Abstract
A newly discovered group of human rhinoviruses (HRVs) has been classified as the HRV-C species based on distinct genomic features. HRV-Cs circulate worldwide, and are important causes of upper and lower respiratory illnesses. Methods to culture and produce these viruses have recently been developed, and should enable identification of unique features of HRV-C replication and biology.
Keywords: Human rhinovirus C, molecular detection, epidemiology, clinical outcomes, genetic diversity, recombination, replication, organ culture, reverse genetics
1. Introduction
Human rhinoviruses (HRVs) are positive-strand RNA viruses of the genus Enterovirus in the family Picornaviridae sharing a common genome organization and virion properties. The first HRV was discovered in 1953 and, by 1987, HRVs were classified into 100 distinct “classical” serotypes based on their antigenic crossreactivity in neutralization tests [1]. These multiple serotypes have also been grouped on the basis of cellular receptor specificity (major, utilizing intercellular adhesion molecule 1 (ICAM-1) and minor, low density lipoprotein receptor (LDLR) groups) and sensitivity to antiviral capsid-binding compounds (A and B groups) [2,3]. More recently, both full-length and partial (mainly VP4/VP2 and VP1 coding regions) genome sequencing of prototype strains and clinical isolates revealed two major HRV genotypes or clades [4–7]. This phylogenetic classification correlated better with the antiviral drug sensitivity than with the receptor grouping, and suggested a fundamental division of HRVs into two species, HRV-A and HRV-B.
The development of highly-sensitive molecular techniques for detecting HRV genome in a variety of clinical specimens led to discovery of novel rhinoviruses designated HRV-C (aka HRV-A2 or HRV-X), that were subsequently shown to meet the species sequence demarcation criteria (less than 70% amino acid identity in the P1, 2C and 3CD regions) in the genus Enterovirus. HRV-C isolates do not grow in standard cell culture (e.g. HeLa or embryonic lung fibroblasts), and this distinguishing feature most likely has postponed their discovery until recently [8–12] using moleculat diagnostics that do not require virus isolation or passaging. Sequencing of the complete reference set of HRV-A and HRV-B at the full-genome level and comparative analysis with available (at the time) complete genomes of seven HRV-Cs confirmed clustering of all strains into three phylogenetically distinct groups [13]. This phylogeny together with the presence of species-specific sequence and structural elements [11,14,15] resulted in recent revision of HRV taxonomic classification by the International Committee on Taxonomy of Viruses to include the third HRV species C [16].
The 99 acknowledged types of HRV-A and HRV-B are quite well studied, and cause more than half of the upper respiratory infections known as the common cold. It is well known that HRV-induced colds are responsible for millions of work hours lost by sniffling patients, but overall the public perception is that HRV infections are generally benign, and, therefore, the more severe consequences of HRV infections are often overlooked. HRVs are detected very frequently, either singly or in combination with other viruses, in patients hospitalized with wheezing or pneumonia [17,18]. One or more HRV are also typically found in the majority of young children and adults with acute symptoms of asthma (reviewed by [19,20]), and HRV infections of all types are additional significant contributors to morbidity associated with exacerbations of other chronic lung diseases such as cystic fibrosis and chronic obstructive pulmonary disease [21,22].
From the dozens of new HRV-C strains described in recent years, it is now recognized that these particular viruses, as well as additional pathogenic varieties of the HRV-A and HRV-B are very widespread and continuously co-circulating on all continents throughout the world [23]. In patients, these virus infections are associated with flu-like symptoms [9], acute low respiratory tract infections, bronchiolitis and pneumonia, especially in children [14,17,18,24–29], and wheezing and asthma exacerbations [30,31]. There is evidence that HRV-C may be more virulent, as HRV-C infections appear to be overrepresented in some studies of children with pneumonia, or acute wheezing and exacerbations of asthma [28,32–34]. Additional population-based studies are required to make definitive conclusions about relative HRV prevalence and virulence.
The molecular typing assays, sequence variability, genomic features and clinical impact of the newly identified HRVs have been recently reviewed in great detail by Arden & Mackay [35] posing, however, a number of important questions on HRV-C biology, evolution and pathogenesis. In the current review, we address some of these questions and discuss current strategies of molecular genotyping, genetic diversity and recombination, recent classification of HRV-C species, as well as first organ culture and reverse genetics system, and our attempts to identify cellular receptor utilized by these viruses.
2. Current strategies for molecular detection and genotyping of HRV-C
Although the first PCR assay for detection of HRV in clinical samples was reported in 1989 [36], PCR-based HRV screening has been more widely used since the discovery of HRV-C known to be refractory to cell culture isolation. Universal diagnostic primers which anneal to highly-conserved motifs in 5’ untranslated region (5’-UTR) provide the most sensitive assay (10–50 molecules per sample) for detection of both prototype strains [36–38] and novel HRV variants in both convential and real-time PCR formats [12,39–41]. Direct sequencing of the relatively short (270 – 390 base pairs (bp)) 5’-UTR amplicons is usually informative enough for strain typing [12,41]; however, species assignment (A, B or C) is sometimes more difficult due to putative recombination events between HRV-A and HRV-C strains in this region resulting in incongruent clustering when compared to that from coding sequence analysis. As it has been noticed previously [35], some HRV-A sequences (e.g. HRV-A51, 65, 71, 12, 45 and 78 types) co-localize with a subset of HRV-C sequences which were consequently designated “HRV-Ca” clade due to their 5’-UTR sequences that resemble those of HRV-A [32].
Besides, both we and others [35,42] sometimes (in about 5% of nasal speciemens tested in our lab) observe non-specific amplification of human genomic DNA (chromosome 6 region 11,152,642– 11,153,065, accession number NT007592) or RNA (large regulator RNA B2, accession number GQ497714) sequences when using 5’-UTR diagnostic primers [12]. Although the non-specific PCR product size is similar to that of HRV-specific amplicon (424 bp vs 390 bp, respectively), DNA sequencing can discriminate false-positive results.
In contrast to 5’-UTR, 1A/1B (VP4/VP2) or 1D (VP1) gene phylogenetic analyses helps to achieve unequivocal HRV species classification [5,7,43]; however, these regions are more variable compared with 5’-UTR that impedes design of universal diagnostic primers. As for genetic typing, a significant proportion of HRV-A and HRV-B prototype sequences still have relatively high pairwise nucleotide identity (from more than 90% to 98% in 1A/1B and 1D) thus complicating the definition of objective thresholds for intra- and intertypic sequence variation. One possible reason for the high levels of identity among distinct serotypes might include incorrect historical serotyping of prototype strains. In addition, mutations in the capsid region could lead to key amino acid changes in neutralization epitopes [44] that could not only affect antigenic properties of genetically closely related strains, but also be associated with increased severity of illness [45].
A preliminary comparison of the relative sensitivity of 10 published HRV-specific PCR primer pairs conducted by testing two panels of genotyped clinical specimens (including all three HRV species) from children with colds, influenza-like illnesses or asthma exacerbations demonstrated that none of the pairs could detect all tested HRVs [42]. Faux et al. did not observe any apparent species-specific bias, but found that specimens with a lower RNA concentration, as determined by real-time PCR assay, were less likely to be detected or typed by using other primer pairs, highlighting the potential problem of inefficient HRV detection in some epidemiological studies.
The combination of molecular diagnostic assays targeting both conserved 5’-UTR (including multiplexed formats such as MassTag PCR [9] or Respiratory MultiCode-PLx Assay [46]) and capsid coding regions allows for rapid, sensitive and comprehensive detection and improved species assignment and strain typing results, and also minimize the risk of missing highly divergent HRV variants. A number of recently published epidemiological studies describing different clinical and phylogenetic aspects of newly identified HRV-C have successfully utilized this approach [31,47–49].
While direct sequencing of PCR products is very fast, simple and cost-efficient way of HRV typing widely used in large epidemiological studies, molecular typing is more challenging when more than one HRV isolate is present in one specimen [32]. Accurate genotyping could be especially important for studying the correlation between clinical outcomes and certain HRV types or species. Based on our experience with 5’-UTR-based diagnostic assay, direct sequencing approach usually allows for typing of more than 90% of HRV-positive samples even in large studies (including more than 1000 specimens), therefore cloning step is required to type less than 10% of dual-HRV-containing samples. We were able to successfully distinguish and type HRV in nasal samples co-infected with different strains of the HRV-A, HRV-B and HRV-C in all possible combinations (Bochkov & Gern, unpublished data).
3. Molecular epidemiology and clinical outcomes associated with HRV-C
HRV-A and HRV-B species appear to differ in terms of prevalence, detection rates and number of known types. HRV-C more closely resembles HRV-A than HRV-B in having high prevalence (at least in children – data in adults is relatively scarce) and high genetic diversity currently comprising more than 60 distinct types. As is the case with other respiratory viruses, the range of clinical outcomes associated with HRV-C varies from mild or even asymptomatic infections to acute lower respiratory tract illnesses including pneumonia, recurrent wheezing and bronchitis. Several first reports suggested that HRV-C tend to induce more severe illnesses in young children than the other two HRV species [11,25,30,31,50]. Cross-sectional studies suggest that HRV-A and HRV-C are the predominant species associated with acute respiratory illnesses in hospitalized children and adults [33,34,51–53], Accordingly, a recent case-control study found that both HRV-A and HRV-C detection rates were significantly higher in young children hospitalized for acute respiratory illnesses (n = 1867) compared with asymptomatic controls (n = 784), while HRV-B viruses were seldom detected in either group [49]. HRV-Cs may be less frequently found than HRV-A and HRV-B in non-hospitalized adults with influenza-like illnesses [48,54].
A recent phylogenetic analysis of 144 HRV-positive samples from lung transplant recipients and hospital patients (both children and adults) with upper and lower respiratory tract infections showed no apparent correlation between a given viral genotype or species and their ability to invade the lower respiratory tract or lead to protracted infection [55]. Notably, protracted infections were found in immunocompromised patients, thus suggesting that host factors rather than the virus genotype could modulate disease outcome. The proportion of HRV-C positive samples in this study was significantly higher in pediatric hospital patients than in adult hospital patients with low or upper respiratory tract infections.
HRV-C appears to show seasonal patterns of infection similarly to the other HRV species demonstrating high incidence peaks in early fall and late spring in most temperate or subtropical countries, and during the rainy season in the tropics [10,11,17,24,54]; however, some species-specific monthly distribution was also observed [34,48,53,56].
Either nasal secretions, bronchoalveolar lavage or nasopharyngeal swabs are typically tested for HRV in different clinical studies. Recently, HRV-C was detected in stool, pericardial fluid, and plasma samples of a 14-month-old child hospitalized with pneumonia and acute pericarditis using real-time PCR suggesting that systemic infection of HRV-C could occur in patients with severe illness [25]. Another case of severe pneumonia was reported in a 3-week old neonate where HRV-C was identified in nasal secretions and bronchoalveolar lavage, as well as in stool and urine samples in the absense of any other viral or bacterial agents [57]. More intriguingly, when serum samples from hospitalized children with severe respiratory illnesses were also tested to determine whether specific HRV species were associated with viremia, more than 12% of them were positive for HRV-A (3%, 4/135) or HRV-C (31%, 26/83) but not for HRV-B (0%, 0/25) suggesting that HRV-C may have a different pathogenicity and can more commonly cause viremia than HRV-A and HRV-B [47]. Population-based longitudinal studies are needed to provide definitive answers about species-specific patterns of virulence and species contribution in disease causality.
4. Distinct genomic features and novel genotypic classification of HRV-C
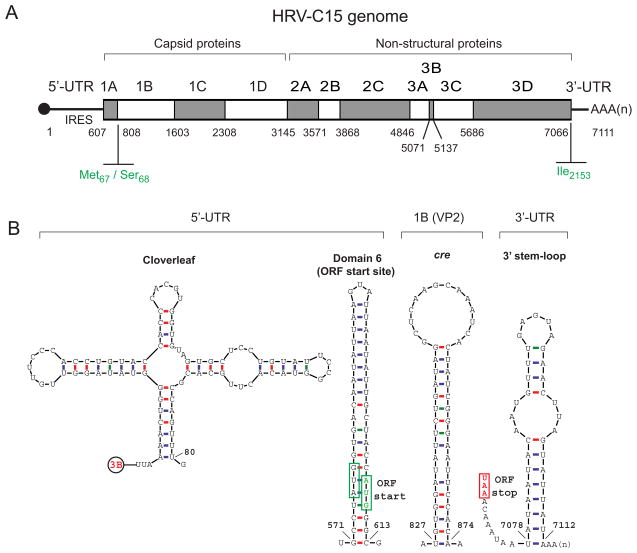
HRV-C shares all basic features of its genomic organization with HRV-A and HRV-B. These features include an RNA genome length of about 7.1 kb containing single open reading frame, 5’-UTR cloverleaf RNA structure necessary for virus replication, type I internal ribosomal entry site (IRES) containing five conserved secondary structure domains (stems) responsible for the cap-independent translation mechanism, and a stem-loop structure in 3’-UTR preceeding poly (A) tract. However, the RNA and deduced polyprotein sequences of HRV-Cs also possess some distinct characteristics supporting its classification as the third HRV species, for example, a putative internal cis-acting replication element (cre) located within 1B (VP2), a unique Met67/Ser68 cleavage site at the VP4/VP2 junction, large species-specific insertions and deletions in the VP1 and some other regions, and an isoleucine at the termination of the 3D polymerase [11,14,58] (Fig. 1).
Fig. 1.
HRV-C genome organization and distinct features. (A) Schematic representation of the HRV-C genome encoding single polyprotein open reading frame, flanked by 5’- and 3’-UTRs. Indicated nucleotide positions correspond to HRV-C15 complete genome sequence (GenBank accession number GU219984). HRV-C-specific genomic features are shown in green. (B) Structural motifs predicted from HRV-C15 genomic RNA and their genomic locations.
Due to the lack of accessible methods for in vitro culture to study cross-neutralization properties, a genetically-based system was proposed for the classification of HRV-C species [59]. Sequence analysis of a large number of HRV-C variants showed clustering of genetic groups into well-defined clades similarly to clustering of HRV-A and HRV-B serotypes, with two well-separated distributions (inter- and intratype) of pairwise distances in structural genes suggesting that genetic types can be defined and demarcated for classification purposes [60]. Since the majority of HRV-C isolates are not yet fully sequenced, this classification is based on sequence identity criteria in two capsid-coding regions (1D and partial 1A/1B); however, the most of the HRV-C published sequence data have been collected for only 5’-UTR and 1A/1B.
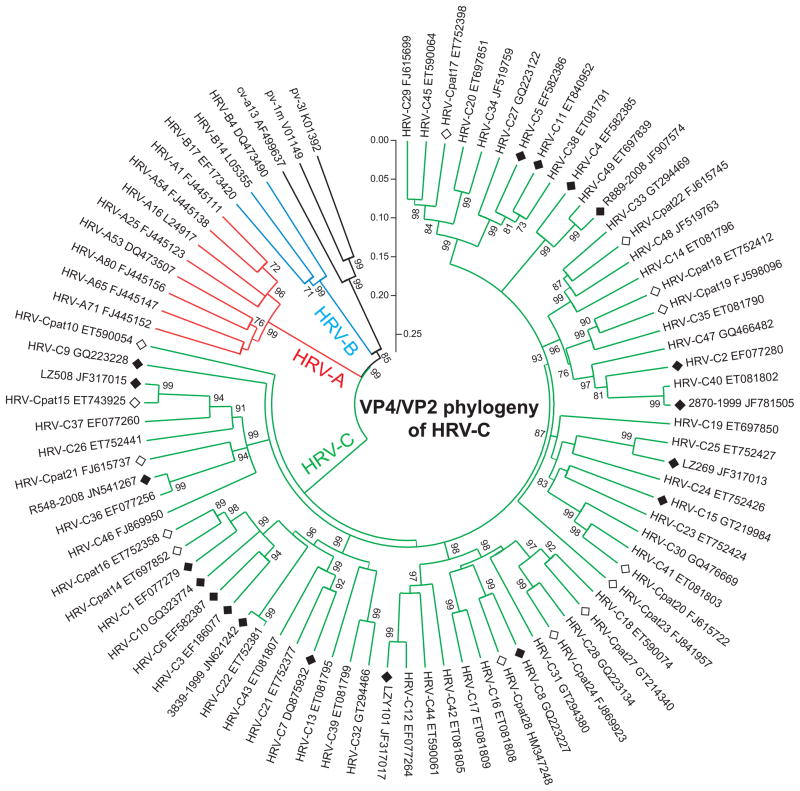
HRV-Cs were classified to 33 types using a threshold of 13% nucleotide divergence in 1D gene, and, additionally, to 28 provisionally assigned types with >10% divergence in 1A/1B (1D sequences are lacking). Therefore, HRV-C variants identified and sequenced by 2010 were assigned into a total of 61 genotypes. This classification has been recently updated by adding additional 1D sequences to the database to include now 49 types (http://www.picornastudygroup.com/types/enterovirus/hrv-c.htm); whereas, 14 provisionally assigned types are still awaiting the 1D or complete genome sequences. (Fig. 2). Either complete genome or complete ORF sequences are currently (November 29, 2011) available for 19 HRV-C types (C1 – 12, 15, 22, 25, 36, 40, 49 and pat15). Indeed, full-genome sequencing of the representatives of each remaining HRV-C type is neccessary for precise strain classification and complete understanding of the virus evolution.
Fig. 2.
Neighbor-joining phylogenetic tree based on partial 1A/1B (VP4/VP2) nucleotide sequences (positions 608–1002 of HRV-C15 complete genome, accession number GU219984) of all sequenced types of HRV-C species, and representatives of major branches of HRV-A and HRV-B species, constructed using MEGA 4.1 software [78]. All major nodes are labeled with bootstrap values (1000 replicates, with its value more than 70%). Branch lengths are proportional to nucleotide similarity (p-distance). Human enteroviruses (polio- and coxsackieviruses) are included as an outgroup. HRV-C types followed by GenBank accession numbers correspond to the recent classification proposal [59]. Fully-sequenced (genomes or ORFs) and provisionally assigned (pat) HRV-C types are shown by filled and open diamonds, respectively.
5. Genetic diversity and recombination in evolution of HRV-C
HRV is a very genetically diverse group compared to other RNA viruses, and the HRV discovery phase is still ongoing rather than complete; however, the newly identified HRV-Cs are not emerging strains, but have most likely been circulating for a long time without detection [23,35]. On the other hand, HRVs often reveal high level of conservation in different genomic regions both among variants of the same strain (e.g. HRV-QPM) detected over relatively short time period [10] and among temporally distant strains of classical HRV-A and HRV-B serotypes which were first detected in 1960–1970s [8,12,35]. Amino acid differences in the structural genes are usually concentrated within predicted neutralization epitopes [45]. In agreement with this, the HRV genome as a whole was proposed to be under purifying selective pressure, with only islands of diversifying pressure in structural genes (1B, 1C and 1D) mapped to the external surface of the virion, and, therefore, potentially interacting with the host immune system, and also in two non-structural genes (3C and 3D) [61]. Analysis of the HRV-A39 genome evolution in experimentally infected human volunteers also confirmed the presence of those mutation hot and cold spots [62]. Overall, HRV-C isolates revealed significantly higher genetic diversity than HRV-A and HRV-B serotypes.
Although underestimated initially as a driving force of HRV evolution [61], recombination has been predicted to occur between certain reference types [13] and circulating clinical strains [40] of HRV-A and HRV-B contributing to wide genetic diversity of these species. Phylogenetic analysis of the partial 5’-UTR and 1A/1B gene sequences revealed incongruent clustering of the 34 Chinese HRV-C strains [32]. They all were initially classified into HRV-C species according to 1A/1B phylogenies showing more than 47% nucleotide difference from HRV-A and HRV-B. However, the majority of them (n = 20) were closely related to some HRV-A strains (HRV-Ca clade) in the phylogenetic tree based on 5’-UTR suggesting interspecies recombination, while the fourteen other strains from this study formed a unique cluster (HRV-Cc) different from the rest of the HRVs and other human enterovirus (HEV) species. Bootscanning and similarity plot analyses in the gene fragment that included partial 5’-UTR and adjacent capsid genes (868 nt) of representative strains of HRV-Ca cluster revealed variable recombination sites all located inside the 5’-UTR rather than in downstream coding sequence for the first time suggesting putative recombination events involving HRV-C [32].
Through sequence comparisons of the 5’-UTR, a total of 175 positive samples identified as HRV-C by 1A/1B screening were classified into 2 groups, HRV-Ca (n = 96) and HRV-Cc (n = 79). The multiple phylogenetic subclades of HRV-Ca in both 5’-UTR and 1A/1B-based phylogenetic trees were consistent with a series of putative interspecies recombination events in their evolutionary history. In agreement with the previous study, there were no significant differences in clinical outcomes or epidemiology between HRV-Ca and HRV-Cc isolates [50].
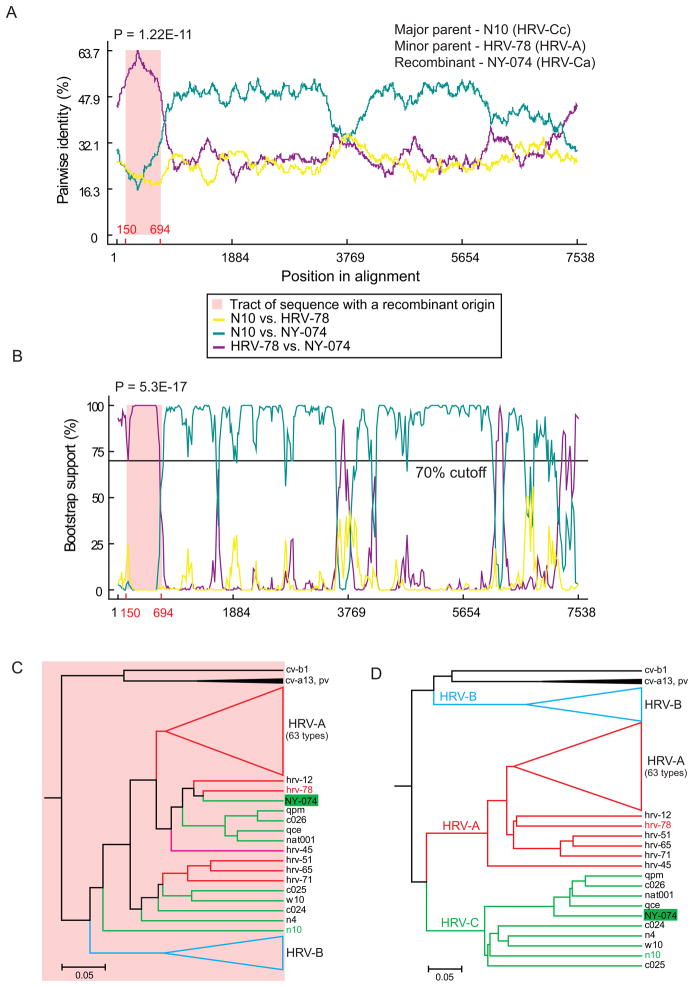
Scanning of our complete genome set of HRV-A, HRV-B and HRV-C sequences augmented with the N10 strain (accession number GQ223228), the only available full-length sequence representing HRV-Cc 5’-UTR cluster, also confirmed potential interspecies recombination events in HRV-C evolution (Fig. 3). Additional full-genome HRV-C sequences are necessary for more accurate and reliable recombination analysis.
Fig. 3.
Putative interspecies recombination in HRV-C 5’-UTR. Similarity plot (A) and bootscanning (B) analyses of the HRV-C7 strain NY-074 (GenBank accession number DQ875932) implemented by the RDP and Bootscan methods (window size 200, step size 20, 1000 replicates), respectively, in RDP3.31 show the predicted recombination crossover site in 5’-UTR. Phylogenetic trees constructed from (C) the putative recombinant sequence (5’-UTR positions 150–594) and (D) the rest of genome reveals incongruent clustering of recombinant strain. Parental strains are shown by red (HRV-A) and green (HRV-C) colors, and the recombinant strain is shaded in green.
More recent analysis of genetic diversity in 89 HRV-C-positive clinical specimens also revealed dual-branch clustering in 5’-UTR (HRV-Ca and HRV-Cc clades) and almost identical tree topologies showing one clearly defined HRV-C branch in different coding regions (e.g. partial 1A/1B, 1D and 3D), suggesting recombination with HRV-A sequences in the 5’-UTR – VP4 junction in more than 60% of analyzed variants [60]. Recombination hot spots were mapped in stem-loop 5 and polypyrimidine tract between stem-loops 5 and 6 of HRV-Ca 5’-UTR, and the corresponding clades were interspersed within the 1A/1B gene tree, indicating several independent recombination events. In addition to the 5’-UTR, the second site of interspecies (A-C) recombination was predicted in the C-terminal domain of 2A protease gene (~ 260 bases) of all HRV-C strains genetically characterized in this region. HRV-C sequences grouped within the HRV-A cluster in a manner similar to that found in the 5’-UTR except N10, which formed distinct HRV-Cc 5’-UTR clade, was embedded within HRV-A clade.
In order to test the interspecies recombination potential in the 5’-UTR among different Enterovirus species, replication and translation efficiences of in vitro generated chimeric genomes containing complete 5’-UTRs from HRV-Ca, HRV-Cc, HRV-B and HEV-A species and the remaining part of the genome from HRV-A16 strain were compared [63]. The results confirmed functional compatibility of all selected 5’-UTRs in the context of HRV-A16 genome resulting in productive translation and replication; however, the wild-type HRV-A16 still retained a replication advantage. Although all of the chimeric viruses were easily propagated in cell culture, the virus progenies were not able to outcompete the well-adapted parental strain. These data support phylogenetic predictions of ancient recombination events among HRV and HEV species.
Overall, recombination analysis of HRV shows dramatic differences from the other Picornaviridae species (e.g. HEV, aphtoviruses and cardioviruses) with extensively documented recombination events in both structural and non-structural regions. It has been proposed that high sequence divergence throughout the coding region of HRV (e.g. more than 30% nucleotide differences between HRV-C types in 3D), in contrast to some other picornaviruses, could decrease the probability of biologically viable recombinants with breakpoints in the nonstructural genes [60]. Although evidence of some evolutionary ancient recombination events at the P1/P2 boundary was found among HRV-A sequences [13] no such evidence has been documented for HRV-C strains yet, probably due to more limited sequence data and even greater sequence divergence between HRV-C groups restricting biological compatibility of recombinants [60]. However, high frequency of recombination was predicted in HRV-C 5’-UTR most likely due to high level of sequence conservation and the presence of similar secondary structure elements (e.g. stem-loops) between the HRV-A and HRV-C species permitting biological compatibility. Multiple occurrence of recombination in HRV-C and the absence of HRV-A with species C 5’-UTR sequences suggest that species A 5’-UTR could be evolutionary advantageous [50].
6. HRV-C pathogenesis: binding, entry and replication
HRV-A and HRV-B enter the host through inoculation of either the eyes or nose and replicate primarily in the airway epithelium. They utilize two major types of cellular receptors to initiate infection, ICAM-1 and LDLR family members including LDLR, very LDLR and LDLR-related protein. Although some major receptor group HRV variants (e.g. HRV-54, 89 and 8) could use another receptor (heparan sulfate) for binding in vitro, the infection is usually less efficient, and these variants preserve their ability to attach to ICAM-1 [64,65].
Our bioinformatic comparisons of the available HRV-C sequences showed unique amino acid composition profiles in the putative ICAM-1 and LDLR receptor footprint locales, inconsistent with all known major and minor group receptor interactions suggesting a unique receptor use. Failure of the existing cell culture systems to grow HRV-C was also consistent with the novel receptor specificity, but could also indicate some other undefined receptor-independent limitation, prohibiting HRV-C replication inside cultured cells. After development of a reverse genetics system for HRV-C, we demonstrated that the full-length viral RNA transcripts synthesized in vitro were infectious when transfected into cell lines resistant to HRV-C infection [66].
HRV-A and HRV-B replication is greatest in the upper airway in most individuals, and high-level replication can also occur in the large and medium-sized airways. In addition to infecting the nasopharynx, conjunctiva and lower airways, classical HRVs have also been recovered in specimens obtained from the middle ear and sinuses [67–69]. After multiple attempts to grow HRV-C isolates from nasal samples in different cell cultures without success, we employed human organ culture of sinus mucosa to propagate in vitro HRV-C15 clinical isolate. HRV-C replication in sinus tissue was limited to the epithelium, and in situ hybridization signals were associated with both non-ciliated and ciliated cells [66].
It has been shown earlier that classical HRVs can replicate in both ciliated cells of nasal polyps and in nonciliated epithelial cells in adenoid fragments infected in vitro [70], as well as in both ciliated and non-ciliated cells in the nasopharyngeal biopsies after experimental HRV infection in humans [71]. The overall frequency of infection was shown to be low, little higher in ciliated then in nonciliated cells. We were not able to confirm HRV-C growth in cultured epithelium from adenoids or nasal polyps. In sinus mucosal organ culture, we observed larger numbers of HRV-C positive epithelial cells than were reported in previous in vitro studies of classical HRVs [70,71], but similar to that detected in sinus biopsy specimens obtained from patients with acute sinusitis [72], and in nasal epithelium biopsies from patients with common colds [73]. Availability of HRV-C-specific antibodies for immunohistochemistry studies might help to confirm specific type of cell in sinus mucosa that supports virus replication.
Thus, HRV-C binds to an unknown receptor(s) that is expressed on epithelial cells in differentiated tissues, but is either absent or underexpressed in many cell lines tested so far, and is clearly distinct from receptors utilized by other HRV species. Notably, replication of HRV-C in sinus organ culture was not affected by antibodies that block attachment of major and minor group HRVs to their receptors (ICAM-1 and LDLR) [66]. It is also possible that expression of HRV-C-specific receptor(s) on epithelial cells may depend on factors found in vivo such as microbial products or interactions with other types of cells also present in human airways. Interestingly, both intestinal microflora and its certain components (e.g. lipopolysaccharide and peptidoglycans) have been shown to interact with poliovirus and promote its replication and transmission in mice [74].
Although differentiated epithelial cells grown at air-liquid interface are more resistant to HRV-A infection compared to undifferentiated monolayers [75], interleukin-13 -induced goblet cell metaplasia lead to greater HRV yield in vitro, suggesting that virus replication is increased in goblet cells [76]. In addition, mechanical damage to well-differentiated cells significantly enhances HRV-A replication in vitro [77]. We tried to utilize these interesting findings to culture HRV-C15 isolate; however, neither interleukin-13 treatment nor mechanical damage helped to make differentiated bronchial epithelial cells (from two donors) susceptible to infection [66]. Additional cultures from larger number of donors and from different airway locales, as well as other treatment options might help to adapt HRV-C to grow in these cultures.
7. Conclusions
The development of highly-sensitive molecular techniques for detecting HRV genome in a variety of clinical specimens has recently resulted in discovery of the novel rhinovirus genotype circulating worldwide, that was further classified as the third HRV species C within the genus Enterovirus. The range of clinical outcomes associated with HRV-C varies from mild or even asymptomatic infections to acute lower respiratory tract illnesses including pneumonia, recurrent wheezing, bronchitis and even more rarely reported cases of systemic infection. The combination of available molecular diagnostic assays targeting both highly conserved motifs in 5’-UTR and more variable capsid-coding regions is necessary for sensitive and comprehensive HRV detection, improved species assignment and strain genotyping results. HRV-C along with HRV-A are typically more prevalent species in most of the epidemiological studies compared to HRV-B which are rarely detected. Current genetically-based classification assigns known HRV-Cs in 63 types using sequence diversity thresholds in 1D and 1A/1B regions; however, the HRV-C discovery phase is not complete yet. A series of both ancient and probably more recent putative interspecies recombination events in 5’-UTR and 2A regions contributed to evolution of a large number of HRV-C strains. HRV-C does not grow in standard cell lines (e.g. HeLa or embryonic lung fibroblasts) or primary airway epithelial cell cultures that readily support infection with HRV-A and HRV-B, but it efficiently replicates in fully-differentiated sinus epithelium cultured in vitro. Unique amino acid composition profiles in the known HRV receptor footprint locales and infectivity of the in vitro synthesized HRV-C RNA transcripts, transfected into cell lines resistant to HRV-C infection, suggest a novel HRV-C receptor binding specificity.
Acknowledgments
This work was supported by NIH grants HHSN272200900052C, P01 HL070831 and U19 AI070503-01.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hamparian VV, Colonno RJ, Cooney MK, Dick EC, Gwaltney JM, Jr, Hughes JH, Jordan WS, Jr, Kapikian AZ, Mogabgab WJ, Monto A, Phillips CA, Rueckert RR, Schieble JH, Stott EJ, Tyrrell DAJ. A collaborative report: rhinoviruses--extension of the numbering system from 89 to 100. Virology. 1987;159:191–192. doi: 10.1016/0042-6822(87)90367-9. [DOI] [PubMed] [Google Scholar]
- 2.Andries K, Dewindt B, Snoeks J, Wouters L, Moereels H, Lewi PJ, Janssen PA. Two groups of rhinoviruses revealed by a panel of antiviral compounds present sequence divergence and differential pathogenicity. J Virol. 1990;64:1117–1123. doi: 10.1128/jvi.64.3.1117-1123.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uncapher CR, DeWitt CM, Colonno RJ. The major and minor group receptor families contain all but one human rhinovirus serotype. Virology. 1991;180:814–817. doi: 10.1016/0042-6822(91)90098-v. [DOI] [PubMed] [Google Scholar]
- 4.Horsnell C, Gama RE, Hughes PJ, Stanway G. Molecular relationships between 21 human rhinovirus serotypes. J Gen Virol. 1995;76:2549–2555. doi: 10.1099/0022-1317-76-10-2549. [DOI] [PubMed] [Google Scholar]
- 5.Savolainen C, Blomqvist S, Mulders MN, Hovi T. Genetic clustering of all 102 human rhinovirus prototype strains: serotype 87 is close to human enterovirus 70. J Gen Virol. 2002;83:333–340. doi: 10.1099/0022-1317-83-2-333. [DOI] [PubMed] [Google Scholar]
- 6.Savolainen C, Mulders MN, Hovi T. Phylogenetic analysis of rhinovirus isolates collected during successive epidemic seasons. Virus Res. 2002;85:41–46. doi: 10.1016/s0168-1702(02)00016-3. [DOI] [PubMed] [Google Scholar]
- 7.Ledford RM, Patel NR, Demenczuk TM, Watanyar A, Herbertz T, Collett MS, Pevear DC. VP1 sequencing of all human rhinovirus serotypes: insights into genus phylogeny and susceptibility to antiviral capsid-binding compounds. J Virol. 2004;78:3663–3674. doi: 10.1128/JVI.78.7.3663-3674.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arden KE, McErlean P, Nissen MD, Sloots TP, Mackay IM. Frequent detection of human rhinoviruses, paramyxoviruses, coronaviruses, and bocavirus during acute respiratory tract infections. J Med Virol. 2006;78:1232–1240. doi: 10.1002/jmv.20689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lamson D, Renwick N, Kapoor V, Liu Z, Palacios G, Ju J, Dean A, St George K, Briese T, Lipkin WI. MassTag polymerase-chain-reaction detection of respiratory pathogens, including a new rhinovirus genotype, that caused influenza-like illness in New York State during 2004-2005. J Infect Dis. 2006;194:1398–1402. doi: 10.1086/508551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McErlean P, Shackelton LA, Lambert SB, Nissen MD, Sloots TP, Mackay IM. Characterisation of a newly identified human rhinovirus, HRV-QPM, discovered in infants with bronchiolitis. J Clin Virol. 2007;39:67–75. doi: 10.1016/j.jcv.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lau SK, Yip CC, Tsoi HW, Lee RA, So LY, Lau YL, Chan KH, Woo PC, Yuen KY. Clinical features and complete genome characterization of a distinct human rhinovirus (HRV) genetic cluster, probably representing a previously undetected HRV species, HRV-C, associated with acute respiratory illness in children. J Clin Microbiol. 2007;45:3655–3664. doi: 10.1128/JCM.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee WM, Kiesner C, Pappas T, Lee I, Grindle K, Jartti T, Jakiela B, Lemanske RF, Jr, Shult PA, Gern JE. A diverse group of previously unrecognized human rhinoviruses are common causes of respiratory illnesses in infants. PLoS One. 2007;2:e966. doi: 10.1371/journal.pone.0000966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palmenberg AC, Spiro D, Kuzmickas R, Wang S, Djikeng A, Rathe JA, Fraser-Liggett CM, Liggett SB. Sequencing and analyses of all known human rhinovirus genomes reveal structure and evolution. Science. 2009;324:55–59. doi: 10.1126/science.1165557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McErlean P, Shackelton LA, Andrews E, Webster DR, Lambert SB, Nissen MD, Sloots TP, Mackay IM. Distinguishing molecular features and clinical characteristics of a putative new rhinovirus species, human rhinovirus C (HRV C) PLoS One. 2008;3:e1847. doi: 10.1371/journal.pone.0001847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arden KE, Faux CE, O'Neill NT, McErlean P, Nitsche A, Lambert SB, Nissen MD, Sloots TP, Mackay IM. Molecular characterization and distinguishing features of a novel human rhinovirus (HRV) C, HRVC-QCE, detected in children with fever, cough and wheeze during 2003. J Clin Virol. 2010;47:219–223. doi: 10.1016/j.jcv.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carstens EB. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2009) Arch Virol. 2010;155:133–146. doi: 10.1007/s00705-009-0547-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lau SK, Yip CC, Lin AW, Lee RA, So LY, Lau YL, Chan KH, Woo PC, Yuen KY. Clinical and molecular epidemiology of human rhinovirus C in children and adults in Hong Kong reveals a possible distinct human rhinovirus C subgroup. J Infect Dis. 2009;200:1096–1103. doi: 10.1086/605697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiang Z, Gonzalez R, Wang Z, Xiao Y, Chen L, Li T, Vernet G, Paranhos-Baccala G, Jin Q, Wang J. Human rhinoviruses in Chinese adults with acute respiratory tract infection. J Infect. 2010;61:289–298. doi: 10.1016/j.jinf.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gern JE, Busse WW. Association of rhinovirus infections with asthma. Clin Microbiol Rev. 1999;12:9–18. doi: 10.1128/cmr.12.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gern JE. The ABCs of rhinoviruses, wheezing, and asthma. J Virol. 2010;84:7418–7426. doi: 10.1128/JVI.02290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wat D, Gelder C, Hibbitts S, Cafferty F, Bowler I, Pierrepoint M, Evans R, Doull I. The role of respiratory viruses in cystic fibrosis. J Cyst Fibros. 2008;7:320–328. doi: 10.1016/j.jcf.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McManus TE, Marley AM, Baxter N, Christie SN, O'Neill HJ, Elborn JS, Coyle PV, Kidney JC. Respiratory viral infection in exacerbations of COPD. Respir Med. 2008;102:1575–1580. doi: 10.1016/j.rmed.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Briese T, Renwick N, Venter M, Jarman RG, Ghosh D, Kondgen S, Shrestha SK, Hoegh AM, Casas I, Adjogoua EV, Akoua-Koffi C, Myint KS, Williams DT, Chidlow G, van den Berg R, Calvo C, Koch O, Palacios G, Kapoor V, Villari J, Dominguez SR, Holmes KV, Harnett G, Smith D, Mackenzie JS, Ellerbrok H, Schweiger B, Schonning K, Chadha MS, Leendertz FH, Mishra AC, Gibbons RV, Holmes EC, Lipkin WI. Global distribution of novel rhinovirus genotype. Emerg Infect Dis. 2008;14:944–947. doi: 10.3201/eid1406.080271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiang Z, Gonzalez R, Xie Z, Xiao Y, Chen L, Li Y, Liu C, Hu Y, Yao Y, Qian S, Geng R, Vernet G, Paranhos-Baccala G, Shen K, Jin Q, Wang J. Human rhinovirus group C infection in children with lower respiratory tract infection. Emerg Infect Dis. 2008;14:1665–1667. doi: 10.3201/eid1410.080545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tapparel C, L'Huillier AG, Rougemont AL, Beghetti M, Barazzone-Argiroffo C, Kaiser L. Pneumonia and pericarditis in a child with HRV-C infection: a case report. J Clin Virol. 2009;45:157–160. doi: 10.1016/j.jcv.2009.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piralla A, Rovida F, Campanini G, Rognoni V, Marchi A, Locatelli F, Gerna G. Clinical severity and molecular typing of human rhinovirus C strains during a fall outbreak affecting hospitalized patients. J Clin Virol. 2009;45:311–317. doi: 10.1016/j.jcv.2009.04.016. [DOI] [PubMed] [Google Scholar]
- 27.Jin Y, Yuan XH, Xie ZP, Gao HC, Song JR, Zhang RF, Xu ZQ, Zheng LS, Hou YD, Duan ZJ. Prevalence and clinical characterization of a newly identified human rhinovirus C species in children with acute respiratory tract infections. J Clin Microbiol. 2009;47:2895–2900. doi: 10.1128/JCM.00745-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Linsuwanon P, Payungporn S, Samransamruajkit R, Posuwan N, Makkoch J, Theanboonlers A, Poovorawan Y. High prevalence of human rhinovirus C infection in Thai children with acute lower respiratory tract disease. J Infect. 2009;59:115–121. doi: 10.1016/j.jinf.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han TH, Chung JY, Hwang ES, Koo JW. Detection of human rhinovirus C in children with acute lower respiratory tract infections in South Korea. Arch Virol. 2009;154:987–991. doi: 10.1007/s00705-009-0383-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khetsuriani N, Lu X, Teague WG, Kazerouni N, Anderson LJ, Erdman DD. Novel human rhinoviruses and exacerbation of asthma in children. Emerg Infect Dis. 2008;14:1793–1796. doi: 10.3201/eid1411.080386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller EK, Khuri-Bulos N, Williams JV, Shehabi AA, Faouri S, Al J, Chen IQ, Heil L, Mohamed Y, Morin LL, Ali A, Halasa NB. Human rhinovirus C associated with wheezing in hospitalised children in the Middle East. J Clin Virol. 2009;46:85–89. doi: 10.1016/j.jcv.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang T, Wang W, Bessaud M, Ren P, Sheng J, Yan H, Zhang J, Lin X, Wang Y, Delpeyroux F, Deubel V. Evidence of recombination and genetic diversity in human rhinoviruses in children with acute respiratory infection. PLoS One. 2009;4:e6355. doi: 10.1371/journal.pone.0006355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mak RK, Tse LY, Lam WY, Wong GW, Chan PK, Leung TF. Clinical spectrum of human rhinovirus infections in hospitalized Hong Kong children. Pediatr Infect Dis J. 2011;30:749–753. doi: 10.1097/INF.0b013e31821b8c71. [DOI] [PubMed] [Google Scholar]
- 34.Smuts HE, Workman LJ, Zar HJ. Human rhinovirus infection in young African children with acute wheezing. BMC Infect Dis. 2011;11:65. doi: 10.1186/1471-2334-11-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arden KE, Mackay IM. Newly identified human rhinoviruses: molecular methods heat up the cold viruses. Rev Med Virol. 2010;20:156–176. doi: 10.1002/rmv.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gama RE, Horsnell PR, Hughes PJ, North C, Bruce CB, al-Nakib W, Stanway G. Amplification of rhinovirus specific nucleic acids from clinical samples using the polymerase chain reaction. J Med Virol. 1989;28:73–77. doi: 10.1002/jmv.1890280204. [DOI] [PubMed] [Google Scholar]
- 37.Ireland DC, Kent J, Nicholson KG. Improved detection of rhinoviruses in nasal and throat swabs by seminested RT-PCR. J Med Virol. 1993;40:96–101. doi: 10.1002/jmv.1890400204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andeweg AC, Bestebroer TM, Huybreghs M, Kimman TG, de Jong JC. Improved detection of rhinoviruses in clinical samples by using a newly developed nested reverse transcription-PCR assay. J Clin Microbiol. 1999;37:524–530. doi: 10.1128/jcm.37.3.524-530.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu X, Holloway B, Dare RK, Kuypers J, Yagi S, Williams JV, Hall CB, Erdman DD. Real-time reverse transcription-PCR assay for comprehensive detection of human rhinoviruses. J Clin Microbiol. 2008;46:533–539. doi: 10.1128/JCM.01739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tapparel C, Junier T, Gerlach D, Van-Belle S, Turin L, Cordey S, Muhlemann K, Regamey N, Aubert JD, Soccal PM, Eigenmann P, Zdobnov E, Kaiser L. New respiratory enterovirus and recombinant rhinoviruses among circulating picornaviruses. Emerg Infect Dis. 2009;15:719–726. doi: 10.3201/eid1505.081286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kiang D, Kalra I, Yagi S, Louie JK, Boushey H, Boothby J, Schnurr DP. Assay for 5' noncoding region analysis of all human rhinovirus prototype strains. J Clin Microbiol. 2008;46:3736–3745. doi: 10.1128/JCM.00674-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Faux CE, Arden KE, Lambert SB, Nissen MD, Nolan TM, Chang AB, Sloots TP, Mackay IM. Usefulness of published PCR primers in detecting human rhinovirus infection. Emerg Infect Dis. 2011;17:296–298. doi: 10.3201/eid1702.101123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laine P, Savolainen C, Blomqvist S, Hovi T. Phylogenetic analysis of human rhinovirus capsid protein VP1 and 2A protease coding sequences confirms shared genus-like relationships with human enteroviruses. J Gen Virol. 2005;86:697–706. doi: 10.1099/vir.0.80445-0. [DOI] [PubMed] [Google Scholar]
- 44.Duechler M, Skern T, Sommergruber W, Neubauer C, Gruendler P, Fogy I, Blaas D, Kuechler E. Evolutionary relationships within the human rhinovirus genus: comparison of serotypes 89, 2, and 14. Proc Natl Acad Sci USA. 1987;84:2605–2609. doi: 10.1073/pnas.84.9.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kiang D, Yagi S, Kantardjieff KA, Kim EJ, Louie JK, Schnurr DP. Molecular characterization of a variant rhinovirus from an outbreak associated with uncommonly high mortality. J Clin Virol. 2007;38:227–237. doi: 10.1016/j.jcv.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 46.Lee WM, Grindle K, Pappas T, Marshall DJ, Moser MJ, Beaty EL, Shult PA, Prudent JR, Gern JE. High-throughput, sensitive, and accurate multiplex PCR-microsphere flow cytometry system for large-scale comprehensive detection of respiratory viruses. J Clin Microbiol. 2007;45:2626–2634. doi: 10.1128/JCM.02501-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fuji N, Suzuki A, Lupisan S, Sombrero L, Galang H, Kamigaki T, Tamaki R, Saito M, Aniceto R, Olveda R, Oshitani H. Detection of human rhinovirus C viral genome in blood among children with severe respiratory infections in the Philippines. PLoS One. 2011;6:e27247. doi: 10.1371/journal.pone.0027247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiang Z, Gonzalez R, Wang Z, Xiao Y, Chen L, Li T, Vernet G, Paranhos-Baccala G, Jin Q, Wang J. Human rhinoviruses in Chinese adults with acute respiratory tract infection. J Infect. 2010;61:289–298. doi: 10.1016/j.jinf.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iwane MK, Prill MM, Lu X, Miller EK, Edwards KM, Hall CB, Griffin MR, Staat MA, Anderson LJ, Williams JV, Weinberg GA, Ali A, Szilagyi PG, Zhu Y, Erdman DD. Human rhinovirus species associated with hospitalizations for acute respiratory illness in young US children. J Infect Dis. 2011;204:1702–1710. doi: 10.1093/infdis/jir634. [DOI] [PubMed] [Google Scholar]
- 50.Wisdom A, Kutkowska AE, McWilliam Leitch EC, Gaunt E, Templeton K, Harvala H, Simmonds P. Genetics, recombination and clinical features of human rhinovirus species C (HRV-C) infections; interactions of HRV-C with other respiratory viruses. PLoS One. 2009;4:e8518. doi: 10.1371/journal.pone.0008518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Piralla A, Baldanti F, Gerna G. Phylogenetic patterns of human respiratory picornavirus species, including the newly identified group C rhinoviruses, during a 1-year surveillance of a hospitalized patient population in Italy. J Clin Microbiol. 2011;49:373–376. doi: 10.1128/JCM.01814-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fry AM, Lu X, Olsen SJ, Chittaganpitch M, Sawatwong P, Chantra S, Baggett HC, Erdman D. Human rhinovirus infections in rural Thailand: epidemiological evidence for rhinovirus as both pathogen and bystander. PLoS One. 2011;6:e17780. doi: 10.1371/journal.pone.0017780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiang Z, Gonzalez R, Xie Z, Xiao Y, Liu J, Chen L, Liu C, Zhang J, Ren L, Vernet G, Paranhos-Baccala G, Shen K, Jin Q, Wang J. Human rhinovirus C infections mirror those of human rhinovirus A in children with community-acquired pneumonia. J Clin Virol. 2010;49:94–99. doi: 10.1016/j.jcv.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watanabe A, Carraro E, Kamikawa J, Leal E, Granato C, Bellei N. Rhinovirus species and their clinical presentation among different risk groups of non-hospitalized patients. J Med Virol. 2010;82:2110–2115. doi: 10.1002/jmv.21914. [DOI] [PubMed] [Google Scholar]
- 55.Tapparel C, Cordey S, Junier T, Farinelli L, Van BS, Soccal PM, Aubert JD, Zdobnov E, Kaiser L. Rhinovirus genome variation during chronic upper and lower respiratory tract infections. PLoS One. 2011;6:e21163. doi: 10.1371/journal.pone.0021163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arakawa M, Okamoto-Nakagawa R, Toda S, Tsukagoshi H, Kobayashi M, Ryo A, Mizuta K, Hasegawa S, Hirano R, Wakiguchi H, Kudo K, Tanaka R, Morita Y, Noda M, Kozawa K, Ichiyama T, Shirabe K, Kimura H. Molecular epidemiological study of human rhinovirus species ABCs from patients with acute respiratory illnesses in Japan. J Med Microbiol. doi: 10.1099/jmm.0.035006-0. (in press) [DOI] [PubMed] [Google Scholar]
- 57.Broberg E, Niemela J, Lahti E, Hyypia T, Ruuskanen O, Waris M. Human rhinovirus C--associated severe pneumonia in a neonate. J Clin Virol. 2011;51:79–82. doi: 10.1016/j.jcv.2011.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cordey S, Gerlach D, Junier T, Zdobnov EM, Kaiser L, Tapparel C. The cis-acting replication elements define human enterovirus and rhinovirus species. RNA. 2008;14:1568–1578. doi: 10.1261/rna.1031408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simmonds P, McIntyre C, Savolainen-Kopra C, Tapparel C, Mackay IM, Hovi T. Proposals for the classification of human rhinovirus species C into genotypically assigned types. J Gen Virol. 2010;91:2409–2419. doi: 10.1099/vir.0.023994-0. [DOI] [PubMed] [Google Scholar]
- 60.McIntyre CL, McWilliam Leitch EC, Savolainen-Kopra C, Hovi T, Simmonds P. Analysis of genetic diversity and sites of recombination in human rhinovirus species C. J Virol. 2010;84:10297–10310. doi: 10.1128/JVI.00962-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kistler AL, Webster DR, Rouskin S, Magrini V, Credle JJ, Schnurr DP, Boushey HA, Mardis ER, Li H, DeRisi JL. Genome-wide diversity and selective pressure in the human rhinovirus. Virol J. 2007;4:40. doi: 10.1186/1743-422X-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cordey S, Junier T, Gerlach D, Gobbini F, Farinelli L, Zdobnov EM, Winther B, Tapparel C, Kaiser L. Rhinovirus genome evolution during experimental human infection. PLoS One. 2010;5:e10588. doi: 10.1371/journal.pone.0010588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schibler M, Gerlach D, Martinez Y, Van BS, Turin L, Kaiser L, Tapparel C. Experimental human rhinovirus and enterovirus interspecies recombination. J Gen Virol. 2012;93:93–101. doi: 10.1099/vir.0.035808-0. [DOI] [PubMed] [Google Scholar]
- 64.Vlasak M, Goesler I, Blaas D. Human rhinovirus type 89 variants use heparan sulfate proteoglycan for cell attachment. J Virol. 2005;79:5963–5970. doi: 10.1128/JVI.79.10.5963-5970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Khan AG, Pichler J, Rosemann A, Blaas D. Human rhinovirus type 54 infection via heparan sulfate is less efficient and strictly dependent on low endosomal pH. J Virol. 2007;81:4625–4632. doi: 10.1128/JVI.02160-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bochkov YA, Palmenberg AC, Lee WM, Rathe JA, Amineva SP, Sun X, Pasic TR, Jarjour NN, Liggett SB, Gern JE. Molecular modeling, organ culture and reverse genetics for a newly identified human rhinovirus C. Nat Med. 2011;17:627–632. doi: 10.1038/nm.2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pitkaranta A, Arruda E, Malmberg H, Hayden FG. Detection of rhinovirus in sinus brushings of patients with acute community-acquired sinusitis by reverse transcription-PCR. J Clin Microbiol. 1997;35:1791–1793. doi: 10.1128/jcm.35.7.1791-1793.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chantzi FM, Papadopoulos NG, Bairamis T, Tsiakou M, Bournousouzis N, Constantopoulos AG, Liapi G, Xatzipsalti M, Kafetzis DA. Human rhinoviruses in otitis media with effusion. Pediatr Allergy Immunol. 2006;17:514–518. doi: 10.1111/j.1399-3038.2006.00448.x. [DOI] [PubMed] [Google Scholar]
- 69.Jang YJ, Kwon HJ, Park HW, Lee BJ. Detection of rhinovirus in turbinate epithelial cells of chronic sinusitis. Am J Rhinol. 2006;20:634–636. doi: 10.2500/ajr.2006.20.2899. [DOI] [PubMed] [Google Scholar]
- 70.de Arruda E, III, Mifflin TE, Gwaltney JM, Jr, Winther B, Hayden FG. Localization of rhinovirus replication in vitro with in situ hybridization. J Med Virol. 1991;34:38–44. doi: 10.1002/jmv.1890340107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Arruda E, Boyle TR, Winther B, Pevear DC, Gwaltney JM, Jr, Hayden FG. Localization of human rhinovirus replication in the upper respiratory tract by in situ hybridization. J Infect Dis. 1995;171:1329–1333. doi: 10.1093/infdis/171.5.1329. [DOI] [PubMed] [Google Scholar]
- 72.Pitkaranta A, Starck M, Savolainen S, Poyry T, Suomalainen I, Hyypia T, Carpen O, Vaheri A. Rhinovirus RNA in the maxillary sinus epithelium of adult patients with acute sinusitis. Clin Infect Dis. 2001;33:909–911. doi: 10.1086/322678. [DOI] [PubMed] [Google Scholar]
- 73.Pitkaranta A, Puhakka T, Makela MJ, Ruuskanen O, Carpen O, Vaheri A. Detection of rhinovirus RNA in middle turbinate of patients with common colds by in situ hybridization. J Med Virol. 2003;70:319–323. doi: 10.1002/jmv.10397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kuss SK, Best GT, Etheredge CA, Pruijssers AJ, Frierson JM, Hooper LV, Dermody TS, Pfeiffer JK. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science. 2011;334:249–252. doi: 10.1126/science.1211057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lopez-Souza N, Dolganov G, Dubin R, Sachs LA, Sassina L, Sporer H, Yagi S, Schnurr D, Boushey HA, Widdicombe JH. Resistance of differentiated human airway epithelium to infection by rhinovirus. Am J Physiol Lung Cell Mol Physiol. 2004;286:L373–L381. doi: 10.1152/ajplung.00300.2003. [DOI] [PubMed] [Google Scholar]
- 76.Lachowicz-Scroggins ME, Boushey HA, Finkbeiner WE, Widdicombe JH. Interleukin-13-induced mucous metaplasia increases susceptibility of human airway epithelium to rhinovirus infection. Am J Respir Cell Mol Biol. 2010;43:652–661. doi: 10.1165/rcmb.2009-0244OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jakiela B, Brockman-Schneider R, Amineva S, Lee WM, Gern JE. Basal cells of differentiated bronchial epithelium are more susceptible to rhinovirus infection. Am J Respir Cell Mol Biol. 2008;38:517–523. doi: 10.1165/rcmb.2007-0050OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]