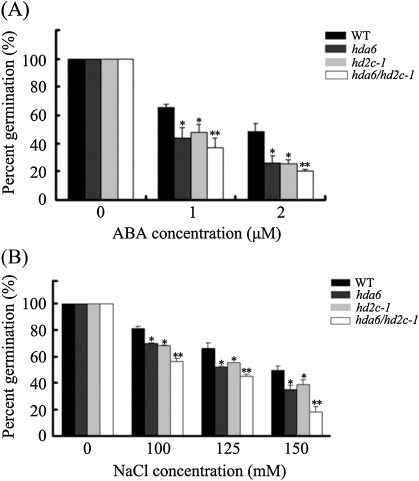
The hda6 mutant axe1-5 is a HDA6 splicing mutant which carries a point mutation in the HDA6 splicing site (Murfett et al., 2001; Wu et al., 2008; Yu et al., 2011). To study the genetic interaction between HDA6 and HD2C, hda6 and hd2c-1 were crossed to generate the hda6/hd2c-1 double mutant. The seed germination rates of hda6, hd2c-1, and hda6/hd2c-1 in response to ABA and NaCl were compared. Without ABA and NaCl treatment, no difference was found in the germination rates among the wild-type and mutant lines. As shown in Fig. 6, hda6, hd2c-1, and hda6/hd2c-1 double mutants displayed lower germination rates compared with the wild type when treated with ABA and NaCl.
Fig. 6.
Seed germination rates of hda6, hd2c-1, and hda6/hd2c-1 plants treated with ABA and NaCl. (A) Germination rates of 2-d-old wild-type (WT), hda6, hd2c-1, and hda6/hd2c-1 seedlings treated with ABA. (B) Germination rates of 2-d-old wild-type (WT), hda6, hd2c-1, and hda6/hd2c-1 seedlings treated with NaCl. Asterisks mark values that are significantly different from the wild type (t test, *P <0.05, **P <0.01). Three biological replicas were performed with three technical replicates for each treatment (n ≥100).
Expression of ABI1, ABI2, and AtERF4 was increased in hda6, hd2c-1, and hda6/hd2c-1 double mutant plants
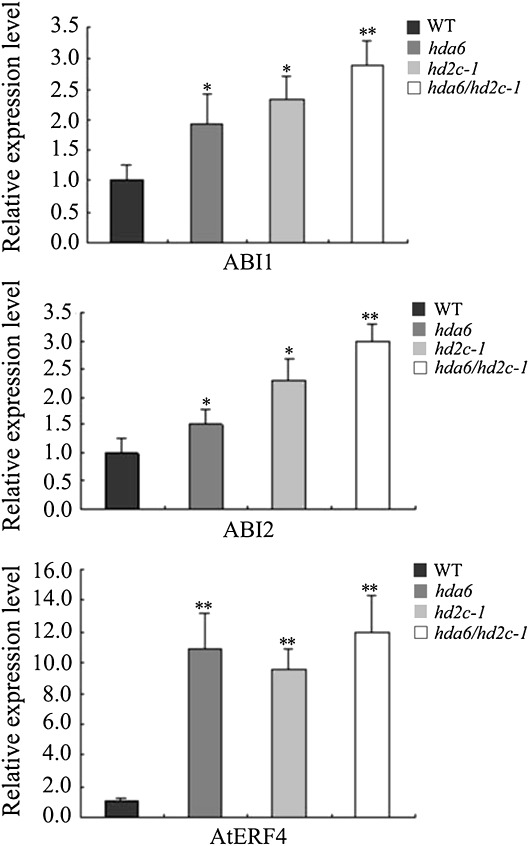
The expression of the ABA-responsive genes, ABI1 and ABI2, in hda6, hd2c-1, and hda6/hd2c-1 double mutant plants was analysed further. ABI1 and ABI2 encode serine/threonine phosphatease 2C (PP2C) that negatively regulate the ABA response (Merlot et al., 2001). As shown in Fig. 7, the expression of ABI1 and ABI2 were higher in hda6, hd2c-1, and hda6/hd2c-1 double mutant plants compared with the wild type. Furthermore, it was found that the AP2-domain transcriptional repressor AtERF4 was also up-regulated in hda6, hd2c-1, and hda6/hd2c-1 double mutant plants. In addition, two transcriptional activators MYB2 and MYC2 were down-regulated in hd2c mutant plants (see Supplementary Fig. S2 at JXB online). Since HDAs are usually associated with the repression of gene expression, the down-regulation of MYB2 and MYC2 in the hd2c mutants suggested that HD2C may affect the expression of MYB2 and MYC2 indirectly.
Fig. 7.
Expression of ABI1, ABI2, and AtERF4 in hda6, hd2c-1, and hda6/hd2c-1 double mutant plants. Total RNA was isolated from leaf tissues and the expression of ABI1, ABI2, and AtERF4 was determined by real-time RT-PCR. Asterisks mark values that are significantly different from the wild type (t test, **P <0.01, *P<0.05). The experiment was repeated three times with similar results.
Histone H3K9K14Ac and H3K9Me2 levels of ABI1, ABI2, and AtERF4 were changed in hda6, hd2c-1, and hda6/hd2c-1 double mutant plants
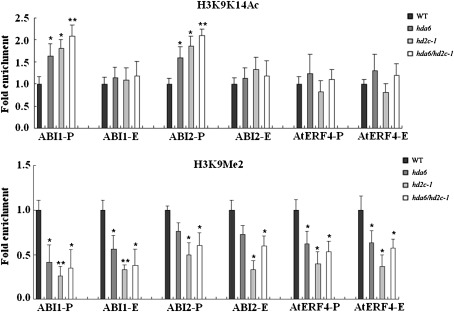
To determine whether the higher expression of ABI1, ABI2, and AtERF4 in hda6, hd2c-1, and hda6/hd2c-1 double mutant plants was related to histone acetylation and methylation in the chromatin, a chromatin immunoprecipitation (ChIP) assay was used to analyse the gene activation mark histone H3K9K14Ac and the gene repression mark H3K9Me2 of the up-regulated genes, ABI1, ABI2, and AtERF4. The levels of histone H3K9K14Ac in the promoter regions of ABI1 and ABI2 in hda6, hd2c-1, and hda6/hd2c-1 double mutant plants were higher than those in the wild-type plants (Fig. 8A; Supplementary Fig. S3 at JXB online). By contrast, no significant changes were found in the levels of histone H3K9K14Ac at AtERF4. These data indicate that HD2C and HDA6 may regulate ABI1 and ABI2 expression by histone acetylation. In addition, our ChIP data also showed that the levels of H3K9Me2 were decreased in the promoter and exon regions of ABI1, ABI2, and AtERF4 in hda6, hd2c-1, and hda6/hd2c-1 double mutant plants (Fig. 8B). These data revealed that the higher expression of ABI1, ABI2, and AtERF4 was associated with increased H3K9K14Ac and/or decreased H3K9Me2.
Fig. 8.
Histone acetylation and methylation of genes up-regulated in hda6, hd2c-1, and hda6/hd2c-1 double mutant plants. Relative levels of H3K9K14Ac (A) and H3K9Me2 (B) in ABI1, ABI2, and AtERF4 promoter and first exon regions were determined. The amounts of DNA after ChIP were quantified and normalized to an internal control ACTIN2 for H3K9K14Ac or Ta3 for H3K9Me2. Error bars represent standard errors. Asterisks mark values that are significantly different from the wild type (t test, **P <0.01, *P<0.05). The experiment was repeated three times with similar results.
HD2C physically associated with histone H3 and affected histone modifications
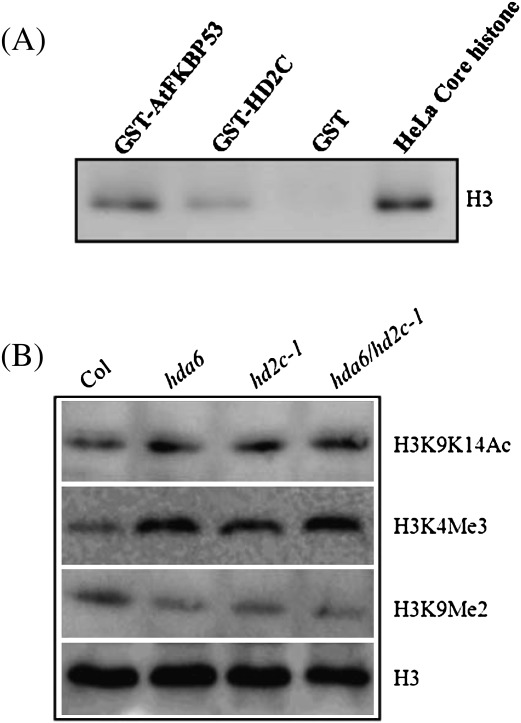
To investigate whether HD2C can interact with histone H3, an in vitro pull-down assay was performed using purified GST-HD2C protein with a HeLa core histone protein. AtFKBP53, a FK506-binding protein that was previously shown to physically interact with histone H3 (Li and Luan, 2010), was taken as a positive control. The result showed that histone H3 can interact with AtHD2C as well as AtFKBP53, but not GST (Fig. 9A).
Fig. 9.
Levels of histone H3K9K14Ac, H3K4Me3, and H3K9Me2 in hda6, hd2c-1, and hda6/hd2c-1 plants. (A) The H3 was pulled down by GST-HD2C and GST-FKBP53 and analysed by Western blotting using anti-H3 antibody. (B) The levels of histone H3K9K14Ac, H3K4Me3, and H3K9Me2 in Col wild-type, hda6, hd2c-1, and hda6/hd2c-1 plants were determined by Western blot analysis. The levels of H3 were shown as a loading control.
To determine whether HD2C can affect histone H3 modifications globally, the levels of histone H3K9K14Ac, H3K4Me3, and H3K9Me2 were analysed in hd2c-1 as well as in hda6 and had6/hdc2c-1 plants. Western blot analysis was performed using Anti-acetyl-Histone H3K9K14, Anti-trimethyl-Histone H3K4, and Anti-dimethyl-Histone H3K9 as the primary antibody. As shown in Fig. 9B, the levels of acetylated histone H3K9K14Ac and H3K4Me2 were increased in hd2c-1, hda6, and had6/hdc2c-1 compared with the wild type (Col-0). By contrast, the level of H3K9Me2 was decreased in hd2c-1, hda6, and had6/hdc2c-1 mutant plants.
Discussion
Histone acetylation is involved in plant abiotic stress responses
The involvement of RPD3-type HDAs, HDA6 and HDA19, in ABA and abiotic stress has been reported (Chen et al., 2010; Chen and Wu, 2010). It was found that hda6 mutant and HDA6 RNA-interference (HDA6-RNAi) plants displayed a phenotype that is more sensitive to ABA and salt stress. Compared with wild-type plants, the expression of ABA and abiotic stress-responsive genes was decreased in hda6 and HDA6-RNAi plants (Chen et al., 2010). Similarly, the Arabidopsis HDA19 T-DNA insertion mutant, hda19-1, was hypersensitive to ABA and salt stress, and the expression of ABA-responsive genes was decreased in hda19-1 plants (Chen and Wu, 2010). Arabidopsis HDA19 may act in a protein complex of AtERF7, a transcription repressor (Song et al., 2005; Yang et al., 2005), to regulate the abiotic stress response genes. Furthermore, AtERF7 interacts with the Arabidopsis homologue of a human global corepressor of transcription, AtSin3, which, in turn, may interact with HDA19. It was proposed that AtERF7, AtSin3, and HDA19 can form a transcriptional repressor complex to regulate ABA and drought response in Arabidopsis (Song et al., 2005).
In this study, it was found that hd2c-1 and hd2c-3 plants displayed increased sensitivity to ABA and NaCl during germination, and decreased tolerance to salt stress during seedling growth. In addition, over-expression of HD2C conferred an ABA-insensitive phenotype and enhanced tolerance to salt and drought stresses in transgenic Arabidopsis plants (Sridha and Wu, 2006). Furthermore, a rice SIR2-related HDA, OsSRT1, could enhance tolerance to oxidative stress when over-expressed in transgenic rice plants (Huang et al., 2007). Taken together, the data described above demonstrate that histone acetylation and deacetylation regulate the plant responses to abiotic stresses, and HDAs could be essential players in such regulation. The observations that both hda6 and hd2c-1 mutants displayed hypersensitivity to ABA and salt suggested that both HD2C and HDA6 are involved in the ABA and salt response of Arabidopsis. Therefore, HD2C may function with an RPD3-type HDA such as HDA6 to regulate gene expression in the ABA and abiotic stress-response pathways.
HD2C regulates the expression of abiotic stress-responsive genes
The HD2C T-DNA knock-out lines, hd2c-1 and hd2c-3, were hypersensitive to ABA and NaCl. hd2c-1 and hd2c-3 plants showed lower germination and survival rates under NaCl treatment. Similarly, Colville et al. (2011) also reported that a hd2c mutant was hypersensitive to ABA and NaCl in seed germination. The expression of ABA-responsive genes, ABI1 and ABI2, which were found to be protein phosphatases (Merlot et al., 2001) that negatively regulate the ABA response (Sheen, 1998), were increased in hda6, hd2c-1, and hda6/hd2c-1 double mutant plants. A previous study reported that the transcription factor AtERF4 is a negative regulator involved in the ABA response (Yang et al., 2005). The expression of AtERF4 was up-regulated in hda6, hd2c-1, and hda6/hd2c-1 double mutant plants. Taken together, our results suggest that HD2C might regulate the expression of abiotic stress-related genes to affect plant stress tolerance. In this study, it was found that the expression of HD2A, HD2B, HD2C, and HD2D can be repressed by ABA and NaCl. Similarly, ABA also represses the expression of HD2-type HDAs, HDT701 and HDT702, in rice (Fu et al., 2007). More recently, Colville et al., (2011) reported that hd2a and hd2c mutants responded differently to ABA and NaCl. It remains to be determined whether HD2 proteins function differently in the abiotic stress response.
The levels of the gene activation mark histone H3K9K14Ac were increased in the promoter regions of ABI1 and ABI2 in hda6, hd2c-1, and hda6/hd2c-1 double mutant plants. In addition, the levels of the gene repression mark H3K9Me2 of ABI1, ABI2, and AtERF4 were decreased in hda6, hd2c-1, and hda6/hd2c-1 double mutant plants. These results revealed that the high expression of ABI1, ABI2, and AtERF4 in the mutants is associated with the increased levels of H3K9K14Ac and/or decreased levels of H3K9Me2. More recently, genome-wide analyses were performed in Arabidopsis, rice, and maize by ChIP-chip or ChIPseq (Lauria and Rossi, 2011). These studies showed that histone H3K9Ac is invariably correlated with transcriptional activation and biased towards the 5′-end of genes, whereas the gene repression mark H3K9me2 was found in repressed genes, spanning both the promoter and the gene body. Similarly, it was found that H3K9K14Ac increased in the promoter regions of ABI1 and ABI2, but H3K9Me2 decreased in the promoter and exon regions of ABI1, ABI2, and AtERF4 in the hda6 and hd2c mutants. These data suggested that HD2C and HDA6 may regulate ABI1, ABI2, and AtERF4 expression through histone modifications.
HD2C affects histone acetylation by interacting with HDA6
HD2-type histone deacetylases were identified as a plant-specific HDACs (Lusser et al., 1997), and sequence analysis revealed that HD2 proteins share some sequence similarities with FKBP family pepridylprolyl cis-trans isomerases (PPIase) and a trypanosomal RNA-binding protein (Aravind, 1998). It has been shown that human FKBP25 shows 17% identity in protein sequence to the maize histone deacetylase HD2, with the most striking homology (22% identity) in residues 15–200 of human FKBP25 and residues 56–283 of maize HD2. FKBP25 physically associates with the histone deacetylases HDAC1 and HDAC2 and with HDA-binding transcriptional regulator YY1 in human cell (Yang et al., 2001). Similarly, it was found that HD2C can interact with HDA6 in Arabidopsis, suggesting that HD2 proteins, like FKBP25 in human cells, might form a complex with RPD3-type HDAs.
Increased histone H3 acetylation was found in hd2c-1 and hda6 plants, suggesting that HD2C may affect the level of histone H3 acetylation by interacting with HDA6. In addition, increased H3K4Me3 and decreased H3K9Me2 were also observed in hd2c-1 and hda6 plants. Modulation of gene expression through cross-talk between histone acetylation and methylation has been reported previously (Lee et al., 2006; Chen et al, 2010). Our recent study indicates that HDA6 can interact with histone demethylase FLD and regulate both histone acetylation and methylation (Yu et al., 2011). The observations that HD2A/HDT1 and HDA6 knock-down plants displayed a similar phenotype with respect to rRNA gene derepression, promoter cytosine methylation, histone modifications, and NOR condensation also support the functional association of HD2 proteins with RPD3-type HDAs (Lawrence et al., 2004; Earley et al., 2006). Our study indicated that HD2C physically associated with histone H3 and affected the levels of histone H3 acetylation and methylation. These results suggest that HD2 proteins may be a part of the chromatin remodelling complexes, including HDAs and other histone modification proteins. Our double mutant analysis indicated that the hda6/hd2c double mutant had enhanced phenotypes compared with the hda6 and hd2c single mutants in ABI1 and ABI2 expression, histone acetylation, and germination, suggesting a synergistic effect. These results indicated that HDA6 and HD2C proteins could indeed cooperate in these responses (Capaldi et al., 2008). Further research is required to determine the molecular mechanisms of HD2C and HDA6 interaction involved in the regulation of ABA-responsive genes and abiotic stress response.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Fig. S1. Negative controls in BiFC assays.
Supplementary Fig. S2. Expression of MYB2 and MYC2 determined by real-time RT-PCR.
Supplementary Fig. S3. Two additional independent analyses of histone acetylation and methylation of ABI1 and ABI2 in hda6 plants.
Supplementary Table S1. Primers used for qRT-PCR analysis.
Supplementary Table S2. Primers used for Chip assay.
Acknowledgments
This work is supported by grants from the National Science Council of Taiwan (99-2321-B-002-027-MY3 and 98-2628-B-002-016-MY3) and the National Taiwan University (10R80917-5), and grants from the Natural Science Foundation of China (No. 30971564, No. 90919038, and No. 31140015).
References
- Alinsug MV, Yu CW, Wu K. Phylogenetic analysis, subcellular localization, and expression patterns of RPD3/HDA1 family histone deacetylases in plants. BMC Plant Biology. 2009;9:37. doi: 10.1186/1471-2229-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L. Second family of histone deacetylases. Science. 1998;280:1167. [Google Scholar]
- Berger SL. Histone modifications in transcriptional regulation. Current Opinion in Genetics and Development. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- Brosch G, Lusser A, Goralik-Schramel M, Loidl P. Purification and characterization of a high molecular weight histone deacetylase complex (HD2) of maize embryos. Biochemistry. 1996;35:15907–15914. doi: 10.1021/bi961294x. [DOI] [PubMed] [Google Scholar]
- Capaldi AP, Kaplan T, Liu Y, Habib N, Regev A, Friedman N, O’Shea EK. Structure and function of a transcriptional network activated by the MAPK Hog1. Nature Genetics. 2008;40:1300–1306. doi: 10.1038/ng.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LT, Luo M, Wang YY, Wu K. Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. Journal of Experimental Botany. 2010;61:3345–3353. doi: 10.1093/jxb/erq154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LT, Wu K. Role of histone deacetylases HDA6 and HDA19 in ABA and abiotic stress response. Plant Signaling and Behavior. 2010;5:1318–1320. doi: 10.4161/psb.5.10.13168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colville A, Alhattab R, Hu M, Labbé H, Xing T, Miki B. Role of HD2 genes in seed germination and early seedling growth in Arabidopsis. Plant Cell Reporter. 2011;30:1969–1979. doi: 10.1007/s00299-011-1105-z. [DOI] [PubMed] [Google Scholar]
- Dangl M, Brosch G, Haas H, Loidl P, Lusser A. Comparative analysis of HD2 type histone deacetylases in higher plants. Planta. 2001;213:280–285. doi: 10.1007/s004250000506. [DOI] [PubMed] [Google Scholar]
- Earley K, Lawrence RJ, Pontes O, Reuther R, Enciso AJ, Silva M, Neves N, Gross M, Viegas W, Pikaard CS. Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes and Development. 2006;20:1283–1293. doi: 10.1101/gad.1417706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Pontvianne F, Wierzbicki AT, Blevins T, Tucker S, Costa-Nunes P, Pontes O, Pikaard CS. Mechanisms of HDA6-mediated rRNA gene silencing: suppression of intergenic Pol II transcription and differential effects on maintenance versus siRNA-directed cytosine methylation. Genes and Development. 2010;24:1119–1132. doi: 10.1101/gad.1914110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W, Wu K, Duan J. Sequence and expression analysis of histone deacetylases in rice. Biochemical and Biophysical Research Communications. 2007;356:843–850. doi: 10.1016/j.bbrc.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Gendrel AV, Lippman Z, Martienssen R, Colot V. Profiling histone modification patterns in plants using genomic tiling microarrays. Nature Methods. 2005;2:213–218. doi: 10.1038/nmeth0305-213. [DOI] [PubMed] [Google Scholar]
- Huang L, Sun Q, Qin F, Li C, Zhao Y, Zhou DX. Down-regulation of a SILENT INFORMATION REGULATOR2-related histone deacetylase gene, OsSRT1, induces DNA fragmentation and cell death in rice. Plant Physiology. 2007;144:1508–1519. doi: 10.1104/pp.107.099473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, To TK, Ishida J, Morosawa T, Kawashima M, Matsui A, Toyoda T, Kimura H, Shinozaki K, Seki M. Alterations of lysine modifications on the histone H3 N-tail under drought stress conditions in Arabidopsis thaliana. Plant and Cell Physiology. 2008;49:1580–1588. doi: 10.1093/pcp/pcn133. [DOI] [PubMed] [Google Scholar]
- Kuzuhara T, Horikoshi M. A nuclear FK506-binding protein is a histone chaperone regulating rDNA silencing. Nature Structural and Molecular Biology. 2004;11:275–283. doi: 10.1038/nsmb733. [DOI] [PubMed] [Google Scholar]
- Lagace M, Chantha SC, Major G, Matton DP. Fertilization induces strong accumulation of a histone deacetylase (HD2) and of other chromatin-remodeling proteins in restricted areas of the ovules. Plant Molecular Biology. 2003;53:759–769. doi: 10.1023/B:PLAN.0000023665.36676.89. [DOI] [PubMed] [Google Scholar]
- Lauria M, Rossi V. Epigenetic control of gene regulation in plants. Biochimica et Biophysica Acta. 2011;1809:369–378. doi: 10.1016/j.bbagrm.2011.03.002. [DOI] [PubMed] [Google Scholar]
- Lawrence RJ, Earley K, Pontes O, Silva M, Chen ZJ, Neves N, Viegas W, Pikaard CS. A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Molecular Cell. 2004;13:599–609. doi: 10.1016/s1097-2765(04)00064-4. [DOI] [PubMed] [Google Scholar]
- Lee MG, Wynder C, Bochar DA, Hakimi MA, Cooch N, Shiekhattar R. Functional interplay between histone demethylase and deacetylase enzymes. Molecular and Cellular Biology. 2006;26:6395–6402. doi: 10.1128/MCB.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Luan S. AtFKBP53 is a histone chaperone required for repression of ribosomal RNA gene expression in Arabidopsis. Cell Research. 2010;20:357–366. doi: 10.1038/cr.2010.22. [DOI] [PubMed] [Google Scholar]
- Lu F, Cui X, Zhang S, Jenuwein T, Cao X. Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nature Genetics. 2011;43:715–719. doi: 10.1038/ng.854. [DOI] [PubMed] [Google Scholar]
- Lu Q, Tang X, Tian G, Wang F, Liu K, Nguyen V, Kohalmi SE, Keller WA, Tsang EWT, Harada JJ. Arabidopsis homolog of the yeast TREX-2 mRNA export complex: components and anchoring nucleoporin. The Plant Journal. 2010;61:259–270. doi: 10.1111/j.1365-313X.2009.04048.x. [DOI] [PubMed] [Google Scholar]
- Lusser A, Brosch G, Loidl A, Haas H, Loidl P. Identification of maize histone deacetylase HD2 as an acidic nucleolar phosphoprotein. Science. 1997;277:88. doi: 10.1126/science.277.5322.88. [DOI] [PubMed] [Google Scholar]
- Lusser A, Kolle D, Loidl P. Histone acetylation: lessons from the plant kingdom. Trends in Plant Science. 2001;6:59–65. doi: 10.1016/s1360-1385(00)01839-2. [DOI] [PubMed] [Google Scholar]
- Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat J. The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. The Plant Journal. 2001;25:295–303. doi: 10.1046/j.1365-313x.2001.00965.x. [DOI] [PubMed] [Google Scholar]
- Murfett J, Wang XJ, Hagen G, Guilfoyle TJ. Identification of Arabidopsis histone deacetylase HDA6 mutants that affect transgene expression. The Plant Cell. 2001;13:1047–1061. doi: 10.1105/tpc.13.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R, Muller A, Napoli CA, Selinger DA, Pikaard CS, Richards EJ, Bender J, Mount DW, Jorgensen RA. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acid Research. 2002;30:5036–5055. doi: 10.1093/nar/gkf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontes O, Lawrence RJ, Silva M, Preuss S, Costa-Nunes P, Earley K, Neves N, Viegas W, Pikaard CS. Postembryonic establishment of megabase-scale gene silencing in nucleolar dominance. PLoS ONE. 2007;2 doi: 10.1371/journal.pone.0001157. e1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. Mutational analysis of protein phosphatase 2C involved in abscisic acid signal transduction in higher plants. Proceedings of the National Academy of Sciences, USA. 1998;95:975–980. doi: 10.1073/pnas.95.3.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol A, Kwiatkowska A, Jerzmanowski A, Prymakowska-Bosak M. Up-regulation of stress-inducible genes in tobacco and Arabidopsis cells in response to abiotic stresses and ABA treatment correlates with dynamic changes in histone H3 and H4 modifications. Planta. 2007;227:245–254. doi: 10.1007/s00425-007-0612-1. [DOI] [PubMed] [Google Scholar]
- Song CP, Agarwal M, Ohta M, Guo Y, Halfter U, Wang P, Zhu JK. Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. The Plant Cell. 2005;17:2384–2396. doi: 10.1105/tpc.105.033043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridha S, Wu K. Identification of AtHD2C as a novel regulator of abscisic acid responses in Arabidopsis. The Plant Journal. 2006;46:124–133. doi: 10.1111/j.1365-313X.2006.02678.x. [DOI] [PubMed] [Google Scholar]
- Ueno Y, Ishikawa T, Watanabe K, Terakura S, Iwakawa H, Okada K, Machida C, Machida Y. Histone deacetylases and ASYMMETRIC LEAVES2 are involved in the establishment of polarity in leaves of Arabidopsis. The Plant Cell. 2007;19:445–457. doi: 10.1105/tpc.106.042325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Tian L, Malik K, Brown D, Miki B. Functional analysis of HD2 histone deacetylase homologues in Arabidopsis thaliana. The Plant Journal. 2000;22:19–27. doi: 10.1046/j.1365-313x.2000.00711.x. [DOI] [PubMed] [Google Scholar]
- Wu K, Tian L, Zhou C, Brown D, Miki B. Repression of gene expression by Arabidopsis HD2 histone deacetylases. The Plant Journal. 2003;34:241–247. doi: 10.1046/j.1365-313x.2003.01714.x. [DOI] [PubMed] [Google Scholar]
- Wu K, Zhang L, Zhou C, Yu CW, Chaikam V. HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. Journal of Experimental Botany. 2008;59:225–234. doi: 10.1093/jxb/erm300. [DOI] [PubMed] [Google Scholar]
- Yang JY, Iwasaki M, Machida C, Machida Y, Zhou X, Chua NH. betaC1, the pathogenicity factor of TYLCCNV, interacts with AS1 to alter leaf development and suppress selective jasmonic acid responses. Genes and Development. 2008;22:2564–2577. doi: 10.1101/gad.1682208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WM, Yao YL, Seto E. The FK506-binding protein 25 functionally associates with histone deacetylases and with transcription factor YY1. The EMBO Journal. 2001;20:4814–4825. doi: 10.1093/emboj/20.17.4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Tian L, Latoszek-Green M, Brown D, Wu K. Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Molecular Biology. 2005;58:585–596. doi: 10.1007/s11103-005-7294-5. [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- Yu CW, Liu X, Luo M, Chen C, Lin X, Tian G, Lu Q, Cui Y, Wu K. HISTONE DEACETYLASE6 interacts with FLOWERING LOCUS D and regulates flowering in Arabidopsis. Plant Physiology. 2011;156:173–184. doi: 10.1104/pp.111.174417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Labbe H, Sridha S, Wang L, Tian L, Latoszek-Green M, Yang Z, Brown D, Miki B, Wu K. Expression and function of HD2-type histone deacetylases in Arabidopsis development. The Plant Journal. 2004;38:715–724. doi: 10.1111/j.1365-313X.2004.02083.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.