Abstract
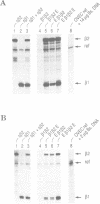
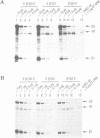
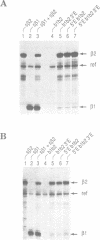
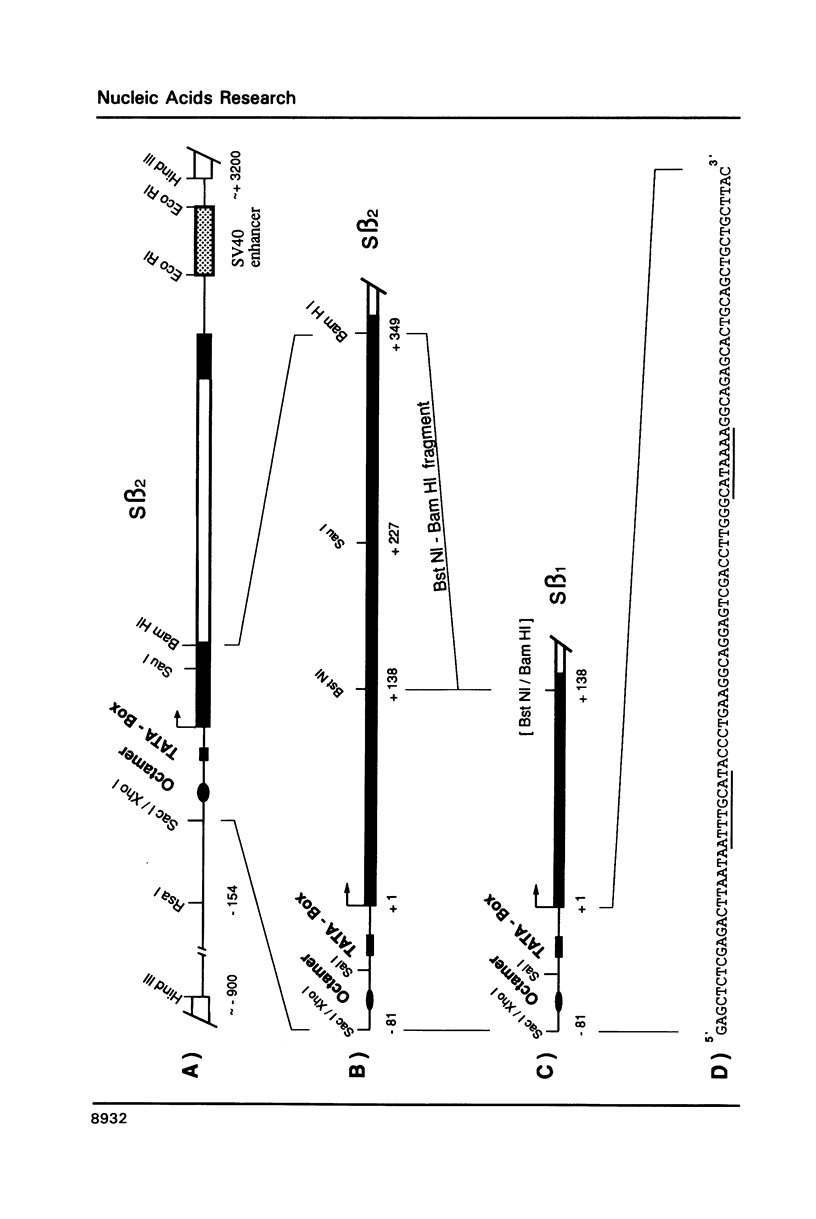
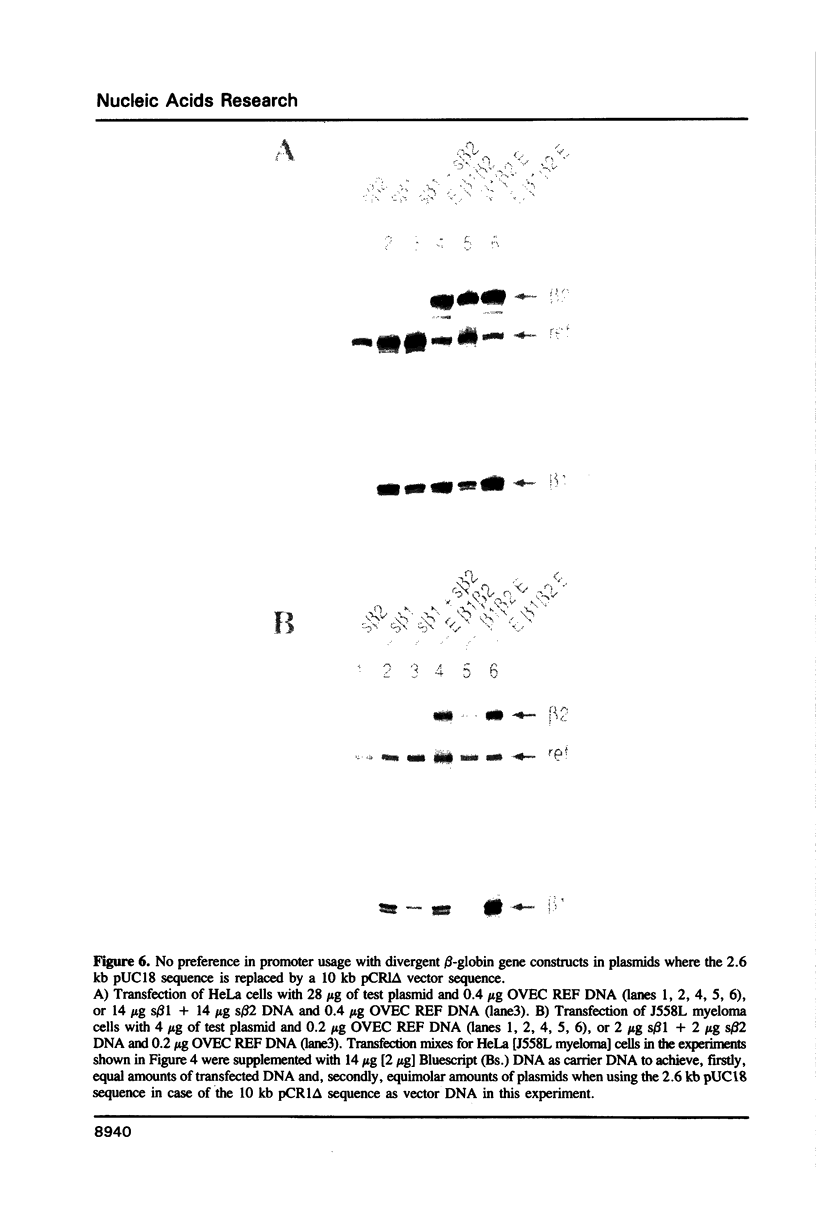
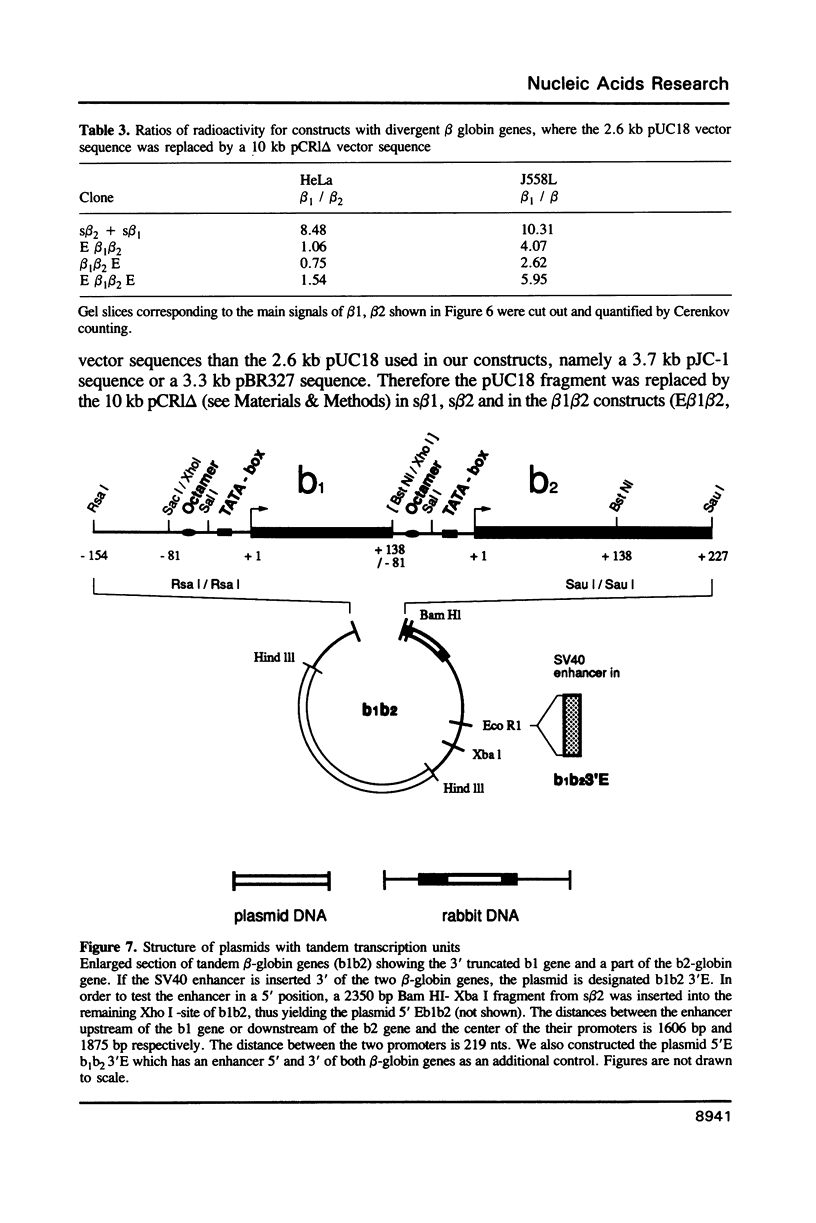
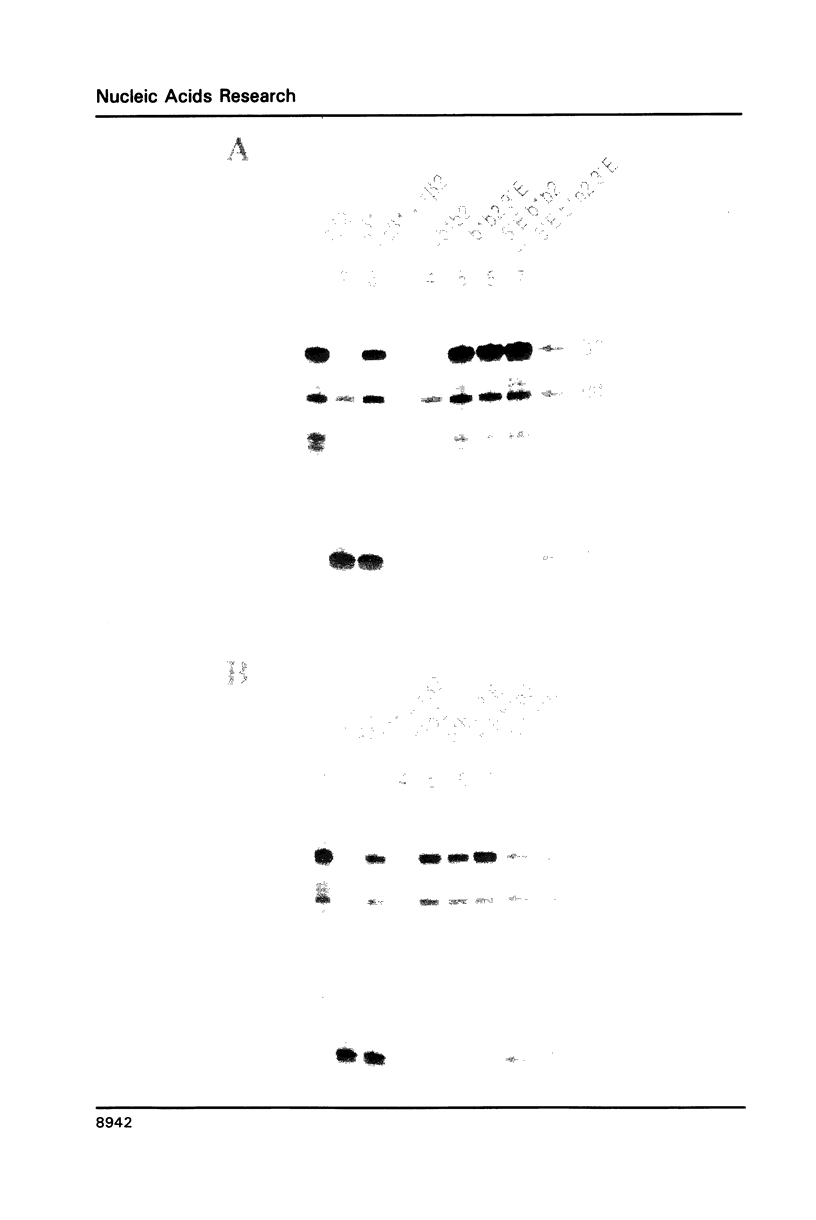
To explain the activation of transcription by a remote enhancer, two models are most often considered, namely looping and scanning. A scanning model, also referred to as 'polymerase entry site' model predicts that for two adjacent promoters the one proximal to an enhancer would be preferentially activated. Preferential activation of the proximal promoter in a tandem promoter arrangement has been found before in several laboratories, including our own, but for technical reasons the data were inconclusive with regards to the enhancer mechanism. In the work presented here, we readdress the question of preferential promoter activation by an enhancer using a more clearly defined system. Two identical promoters were kept closeby in a divergent, or directly repeated orientation. The SV40 enhancer was placed at a great distance on one or the other side of the two promoters, to see whether the enhancer position influenced the relative efficiency of the two promoters in transfected cells. Our finding that the promoter usage is virtually unaffected by the enhancer position does not favor a scanning model, but is compatible with a looping model of enhancer action.
Full text
PDF

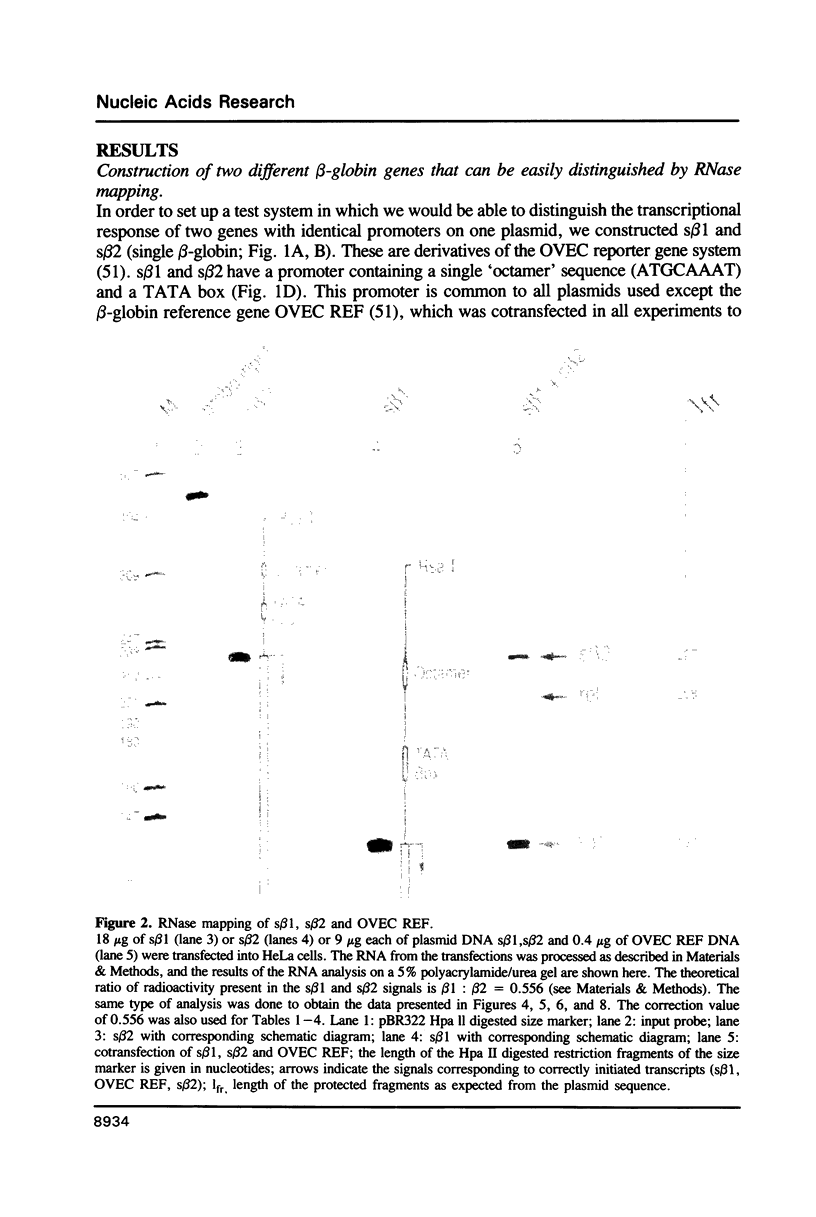
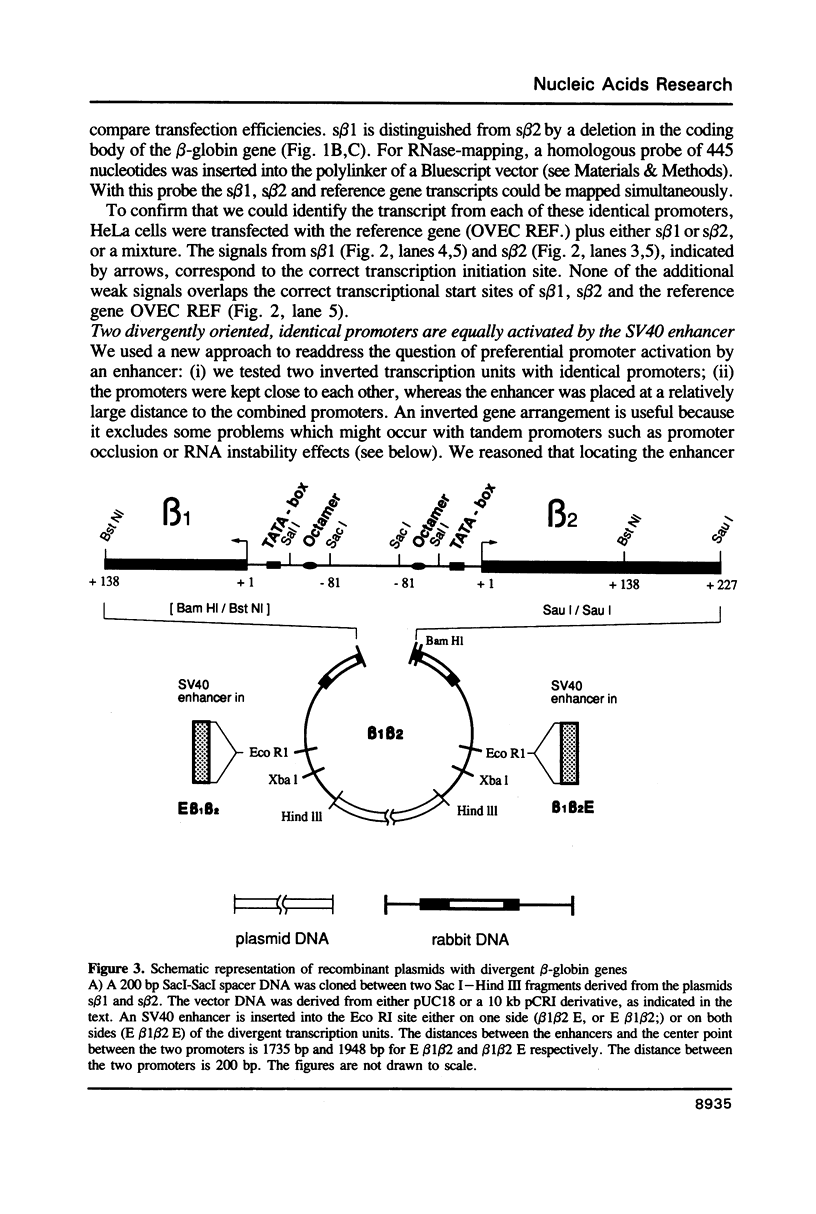
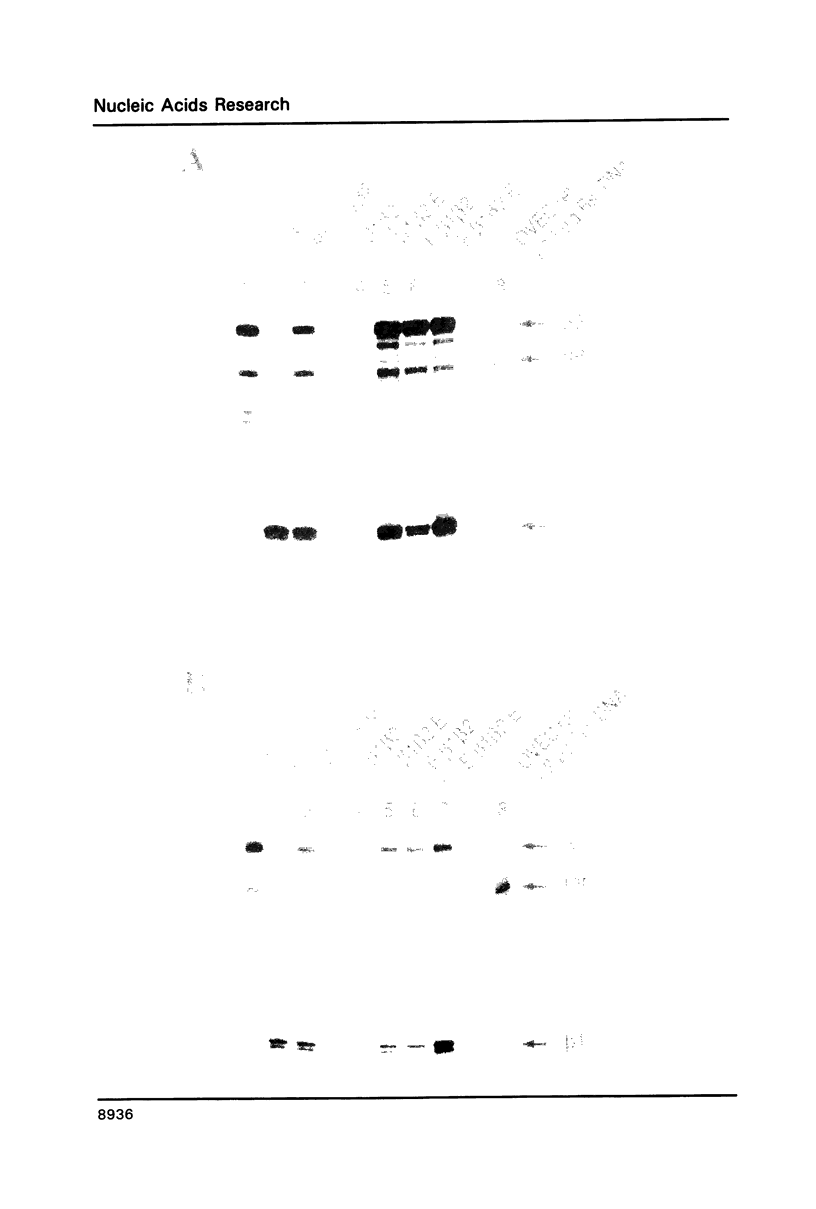







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armstrong K. A., Hershfield V., Helinski D. R. Gene cloning and containment properties of plasmid Col E1 and its derivatives. Science. 1977 Apr 8;196(4286):172–174. doi: 10.1126/science.322277. [DOI] [PubMed] [Google Scholar]
- Atchison M. L., Perry R. P. Tandem kappa immunoglobulin promoters are equally active in the presence of the kappa enhancer: implications for models of enhancer function. Cell. 1986 Jul 18;46(2):253–262. doi: 10.1016/0092-8674(86)90742-7. [DOI] [PubMed] [Google Scholar]
- Baker S. M., Platt T. Pol I transcription: which comes first, the end or the beginning? Cell. 1986 Dec 26;47(6):839–840. doi: 10.1016/0092-8674(86)90795-6. [DOI] [PubMed] [Google Scholar]
- Banerji J., Rusconi S., Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981 Dec;27(2 Pt 1):299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- Brent R., Ptashne M. A bacterial repressor protein or a yeast transcriptional terminator can block upstream activation of a yeast gene. Nature. 1984 Dec 13;312(5995):612–615. doi: 10.1038/312612a0. [DOI] [PubMed] [Google Scholar]
- Chodosh L. A., Carthew R. W., Morgan J. G., Crabtree G. R., Sharp P. A. The adenovirus major late transcription factor activates the rat gamma-fibrinogen promoter. Science. 1987 Oct 30;238(4827):684–688. doi: 10.1126/science.3672119. [DOI] [PubMed] [Google Scholar]
- Cohen R. S., Meselson M. Periodic interactions of heat shock transcriptional elements. Nature. 1988 Apr 28;332(6167):856–858. doi: 10.1038/332856a0. [DOI] [PubMed] [Google Scholar]
- Courey A. J., Plon S. E., Wang J. C. The use of psoralen-modified DNA to probe the mechanism of enhancer action. Cell. 1986 May 23;45(4):567–574. doi: 10.1016/0092-8674(86)90288-6. [DOI] [PubMed] [Google Scholar]
- Covey C., Richardson D., Carbon J. A method for the deletion of restriction sites in bacterial plasmid deoxyribonucleic acid. Mol Gen Genet. 1976 May 7;145(2):155–158. doi: 10.1007/BF00269587. [DOI] [PubMed] [Google Scholar]
- Dunn T. M., Hahn S., Ogden S., Schleif R. F. An operator at -280 base pairs that is required for repression of araBAD operon promoter: addition of DNA helical turns between the operator and promoter cyclically hinders repression. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5017–5020. doi: 10.1073/pnas.81.16.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynan W. S. Modularity in promoters and enhancers. Cell. 1989 Jul 14;58(1):1–4. doi: 10.1016/0092-8674(89)90393-0. [DOI] [PubMed] [Google Scholar]
- Gellert M., Nash H. Communication between segments of DNA during site-specific recombination. 1987 Jan 29-Feb 4Nature. 325(6103):401–404. doi: 10.1038/325401a0. [DOI] [PubMed] [Google Scholar]
- Gralla J. D. Bacterial gene regulation from distant DNA sites. Cell. 1989 Apr 21;57(2):193–195. doi: 10.1016/0092-8674(89)90955-0. [DOI] [PubMed] [Google Scholar]
- Griffith J., Hochschild A., Ptashne M. DNA loops induced by cooperative binding of lambda repressor. Nature. 1986 Aug 21;322(6081):750–752. doi: 10.1038/322750a0. [DOI] [PubMed] [Google Scholar]
- Hahn S., Hendrickson W., Schleif R. Transcription of Escherichia coli ara in vitro. The cyclic AMP receptor protein requirement for PBAD induction that depends on the presence and orientation of the araO2 site. J Mol Biol. 1986 Apr 5;188(3):355–367. doi: 10.1016/0022-2836(86)90160-9. [DOI] [PubMed] [Google Scholar]
- Hochschild A., Ptashne M. Cooperative binding of lambda repressors to sites separated by integral turns of the DNA helix. Cell. 1986 Mar 14;44(5):681–687. doi: 10.1016/0092-8674(86)90833-0. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986 Sep 12;46(6):885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- Kadesch T., Berg P. Effects of the position of the simian virus 40 enhancer on expression of multiple transcription units in a single plasmid. Mol Cell Biol. 1986 Jul;6(7):2593–2601. doi: 10.1128/mcb.6.7.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer H., Niemöller M., Amouyal M., Revet B., von Wilcken-Bergmann B., Müller-Hill B. lac repressor forms loops with linear DNA carrying two suitably spaced lac operators. EMBO J. 1987 May;6(5):1481–1491. doi: 10.1002/j.1460-2075.1987.tb02390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987 Mar 13;48(5):847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Goodbourn S., Fischer J. A. Regulation of inducible and tissue-specific gene expression. Science. 1987 Jun 5;236(4806):1237–1245. doi: 10.1126/science.3296191. [DOI] [PubMed] [Google Scholar]
- Martin K., Huo L., Schleif R. F. The DNA loop model for ara repression: AraC protein occupies the proposed loop sites in vivo and repression-negative mutations lie in these same sites. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3654–3658. doi: 10.1073/pnas.83.11.3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau P., Hen R., Wasylyk B., Everett R., Gaub M. P., Chambon P. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res. 1981 Nov 25;9(22):6047–6068. doi: 10.1093/nar/9.22.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Erickson H., Bastia D. Detection of DNA looping due to simultaneous interaction of a DNA-binding protein with two spatially separated binding sites on DNA. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6287–6291. doi: 10.1073/pnas.85.17.6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müeller-Storm H. P., Sogo J. M., Schaffner W. An enhancer stimulates transcription in trans when attached to the promoter via a protein bridge. Cell. 1989 Aug 25;58(4):767–777. doi: 10.1016/0092-8674(89)90110-4. [DOI] [PubMed] [Google Scholar]
- Müller M. M., Gerster T., Schaffner W. Enhancer sequences and the regulation of gene transcription. Eur J Biochem. 1988 Oct 1;176(3):485–495. doi: 10.1111/j.1432-1033.1988.tb14306.x. [DOI] [PubMed] [Google Scholar]
- Ondek B., Gloss L., Herr W. The SV40 enhancer contains two distinct levels of organization. Nature. 1988 May 5;333(6168):40–45. doi: 10.1038/333040a0. [DOI] [PubMed] [Google Scholar]
- Ondek B., Shepard A., Herr W. Discrete elements within the SV40 enhancer region display different cell-specific enhancer activities. EMBO J. 1987 Apr;6(4):1017–1025. doi: 10.1002/j.1460-2075.1987.tb04854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard D. Viral and cellular transcription enhancers. Oxf Surv Eukaryot Genes. 1985;2:24–48. [PubMed] [Google Scholar]
- Pierce J. W., Lenardo M., Baltimore D. Oligonucleotide that binds nuclear factor NF-kappa B acts as a lymphoid-specific and inducible enhancer element. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1482–1486. doi: 10.1073/pnas.85.5.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plon S. E., Wang J. C. Transcription of the human beta-globin gene is stimulated by an SV40 enhancer to which it is physically linked but topologically uncoupled. Cell. 1986 May 23;45(4):575–580. doi: 10.1016/0092-8674(86)90289-8. [DOI] [PubMed] [Google Scholar]
- Ptashne M. Gene regulation by proteins acting nearby and at a distance. Nature. 1986 Aug 21;322(6081):697–701. doi: 10.1038/322697a0. [DOI] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988 Oct 20;335(6192):683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Ruden D. M., Ma J., Ptashne M. No strict alignment is required between a transcriptional activator binding site and the "TATA box" of a yeast gene. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4262–4266. doi: 10.1073/pnas.85.12.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirm S., Jiricny J., Schaffner W. The SV40 enhancer can be dissected into multiple segments, each with a different cell type specificity. Genes Dev. 1987 Mar;1(1):65–74. doi: 10.1101/gad.1.1.65. [DOI] [PubMed] [Google Scholar]
- Schleif R. DNA looping. Science. 1988 Apr 8;240(4849):127–128. doi: 10.1126/science.3353710. [DOI] [PubMed] [Google Scholar]
- Schleif R. Gene regulation: why should DNA loop? Nature. 1987 Jun 4;327(6121):369–370. doi: 10.1038/327369a0. [DOI] [PubMed] [Google Scholar]
- Schüle R., Muller M., Kaltschmidt C., Renkawitz R. Many transcription factors interact synergistically with steroid receptors. Science. 1988 Dec 9;242(4884):1418–1420. doi: 10.1126/science.3201230. [DOI] [PubMed] [Google Scholar]
- Schüle R., Muller M., Otsuka-Murakami H., Renkawitz R. Cooperativity of the glucocorticoid receptor and the CACCC-box binding factor. Nature. 1988 Mar 3;332(6159):87–90. doi: 10.1038/332087a0. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Vigneron M., Matthes H., Wildeman A., Zenke M., Chambon P. Requirement of stereospecific alignments for initiation from the simian virus 40 early promoter. Nature. 1986 Jan 9;319(6049):121–126. doi: 10.1038/319121a0. [DOI] [PubMed] [Google Scholar]
- Théveny B., Bailly A., Rauch C., Rauch M., Delain E., Milgrom E. Association of DNA-bound progesterone receptors. Nature. 1987 Sep 3;329(6134):79–81. doi: 10.1038/329079a0. [DOI] [PubMed] [Google Scholar]
- Treisman R., Maniatis T. Simian virus 40 enhancer increases number of RNA polymerase II molecules on linked DNA. Nature. 1985 May 2;315(6014):73–75. doi: 10.1038/315072a0. [DOI] [PubMed] [Google Scholar]
- Veldman G. M., Lupton S., Kamen R. Polyomavirus enhancer contains multiple redundant sequence elements that activate both DNA replication and gene expression. Mol Cell Biol. 1985 Apr;5(4):649–658. doi: 10.1128/mcb.5.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. C., Giaever G. N. Action at a distance along a DNA. Science. 1988 Apr 15;240(4850):300–304. doi: 10.1126/science.3281259. [DOI] [PubMed] [Google Scholar]
- Wang X. F., Calame K. The endogenous immunoglobulin heavy chain enhancer can activate tandem VH promoters separated by a large distance. Cell. 1985 Dec;43(3 Pt 2):659–665. doi: 10.1016/0092-8674(85)90238-7. [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Wasylyk C., Augereau P., Chambon P. The SV40 72 bp repeat preferentially potentiates transcription starting from proximal natural or substitute promoter elements. Cell. 1983 Feb;32(2):503–514. doi: 10.1016/0092-8674(83)90470-1. [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Wasylyk C., Chambon P. Short and long range activation by the SV40 enhancer. Nucleic Acids Res. 1984 Jul 25;12(14):5589–5608. doi: 10.1093/nar/12.14.5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber F., Schaffner W. Simian virus 40 enhancer increases RNA polymerase density within the linked gene. Nature. 1985 May 2;315(6014):75–77. doi: 10.1038/315075a0. [DOI] [PubMed] [Google Scholar]
- Westin G., Gerster T., Müller M. M., Schaffner G., Schaffner W. OVEC, a versatile system to study transcription in mammalian cells and cell-free extracts. Nucleic Acids Res. 1987 Sep 11;15(17):6787–6798. doi: 10.1093/nar/15.17.6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth T., Staudt L., Baltimore D. An octamer oligonucleotide upstream of a TATA motif is sufficient for lymphoid-specific promoter activity. Nature. 1987 Sep 10;329(6135):174–178. doi: 10.1038/329174a0. [DOI] [PubMed] [Google Scholar]
- Wu L., Berk A. Constraints on spacing between transcription factor binding sites in a simple adenovirus promoter. Genes Dev. 1988 Apr;2(4):403–411. doi: 10.1101/gad.2.4.403. [DOI] [PubMed] [Google Scholar]
- de Villiers J., Olson L., Banerji J., Schaffner W. Analysis of the transcriptional enhancer effect. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):911–919. doi: 10.1101/sqb.1983.047.01.105. [DOI] [PubMed] [Google Scholar]