Abstract
The patatin-related phospholipase A (pPLA) hydrolyzes membrane glycerolipids to produce monoacyl compounds and free fatty acids. Phospholipids are cleaved by pPLAIIα at the sn-1 and sn-2 positions, and galactolipids, including those containing oxophytodienoic acids, can also serve as substrates. Ablation of pPLAIIα decreased lysophosphatidylcholine and lysophosphatidylethanolamine levels, but increased free linolenic acid. pPLAIIα-deficient plants displayed a higher level of jasmonic acid and methyl jasmonate, as well as the oxylipin-biosynthetic intermediates 13-hydroperoxylinolenic acid and 12-oxophytodienoic acid than wild-type (WT) plants. The expression of genes involved in oxylipin production was also higher in the pPLAIIα-deficient mutant than in WT plants. The mutant plants lost water more quickly than WT plants. The stomata of WT and mutant plants responded similarly to abscisic acid. In response to desiccation, the mutant and WT leaves produced abscisic acid at the same rate, but, after 4 h of desiccation, the jasmonic acid level was much higher in mutant than WT leaves. These results indicate that pPLAIIα negatively regulates oxylipin production and suggest a role in the removal of oxidatively modified fatty acids from membranes.
Keywords: patatin-related phospholipase A, oxidative modified lipids, jasmonate synthesis, water loss, Arabidopsis thaliana
INTRODUCTION
The release of fatty acids from cellular membrane glycerolipids has been implicated in various cellular processes including the production of signaling messengers, membrane remodeling, membrane deterioration during senescence, and stress damage (Wang, 2004; Scherer et al., 2010). The patatin-related phospholipase A (pPLA) is a major family of enzymes that hydrolyze membrane glycerolipids to produce lysolipids and free fatty acids. Patatin refers to a group of storage glycoproteins that were originally discovered in potato tubers (Galliard, 1971); patatin-related proteins were later found in other plant species and tissues, as well as in animal cells (Holk et al., 2002). The well-characterized mammalian cytosolic PLA2s contain a patatin domain that serves as catalytic site (Schrag and Cygler, 1997; Hirschberg et al., 2001). The Arabidopsis pPLA family consists of 10 genes that are grouped into three subfamilies, pPLAI, pPLAII (α, β, γ, δ, ε), and pPLAIII (α, β, γ, δ), based on gene structures and deduced protein sequences (Scherer et al., 2010). pPLAI contains 1257 amino acid residues whereas the size of pPLAIIs and IIIs ranges from 382 to 526 amino acid residues. pPLAs are involved in various processes, including fungal and bacterial pathogen infection, phosphate deprivation, auxin response, cellulose deposition, cell elongation, and grain yield (La Camera et al., 2005; Yang et al., 2007; Rietz et al., 2010; Li et al., 2011). However, the metabolic and cellular mechanism by which pPLAs affect plant growth and stress response remain elusive.
One of potential mechanisms of action for pPLAs starts with the hydrolysis of membrane lipids, releasing polyunsaturated fatty acids (PUFAs), such as linolenic acid that serves as a substrate for the synthesis of jasmonic acid (JA) in response to stress. JA and other cyclopentenone derivatives are synthesized via the octadecanoic pathway from linolenic acid in plants. These oxylipins modulate plant response to various stresses, including pathogen infection, insect attack, and drought (Staswick, 1992; Wasternack and Parthier, 1997; Reymond and Farmer, 1998; Farmer, 2001). One PLA1 was identified as being involved in JA production, pollen maturation, and anther dehiscence in Arabidopsis (Ishiguro et al., 2001), but the identity of the enzyme(s) involved in stress-induced production of PUFAs for oxylipin production it is still unclear. pPLAI has been implicated in the production of basal levels of JA, but not pathogen or wounding-induced JA production (Yang et al., 2007). pPLAI plays a positive role in Arabidopsis resistance to Botrytis cinerea (Yang et al., 2007), whereas the suppression of pPLAIIα was reported to render Arabidopsis plants more resistant to fungal and bacterial infection (La Camera et al., 2005). This negative effect of pPLAIIα on plant–pathogen interactions appears to argue against a role of pPLAIIα in promoting JA production. The expression of pPLAIIα is induced in response to various biotic and abiotic stresses, including pathogens, low temperature, high salinity, abscisic acid, salicylic acid, methyl jasmonate, ethylene, iron, and phosphate deficiency (Narusaka et al., 2003; Rietz et al., 2004). These observations raise intriguing questions regarding the role of pPLAIIα in JA production and plant stress responses.
Recent studies have shown that complex membrane lipids, such as monogalactosyldiacylglycerol (MGDG), digalactosyldiacylglycerol (DGDG), and phospholipids, are oxidatively modified (Andersson et al., 2006; Buseman et al., 2006). Oxidized (ox-) MGDG and DGDG containing 12-oxophytodienoic acid (OPDA) and dinor-oxophytodienoic acid (dnOPDA), including Arabidopside A (OPDA/dnOPDA MGDG), B (OPDA/OPDA MGDG), C (OPDA/dnOPDA DGDG), and D (OPDA/OPDA DGDG), are produced in response to wounding, bacterial and fungal infection, dark, aging, and osmotic stress (Andersson et al., 2006; Buseman et al., 2006; Kourtchenko et al., 2007; Glauser et al., 2008; Maeda et al., 2008; Xiao et al., 2010). pPLAI has been shown to use arabidopsides as substrates, and to prefer them as substrates to unoxidized, normal MGDG and DGDG (Yang et al., 2007). In this study, we analyzed pPLAIIα’s lipid-hydrolyzing activity and effect on JA formation. The results suggest that this enzyme negatively regulates oxylipin production, possibly via participating in membrane repair that includes removal of oxidatively modified lipids.
RESULTS
pPLAIIα Hydrolyzes Glycerolipids at the sn-1 and sn-2 Positions
The full-length cDNA of pPLAIIα gene encodes a protein of 407 amino acids with a predicted pI of 5.57 and molecular weight of 44 239. pPLAIIα contains the conserved serine hydrolase motif GXSXG at residues 64–68, and a conserved Asp residue at 215 within the patatin domain (residues 21–228). The serine and aspartic acid residues constitute a catalytic dyad for its acyl hydrolase activity (Hirschberg et al., 2001; Rydel et al., 2003). The pPLAIIα cDNA was expressed in E. coli and a protein of 44 kDa corresponding to the predicted size of pPLAIIα was purified to apparent homogeneity for determination the enzymatic properties of pPLAIIα (Figure 1A).
Figure 1.
pPLAIIα Activity on Labeled Phospholipid Substrates.
(A) Coomassie blue staining of an 8% SDS–PAGE gel loaded with affinity purified pPLAIIα from E. coli. M, protein marker; P, purified protein.
(B) Fluorescent fatty acid and lysophospholipid released by pPLAIIα when NBD-phospholipids were used as substrates.
(C) Production of radioactive fatty acid and lysoPI by pPLAIIα when 1-stearoyl-2-[1-14C]arachidonyl-PI was used as substrate. Values are mean ± SD (n = 3).
Purified pPLAIIα was assayed for its ability to hydrolyze various phospholipids, using fluorescently [12-(7-nitro-2–1,3-benzoxadiazol-4-yl)amino]-dodecanoyl (NBD)-labeled phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylglycerol (PG), phosphatidic acid (PA), and phosphatidylserine (PS) as substrates. The pPLAIIα reaction produced both NBD-labeled free fatty acids and NBD-lysophospholipids (Figure 1B). Because the fluorescent label was at the sn-2 position, the detection of both the NBD-lysophospholipids and NBD-free fatty acid indicates that the enzyme releases fatty acids at both the sn-1 and sn-2 positions. The relative amount of NBD-fatty acids versus NBD-lysophospholipids formed indicates the relative hydrolysis at the sn-2 versus sn-1 position by the enzyme. pPLAIIα preferred releasing the fatty acid at the sn-1 to sn-2 position when PC or PE was the substrate whereas it favors the sn-2 position when PG, PA, or PS was the substrate (Figure 1B and 1C). The enzyme was less active towards PS and PA in comparison to the other phospholipids used in the assay (Figure 1B and 1C). Radioactive 1-stearoyl-2-(1-[14C]arachidonyl)-phosphatidylinositol (PI) was used to determine the hydrolytic activity towards PI. Consistently with results from the fluorescent lipid assays, both radioactive free fatty acid and lysoPI were generated (Figure 1C). The amount of [14C]-fatty acid formed was about fivefold higher than that of [14C]-lysoPI, suggesting that the enzyme prefers hydrolysis of PI at the sn-2 to sn-1 position.
pPLAIIα Act on Oxidized and Unoxidized Glycerolipids
To determine the substrate selectivity, total lipids extracted from leaves of a wounded plant were used as substrates for the purified pPLAIIα. As a control, the same number of leaf lipids was incubated with empty-vector bacterial proteins that were subjected to the same purification process as pPLAIIα. After reaction, phospholipid and galactolipid species in the pPLAIIα and control reactions were measured. Addition of pPLAIIα decreased PC, PE, PI but little PS and PA species (Figure 2A and 2B). Lyso-phospholipid levels rose (LPC and LPE determined) (Figure 2C). pPLAIIα also acted on nearly every species of MGDG and DGDG (Figure 3A), including those containing OPDA (Figure 3B), resulting in the production of corresponding lysogalactolipids monogalactosyl monoacylglyceride and digalactosyl monoacylglyceride (Figure 3C). Taken together, the data show that the enzyme is capable of acting on nearly every membrane lipid substrate determined in wounded Arabidopsis leaves.
Figure 2.
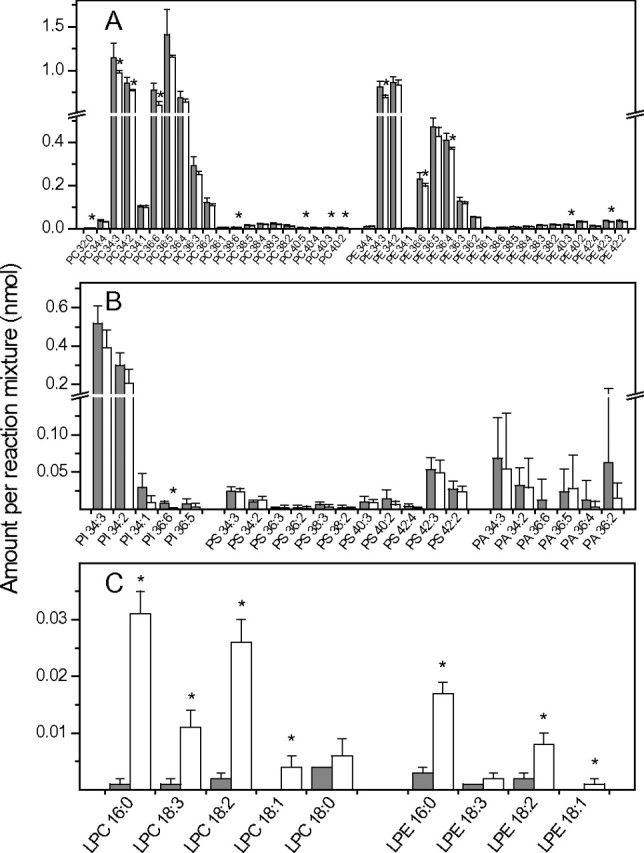
Hydrolysis of Phospholipids by pPLAIIα Using Total Lipids from Mature Wounded Arabidopsis Leaves.
Equal amounts of total Arabidopsis lipids were incubated with purified pPLAIIα (+ pPLAII, open bars) or protein from empty-vector control (– pPLAII, gray bars) at 30°C for 1 h.
(A) Amounts of PC and PE molecular species present in the + pPLAII and – pPLAII mixture after reactions.
(B) Amounts of PI, PS, and PA molecular species present in the + pPLAII and – pPLAII mixture after reactions.
(C) Lysophospholipid species formed in the + pPLAII and – pPLAII mixtures after reactions. Values are means ± SD (n = 5). Asterisks mark differences between the control and + pPLAIIα reactions at p < 0.05 according to Student’s t-test.
Figure 3.

Hydrolysis of Galactolipids by pPLAIIα Using Total Lipids from Mature Wounded Arabidopsis Leaves.
Equal amounts of total Arabidopsis lipids were incubated with purified pPLAIIα (+ pPLAII, open bars) or protein from empty-vector control (– pPLAII, gray bars) at 30°C for 1 h.
(A) Amounts of normal-chain DGDG, MGDG, and PG molecular species present in the + pPLAII and – pPLAII mixture after reactions.
(B) Amounts of OPDA-containing DGDG, MGDG, and PG molecular species in the + pPLAII and – pPLAII mixture after reactions.
(C) MGMG and DGMG species formed in the + pPLAII and – pPLAII mixtures after reactions. Values are means ± SD (n = 5). Asterisks mark differences between the control and + pPLAIIα reactions at P < 0.05 according to Student’s t-test. One unit is the amount of signal produced by one nmol of internal standard (panels A and C) or one unit of 34:6 MGDG signal (panel B) as described in ‘Methods’. Asterisks mark differences between the control and + pPLAIIα reactions at p < 0.05 according to Student’s t-test.
JA Production Is Up-Regulated in pPLAIIα-Deficient Leaves
A T-DNA insertion knockout (KO) mutant was isolated and the mutant lost the expression of pPLAIIα, as indicated by real-time quantitative PCR (Figure 4A). pPLAIIα KO leaves contained approximately 30 and 15% less LPC and LPE, respectively, but the level of LPG was the same in the KO and wild-type (WT) leaves (Figure 4B). The decrease occurred with LPC species with 16:0, 18:2, or 18:0, but not with 18:3, whereas only 16:0-LPE was significantly decreased in the KO leaves (Figure 4C).
Figure 4.
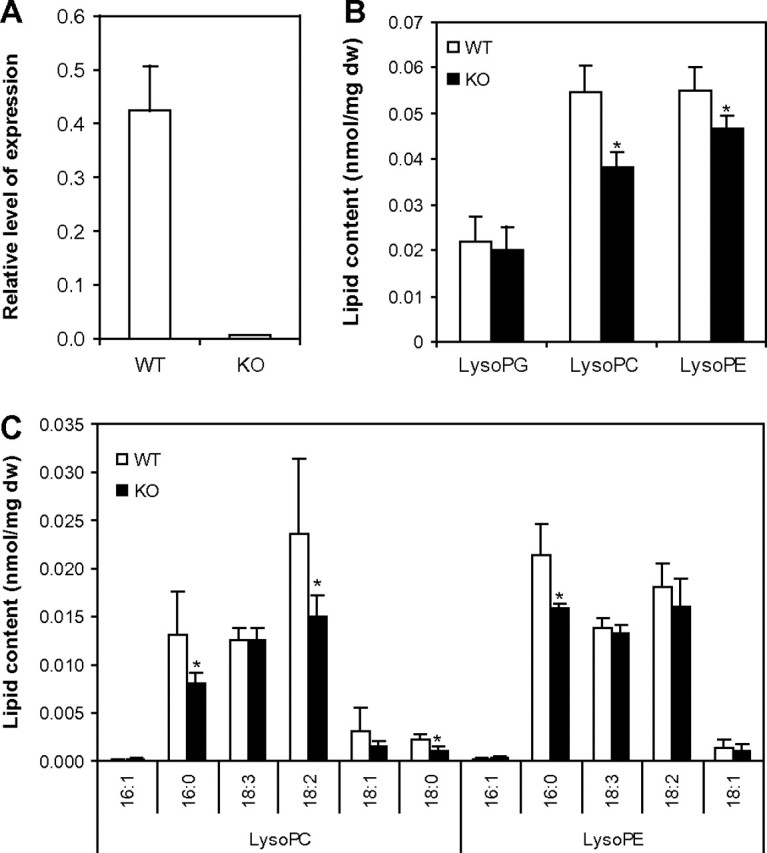
Lysophospholipid Level in pPLAIIα-KO and WT Leaves.
(A) Relative level of pPLAIIα gene expression in wild-type and mutant plants was checked with real-time quantitative PCR.
(B) Profiles of total LPC, LPE, and LPG in WT and mutants.
(C) Profiles of LPC and LPE species in WT and mutant plants. Values are means ± SD (n = 5). Statistically significant differences between the wild-type and mutant line are marked with asterisks (p < 0.05, according to Student’s t-test).
In contrast to the change in lysophospholipids, the level of free linolenic acid in pPLAIIα-KO leaves was significantly higher than that of WT (Figure 5A). The KO leaves produced approximately twice the amount of methyl jasmonate (MeJA) and JA of WT plants (Figure 5A). Measurements of the potential intermediates in the oxylipin biosynthetic pathway showed that the levels of 13-hydroperoxylinolenic acid (13-HPOT) and OPDA in pPLAIIα-KO leaves were significantly higher than those of WT plants (Figure 5A).
Figure 5.

Contents of Intermediate Compounds and the Expression Levels of Several Genes in the Oxlipin Pathway in WT and KO Plants.
(A) The amount of jasmonates in the oxylipin pathway in WT and mutants. 18:3, linolenic acid; 13-HPOT, 13-hydroperoxylinolenic acid; OPDA, 12-oxo-phytodienoic acid; JA, jasmonic acid; MeJA, methyl jasmonic acid. Values are means ± SD (n = 5).
(B) Expression of several genes in the oxylipin pathway. PLAIIA, pPLAIIα; LOX1, lipoxygenase 1; LOX2, lipoxygenase 2; LOX3, lipoxygenase 3; AOS, allene oxide synthase; HPL, hydroperoxide lyase; AOC1, allene oxide cyclase 1; AOC2, allene oxide cyclase 2; MJT, jasmonic acid carboxyl methyltransferase; OPR, 12-oxo-phytodienoic acid (OPDA) reductase 3; VSP2, vegetative storage protein 2.
The expression of the genes for the enzymes in the oxylipin synthesis and responsive processes was monitored by quantitative real-time PCR (Figure 5B). pPLAIIα-KO plants displayed higher levels of lipoxygenase 1, 3 (LOX1, LOX3), allene oxide synthase (AOS), allene oxide cyclase 1 and 2 (AOC1 and AOC2), and carboxyl methyltransferase (MJT). The fold increases were 2.4, 65, 1.6, 2, 5, and 2 for LOX1, LOX3, AOS, AOC1, AOC2, and MJT, respectively. The levels of expression for LOX2, hydrogen peroxide lyase (HPL), OPDA reductase 3 (OPR3), and vegetative storage protein 2 (VSP2) were comparable between KO and WT plants (Figure 5B). These results indicate that an oxylipin pathway in pPLAIIα-KO plants is up-regulated at the level of both gene expression and the accumulation of the metabolic products.
Knockout of pPLAIIα Increases Water Loss and Drought Sensitivity
During growth and handling of transgenic plants, it was noted that the pPLAIIα-KO plants became dehydrated more quickly than WT plants. When fresh weights of detached leaves were measured, pPLAIIα-KO leaves lost 25% more water than WT leaves plants 2 h after detachment (Figure 6A). When plants were left un-watered, the mutant plants became desiccated and died more quickly than WT plants (Figure 6B). When the mutant was complemented with the genomic gene of pPLAIIα, the rate of water loss and the sensitivity to drought were restored to those of WT (Figure 6), indicating that the accelerated desiccation is caused by the loss of the pPLAIIα function.
Figure 6.
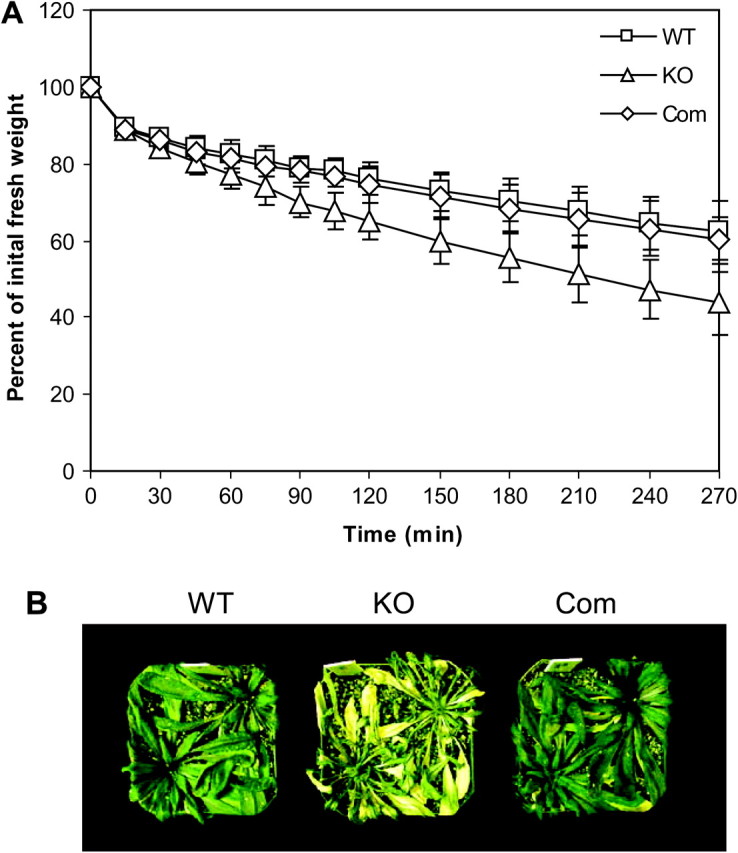
Water Loss and Phenotype of pPLAIIα-KO and WT Plants.
(A) Water loss as measured by decrease in fresh weight in detached leaves from WT, pPLAIIa-KO, and the complemented plants (Com). Values are means ± SD (n = 5).
(B) Phenotype of drought sensitivity in WT, pPLAIIα-KO, and complemented plants. Five-week-old plants grown in a growth chamber were left without watering for 10 d under 12/12-h light/dark period, 150 μmol m−2 s−1 and 30% humidity in a growth chamber.
Water loss in terrestrial plants results mostly from transpiration via stomata, and the plant hormone abscisic acid (ABA) promotes stomatal closure and reduces water loss. Thus, the stomatal response to ABA was compared between pPLAIIα-KO and WT leaves. ABA in different concentrations caused the similar closure in stomata in WT and pPLAIIα-KO leaves (Figure 7). This result suggests that the loss of pPLAIIα does not alter stomatal response to ABA. To determine how the levels of ABA and JA change during dehydration in pPLAIIα-KO and WT plants, the hormone levels were measured at different time intervals in detached leaves. ABA level increased continuously in both WT and mutant plants during the 4-h period tested, but no significant difference was found between the two genotypes (Figure 8A). The JA levels increased in the first hour after detachment in both WT and KO leaves. The JA levels displayed no further increase at the second and third hours in both genotypes. At 4 h of treatment, JA level in the mutant became almost threefold higher than that in WT (Figure 8B).
Figure 7.
Response of Stomatal Aperture to ABA.
Leaf epidermal peels were incubated with different levels of ABA under light for 2.5 h before stomatal aperture was measured. Values are the means ± SD (n > 30).
Figure 8.

Changes in ABA and JA Levels in Detached Leaves of WT and pPLAIIα-KO Mutants.
(A) ABA change in leaves of WT and mutant plants upon desiccation treatment.
(B) JA change in leaves of WT and mutant plants upon desiccation treatment. Values are means ± SD (n = 5). Statistically significant differences between the wild-type and mutant line are marked with asterisks (p < 0.05, according to Student’s t-test).
DISCUSSION
The present study shows that pPLAIIα hydrolyzes various membrane glycerolipids. Comparison of pPLAIIα activity with pPLAI and III reveals differences in substrate usage. pPLAI hydrolyzes MGDG four times greater than PG (Yang et al., 2007), whereas pPLAIIIβ uses PG four times greater than MGDG (Li et al., 2011). pPLAIIs have similar activities toward MGDG and PG (Rietz et al., 2010). The specific activity of pPLAIIα is much higher than that of pPLAIIIβ (Li et al., 2011). These differences suggest that activation of individual pPLAs in the cell may selectively hydrolyze different classes of membrane lipids.
The pPLAIIα-deficient plants displayed increased basal levels of JA and methyl JA. The result indicates that pPLAIIα is not involved in providing free fatty acids for JA biosynthesis. On the contrary, pPLAIIα acts more like as a repressor of the JA production. The enhanced JA production in the pPLAIIα-deficient plants is further supported by the increase in several major intermediates, linolenic acids, 13-HPOT, OPDA, and increased expression of genes involved in the JA biosynthesis. The effect of pPLAIIα-KO on the increased basal JA production is opposite to that of pPLAI-KO that shows a decreased basal level of JA (Yang et al., 2007). These results indicate that individual pPLAs have distinctly different functions. In addition, the increase in LOX1, a 9-LOX that is not directly involved in the JA production, suggests that the production of other oxylipins besides JA is increased in pPLAIIα-knockout plants. pPLAIIα might be involved in the removal of oxidatively modified fatty acids from membranes in membrane remodeling and repair, and the loss of pPLAIIα might impair the membrane repair, resulting in increases in oxylipin production.
The increased water loss in pPLAIIα-deficient plants suggests that pPLAIIα plays a role in modulating leaf water status. ABA is a key water stress hormone that triggers stomatal closure and reduces transpirational water loss (Kim et al., 2010). However, ablation of pPLAIIα does not impact on stomatal response to ABA or ABA production in response to water deficits. This result indicates that pPLAIIα modulates plant response to water deficits in an ABA-independent manner. JA may be involved in the altered response, as it was increased significantly more in KO than WT leaves 4 h after desiccation. On the other hand, JA has been reported to induce stomatal closure and its effect depends on the cytosolic Ca2+ concentration involving a calcium-dependent protein kinase in guard cells (Munemasa et al., 2011). How JA participates in pPLAIIα modulation of plant response to water deficits without affecting ABA is not known. Further study is needed to determine whether and how the increased oxylipin level may play a role in increasing water loss in the pPLAIIα-deficient plants.
METHODS
cDNA Cloning and Expression of Recombinant pPLAIIα Protein in E. coli
The full-length cDNA of pPLAIIα was obtained with PCR using a cDNA clone from the Arabidopsis Biological Resource Center at Columbus, Ohio, using primers of forward 5'-TCCGGATCCATGCAAATGGACAGCCCCA-3' and reverse 5'-ATCGTCGACGATCCTAATTGGAGCTTTTGCATG-3'. The coding sequence of the cDNA (1221 bp) was cloned into the BamHI and SalI sites of pET28a(+)(Novagen) and both strands were sequenced. The construct was introduced into E. coli strain BL-21(DE3). The cells were grown to an OD600 of 0.7 and induced with 0.4 mM IPTG for 12 h at 22°C. The purification of the protein was performed as described previously (Pappan et al., 2004) and the protein concentration was determined with the Bradford method (Bradford, 1976). The purity of the protein was analyzed by 8% SDS–polyacylamide gel electrophoresis (PAGE), followed by Coomasie blue staining.
Isolation of pPLAIIα-KO Mutant and Genetic Complementation
Arabidopsis thaliana plants (ecotype Columbia) were grown in growth chambers under a day/night regimen of 23/18°C and 12/12-h photoperiod under white light of 150 μmol m−2 s−1. A T-DNA insertion mutant at the pPLAIIα locus At2g26560 was isolated by a screening T-DNA insertion line Salk_059119. The T-DNA was inserted at the last intron and the site of T-DNA insertion was confirmed by sequencing. The loss of pPLAIIα expression was verified by quantitative real-time PCR. The mutant gene co-segregated with kanamycin resistance in a 3:1 ratio and the homozygous knockout plants were isolated and used in the study. A 3.34-kb genomic fragment of DNA containing the coding region of pPLAIIα and 1.10 kb 5' and 0.45 kb 3' untranslated regions was amplified by PCR. The PCR product was cloned into the AscI site of pEC291 binary vector. The resulting vector was introduced into the C58C1 strain of Agrobacterium tumefaciens, and the pPLAIIα mutant plants were transformed as described (Clough and Bent, 1998).
Quantitative Real-Time PCR
RNA was isolated from liquid nitrogen-frozen plant leaves using a cetyltrimethylammonium bromide extraction method as described previously (Fan et al., 1997). Ten micrograms of RNA were reverse-transcribed using the iScript™ cDNA Synthesis Kit (Bio-Rad) containing a blend of oligo (dT) and random hexamer primers. Quantitative RT–PCR was performed on 50 ng cDNA with the iQ™ SYBR Green Supermix (Bio-Rad). Amplification of Arabidopsis polyubiquitin gene (UBQ10) transcripts derived from gene At4g05320 was used as reference. The gene specific primer pairs used for the PCR are (from 5′ to 3′): pPLAIIα (At2g26560): forward CTTGCCAAGACAGGAGATGAACTAC, reverse GCATGAGGTGA ACGAATGTCTCGGAT; lipoxygenase1 (LOX1, At1g55020): forward ACCATCACA GCTCAGCTTCAGACA, reverse TCCAAACTTCTCGAACGCCTCCAA; lipoxygenase2 (LOX2, At3g45140): forward AGAGCTGGA GCTGGTGTTGTTAAG, reverse GAACACCCATTCCGGTAACACCAT; lipoxygenase 3 (LOX3, At1g17420): forward TATGGATTTGCGGCA GAGATCGGA, reverse AGGCTCAGAACTCGGAACCAACAA; allene oxide synthase (AOS, At5g42650): forward TGGTGGCGAGGTTGTTTGTGATTG, reverse ATTAACGGAGCTTCCTAACGGCGA; allene oxide cyclase 1 (AOC1, At3g25760): forward CTTAAGCCCAGTGGAGTTGTAAGC, reverse TATACAGGACACGAG AAAGATAAGACT; allene oxide cyclase 2 (AOC2, At3g25780): forward GCACTGGAGCCTAGCGGAGTTATAAGT, reverse TATATCATTACACAGCGATACGAGAAAC; OPDA reductase (OPR, At2g06050): forward CGGATTTGGTTTCGCGGTTCAAGA, reverse TCCGTGTAGCCAACAACTGGATCT; jasmonic acid carboxyl methyltransferase (MJT, At1g19640): forward TATGTAAGCTCGCCACGATACGCT, reverse AACACGATCAACCGGCTCTAACGA; vegetative storage protein 2 (VSP2, At5g24780): forward GCAAGTGGTGTACAAGTCAAAGGT, reverse ATCCTCAACCAAATCAGCCCATTG; polyubiquitin (UBQ10, At4g05320): forward CACACTCCACTTGGTCTTGCGT, reverse TGGTCTTTCCGGTGAGAGTCTTCA. The PCR amplification was performed with a MyiQ™ Single Color Real-Time PCR Detection System (Bio-Rad) according to the manufacturer’s instructions.
Assaying pPLAIIα Activity Using Individual Phospholipids as Substrates
NBD-PC, NBD-PE, NBD-PG, and NBD-PA were obtained from Avanti Polar Lipids, Inc. (Alabaster, AL, USA). Individual classes of NBD-lipids in chloroform were dried under a nitrogen stream and emulsified in distilled water containing 0.05% Triton X-100 by sonication. Enzymatic activities were assayed in a reaction mixture containing 50 mM Tris-HCl, pH 8.0, 10 mM CaCl2, and 0.05% Triton X-100. Sixty μM of NBD-lipids was used as substrate and 3 μg of purified protein was added to the mixture in a final volume of 200 μl. The reaction was incubated at 30°C for 60 min and was stopped by adding 700 μl chloroform/methanol (2:1, v/v) and 200 μl of 2 M KCl. After vortexing and centrifugation, the aqueous phase was removed and remaining chloroform was dried under a stream of nitrogen. Lipid was dissolved in 10 μl chloroform and spotted on a thin layer chromatography (TLC) plate (silica gel 60, Merck, Darmstadt, Germany). The TLC plate was developed in chloroform/methanol/ammonium hydroxide/water (65:39:4:4, v/v/v/v). Lipids on the plate were visualized under ultraviolet light. The spots corresponding to free fatty acids and lysophospholipids were scraped into vials. The lipid in the vials was extracted with methanol and its fluorescence was measured at 460 nm (excitation) and 534 nm (emission). For activity assays using radioactive PI as a substrate, 0.6 μCi 1-stearoyl-2-[1-14C arachidonyl]-PI (New England Nuclear) and 3 μmol of unlabeled soy PI (Sigma, St Louis, MO) were mixed in chloroform. The solvent was evaporated under a nitrogen stream, and the lipid was emulsified in 0.5 ml H2O containing 0.05% Triton X-100 by sonication. The enzyme assay was performed as described above, using 20 μl of the radioactive substrate. After separation on a TLC plate, the lipids on the plate were visualized by exposure to iodine vapor. The spots corresponding to the lipid standards were scraped into vials, and their radioactivity was determined by liquid scintillation counting.
Assaying pPLAIIα Enzymatic Activity Using Mixed Plant Lipids as Substrates
To use plant lipids as substrates, fully expanded Arabidopsis leaves were wounded mechanically with a forceps and total lipids were extracted 45 min after wounding as described (Zien et al., 2001). Solvents of the lipid extracts were evaporated under a stream of nitrogen, emulsified in distilled water containing 0.05% Triton X-100 by sonication, and used as substrates. Thirty μg of the lipids (1 μg μl−1) and 3 μg purified pPLAIIα were added to each reaction in a final volume of 200 μl. Control reactions contained the same amount of lipids and empty-vector bacterial proteins and were subjected to the same purification process as those reactions containing pPLAIIα. The other components in the assay and the reaction condition were the same as described above. After the reaction at 30°C for 60 min, the resulting lipids were extracted and the organic phase was analyzed by the triple quadrupole mass spectrometry.
Lipid Analysis by Mass Spectrometry
Lipid extraction, lipid analysis, and lipid quantification of all lipids except MGMG and DGMG were performed as described by Xiao et al. (2010), except that negative-ion precursor scan mass spectral data for OPDA-containing lipids were normalized to amounts of 34:6 MGDG in the samples. This was done by multiplying the negative ion (Pre 291.2, i.e. OPDA) signals by the amount (nmol) of 34:6 MGDG determined by Neutral Loss scanning for the MGDG head group (NL 179.1) in positive ion mode and dividing by the signal for 34:6 MGDG in negative ion mode (Pre 277.2, i.e. 18:3). MGMG and DGMG were quantified similarly to MGDG and DGDG as [M + NH4]+ in positive ion mode with NL179.1 and NL 341.1, except that the internal standards were 18:0 MGMG and 18:0 DGMG. Five replicates of each treatment for each phenotype were processed and analyzed. Paired values were subjected to the Student’s t-test to determine the statistical significance.
Measurements of Jasmonates and ABA
The JA and ABA analysis was performed as described previously (Pan et al., 2008). Briefly, approximately 50–100 mg of fresh tissue was sealed in 1.5-ml snap-cap vials. After being frozen in liquid nitrogen, the leaves were ground to powder, and 500 μl of 1-propanol/H2O/concentrated HCl (2:1:0.002, v/v/v) with internal standards were added, followed by agitation for 30 min at 4°C. Dichloromethane (1 ml) was added, followed by agitation for another 30 min and then centrifugation at 13 000 g for 1 min. The bottom layer was used for jasmonate and ABA analysis. Dihydrojasmonic acid (H2JA) and heptadecanoic acid (C17:0) were used as internal standards for JA and linolenic acid quantification, respectively. Plant extracts were first separated by HPLC, equipped with a reversed-phase column (C18 Gemini 5 μ, 150 × 2.00 mm, Phenomenex, CA, USA), using a binary solvent system composed of water with 0.1% formic acid and methanol with 0.1% formic acid as a mobile phase at a flow rate of 0.3 ml min−1 and a gradient of linearly increasing methanol content from 30 to 100% at 30 min. A hybrid triple quadrupole/linear ion trap mass spectrometer (ABI 4000 Q-Trap, Applied Biosystems, Foster City, CA) outfitted with an electrospray ion source was used in multiple reaction monitoring (MRM) mode. JA, H2JA, linolenic acid, 13-hydroperoxyoctadeca-9,11,15-trienoic acid (13-HPOT), and oxophytodienoic acid (OPDA) were measured according to the procedure described (Pan et al., 2010).
Measurements of Water Loss and Stomatal Aperture
Arabidopsis plants were grown in growth chambers under a day/night regimen of 23/18°C and 12/12-h photoperiod under white light of 150 μmol m−2 s−1. Leaves from 4-week-old plants were detached and placed on a bench at room temperature and under white light of 100 μmol m−2 s−1. The weight of the leaf was recorded at different time points after detachment, and the percentage over initial weight was calculated for the water loss rate of the leaf. Stomatal responses to ABA and JA were analyzed in 4-week-old plants based on the procedure described by Kuhn et al. (2006). Briefly, epidermal layers of leaves from 4-week-old plants grown in the growth chamber were peeled and floated in stomata opening buffer. After incubation at room temperature under light of 200 μmol m−2 s−1 for 2.5 h, ABA or JA was added to the solution at various concentrations and the peels were incubated under light for another 2.5 h before the stomata were photographed under a microscope and the stomatal aperture was measured. For measurement of JA and ABA in response to desiccation, leaves from 4-week-old plants were detached and placed on a bench at room temperature under white light of 100 μmol m−2 s−1. At different time points after desiccation, the leaves were placed in liquid nitrogen for hormone preparation and measurement (Pan et al., 2008).
FUNDING
This work was supported by grants from the National Science Foundation (MCB-0920681; IOS-0818740) and the US Department of Energy (DE-SC0001295). The Kansas Lipidomics Research Center’s research was supported by grants from the National Science Foundation (MCB-0920663, DBI-0521587, and Kansas Experimental Program to Stimulate Competitive Research Award EPS-0236913), with support from the State of Kansas through the Kansas Technology Enterprise Corporation and Kansas State University, as well from US Public Health Service Grant P20 RR-016475 from the IDeA Network of Biomedical Research Excellence program of the National Center for Research Resources. No conflict of interest declared.
References
- Andersson MX, Kourtchenko O, Dangl JL, Mackey D, Ellerström M. Phospholipase-dependent signalling during the AvrRpm1- and AvrRpt2-induced disease resistance responses in Arabidopsis thaliana. Plant J. 2006;47:947–959. doi: 10.1111/j.1365-313X.2006.02844.x. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Buseman CM, et al. Wounding stimulates the accumulation of glycerolipids containing oxophytodienoic acid and dinor-oxophytodienoic acid in Arabidopsis leaves. Plant Physiol. 2006;142:28–39. doi: 10.1104/pp.106.082115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent A. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Fan L, Zheng S, Wang X. Antisense suppression of phospholipase D alpha retards abscisic acid- and ethylene-promoted senescence of postharvest Arabidopsis leaves. Plant Cell. 1997;9:2183–2196. doi: 10.1105/tpc.9.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer EE. Surface-to-air signals. Nature. 2001;411:854–856. doi: 10.1038/35081189. [DOI] [PubMed] [Google Scholar]
- Galliard T. Enzymic deacylation of lipids in plants: the effects of free fatty acids on the hydrolysis of phospholipids by the lipolytic acyl hydrolase of potato tubers. Eur. J. Biochem. 1971;21:90–98. doi: 10.1111/j.1432-1033.1971.tb01444.x. [DOI] [PubMed] [Google Scholar]
- Glauser G, Grata E, Dubugnon L, Rudaz S, Farmer EE, Wolfender JL. Spatial and temporal dynamics of jasmonate synthesis and accumulation in Arabidopsis in response to wounding. J. Biol. Chem. 2008;283:16400–16407. doi: 10.1074/jbc.M801760200. [DOI] [PubMed] [Google Scholar]
- Hirschberg HJHB, Simons J-WFA, Dekkar N, Egmond MR. Cloning, expression, purification and characterization of patatin, a novel phospholipase A. Eur. J. Biochem. 2001;268:5037–5044. doi: 10.1046/j.0014-2956.2001.02411.x. [DOI] [PubMed] [Google Scholar]
- Holk A, Rietz S, Zahn M, Quader H, Scherer GF. Molecular identification of cytosolic, patatin-related phospholipases A from Arabidopsis with potential functions in plant signal transduction. Plant Physiol. 2002;130:90–101. doi: 10.1104/pp.006288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro S, Kawai-Oda A, Ueda J, Nishida I, Okada K. The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell. 2001;13:2191–2209. doi: 10.1105/tpc.010192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Böhmer M, Hu H, Nishimura N, Schroeder JI. Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu. Rev. Plant Biol. 2010;61:561–591. doi: 10.1146/annurev-arplant-042809-112226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtchenko O, Andersson MX, Hamberg M, Brunnström A, Göbel C, McPhail KL, Gerwick WH, Feussner I, Ellerström M. Oxo-phytodienoic acid-containing galactolipids in Arabidopsis: jasmonate signaling dependence. Plant Physiol. 2007;145:1658–1669. doi: 10.1104/pp.107.104752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JM, Boisson-Dernier A, Dizon MB, Maktabi MH, Schroeder JI. The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiol. 2006;140:127–139. doi: 10.1104/pp.105.070318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Camera S, Geoffroy P, Samaha H, Ndiaye A, Rahim G, Legrand M, Heitz T. A pathogen-inducible patatin-like lipid acyl hydrolase facilitates fungal and bacterial host colonization in Arabidopsis. Plant J. 2005;44:810–825. doi: 10.1111/j.1365-313X.2005.02578.x. [DOI] [PubMed] [Google Scholar]
- Li M, Bahn SC, Guo L, Musgrave W, Berg H, Welti R, Wang X. Patatin-related phospholipase pPLAIIIβ-induced changes in lipid metabolism alter cellulose content and cell elongation in Arabidopsis. Plant Cell. 2011;23:1107–1123. doi: 10.1105/tpc.110.081240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H, Sage TL, Isaac G, Welti R, Dellapenna D. Tocopherols modulate extraplastidic polyunsaturated fatty acid metabolism in Arabidopsis at low temperature. Plant Cell. 2008;20:452–470. doi: 10.1105/tpc.107.054718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munemasa S, Hossain MA, Nakamura Y, Mori IC, Murata Y. The Arabidopsis calcium-dependent protein kinase, CPK6, functions as a positive regulator of methyl jasmonate signaling in guard cells. Plant Physiol. 2011;155:553–561. doi: 10.1104/pp.110.162750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narusaka Y, et al. Expression profiles of Arabidopsis phospholipase A IIA gene in response to biotic and abiotic stresses. Plant Cell Physiol. 2003;44:1246–1252. doi: 10.1093/pcp/pcg138. [DOI] [PubMed] [Google Scholar]
- Pan X, Welti R, Wang X. Simultaneous quantification of plant hormones by high-performance liquid-chromatography electrospray tandem mass spectrometry. Phytochemistry. 2008;69:1773–1781. doi: 10.1016/j.phytochem.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Pan X, Welti R, Wang X. Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography–mass spectrometry. Nature Protocols. 2010;5:986–992. doi: 10.1038/nprot.2010.37. [DOI] [PubMed] [Google Scholar]
- Pappan K, Zheng L, Krishnamoorthi R, Wang X. Evidence for and characterization of Ca2+ binding to the catalytic region of Arabidopsis thaliana phospholipase Dβ. J. Biol. Chem. 2004;279:47833–47839. doi: 10.1074/jbc.M402789200. [DOI] [PubMed] [Google Scholar]
- Reymond P, Farmer EE. Jasmonate and salicylate as global signals for defense gene expression. Curr. Opin. Plant Biol. 1998;1:404–411. doi: 10.1016/s1369-5266(98)80264-1. [DOI] [PubMed] [Google Scholar]
- Rietz S, Dermendjiev G, Oppermann E, Tafesse FG, Effendi Y, Holk A, Parker JE, Teige M, Scherer GF. Roles of Arabidopsis patatin-related phospholipases a in root development are related to auxin responses and phosphate deficiency. Mol. Plant. 2010;3:524–538. doi: 10.1093/mp/ssp109. [DOI] [PubMed] [Google Scholar]
- Rietz S, Holk A, Scherer GF. Expression of the patatin-related phospholipase A gene AtPLA IIA in Arabidopsis thaliana is up-regulated by salicylic acid, wounding, ethylene, and iron and phosphate deficiency. Planta. 2004;219:743–753. doi: 10.1007/s00425-004-1275-9. [DOI] [PubMed] [Google Scholar]
- Rydel TJ, Williams JM, Krieger E, Moshiri F, Stallings WC, Brown SM, Pershing JC, Purcell JP, Alibhai MF. The crystal structure, mutagenesis, and activity studies reveal that patatin is a lipid acyl hydrolase with a Ser-Asp catalytic dyad. Biochemistry. 2003;42:6696–6708. doi: 10.1021/bi027156r. [DOI] [PubMed] [Google Scholar]
- Scherer GF, Ryu SB, Wang X, Matos AR, Heitz T. Patatin-related phospholipase A: nomenclature, subfamilies and functions in plants. Trends Plant Sci. 2010;15:693–700. doi: 10.1016/j.tplants.2010.09.005. [DOI] [PubMed] [Google Scholar]
- Schrag JD, Cygler M. Lipases and alpha/beta hydrolase fold. Methods Enzymol. 1997;284:85–107. doi: 10.1016/s0076-6879(97)84006-2. [DOI] [PubMed] [Google Scholar]
- Staswick PE. Jasmonate, genes, and fragrant signals. Plant Physiol. 1992;99:804–807. doi: 10.1104/pp.99.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. Lipid signaling. Curr. Opin Plant Biol. 2004;7:329–336. doi: 10.1016/j.pbi.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Wasternack C, Parthier B. Jasmonate signalled plant gene expression. Trends Plant Sci. 1997;2:302–307. [Google Scholar]
- Xiao S, Gao W, Chen QF, Chan SW, Zheng SX, Ma J, Wang M, Welti R, Chye ML. Overexpression of Arabidopsis acyl-CoA binding protein ACBP3 promotes starvation-induced and age-dependent leaf senescence. Plant Cell. 2010;22:1463–1482. doi: 10.1105/tpc.110.075333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Shivakumar PD, Pan X, Giorgis I, Welti R, Wang X. AtPLAI is an acyl hydrolase involved in basal jasmonic acid production and Arabidopsis resistance to Botrytis cinerea. J. Biol. Chem. 2007;282:18116–18128. doi: 10.1074/jbc.M700405200. [DOI] [PubMed] [Google Scholar]
- Zien CA, Wang C, Wang X, Welti R. In vivo substrates and the contribution of the common phospholipase D, PLDalpha, to wound-induced metabolism of lipids in Arabidopsis. Biochim. Biophys. Acta. 2001;1530:236–248. doi: 10.1016/s1388-1981(01)00091-9. [DOI] [PubMed] [Google Scholar]




