Abstract
Common variable immunodeficiency (CVID) is a heterogeneous disorder of B cell differentiation or function with inadequate antibody production. Our laboratory studies a natural form of CVID in horses characterized by late-onset B cell lymphopenia due to impaired B cell production in the bone marrow. This study was undertaken to assess the status of B cell differentiation in the bone marrow of CVID-affected horses by measuring the expression of genes essential for early B cell commitment and development. Standard RT-PCR revealed that most of the transcription factors and key signaling molecules that directly regulate B cell differentiation in the bone marrow and precede PAX5 are expressed in the affected horses. Yet, the expression of PAX5 and relevant target genes was variable. Quantitative RT-PCR analysis confirmed that the mRNA expression of E2A, PAX5, CD19, and IGHD was significantly reduced in equine CVID patients when compared to healthy horses (p < 0.05). In addition, the PAX5/EBF1 and PAX5/B220 ratios were significantly reduced in CVID patients (p < 0.01). Immunohistochemical analysis confirmed the absence of PAX5-BSAP expression in the bone marrow of affected horses. Our data suggest that B cell development seems to be impaired at the transition between pre-pro-B cells and pro-B cells in equine CVID patients.
Keywords: CVID, humoral immunodeficiency, B cell development
1. Introduction
Common variable immunodeficiency (CVID), described more than 50 years ago, is the most frequent clinically relevant primary immunodeficiency in human patients. CVID has challenged the field of clinical immunology in regards to etiology and, consequently, therapeutic intervention. CVID in human patients is a clinically and immunologically heterogeneous primary immunodeficiency, characterized by hypogammaglobulinemia, recurrent infections and other complications, such as autoimmunity, granulomatous infiltrations, lymphoid hypertrophy, and non-Hodgkin’s lymphoma (Cunningham-Rundles and Knight 2007). In human patients with CVID, B cell numbers may be normal or reduced, and immunoglobulin production is impaired likely through different mechanisms. Two striking aspects of this disorder include manifestation later in life, and single-familial cases; consequently, these elements complicate the study of putative genetic causes. Only a small percentage of patients have been diagnosed with a genetic disorder, and variations in TACI, MSH5, ICOS, CD19, and BAFF-R genes have been described among CVID patients (Sekine et al. 2007; Park et al. 2008; Salzer et al. 2008; Warnatz et al. 2009; Wilmott 2009; Yong et al. 2011). In addition to the clinical parameters listed above, the diagnosis of CVID is also based on the negative results of the genetic-based tests available for Bruton’s X-linked agammaglobulinemia (XLA), X-linked hyper IgM syndrome, X-linked lymphoproliferative disease (XLP), adenosine deaminase deficiency, caspase 8 deficiency, or activation-induced cytidine deaminase (AID). The incidence of CVID in the human population may be underestimated due to availability or limitations of immunologic and genetic diagnostics (Chapel and Cunningham-Rundles 2009).
Our laboratory diagnosed the index case and further characterized CVID in horses, which manifests as late-onset recurrent bacterial infections, hypo- or agammaglobulinemia, progressive B cell lymphopenia or depletion, and poor response to protein (tetanus toxoid) vaccination (Flaminio et al. 2002; Pellegrini-Masini et al. 2005; Flaminio et al. 2009; Tennent-Brown et al. 2010). Affected patients are unrelated adult horses (average age 10.7 ± 4.4 years) of both sexes, many different breeds, and living in distinct parts of the country. Clinical signs most commonly include recurrent fevers and pneumonia, but also meningitis and/or neurologic disorders, gingivitis, sinusitis, hepatitis, diarrhea, and skin abscesses. The majority of the affected horses are submitted to euthanasia due to severe pneumonia, meningitis or septicemia. When affordable, a few horses are managed for 1 to 5 years on continuous or intermittent antibiotic and immunoglobulin therapies. By the time of diagnosis of CVID in horses, B cells are extremely rare in primary and secondary lymphoid tissues. Our previous work indicated that CD19+IgM+ B cells were absent in the bone marrow of CVID horses (Flaminio et al. 2002; Pellegrini-Masini et al. 2005). Because B cells are continuously generated over an individual’s lifetime from hematopoietic stem cells in the bone marrow, this study was undertaken to determine the extent of ongoing B cell differentiation in CVID horses (Medina and Singh 2005).
B cell differentiation and development is regulated by a network of transcription factors and demethylation of CpG dinucleotides at promoter sites (Maier et al. 2004; Hagman and Lukin 2006). The combinatorial expression of transcription factors PU.1, IKAROS, E2A, BCL11A, EBF1, and PAX5, and cytokine receptors FLT3 and IL7R are critical in the differentiation of common lymphoid progenitors (CLP) into pro-B cells (DeKoter et al. 2002; Singh et al. 2005; Ramirez et al. 2010). In the mouse, targeted inactivation of FLT3 and IL7R results in severe deficiency in the generation of B lineage progenitors and common lymphoid progenitors (Mackarehtschian et al. 1995; Sitnicka et al. 2003; Sitnicka et al. 2007). B cell lineage commitment is promoted by the transcription factor IRF8, which represses transcription of PU.1 while activating transcription of EBF1 and BCL11A (Liu et al. 2003; Wang et al. 2008). The activation of FLT3 and IL7R also promotes the expression of transcriptional regulators E2A and EBF1, which are considered primary B cell fate determinants with synergistic function. Together, they regulate the expression of a secondary B cell fate determinant PAX5, for subsequent early program B lineage gene expression. PAX5 expands the early B cell lineage program by inducing CD79A, CD19, IGHM and BLNK, which all encode for antigen receptor or co-receptor signaling components (Fuxa and Skok 2007). CD19 is a co-receptor for B cells that determines signaling thresholds in B cells by regulating Lyn kinase activity, and a commonly used marker for B cells that have matured beyond the pro-B cell development stage.
Based on the fact that immunohistochemical analysis of bone marrow samples from a population of horses with CVID indicated the lack of positive CD19 and IgM cells, our hypothesis was that the development of B cells in these horses is detained at least at the pro-B cell stage (Flaminio et al. 2009). This study measured the expression of the essential B cell genes described above to determine missing differentiation steps in early B cell commitment and development. RT-PCR was utilized to screen several transcription factors important in early B cell development stages, and real-time RT-PCR was subsequently used to quantify genes of particular interest. Gene sequence and immunohistochemical analysis further defined the genes studied.
2. Material and Methods
2.1 Samples from horses with CVID and control horses
This study was conducted following a protocol approved by Cornell University Center for Animal Resources and Education and regulations mandated by the Institutional Animal Care and Use Committee. CVID horses included in this study (n = 8) presented homogeneously with clinical signs of recurrent bacterial infections, abnormal ante-mortem immunologic testing (severe hypogammaglobulinemia, B cell lymphopenia, and no response to tetanus toxoid vaccination) and histopathology (lack of germinal centers and plasma cells, depletion of CD19+IgM+ B cells in bone marrow and spleen, occasional CD19+IgM+ B cells in lymph nodes) (Flaminio et al. 2009). Frozen bone marrow samples collected during necropsy of horses with CVID were collected through the Cornell University Veterinary Pathology service or forwarded overnight on dry ice to our laboratory, and stored at minus 80°C. Control bone marrow samples from adult horses (n = 3) with no clinical history of infections were archived at minus 80°C and were available for this study from research studies performed by other investigators over the years at Cornell University College of Veterinary Medicine.
2.2 Reverse transcriptase polymerase chain reaction (RT-PCR)
RNA was isolated from snap frozen tissues following homogenization by QIAshredder (Qiagen, Valencia, CA) as directed by the RNeasy kit (Qiagen, Valencia, CA), and quantified with a biophotometer. One half microgram of RNA was treated with DNase I (Invitrogen, Carlsbad, CA or Ambion Inc., Austin, TX) to degrade any contaminating genomic DNA. For reverse transcription, cDNA synthesis reactions contained 1X M-MuLV RT buffer, 5.5 mM MgCl2, 0.5 mM dNTPs, 2.5 μM oligod(T) (Applied Biosystems, Foster City, CA), 0.4 U RNasin Ribonuclease Inhibitor (Promega, Madison, WI), and 1 U Moloney Murine Leukemia Virus Reverse Transcriptase (M-MuLV RT, Applied Biosystems, Foster City, CA). To test for residual genomic DNA contamination, control samples did not receive M-MuLV RT. Amplification reactions contained 1X PCR buffer, 1.5 mM MgCl2, 0.25 mM dNTPs, 0.5 μM forward and reverse primer (Table 1, Integrated DNA Technologies, Coralville, IA), and 2 U Taq (Invitrogen, Carlsbad, CA). Thermal cycling parameters were 95 °C for 5 minutes; 35 cycles of 95 °C for 60 seconds, 58 °C for 60 seconds, 72 °C for 30 seconds; and a final extension of 72°C for 10 minutes. Amplification products were run on 1% agarose gels and stained with GelGreen Nucleic Acid Stain (Phenix Research Products, Candler, NC) for visualization.
Table 1.
Equine B cell development gene primers for RT-PCR.
| Gene | Forward primer (5′-3′) | Reverse primer (5′-3′) | Product | Sequence database IDa |
|---|---|---|---|---|
| ACTB | TCCCTGGAGAAGAGCTACGA | GTGGACAATGAGGCCACAAT | 350 | NM_001081838.1 |
| ARNT | TGGACAAACTTCGTGAGCAG | TGCTTTGATGTAGCCTGTGC | 290 | XM_001916097.1 |
| B220 | TCGAAGTGACCCCTTACCTG | CTGCTGGGTGTCTGAGTGTC | 315 | EU761588 |
| BLK | TCACCAAGGAGCCCATCTAC | CGATTTTGCAGCACAAGGTC | 222 | XM_001497947 |
| BLNK | TGCCTGTGATCGAAAGTCTG | TCTTCCTGCCCAAAGCATAC | 191 | XM_001500520 |
| BTK | CTGGAGAGCATCTTTCTGAAG | TCTTCAGTTGGGGAGAAGAC | 341 | XM_001493218.3 |
| CD19 | CCAGTCACCAGGACAACAGA | GGCTGAAGTTTCGCTCATGT | 396 | JN979558 |
| CD43 | ACAGCAACAGCAGGAGTGTC | CTCCTCCTCTCCCTTTAGGC | 177 | XM_001496248 |
| CD79A | CCTCCAGTGCAAACACAATG | AGTTCTGCCATCGTTTCCTG | 369 | XM_001916779 |
| CD79B | GAGCCTCCATCCTTCCTCGTCAC | GATGATGAGCAGGGTCTGGATCA | 544 | CD470799 |
| DNTT | TGGAGAAGAAAATGGGAACC | TTCAGAACTTTCTCCATCTTCAA | 590 | EU761589 |
| E2A | GCTGCACCTCAACAGTGAGA | CCGACACCTTCTCCTCTTCC | 152 | XM_001915704 |
| EBF1 | GCGATATCTGGCATGATTGTT | TCACCACTTCATTCTCCCTTT | 446 | CX601479 |
| FCRL1 | GAAGAATGCTCCTCCTCCAG | GTCTCGCCCTGTCCTCAC | 172 | ENSECAT00000002770 |
| FLT3LG | GGCCGAGATGATAGTGCTG | AGGTGGGAGATGTTGGTCTG | 402 | CX597567.1 |
| IGHD | TCCATGTCTTCTCCCTGACC | CTTCTGGGTTTCTGGCTGAG | 227 | AY631942 |
| IGHJ@ | TGTGTCCGGGTTACTTCCAG | TGGTTCTCACTGGTGAGGTG | 136 | XM_003364702.1 |
| IGHM | CTTCACTACGGAAGAGGTGC | ACTCAGGCTGTCATAGGTGC | 295 | L49414 |
| IGKC | TTCATCTTCCCACCGTCTTC | GAGACCTCGCAGGCATAGAC | 245 | XM_003362951 |
| IL7 | ATGTTCCATGTTTCTTTTAGG | TCAGTGTTCTTTAGCGTCCC | 531 | CX592622 |
| IL7R | ACATGCCTGCTCTGGTCTCT | TCACGTGCATCCAATCATTT | 580 | CD469725 |
| IRF8 | ACGTGGTGGTCAAGGTCTTC | AGCTCTTCCCAGCTTCTTCC | 207 | XM_001502568 |
| NOTCH1 | TCCTTCCTGACCTGGATGAC | CCATGTTGTCCTGGATGTTG | 393 | XM_001498582 |
| PAX5 | CATCAAGCCTGGGGTAATTG | CACGGTGTCATTGTCACACA | 157 | XM_001504306 |
| PU.1 | GAGACCATCCAGCTCCAGAC | CAGCTCGGTGAAGTGGTTCT | 315 | JN979560 |
| RUNX1 | ATGACCTCAGGTTTGTCGGT | CCACCGTGATTTTGATGGCT | 117 | XM_001916234 |
All sequence database identifiers refer to GenBank except FCRL1, which is refers to Ensembl
2.3 Real-time RT-PCR reactions for selected B cell development genes
RNA was quantified on a NanoDrop (Thermo Scientific, Wilmington, DE), and 50 nanograms of RNA were used per qRT-PCR reaction with 500 nM of primer and iScript One-Step RT-PCR kit with SYBR Green mix (Bio-Rad, Hercules, CA) in a CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA). Reactions were performed in triplicate. Cycling parameters were: 1 cycle of 50°C for 10 minutes, 1 cycle of 95°C for 5 minutes, 40 cycles of 95°C for 10 seconds then 60°C for 30 seconds, followed by melt curve analysis. Two exceptions to the above protocols were 1) for CD19 the annealing temperature was 58°C and 2) for CD79A 300 nM of primer were used to avoid primer dimers. SYBR primers spanning intron/exon boundaries were designed with Beacon Designer 7.91 software (PREMIER Biosoft International, Palo Alto, CA), and were synthesized by Eurofins MWG Operon, Huntsville, AL (Table 1).
RNA standard curves were prepared for each gene. First, a region of each target gene was amplified from healthy adult bone marrow cDNA (primers in Table 2; iProof polymerase, Bio-Rad, Hercules, CA), cloned (pJET1.2 vector, Fermentas, Glen Burnie, MD), and sequenced (Cornell University Life Sciences Core Laboratories Center Genomics Facility, Ithaca, NY). RNA transcription was performed on linearized plasmid DNA from the T7 site on the vector (Fermentas, Glen Burnie, MD). In vitro transcribed RNA was purified (Zymo Research Corporation, Irvine, CA) and quantified (NanoDrop, Thermo Scientific, Wilmington, DE). Ten-fold serial dilutions were made for the standard curve. SYBR primers were validated on the RNA standard curve. Reaction efficiency was between 90 and 110% (the slope of the curve was between −3.1 and −3.6) and no primer dimers were observed on the melt curve analysis. Absolute quantification of mRNA transcript numbers was determined from the RNA standard curve with CFX Manager software (Bio-Rad, Hercules, CA) and box plots were generated with KaleidaGraph (Synergy Software, Reading, PA). For most genes, the number of transcripts was not normally distributed as determined by the Shapiro Wilk test. Therefore, significance was determined with the Wilcoxon-Mann-Whitney Rank Sum test.
Table 2.
Sequence of equine B cell gene primers used to generate clones for standard curves and SYBR real-time RT-PCR.
| Gene, GenBank ID | Primer for clone (5′ – 3′) | SYBR primer (5′ – 3′) | |
|---|---|---|---|
| ACTB | F | GGGACCTGACGGACTACCTC | GATGCAGAAGGAGATCACAGC |
| NM_001081838.1 | R | GTGGACAATGAGGCCACAAT | GAGCCGCCGATCCATACG |
| B220 | F | TCGAAGTGACCCCTTACCTG | CACTGAACTCTTTGGATAATGC |
| EU761588 | R | CTGCTGGGTGTCTGAGTGTC | GGCGACACCTGTTACATT |
| BLNK | F | CTGCCAAGATTTCCAGAAGG | TGCCAAGATTTCCAGAAGGG |
| XM_001500520 | R | TCTTCCTGCCCAAAGCATAC | CACAGGCACCAGCATACC |
| CD19 | F | GGAGTCTGGCCACCATGC | TCCTCTCCTCCTCTTCTTC |
| JN979558 | R | AGGCTGAAGGGAATCACCTG | AGCATTGTCTCCCTCTTTAG |
| CD79A | F | CCTCCAGTGCAAACACAATG | GCTGCTGCTGTTCAGGAA |
| XM_001916779 | R | TCGTACATGGAACAGTCATCG | CATGGAACAGTCATCGAGGTT |
| CXCL12 | F | CAGCCTGAGCTACAGATGTCC | GAGAGCCACGTTGCCAGAG |
| XM_001489644 | R | CCTTTTCTGAGCAGCCTTTC | TTCAGCCTTGCCACGATCT |
| E2A | F | GTGTTGGTGGCTTGACTCAG | GCCCAAGAAAGTCCGAAA |
| XM_001915704 | R | CCTGCCGTAGTCGTCACC | CCGTAGTCGTCACCTGAG |
| EBF1 | F | GGAGATTTTCCACAAGAAAAGG | GGGTTCGTGGAGAAGGAA |
| CX601479 | R | CAACTCACTCCAGACCAGCA | AGCAAGACTCGGCACATT |
| IGHD | F | TCCATGTCTTCTCCCTGACC | CACAGTTCACCACAAAGC |
| AY631942 | R | CTTCTGGGTTTCTGGCTGAG | CTTGGATGTCACAGGTAATG |
| IGHM | F | CTTCACTACGGAAGAGGTGC | CATCCCTCCCTCCTTTGC |
| NW_001876796 | R | ACTCAGGCTGTCATAGGTGC | TCATAGGTGCCCAGGTTTG |
| PAX5 | F | CATCAAGCCTGGGGTAATTG | GCTGAGTATAAACGCCAAA |
| XM_001504306 | R | CATCTCTCTTGCGCTTGTTG | CGGATGATCCTGTTGATG |
| PU.1 | F | GAGACCATCCAGCTCCAGAC | GTATCTCAGCAGTGATGGA |
| JN979560 | R | CAGCTCGGTGAAGTGGTTCT | CTCGGTGAAGTGGTTCTC |
2.4 PAX5 sequencing
The PAX5 coding sequence was determined from bone marrow RNA isolated from CVID horse 7 because clinical and immunologic data were collected for 6 years, and a complete necropsy was obtained. Gene-specific reverse transcription was performed with reverse primer 5′ GGTCAGTGGCGGTCGTAG 3′ (Integrated DNA Technologies, Coralville, IA) and SuperScript III First-Strand synthesis (Invitrogen, Carlsbad, CA). Amplification of the PAX5 transcript was performed with primers 5′ GTCCATTCCATCAAGTTCTGA 3′ and 5′ GGTCAGTGGCGGTCGTAG 3′ (Integrated DNA Technologies, Coralville, IA), and iProof high-fidelity polymerase (Bio-Rad Laboratories, Hercules, CA). PCR products were cloned with the CloneJET PCR cloning kit (Fermentas, Glen Burnie, MD). Plasmid DNA was purified with the GeneJET plasmid miniprep kit (Fermentas, Glen Burnie, MD) and multiple clones were sequenced from each direction at the Cornell University Life Sciences Core DNA Sequence and Genotyping Laboratory (Ithaca, NY). Sequences were analyzed with Geneious Pro 4.8.4 software (Biomatters Ltd, Auckland, New Zealand) (Drummond 2009).
2.5 Immunohistochemistry of tissue sections
Bone marrow and mesenteric lymph node tissue sections were collected during necropsy, preserved in optimal cutting temperature compound (O.C.T. Tissue Tek, Sakura Finetek Inc., Torrance, CA), and stored at minus 80° C. Five micron serial sections were obtained using a cryotome, and tissues were fixed in acetone for 10 minutes. Blocking steps included individual 15 minute incubations with a) 10% normal goat serum in TBS; and b) 0.1% sodium azide (Sigma, St. Louis, MO), and 0.3% hydrogen peroxide (Fisher Scientific, Fair Lawn, NJ) in Tris buffered saline. Primary antibody staining [irrelevant monoclonal antibody (negative control); anti-horse CD19 (clone cz2.1, hybridoma kindly provided by Dr. Douglas Antczak at Cornell University); anti-human IgM (clone CM7, AbD Serotec, Raleigh, NC)]. Peroxidase-conjugated goat anti-mouse IgG monoclonal antibody (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) was separately incubated for 30 minutes. The substrate solution was prepared with 3-amino-9-ethylcarbazole, peroxide and acetate buffer (A.E.C., Sigma, St. Louis, MO), and applied on tissue sections for 50 minutes. The PAX5-BSAP staining was performed at the at the New York State Diagnostic Laboratory at Cornell University using the anti-human PAX5-BSAP (clone 24, Invitrogen Corporation, Camarillo, CA), and irrelevant antibody control. The tissues sections were fixed in 10% paraformaldehyde. The primary antibody incubation period was 90 minutes, and the secondary antibody was a biotinylated goat anti-mouse (Invitrogen Corporation, Camarillo, CA) applied to the slides for 20 minutes. The streptavidin-peroxidase conjugate (Invitrogen Corporation, Camarillo, CA) was applied for 10 minutes. The chromogen, 3,3-diaminobenzidine-tetra hydrochloride (DAB from Dakocytomation) was added to the slides for 1 minute. Counterstaining was performed using hematoxylin for 2 minutes, and finished slides were coverslipped using mounting media.
3. Results
3.1 mRNA expression of genes essential for B cell differentiation
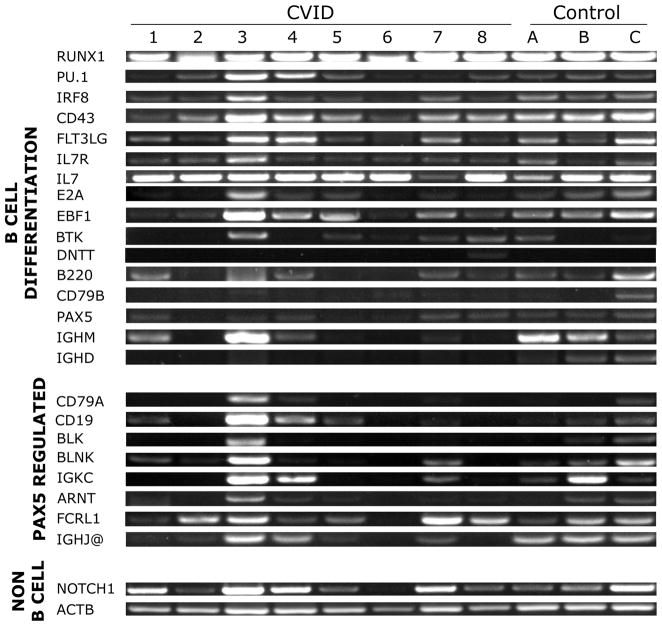
The mRNA expression of essential genes involved in B cell differentiation was tested in bone marrow samples of horses with CVID and control healthy horses (Figure 1). Standard RT-PCR revealed that all equine CVID patients and control healthy horses expressed RUNX1, PU.1, IRF8, CD43, FLT3LG, IL7R, IL7, EBF1, DNTT, and B220, which are transcription factors and key signaling molecules that directly regulate B cell differentiation up to PAX5 expression. The mRNA expression of BTK and CD79B was variable in both groups of horses. Because the standard RT-PCR prevented the consistent detection of DNTT expression even in the control horses, we performed real-time quantitative RT-PCR for this gene, and results were consistent among all samples, including control healthy horses (average CT value 36.8, data not shown). Yet, the expression of E2A, PAX5 and its target genes CD79A, BLK, BLNK, IGKC, CD19, and IGHM was variable, with some patients missing the expression of those genes in their bone marrow. The expression of other PAX5 target genes (ARNT, FCRL1, and IGHJ@) was present in the affected horses. Control genes, such as β-actin (ACTB) and a transcription factor for T cell development, NOTCH1, were present in all samples tested.
Figure 1. RT-PCR of genes involved in bone marrow B cell differentiation in horses with CVID and healthy horses.
The mRNA expression of essential genes involved in B cell differentiation was tested in bone marrow samples of horses with CVID (1 to 8) and control healthy horses (A, B, C). Standard RT-PCR revealed that all equine CVID patients expressed RUNX1, PU.1, IRF8, CD43, FLT3LG, IL7R, IL7, B220, and EBF1, which are transcription factors and key signaling molecules that directly regulate B cell differentiation up to PAX5 expression. Yet, the expression of E2A, PAX5 and its target genes CD79A, BLK, BLNK, IGKC CD19 and IGHM was variable, with some patients missing the expression of those genes in their bone marrow.
3.2 Quantitation of selected genes essential for B cell differentiation
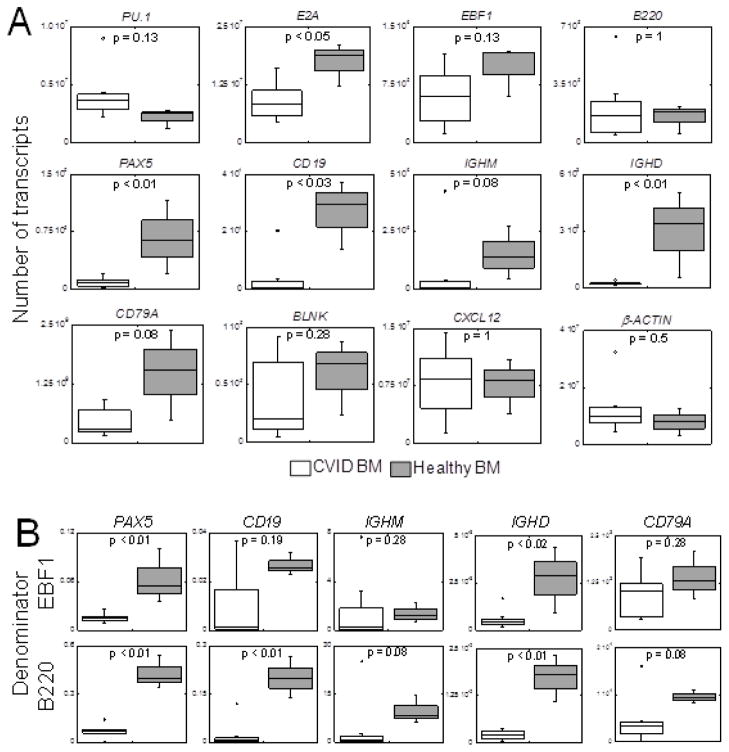
Real-time quantitative RT-PCR revealed comparable (p = 0.1) numbers of PU.1 transcripts in CVID horses (Figure 2A), which is representative of multipotential progenitor and common lymphoid progenitor stages of differentiation. B cell specification is established with expression of E2A and EBF1. Numbers of EBF1 transcripts in equine CVID patients and healthy horses were comparable (p = 0.1), whereas the number of E2A transcripts (both E12 and E47 isoforms) was significantly decreased (p = 0.048) in affected horses. Expression of B220 was equivalent (p = 1) between CVID and healthy horses. Transcript numbers of PAX5 (p = 0.01) and CD19 (p = 0.02) were significantly reduced in equine CVID patients. The number of IGHM transcripts was markedly reduced in 7 out of 8 equine CVID patients compared to healthy controls; however, 1 CVID horse expressed higher numbers of IGHM transcripts than healthy horses, resulting in a p-value = 0.08. This finding was confirmed by an independent experiment. The number of IGHD transcripts was significantly decreased in all 8 equine CVID patients (p = 0.01). Expression of PAX5 downstream targets, such as CD79A (p = 0.08) and BLNK (p = 0.3), were not significantly decreased. CXCL12 mRNA expression was not different (p = 1) between CVID and healthy horses. Similarly, β-actin (p = 0.5) did not differ between the groups. To better understand the relevance of these findings in the absence of B cell production, ratio values were created for each horse using B220 or EBF1 transcript numbers as a denominator because these early B cell differentiation genes were expressed comparably between affected and control horses. In equine CVID patients, the median PAX5/B220 (p < 0.01), CD19/B220 (p < 0.01), and PAX5/EBF1 (p = 0.01) ratios were significantly reduced in comparison to healthy horses (Figure 2B). The ratios of IGHD/B220 and IGHD/EBF1 were also significantly decreased in the bone marrow of CVID horses.
Figure 2. Quantitative RT-PCR of essential B cell differentiation genes in the bone marrow of horses with CVID and healthy horses.
A) Real-time quantitative RT-PCR confirmed that the number of E2A, PAX5, CD19, and IGHD mRNA transcripts (y-axis) was significantly reduced in equine CVID patients (n = 8, open boxes) when compared to healthy horses (n = 3, shaded boxes). Outliers were due to three different horses. B) Gene expression levels were assessed in the context of B cell differentiation by calculating ratios using a PAX5 upstream B cell gene (EBF1 or B220) as a denominator. A median ratio value was calculated using the individual horse ratios. The median ratios of PAX5/EBF1, PAX5/B220, CD19/B220, IGHD/EBF1, and IGHD/B220 were significantly decreased (* p < 0.02) in the bone marrow of horses with CVID. Ratios for CD79A and IGHM were not significantly different between the groups (p > 0.05).
3.3 PAX5 sequencing from an equine CVID patient
Because of the variable expression of PAX5 in affected horses, we conducted gene amplification, cloning and sequencing using bone marrow samples of an affected horse. The nucleotide alignment was identical between all PAX5 clones from a horse with CVID and the equine PAX5 predicted gene in National Center for Biotechnology Information (NCBI, XM_001504306).
3.4 Immunohistochemical analysis of B cell markers in lymphoid tissues
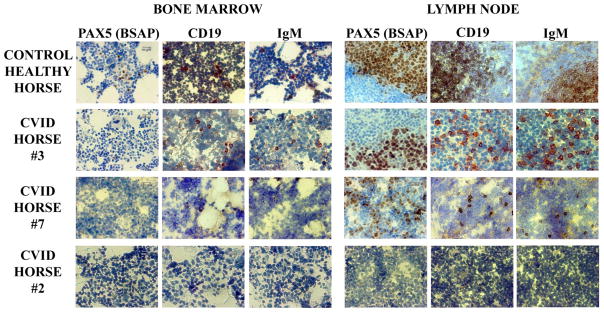
To further evaluate the expression of PAX5 protein (BSAP) in the lymphoid tissues of affected horses, and assess the implications of our gene data, we performed immunohistochemical analysis of B cell markers in the bone marrow and mesenteric lymph node of affected and control healthy horses (Figure 3). In the bone marrow of affected horses, there were no positive cells for the PAX5-BSAP molecule. Occasional, single positive B cells (PAX5-BSAP, CD19 and IgM positive) were identified in the mesenteric lymph nodes of affected horses. Secondary lymphoid tissues of horses with CVID lacked germinal centers and presented overall paucity of cells (also confirmed by histology). In contrast, healthy horses had normal germinal centers, and normal distribution of B cells (PAX5-BSAP, CD19 and IgM positive) in the bone marrow and secondary lymphoid tissues. Tissues from three CVID-affected horses are shown, selected based on high (CVID horse #3), moderate (CVID horse #7), or low (CVID horse #2) numbers of B cell gene transcripts from Figure 2A. Only CVID horse #3 had rare CD19 and IgM positive cells in the bone marrow, and no PAX-BSAP positive cells were detected; nevertheless, this same horse had the most abundant number of CD19, IgM and PAX5-BSAP positive cells in the lymph node. In contrast to the other horses, affected horse #2 did not have any detectable B cells in bone marrow or lymph node.
Figure 3. Immunohistochemical analysis of B cells in primary and secondary lymphoid tissues of horses with CVID and healthy horses.
Secondary lymphoid tissues of horses with CVID (n = 8; 3 examples shown) lacked germinal centers and presented overall paucity of cells. Note negative staining for PAX5-BSAP and few CD19+ or IgM+ cells in the bone marrow, and single positive B cells in the mesenteric lymph node of affected horses. In contrast, healthy horses had normal germinal centers and normal distribution of B cells (PAX5-BSAP, CD19, and IgM positive) in the bone marrow and lymph node. Intended for color reproduction on the Web and in print.
4. Discussion
Using the information available for the regulation of B cell differentiation in mouse and human bone marrow, we measured the expression of the major genes dictating B cell differentiation in samples from horses with CVID and control healthy horses. Most of the transcription factors and key signaling molecules that directly regulate B cell differentiation and precede PAX5 were expressed in the affected horses. Nevertheless, quantitative analysis revealed that the mRNA expression of E2A, PAX5, CD19, and IGHD was significantly reduced in the affected horses when compared to healthy horses, in contrast to other genes important for B cell commitment, such as EBF1 and B220. Expression of major PAX5 target genes, including CD79A, BLK, BLNK, and IGKC was variable among horses with CVID. Immunohistochemical analysis supported our gene expression findings: expression of PAX5-BSAP molecules were not detected in the bone marrow of affected horses. Altogether, these data suggest that CVID in horses is caused by impairment of B cell development in the bone marrow at the transition between pre-pro-B cells and pro-B cells.
The significant reduction in E2A expression but not EBF1 is unexpected as it has been shown that E2A triggers EBF1 transcription (Roessler et al. 2007). The mean transcript number of both genes is reduced approximately 50% in the bone marrow of CVID horses, but more expression variability is present for EBF1, perhaps obscuring statistical difference. Because CVID seems to be a late-onset and progressive disease, the patients in this population were likely at different stages of the disease course, and with variation in gene expression levels, despite a confirmed clinical and immunologic B cell deficiency. To date, direct regulation of PAX5 by E2A has not been reported (Kwon et al. 2008).
As an example of the dynamic interaction of these genes, E2A expression is also modified by EBF1 at transcriptional and post-translational levels (Thal et al. 2009; Treiber et al. 2010). In mouse models, targeted inactivation of E2A or EBF1 results in a block in B cell development at the pre-pro-B cell stage, before the onset of early B lineage gene expression (i.e. PAX5) (Bain et al. 1994; Zhuang et al. 1994; Kwon et al. 2008; Zandi et al. 2008). When EBF1 is missing in mice, the bone marrow lacks D-J rearrangements and pre-B cell restricted-genes, but CD43+ and B220+ cells are present, suggesting a B cell commitment but pro-B cell arrest (Lin and Grosschedl 1995). This sustained expression of B220 is consistent with the data presented herein, in which CVID horses express comparable levels of B220 to healthy horses. EBF1 and PAX5 are also involved in an autoregulatory loop as PAX5 upregulates transcription of EBF1 through its proximal promoter (Roessler et al. 2007). Therefore, given the near complete loss of PAX5 mRNA and protein observed in the bone marrow of CVID horses, we expected to observe a corresponding decrease in EBF1.
The loss of PAX5 expression in the bone marrow of CVID horses is especially intriguing because this gene is essential for B lineage commitment and development (Michaelson et al. 1996; Nutt et al. 1998; Horcher et al., 2001). PAX5 is regulated by EBF1 in the promoter region and a potent enhancer has been identified in intron 5 (Decker et al. 2009). Importantly, EBF1 and PAX5 are continuously expressed from the earliest B lymphoid progenitor through all B cell states, except plasma cells. Therefore, EBF1 and PAX5 are required not only for commitment to the B lineage and suppressing alternative cell fates, but are essential for B cell identity and survival in the periphery (Hao and Rajewsky 2001). PAX5 directly induces transcription favorable chromatin structure at promoters or putative enhancers of target genes, and promotes the expression of B lymphoid-specific genes. After a hierarchical dependence on EBF1 and E2A function, PAX5 regulates CD19, CD79A, LAMBDA5, VpreB, Rag-1,2, B cell-specific tyrosine kinase BLK, LEF1, BCLX, N-MYC, and facilitates immunoglobulin variable-diversity-joining rearrangements of the IgH locus (Zwollo and Desiderio 1994; Nutt et al. 1997; Morrison et al. 1998; Ebert et al. 2011). Simultaneously, PAX5 represses the transcription of non-B lineage genes (e.g. down-regulating the expression of NOTCH 1, required for T cell development; and macrophage colony-stimulating factor receptor M-CSFR, required for myeloid differentiation) (Nutt et al. 1998; Cobaleda et al. 2007). Recently, an additional 170 genes regulated by PAX5 have been identified, and some activated genes are the immunoglobulin light chain kappa (IGKC), the nuclear protein ARNTI (ARNT), and the Fc receptor-like protein 1 (FCRL1) (Schebesta et al. 2007). The immunoglobulin J-chain (IGHJ@) was identified as a direct repression target in the same study. B cell development is arrested at the pro-B cell stage in the bone marrow of PAX5 mutant mice (Urbanek et al. 1994).
Expression of several PAX5 direct and indirect targets was significantly reduced in CVID-affected horses (E2A, CD19, IGHM in 7 of 8 CVID horses, IGHD), but expression of other PAX5 targets (CD79A, BLNK, FCRL1) was more variable and/or maintained at levels overlapping that of healthy horses. PAX5 −/− pro-B lines express early B lineage genes (LAMBDA5, VpreB, including CD79A) and undergo V-D-J gene rearrangements, suggesting the role of additional transcription factors in their regulation (Nutt et al. 1997). Alternatively, PAX5 downstream genes detected in the CVID-affected horses may result from a small number of residual plasma cells in the bone marrow, as CD79A, BLNK, and FCRL1 are expressed in plasma cells independently of PAX5 but E2A, PAX5, and CD19 are dispensable for plasma cells. However, the latter is not well supported by the fact that histology of tissues from CVID-affected horses revealed the absence of plasma cells in the intestine and bone marrow (Schebesta et al. 2007).
Our immunohistochemical results revealed absent B cells [PAX5-BSAP] in the bone marrow of affected horses but occasional B cells in the lymph node, dispersed in the tissue, but not forming germinal centers. It is likely that B cell production was sustained in these patients for a period of time before the diagnosis of CVID, and secondary lymphoid tissues were then populated with B cells, including memory cell development; however, a late-onset event impaired B cell development in the bone marrow, and the continuous population of peripheral tissues could not be sustained. The PAX5 expression in affected horses was significantly reduced in comparison to control horses. The CVID horse with the least number of PAX5 transcripts was most depleted of B cells in the primary and secondary lymphoid tissues. An example of a CVID horse with a moderate number of PAX5 transcripts did not have positive B cells in the bone marrow, but PAX5-BSAP, CD19, and IgM positive B cells were detected in the lymph node. The CVID horse with the highest number of PAX5 transcripts had detectable CD19 and IgM positive cells in the bone marrow, and larger numbers of PAX5-BSAP, CD19, and IgM positive in the lymph node.
Collectively, our data suggest that B cell development in the affected horses seems to be abrogated at the transition between pre-pro-B cells and pro-B cells. Based on our data, the loss of PAX5 mRNA expression does not seem to be due to mutation of the PAX5 coding sequence, rather this event may involve mechanisms or regulatory genes that control PAX5 expression in the bone marrow. We are further investigating genetic, regulatory (promoter and intron 5 enhancer regions), and epigenetic mechanisms that could be involved in silencing of PAX5 and other early B cell development genes in horses with CVID. Our current studies focus on the genetic and epigenetic regulatory mechanisms for the expression of E2A, EBF1, and PAX5 in affected and control horses, and their immediate target genes (Zandi et al. 2008; Treiber et al. 2010). In addition, we are searching for reliable reagents that can measure the protein expression of equine essential B cell molecules (e.g. E2A, EBF1).
In summary, we present herein a natural disturbance in B cell development at the level of pre-pro and pro-B cells in a population of horses with CVID. Our data indicates that the expression of the E2A and PAX5 commitment genes are severely impaired, which directly affects B cell fate and development. It is yet to be determined if all cases of CVID in horses involve a single genetic or epigenetic faulty mechanism, or if the disease manifests heterogeneously as in human patients (Yong et al. 2011). Nevertheless, the equine condition brings a valuable opportunity to study impaired B cell development and its clinical consequences; and it serves as a model for the study of primary immunodeficiency etiology in a natural setting, with hopes to improve diagnostics and understand mechanisms of disease in human patients. Additionally, it provides a unique opportunity to further understand regulatory mechanisms of B cell differentiation and survival.
Highlights.
E2A, PAX5, CD19, and IGHD mRNA expressions were reduced in equine CVID patients.
No mutations in the PAX5 gene were identified in equine CVID patients.
In equine CVID patients, the pre-pro-B to pro-B cell transition appears impaired.
Acknowledgments
The authors would like to thank all the veterinarians and owners of horses with CVID for their efforts in providing clinical data, blood and tissue samples for the study of this disease. The authors also thank Ms. Rebecca A. King for technical assistance, and Stephen Nutt at the The Walter and Eliza Hall Institute of Medical Research, Victoria, Australia for his guidance with PAX5 immunohistochemical staining. This study was support by the National Institutes of Health grant R03AI079796 and New Director’s Innovator Award DP2OD007216.
Abbreviations
- cDNA
complementary DNA
- CT
cycle threshold
- CVID
common variable immunodeficiency
- DNA
deoxyribonucleic acid
- EDTA
ethylenediaminetetraacetic acid
- mRNA
messenger ribonucleic acid
- RNA
ribonucleic acid
- RT-PCR
reverse transcriptase polymerase chain reaction
- TBS
Tris buffered saline
Glossary
- Hypo- or agammaglobulinemia
a condition in which an individual produces few or no antibodies (gamma globulins)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
R.L. Tallmadge, Email: rlt8@cornell.edu.
K.A. Such, Email: kimberlyasuch@gmail.com.
K.C. Miller, Email: kcm9@cornell.edu.
M.B. Matychak, Email: mbm10@cornell.edu.
M.J.B. Felippe, Email: mbf6@cornell.edu.
References
- Bain G, Maandag EC, Izon DJ, Amsen D, Kruisbeek AM, Weintraub BC, Krop I, Schlissel MS, Feeney AJ, van Roon M, et al. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 1994;79(5):885–92. doi: 10.1016/0092-8674(94)90077-9. [DOI] [PubMed] [Google Scholar]
- Chapel H, Cunningham-Rundles C. Update in understanding common variable immunodeficiency disorders (CVIDs) and the management of patients with these conditions. Br J Haematol. 2009;145(6):709–27. doi: 10.1111/j.1365-2141.2009.07669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nat Immunol. 2007;8(5):463–70. doi: 10.1038/ni1454. [DOI] [PubMed] [Google Scholar]
- Cunningham-Rundles C, Knight AK. Common variable immune deficiency: reviews, continued puzzles, and a new registry. Immunol Res. 2007;38(1–3):78–86. doi: 10.1007/s12026-007-0024-0. [DOI] [PubMed] [Google Scholar]
- Decker T, Pasca di Magliano M, McManus S, Sun Q, Bonifer C, Tagoh H, Busslinger M. Stepwise activation of enhancer and promoter regions of the B cell commitment gene Pax5 in early lymphopoiesis. Immunity. 2009;30(4):508–20. doi: 10.1016/j.immuni.2009.01.012. [DOI] [PubMed] [Google Scholar]
- DeKoter RP, Lee HJ, Singh H. PU.1 regulates expression of the interleukin-7 receptor in lymphoid progenitors. Immunity. 2002;16(2):297–309. doi: 10.1016/s1074-7613(02)00269-8. [DOI] [PubMed] [Google Scholar]
- Drummond A, Ashton B, Cheung M, Heled J, Kearse M, Moir R, Stones-Havas S, Thierer T, Wilson A. Geneious v4.7. 2009 Available from http://www.geneious.com/
- Ebert A, McManus S, Tagoh H, Medvedovic J, Salvagiotto G, Novatchkova M, Tamir I, Sommer A, Jaritz M, Busslinger M. The distal V(H) gene cluster of the Igh locus contains distinct regulatory elements with Pax5 transcription factor-dependent activity in pro-B cells. Immunity. 2011;34:175–87. doi: 10.1016/j.immuni.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Flaminio MJ, LaCombe V, Kohn CW, Antczak DF. Common variable immunodeficiency in a horse. J Am Vet Med Assoc. 2002;221(9):1296–302. 67. doi: 10.2460/javma.2002.221.1296. [DOI] [PubMed] [Google Scholar]
- Flaminio MJ, Tallmadge RL, Salles-Gomes CO, Matychak MB. Common variable immunodeficiency in horses is characterized by B cell depletion in primary and secondary lymphoid tissues. J Clin Immunol. 2009;29(1):107–16. doi: 10.1007/s10875-008-9221-4. [DOI] [PubMed] [Google Scholar]
- Fuxa M, Skok JA. Transcriptional regulation in early B cell development. Curr Opin Immunol. 2007;19(2):129–36. doi: 10.1016/j.coi.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Hagman J, Lukin K. Transcription factors drive B cell development. Curr Opin Immunol. 2006;18(2):127–34. doi: 10.1016/j.coi.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Hao Z, Rajewsky K. Homeostasis of peripheral B cells in the absence of B cell influx from the bone marrow. J Exp Med. 2001;194(8):1151–64. doi: 10.1084/jem.194.8.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horcher M, Souabni A, Busslinger M. Pax5/BSAP maintains the identity of B cells in late B lymphopoiesis. Immunity. 2001;14(6):779–90. doi: 10.1016/s1074-7613(01)00153-4. [DOI] [PubMed] [Google Scholar]
- Kwon K, Hutter C, Sun Q, Bilic I, Cobaleda C, Malin S, Busslinger M. Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity. 2008;28(6):751–62. doi: 10.1016/j.immuni.2008.04.014. [DOI] [PubMed] [Google Scholar]
- Liu P, Keller JR, Ortiz M, Tessarollo L, Rachel RA, Nakamura T, Jenkins NA, Copeland NG. Bcl11a is essential for normal lymphoid development. Nat Immunol. 2003;4(6):525–32. doi: 10.1038/ni925. [DOI] [PubMed] [Google Scholar]
- Mackarehtschian K, Hardin JD, Moore KA, Boast S, Goff SP, Lemischka IR. Targeted disruption of the flk2/flt3 gene leads to deficiencies in primitive hematopoietic progenitors. Immunity. 1995;3(1):147–61. doi: 10.1016/1074-7613(95)90167-1. [DOI] [PubMed] [Google Scholar]
- Maier H, Ostraat R, Gao H, Fields S, Shinton SA, Medina KL, Ikawa T, Murre C, Singh H, Hardy RR, Hagman J. Early B cell factor cooperates with Runx1 and mediates epigenetic changes associated with mb-1 transcription. Nat Immunol. 2004;5(10):1069–77. doi: 10.1038/ni1119. [DOI] [PubMed] [Google Scholar]
- Medina KL, Singh H. Gene regulatory networks orchestrating B cell fate specification, commitment, and differentiation. Curr Top Microbiol Immunol. 2005;290:1–14. doi: 10.1007/3-540-26363-2_1. [DOI] [PubMed] [Google Scholar]
- Michaelson JS, Singh M, Birshtein BK. B cell lineage-specific activator protein (BSAP). A player at multiple stages of B cell development. J Immunol. 1996;156(7):2349–51. [PubMed] [Google Scholar]
- Morrison AM, Nutt SL, Thevenin C, Rolink A, Busslinger M. Loss- and gain-of-function mutations reveal an important role of BSAP (Pax-5) at the start and end of B cell differentiation. Semin Immunol. 1998;10(2):133–42. doi: 10.1006/smim.1998.0115. [DOI] [PubMed] [Google Scholar]
- Nutt SL, Morrison AM, Dorfler P, Rolink A, Busslinger M. Identification of BSAP (Pax-5) target genes in early B-cell development by loss- and gain-of-function experiments. Embo J. 1998;17(8):2319–33. doi: 10.1093/emboj/17.8.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt SL, Urbanek P, Rolink A, Busslinger M. Essential functions of Pax5 (BSAP) in pro-B cell development: difference between fetal and adult B lymphopoiesis and reduced V-to-DJ recombination at the IgH locus. Genes Dev. 1997;11(4):476–91. doi: 10.1101/gad.11.4.476. [DOI] [PubMed] [Google Scholar]
- Park MA, Li JT, Hagan JB, Maddox DE, Abraham RS. Common variable immunodeficiency: a new look at an old disease. Lancet. 2008;372(9637):489–502. doi: 10.1016/S0140-6736(08)61199-X. [DOI] [PubMed] [Google Scholar]
- Pellegrini-Masini A, Bentz AI, Johns IC, Parsons CS, Beech J, Whitlock RH, Flaminio MJ. Common variable immunodeficiency in three horses with presumptive bacterial meningitis. J Am Vet Med Assoc. 2005;227(1):114–22. 87. doi: 10.2460/javma.2005.227.114. [DOI] [PubMed] [Google Scholar]
- Ramirez J, Lukin K, Hagman J. From hematopoietic progenitors to B cells: mechanisms of lineage restriction and commitment. Curr Opin Immunol. 2010;22(2):177–84. doi: 10.1016/j.coi.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler S, Gyory I, Imhof S, Spivakov M, Williams RR, Busslinger M, Fisher AG, Grosschedl R. Distinct promoters mediate the regulation of Ebf1 gene expression by interleukin-7 and Pax5. Mol Cell Biol. 2007;27(2):579–94. doi: 10.1128/MCB.01192-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzer U, Neumann C, Thiel J, Woellner C, Pan-Hammarstrom Q, Lougaris V, Hagena T, Jung J, Birmelin J, Du L, Metin A, Webster DA, Plebani A, Moschese V, Hammarstrom L, Schaffer AA, Grimbacher B. Screening of functional and positional candidate genes in families with common variable immunodeficiency. BMC Immunol. 2008;9:3. doi: 10.1186/1471-2172-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schebesta A, McManus S, Salvagiotto G, Delogu A, Busslinger GA, Busslinger M. Transcription factor Pax5 activates the chromatin of key genes involved in B cell signaling, adhesion, migration, and immune function. Immunity. 2007;27(1):49–63. doi: 10.1016/j.immuni.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Sekine H, Ferreira RC, Pan-Hammarstrom Q, Graham RR, Ziemba B, de Vries SS, Liu J, Hippen K, Koeuth T, Ortmann W, Iwahori A, Elliott MK, Offer S, Skon C, Du L, Novitzke J, Lee AT, Zhao N, Tompkins JD, Altshuler D, Gregersen PK, Cunningham-Rundles C, Harris RS, Her C, Nelson DL, Hammarstrom L, Gilkeson GS, Behrens TW. Role for Msh5 in the regulation of Ig class switch recombination. Proc Natl Acad Sci U S A. 2007;104(17):7193–8. doi: 10.1073/pnas.0700815104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H, Medina KL, Pongubala JM. Contingent gene regulatory networks and B cell fate specification. Proc Natl Acad Sci U S A. 2005;102(14):4949–53. doi: 10.1073/pnas.0500480102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitnicka E, Brakebusch C, Martensson IL, Svensson M, Agace WW, Sigvardsson M, Buza-Vidas N, Bryder D, Cilio CM, Ahlenius H, Maraskovsky E, Peschon JJ, Jacobsen SE. Complementary signaling through flt3 and interleukin-7 receptor alpha is indispensable for fetal and adult B cell genesis. J Exp Med. 2003;198(10):1495–506. doi: 10.1084/jem.20031152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitnicka E, Buza-Vidas N, Ahlenius H, Cilio CM, Gekas C, Nygren JM, Mansson R, Cheng M, Jensen CT, Svensson M, Leandersson K, Agace WW, Sigvardsson M, Jacobsen SE. Critical role of FLT3 ligand in IL-7 receptor independent T lymphopoiesis and regulation of lymphoid-primed multipotent progenitors. Blood. 2007;110(8):2955–64. doi: 10.1182/blood-2006-10-054726. [DOI] [PubMed] [Google Scholar]
- Tennent-Brown BS, Navas de Solis C, Foreman JH, Goetz TE, Fredrickson RL, Borst LB, Flaminio MJBF. Common variable immunodeficiency in a horse with chronic peritonitis. Equine vet Educ. 2010;22(8):393–9. [Google Scholar]
- Thal MA, Carvalho TL, He T, Kim HG, Gao H, Hagman J, Klug CA. Ebf1-mediated down-regulation of Id2 and Id3 is essential for specification of the B cell lineage. Proc Natl Acad Sci U S A. 2009;106(2):552–7. doi: 10.1073/pnas.0802550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiber T, Mandel EM, Pott S, Gyory I, Firner S, Liu ET, Grosschedl R. Early B cell factor 1 regulates B cell gene networks by activation, repression, and transcription- independent poising of chromatin. Immunity. 2010;32(5):714–25. doi: 10.1016/j.immuni.2010.04.013. [DOI] [PubMed] [Google Scholar]
- Urbanek P, Wang ZQ, Fetka I, Wagner EF, Busslinger M. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell. 1994;79(5):901–12. doi: 10.1016/0092-8674(94)90079-5. [DOI] [PubMed] [Google Scholar]
- Wang H, Lee CH, Qi C, Tailor P, Feng J, Abbasi S, Atsumi T, Morse HC., 3rd IRF8 regulates B-cell lineage specification, commitment, and differentiation. Blood. 2008;112(10):4028–38. doi: 10.1182/blood-2008-01-129049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnatz K, Salzer U, Rizzi M, Fischer B, Gutenberger S, Bohm J, Kienzler AK, Pan-Hammarstrom Q, Hammarstrom L, Rakhmanov M, Schlesier M, Grimbacher B, Peter HH, Eibel H. B-cell activating factor receptor deficiency is associated with an adult-onset antibody deficiency syndrome in humans. Proc Natl Acad Sci U S A. 2009;106(33):13945–50. doi: 10.1073/pnas.0903543106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmott RW. Delayed diagnosis in common variable immunodeficiency. J Pediatr. 2009;154(6):A1. doi: 10.1016/j.jpeds.2009.04.019. [DOI] [PubMed] [Google Scholar]
- Yong PF, Thaventhiran JE, Grimbacher B. “A Rose is a Rose is a Rose,” but CVID is Not CVID Common Variable Immune Deficiency (CVID), What do we Know in 2011? Adv Immunol. 2011;111:47–107. doi: 10.1016/B978-0-12-385991-4.00002-7. [DOI] [PubMed] [Google Scholar]
- Zandi S, Mansson R, Tsapogas P, Zetterblad J, Bryder D, Sigvardsson M. EBF1 is essential for B-lineage priming and establishment of a transcription factor network in common lymphoid progenitors. J Immunol. 2008;181(5):3364–72. doi: 10.4049/jimmunol.181.5.3364. [DOI] [PubMed] [Google Scholar]
- Zhuang Y, Soriano P, Weintraub H. The helix-loop-helix gene E2A is required for B cell formation. Cell. 1994;79(5):875–84. doi: 10.1016/0092-8674(94)90076-0. [DOI] [PubMed] [Google Scholar]
- Zwollo P, Desiderio S. Specific recognition of the blk promoter by the B-lymphoid transcription factor B-cell-specific activator protein. J Biol Chem. 1994;269(21):15310–7. [PubMed] [Google Scholar]