Abstract
Purpose of review
To provide an update on the origin of the HIV epidemic and insights into how the immune response is shaping virus evolution.
Recent findings
Characterization of archival samples showed that by the 1960s, HIV had already diverged within humans. It is now estimated that HIV has been in humans since at least the early 1900s. However, despite the potential for different divergent viruses to spread, surprisingly few viruses successfully expanded to cause the global epidemic. In approximately 80% of cases, productive infection is the result of infection with only a single virus or single virus-infected cell. After transmission, HIV evolves at a rapid rate driven by the immune pressure until the virus reaches a delicate survival balance: on one hand avoiding elimination through the development of cytotoxic T-cell immune escape mutations, and on the other sacrificing replication fitness as these mutations may come with a severe fitness cost to the virus. People infected with these ‘attenuated’ cytotoxic T-cell escape viruses can have a survival advantage. Cytotoxic T-cell responses are molding HIV diversity at a population level resulting in a loss of some of the common immune epitopes.
Summary
Insights into the origin of HIV and its evolution between populations and within individuals is essential to understanding HIV pathogenesis and imperative for the design of effective biomedical interventions such as vaccines.
Keywords: CTL escape, HIV evolution, origin, pathogenesis, reversion, transmission
Introduction
The rapid evolution of HIV is a hallmark of this virus and high diversity is one of the key reasons for the success of this pathogen. In order to meaningfully intervene through vaccines, microbicides or antiretroviral therapy, scientists need to develop a holistic and precise understanding of virus evolution in the context of host responses.
HIV origin
HIV-1 is thought to have crossed into humans three times to result in three distinct phylogenetic lineages (M, N and O) with group M responsible for the HIV pandemic [1]. A few years ago an elegant study was published whereby investigators analyzed primate feces from the jungles of central Africa. This study not only definitively identified the chimpanzee (Pan troglodyte troglodyte) as the natural host of the group M and N viruses but also determined that the likely geographical location of the epidemic was in west-central Africa [1]. Until recently, however, the natural host of the group O viruses remained inconclusive. These group O viruses are most closely related to viruses infecting gorillas, and new evidence suggests that chimpanzees are also the origin of infection of these gorillas [2]. This suggests that chimpanzees could be the common source of all three HIV lineages. However, one cannot rule out that the group O viruses may have been introduced into humans via a gorilla intermediate [2,3].
Using estimated rates of viral diversification it is possible to date the common ancestor for the HIV M group, and to estimate the latest time at which the transfer from non-human primates occurred. The identification of a second HIV-positive specimen collected in 1960 from the Democratic Republic of the Congo (DRC, previously Zaire), together with the information on the earliest HIV sample characterized from 1959 [4], showed that even at this time the group M viruses had diversified significantly (~12%) suggesting that it had been evolving in humans for longer than previously thought [5••]. It is now estimated that HIV has been in humans since at least 1902–1921. Interestingly, this timing coincides with urbanization when the first towns were emerging in this region of Africa.
Global spread
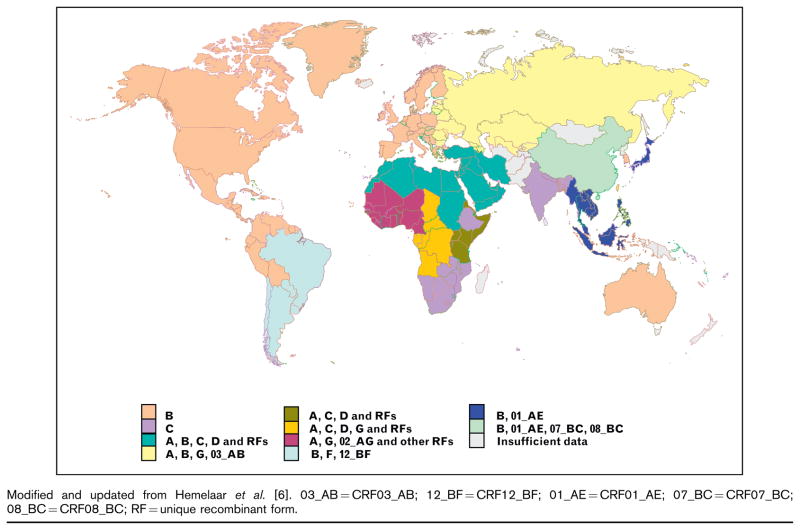
Since the introduction of HIV into humans, the virus has evolved into distinct phylogenetic groups or subtypes labeled A1, A2, B, C, D, F1, F2, G, H, J and K (Fig. 1) [6]. In addition, there are now at least 43 mosaic viruses circulating (called circulating recombinant forms) which comprise of components of more than one subtype [7]. The highest diversity of HIV remains in the region where HIV originated in west-central Africa and despite the potential for divergent viruses to spread, surprisingly few viruses have successfully expanded with 90% of the epidemic comprising of four subtypes (A, B, C and D) and two circulating recombinant forms (CRFs) (CRF01_AE and CRF02_AG). Historically, many more viruses undoubtedly emerged from Africa; however, these strains did not establish themselves within transmission networks, or were of lower fitness which limited their dispersal. Founder effects can probably account for most of the current dominant epidemics whereby a single chance introduction of an ancestral virus resulted in major spread. This is clearly illustrated in a study by Gilbert et al. [8] who investigated the spread of subtype B to Haiti and the US. They demonstrated that the epidemic originated in Haiti around 1966 and within 5 years a single transmission event occurred that culminated in the subtype B epidemic in the US. HIV was evidently smoldering in North America for approximately 10 years prior to its discovery in 1981. This subtype has become massively successful accounting for most infections in the industrialized world (Fig. 1).
Figure 1. Regional HIV-1 subtype and circulating recombinant form (CRF) distribution.
Modified and updated from Hemelaar et al. [6]. 03_AB = CRF03_AB; 12_BF = CRF12_BF; 01_AE = CRF01_AE; 07_BC = CRF07_BC; 08_BC = CRF08_BC; RF = unique recombinant form.
Population bottlenecks result in a drastic reduction of population size with diminished genetic variability and, in RNA viruses, are usually associated with reduction in viral fitness [9]. The question has been asked as to whether HIV replication fitness has changed over the course of the epidemic. Ex-vivo replication fitness assays have been developed and early cross-sectional studies comparing fitness in samples collected in the late 1980s compared to the early 2000 suggested that HIV was becoming less pathogenic over time [10]. However, viral fitness can increase with disease progression and a similar study which controlled for time postinfection found the opposite results in that there was a trend for viruses to gain in fitness over the course of the epidemic in Amsterdam [11]. Similarly, conflicting results were obtained in cohort studies. A comparison of early disease progression markers in large cohorts spanning Europe, Canada and Australia showed trends of decreasing CD4 cell counts and increasing viral loads from 1985 to 2002 [12], whereas no change in disease progression was observed in the US from 1984 to 2005 [13]. One of the major drawbacks of comparing historical to more recent data is that advances in diagnostic techniques may change the readout and thus bias results. These studies are also difficult to interpret within the context of host genotype, subtype, nutritional status, access to healthcare – all of which might impact on disease progression. Whether HIV replicative fitness has changed since the start of the pandemic, remains inconclusive.
Transmission
As vaccines and microbicides target transmitted viruses there is intense interest in understanding the genetic characteristics of these variants, which may differ from those of chronic infection [14]. It has been shown that despite a swarm of closely related viruses present in the donor, there is a genetic bottleneck associated with transmission so that only a limited number of variants get transmitted to the recipient [14–17]. Recent studies have significantly advanced our understanding of transmission: first, by investigating a large number of individuals in very early stage of infection, many of whom were HIV RNA-positive antibody-negative; and second, by applying more quantitative methods using single-genome amplification (SGA) which has enabled investigators to infer the sequence of the infecting virus and thus enumerate the number of infectious units [18••,19••,20,21••]. This more standardized approach has also enabled us to directly compare results between cohorts.
The largest of these studies have been done in 102 subtype B-infected individuals infected via men who have sex with men (MSM) route or heterosexually [19••], and in 69 subtype C heterosexually infected men and women [21••]. Interestingly, both studies estimated that approximately 80% of productive infections were the result of a single virus or a single virus-infected cell. The transmission of a single infectious unit provides a window of opportunity for vaccines which, at least in the majority of cases, would need to protect against a very small incoming viral dose of limited diversity. It is still an unresolved question whether these viruses have distinctive features associated with sexual transmission. However, a recent study of vertical transmission in seven mother–infant pairs showed that replicationally fit viruses were selectively transmitted to the infant in the presence of less fit variants in the mother [22•].
Approximately 20% of people were infected with multiple viruses (between two and six variants) [19••,21••] and there is interest in understanding factors that disrupt this bottleneck not only because it has vaccine implications but also because high diversity following transmission has been associated with more rapid disease progression [23–25]. Studies in macaques have shown that there is a relationship between dose and frequency of multiple variant transmission suggesting that factors that increase HIV transmission rates will also increase frequency of multiple variant transmission [26]. Similarly these macaques studies showed that whereas intravenous inoculation resulted in multiple variant transmission, mucosal routes of inoculation recapitulated human studies, implying that it is the mucosal barrier that is responsible for the population bottleneck during transmission. This is further supported by recent studies confirming that sexually transmitted infections, which disrupt the mucosal barrier, are associated with increased frequency of multiple variant transmission [18••]. Lastly, mode of transmission may also be important with increased frequency of multiple variant transmissions reported in MSM [27].
HIV adaptation to humans
As HIV is passaged through populations, the virus is continually adapting to avoid immune responses. There is evidence to support that mutations associated with immune evasion are accumulating at a population level [28,29•,30••]. This imprinting on the viral genome appears to be due to cytotoxic T-cell (CTL) responses which recognize infected cells through the presentation of short peptides by the human leukocyte antigen (HLA). The virus escapes these responses through mutations which interfere with epitope processing, binding or recognition. A study of 2800 patients spanning five continents showed that HLA frequency is correlated with the persistence of certain polymorphisms in the viral genome [30••]. HLA alleles which occur at high frequency in certain populations, such as HLA-B*51, HLA-B*27 and HLA-B*57, appear to be driving the fixation of certain CTL escape mutations within viruses in a given population. We do not yet know if infection of individuals bearing these HLAs with these ‘less immunogenic viruses’ will affect their disease progression; however, presumably they would elicit CTL responses to other subdominant epitopes.
These escape mutations are not necessarily ‘bad news’ to the host as some of them come with a fitness cost to the virus [31,32]. One group of HIV-1-infected individuals, the elite controllers, is able to control viremia to below 50 copies/ml and many different mechanisms have been suggested to be associated with this control including host genetics and immune responses [33–36]. Viruses infecting elite controllers are enriched for HLA-associated polymorphisms and one virological mechanism which is thought to contribute to this viremic control is HLA-mediated attenuation of viruses [37,38•,39]. Mutations that carry a fitness cost are also known to revert to wild type in the absence of immune pressure and the speed at which this happens is related to the fitness cost of the mutation [29•]. However, the rate or timing of reversion is affected by the presence of compensatory mutations that partially restore replication fitness [40]. In fact the selection of one CTL escape mutation above others that are more effective at disrupting epitope binding is influenced by the ability of some CTL escape mutations to occur in the presence of compensatory mutations [41]. Therefore, the persistence of variants carrying escape mutations depends on whether the fitness cost of the escape mutation outweighs the selective advantage of immune evasion [31].
Certain HLAs such as B*57/5801 appear to drive HIV to a more attenuated form and two studies have shown that individuals who do not have these protective HLAs can also benefit. We have shown that transmission of viral variants carrying HLA-B*57/5801-associated Gag escape mutations to HLA-mismatched recipients was associated with lower viral loads and higher CD4 cell counts during early infection [42••]. A similar study of HIV-infected donor and recipient couples demonstrated that transmission of variants carrying a high number of CTL escape mutations within Gag was significantly associated with reduced viremia in the recipients 6 months after infection [43••]. Together these results suggest that infection with viruses containing CTL mutations that come with high fitness costs may ameliorate infection in these individuals. Vaccine immunogens that elicit CTL responses that drive the virus into an attenuated form could provide a survival advantage in vaccine recipients who become infected despite vaccination. However, the potential loss of epitopes in circulating viruses [30••] does imply that vaccine immunogens would need to be reviewed over time to ensure they reflect the evolving diversity.
Conclusion
HIV has been evolving in humans since at least the early 1900s. Despite high diversity in the west-central African geographical origin of the current pandemic, a limited number of viruses have spread with only four subtypes and two circulating recombinant forms responsible for 90% of infections. Even though a number of studies previously described genetic complexity of transmitted variants, it is only in the past year when it was definitively shown that 80% of infections were the result of single virus or single virus-infected cell in both subtypes B and C, the major contributors of the global pandemic. Similar figures have been shown for other subtypes and cohorts [18••]. There is accumulating evidence that HIV is adapting to human populations resulting in an increased number of viruses carrying mutations which make them less recognizable by individuals bearing certain common HLAs [30••,44]. Some of these CTL escape mutations come with a fitness cost to the virus and infection with these attenuated CTL escape viruses have been associated with lower viral loads. Transmission of these CTL escape viruses to HLA-mismatched individuals has also been associated with lower viral loads and high CD4+ cell counts. However, how the circulation of these attenuated viruses will impact on pathogenesis and the epidemic remains to be defined. Understanding viral evolution will provide vital information to understanding HIV transmission and pathogenesis, which are key research areas needed to design more effective HIV vaccines – the greatest challenges facing HIV research.
Acknowledgments
We would like to acknowledge Debbie Stewart for her assistance in gathering the research material and generating the figure. These authors receive funding from the National Institute of Allergy and Infectious Disease (NIAID), National Institutes of Health (NIH), and the US Department of Health and Human Services (DHHS) (# AI51794, CAPRISA and U19-AI067854-03, CHAVI), as well as by the National Research Foundation, SA (# 67385), the Bill and Melinda Gates Foundation, and the South African AIDS Vaccine Initiative. Z.W. is a Sidney Brenner Postdoctoral Fellow.
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest Additional references related to this topic can also be found in the Current World Literature section in this issue (pp. 335–336).
- 1.Keele BF, Van Heuverswyn F, Li Y, et al. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006;313:523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takehisa J, Kraus MH, Ayouba A, et al. Origin and biology of simian immunodeficiency virus in wild-living western gorillas. J Virol. 2009;83:1635–1648. doi: 10.1128/JVI.02311-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Heuverswyn F, Li Y, Neel C, et al. Human immunodeficiency viruses: SIV infection in wild gorillas. Nature. 2006;444:164. doi: 10.1038/444164a. [DOI] [PubMed] [Google Scholar]
- 4.Zhu TF, Korber BT, Nahmias AJ, et al. An African HIV-1 sequence from 1959 and implications for the origin of the epidemic. Nature. 1998;391:594–597. doi: 10.1038/35400. [DOI] [PubMed] [Google Scholar]
- 5••.Worobey M, Gemmel M, Teuwen DE, et al. Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature. 2008;455:661–757. doi: 10.1038/nature07390. This study reports the identification of an HIV sequence from a Bouin’s-fixed paraffin-embedded lymph node biopsy specimen obtained in 1960 from an adult female in Leopoldville, Belgian Congo [now Kinshasa, Democratic Republic of the Congo (DRC)], that predates the recognition of AIDS. Phylogenetic comparison of this sequence with that of another early sequence from 1959 showed that there was extensive diversification between these two sequences suggesting that HIV had been evolving in Kinshasa long before 1960. The most common ancestor of the M group is now estimated to be in the early 1900s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hemelaar J, Gouws E, Ghys PD, Osmanov S. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS. 2006;20:W13–W23. doi: 10.1097/01.aids.0000247564.73009.bc. [DOI] [PubMed] [Google Scholar]
- 7.Los Alamos National Laboratory database. 2009 www.hiv.lanl.gov. [Ref. type: Internet Communication]
- 8.Gilbert MTP, Rambaut A, Wlasiuk G, et al. The emergence of HIV/AIDS in the Americas and beyond. Proc Natl Acad Sci U S A. 2007;104:18566–18570. doi: 10.1073/pnas.0705329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clarke DK, Duarte EA, Moya A, et al. Genetic bottlenecks and population passages cause profound fitness differences in RNA viruses. J Virol. 1993;67:222–228. doi: 10.1128/jvi.67.1.222-228.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arien KK, Troyer RA, Gali Y, et al. Replicative fitness of historical and recent HIV-1 isolates suggests HIV-1 attenuation over time. AIDS. 2005;19:1555–1564. doi: 10.1097/01.aids.0000185989.16477.91. [DOI] [PubMed] [Google Scholar]
- 11.Gali Y, Berkhout B, Vanham G, et al. Survey of the temporal changes in HIV-1 replicative fitness in the Amsterdam cohort. Virology. 2007;364:140–146. doi: 10.1016/j.virol.2007.02.021. [DOI] [PubMed] [Google Scholar]
- 12.Dorrucci M, Rezza G, Porter K, Phillips A. Temporal trends in postseroconversion CD4 cell count and HIV load: the concerted action on seroconversion to AIDS and death in Europe collaboration, 1985–2002. J Infect Dis. 2007;195:525–534. doi: 10.1086/510911. [DOI] [PubMed] [Google Scholar]
- 13.Herbeck JT, Lyagoba F, Moore SW, et al. Prevalence and genetic diversity of HIV type 1 subtypes A and D in women attending antenatal clinics in Uganda. AIDS Res Hum Retroviruses. 2007;23:755–760. doi: 10.1089/aid.2006.0237.A. [DOI] [PubMed] [Google Scholar]
- 14.Derdeyn CA, Decker JM, Bibollet-Ruche F, et al. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science. 2004;303:2019–2022. doi: 10.1126/science.1093137. [DOI] [PubMed] [Google Scholar]
- 15.Wolfs TFW, Zwart G, Bakker M, Goudsmit J. HIV-1 genomic RNA diversification following sexual and parenteral virus transmission. Virology. 1992;189:103–110. doi: 10.1016/0042-6822(92)90685-i. [DOI] [PubMed] [Google Scholar]
- 16.Wolinsky SM, Wike CM, Korber BTM, et al. Selective transmission of human-immunodeficiency-virus type-1 variants from mothers to infants. Science. 1992;255:1134–1137. doi: 10.1126/science.1546316. [DOI] [PubMed] [Google Scholar]
- 17.Ritola K, Pilcher CD, Fiscus SA, et al. Multiple V1/V2 ENV variants are frequently present during primary infection with human immunodeficiency virus type. J Virol. 2004;78:11208–11218. doi: 10.1128/JVI.78.20.11208-11218.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18••.Haaland RE, Hawkins PA, Salazar-Gonzalez J, et al. Inflammatory genital infections mitigate a severe genetic bottleneck in heterosexual transmission of subtype A and C HIV-1. PLoS Pathogens. 2009;5:e1000274. doi: 10.1371/journal.ppat.1000274. This study investigated the genetic bottleneck during transmission using single-genome amplified viral sequences derived from 20 epidemiologically linked subtype C and subtype A transmission pairs very early after infection. They showed that 90% of the transmission pairs became infected with single infectious units and that there was an association between sexually transmitted infections and infection by multiple variants, highlighting the role of the mucosal barrier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19••.Keele BF, Giorgi EE, Salazar-Gonzalez JF, et al. Identification and characterisation of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. This study used single genome amplification to generate 3449 envelope sequences from subtype B variants from 102 individuals in acute stage of HIV infection. Seventy-six percentage of the individuals were infected with a single infectious unit suggesting that vaccines might have a small window of opportunity when they would need to act against a viral population with very low diversity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salazar-Gonzalez JF, Bailes E, Pham KT, et al. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol. 2008;82:3952–3970. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21••.Abrahams MR, Anderson JA, Giorgi EE, et al. for the CAPRISA Acute Infection Study Team and the Center for HIV-AIDS Vaccine Immunology Consortium. Quantitating the multiplicity of infection with HIV-1 subtype C reveals a non-Poisson distribution of transmitted variants. J Virol. 2009 doi: 10.1128/JVI.02132-08. [Epub ahead of print] This study uses single genome amplification to enumerate the number of variants responsible for infection in 69 HIV-1 subtype C heterosexually infected individuals. Analysis of 1505 envelope sequences showed that 78% of infections involved single-variant transmission and 22% multiple-variant transmission (median of three transmitted variants). It also demonstrated that the transmission of multiple variants were not independent events. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22•.Kong XH, West JT, Zhang H, et al. The human immunodeficiency virus type 1 envelope confers higher rates of replicative fitness to perinatally transmitted viruses than to nontransmitted viruses. J Virol. 2008;82:11609–11618. doi: 10.1128/JVI.00952-08. This study investigated seven mother–infant pairs and demonstrated that the variants that were transmitted to the infants had higher replication fitness than the variants that were not transmitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gottlieb GS, Nickle DC, Jensen MA, et al. Dual HIV-1 infection associated with rapid disease progression. Lancet. 2004;363:619–622. doi: 10.1016/S0140-6736(04)15596-7. [DOI] [PubMed] [Google Scholar]
- 24.Sagar M, Lavreys L, Baeten JM, et al. Identification of modifiable factors that affect the genetic diversity of the transmitted HIV-1 population. AIDS. 2004;18:615–619. doi: 10.1097/00002030-200403050-00005. [DOI] [PubMed] [Google Scholar]
- 25.Grobler J, Gray CM, Rademeyer C, et al. Incidence of HIV-1 dual infection and its association with increased viral load set point in a cohort of HIV-1 subtype C-infected female sex workers. J Infect Dis. 2004;190:1355–1359. doi: 10.1086/423940. [DOI] [PubMed] [Google Scholar]
- 26.Rectal or vaginal inoculation of rhesus macaques by SIVsmE660 or SIVmac251 recapitulates features of mucosal human infection by HIV-1. 16th Conference on Retroviruses and Opportunistic Infections; 2009. [Google Scholar]
- 27.Investigation by single genome amplification of HIV-1 transmission and envelope diversity among men who have sex with men. 16th Conference on Retroviruses and Opportunistic Infections; 2009. [Google Scholar]
- 28.Bhattacharya T, Daniels M, Heckerman D, et al. Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science. 2007;315:1583–1586. doi: 10.1126/science.1131528. [DOI] [PubMed] [Google Scholar]
- 29•.Brumme ZL, Brumme CJ, Carlson J, et al. Marked epitope- and allele-specific differences in rates of mutation in human immunodeficiency type 1 (HIV-1) Gag, Pol, and Nef cytotoxic T-lymphocyte epitopes in acute/early HIV-1 infection. J Virol. 2008;82:9216–9227. doi: 10.1128/JVI.01041-08. This study investigated the HLA-associated viral adaptation during acute subtype B infection by following 98 HIV-infected individuals for 12 months. They determined that the rate of both escape and reversion varied across codons, genes and epitopes and that this formed a large percentage of HIV adaptation during early infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30••.Adaptation of HIV-1 to human leukocyte antigen class I. Nature. 2009 doi: 10.1038/nature07746. [Epub ahead of print] This study demonstrated that the frequency of escape mutations to CTL pressure in a given population was correlated with the prevalence of the relevant HLA allele in that population by investigating nine cohorts from North America, the Caribbean, Europe, sub-Saharan Africa, Australia and Japan. Therefore, the accumulation and fixation of escape mutations at a population level suggests that viruses are becoming resistant to CTL responses and that previously identified HLAs (B*57, B351 and B*27) might lose their protective capacity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y, McNevin J, Zhao H, et al. Evolution of human immunodeficiency virus type I cytotoxic T-lymphocyte epitopes: fitness-balanced escape. J Virol. 2007;81:12179–12188. doi: 10.1128/JVI.01277-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martinez-Picado J, Prado JG, Fry EE, et al. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J Virol. 2006;80:3617–3623. doi: 10.1128/JVI.80.7.3617-3623.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deeks SG, Walker BD. Human immunodeficiency virus controllers: mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity. 2007;27:406–416. doi: 10.1016/j.immuni.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 34.Emu B, Sinclair E, Hatano H, et al. HLA class I-restricted T-cell responses may contribute to the control of human immunodeficiency virus infection, but such responses are not always necessary for long-term virus control. J Virol. 2008;82:5398–5407. doi: 10.1128/JVI.02176-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miura T, Brumme CJ, Brockman MA, et al. HLA-associated viral mutations are common in human immunodeficiency virus type 1 elite controllers. J Virol. 2009;83:3407–3412. doi: 10.1128/JVI.02459-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pereyra F, Addo MM, Kaufmann DE, et al. Genetic and immunologic heterogeneity among persons who control HIV infection in the absence of therapy. J Infect Dis. 2008;197:563–571. doi: 10.1086/526786. [DOI] [PubMed] [Google Scholar]
- 37.Miura T, Brumme C, Brockman MA, et al. HLA-associated mutations occur less frequently in HIV elite controllers than in chronically infected individuals. AIDS Res Hum Retroviruses. 2008;24:77–78. [Google Scholar]
- 38•.Miura T, Brockman MA, Brumme ZL, et al. HLA-associated alterations in replication capacity of chimeric NL4-3 viruses carrying gag-protease from elite controllers of human immunodeficiency virus type 1. J Virol. 2009;83:140–149. doi: 10.1128/JVI.01471-08. This study focused on elite controllers who are HIV-infected individuals that are able to control viremia to below 50 copies/ml. These findings indicate that cytotoxic T-lymphocyte (CTL) selection pressure on gag-protease alters viral replication fitness, and HIV-specific CTLs that induce escape mutations with a fitness cost in this region may be important for strict viremic control in elite controllers of HIV. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miura T, Brockman MA, Schneidewind A, et al. HLA-B57/B*5801 human immunodeficiency virus type 1 elite controllers select for rare gag variants associated with reduced viral replication capacity and strong cytotoxic T-lymphotye recognition. J Virol. 2009;83:2743–2755. doi: 10.1128/JVI.02265-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crawford H, Prado JG, Leslie A, et al. Compensatory mutation partially restores fitness and delays reversion of escape mutation within the immunodominant HLA-B*5703-restricted Gag epitope in chronic human immunodeficiency virus type 1 infection. J Virol. 2007;81:8346–8351. doi: 10.1128/JVI.00465-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schneidewind A, Brockman MA, Yang R, et al. Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J Virol. 2007;81:12382–12393. doi: 10.1128/JVI.01543-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42••.Chopera DR, Woodman Z, Mlisana K, et al. Transmission of HIV-1CTL escape variants provides HLA-mismatched recipients with a survival advantage. PLoS Pathogens. 2008;4:e1000033. doi: 10.1371/journal.ppat.1000033. This study showed that 9 out of 21 HLA-B*27, HLA-B*57 and HLA-B*5801-negative individuals that were infected with variants carrying the relevant HLA escape mutations, T242N and A146X, had lower viral loads and higher CD4 cell counts at both 3 and 12 months postinfection when compared to the 12 individuals infected with viruses not carrying the mutations. This suggested that CTL escape mutations that came with a fitness cost not only benefited the donor but also remained beneficial in HLA-mismatched individuals, at least in early infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43 ••.Goepfert PA, Lumm W, Farmer P, et al. Transmission of HIV-1 Gag immuneescape mutations is associated with reduced viral load in linked recipients. J Experiment Med. 2008;205:1009–1017. doi: 10.1084/jem.20072457. This study demonstrated in 114 epidemiologically linked transmission pairs that the higher the number of Gag CTL escape mutations carried by transmitted variants, the lower the viral loads in the recipient. Therefore, CTL escape mutations that resulted in a loss of replication fitness were beneficial not only for the donor, but also the recipient. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matthews PC, Prendergast A, Leslie A, et al. Central role of reverting mutations in HLA associations with human immunodeficiency virus set point. J Virol. 2008;82:8548–8559. doi: 10.1128/JVI.00580-08. [DOI] [PMC free article] [PubMed] [Google Scholar]