Abstract
Discoveries made over the past 20 years highlight the importance of mRNA decay as a means to modulate gene expression and thereby protein production. Up until recently, studies focused largely on identifying cis-acting sequences that serve as mRNA stability or instability elements, the proteins that bind these elements, how the process of translation influences mRNA decay, and the ribonucleases that catalyze decay. Now, current studies have begun to elucidate how the decay process is regulated. This review examines our current understanding of how mammalian-cell mRNA decay is controlled by different signaling pathways and lays out a framework for future research.
Introduction
mRNAs exist largely to produce proteins. The amount of protein generated from any given mRNA depends not only on the rate of mRNA translation but also on the rates of mRNA synthesis and decay. mRNA levels are influenced by the efficiencies and nucleotide positions at which gene transcription, pre-mRNA splicing, pre-mRNA 3′-end formation, and other post-transcriptional modifications such as A-to-I editing occur, as well as the efficiency of mRNA export from the nucleus to the cytoplasm; remarkably, cytoplasmic mRNA decay can be affected by these same nuclear processes, which have the potential to alter mRNA metabolism by altering mRNA sequences and/or mRNP composition, as well as the translational status of the mRNA1.
While this review will focus on the regulation of mRNA decay in mammalian cells, the paradigms, if not the specifics, generally apply to other organisms. mRNA decay can be divided broadly into two classes: mechanisms of quality control that eliminate the production of potentially toxic proteins, and mechanisms that lengthen or shorten mRNA half-life for the purpose of changing the abundance of functional proteins. The cytoplasmic decay machinery consists of five types of ribonucleolytic activities, the combinatorial and ordered use of which varies depending on the mRNA substrate and cellular conditions. These five activities mediate decapping, 5′-to-3′ exonucleolytic decay, deadenylation, 3′-to-5′ exonucleolytic decay, or endonucleolytic cleavage2–4 (Box 1). Each activity may gain access to an mRNA depending on the proteins and, possibly, antisense noncoding (nc)RNA(s) that are bound to the mRNA.
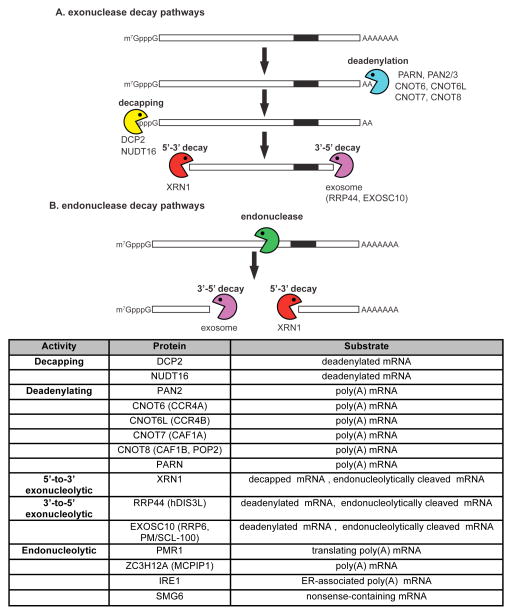
BOX 1. The major mRNA decay pathways.
Most mRNAs and all of the ARE-containing mRNAs are degraded by exonucleases acting at both ends of the molecule. Exonuclease-mediated decay (panel a) begins with shortening of the poly(A) tail by the deadenylases poly(A) ribonuclease (PARN), the complex of PAB-specific ribonuclease 2 and 3 (PAN2/3), or by a multisubunit complex containing the CNOT6, CNOT6L, CNOT7 or CNOT8 deadenylases (reviewed in2). The protein constituents of the mRNP determine which deadenylase is selected, but the details of this process are poorly understood. Next the Nudix domain proteins DCP2 or NUDT16 hydrolyze the cap, and the body of the mRNA is degraded with 5′-to-3′ polarity by XRN13 and/or 3′-to-5′ polarity by the exosome-associated exonucleases RRP44 or EXOSC10 (PM/SCL-100)101, although the latter can occur without prior decapping43. For some mRNAs, decay is initiated by endonuclease cleavage within their body, followed by degradation of the upstream cleavage product by the exosome and degradation of the downstream cleavage product by XRN1 (reviewed in4) (panel b). The mRNA degradative enzymes that are relevant to this review, their activities and substrates are listed in panel c. It should be noted that mammalian cells have 2 forms of RRP44, termed hDIS3 and hDIS3L, which are found in the nucleus and cytoplasm, respectively. Although hDIS3 has both exonuclease and endonuclease activity there is no evidence that its endonuclease activity functions in vertebrate mRNA decay.
Box 1.
mRNA-associated proteins and ncRNAs can influence the rate of mRNA decay directly by promoting or precluding decay-factor binding, indirectly by influencing the cellular location and/or translational status of the mRNA, or all of the above. mRNP constituents provide a plethora of possible fates for an mRNA as they associate or dissociate from mRNA in response to cellular conditions, such as the cell cycle or external stimuli, and these conditions can also regulate the abundance, cellular location and activity of mRNP constituents. Thus, a complicated intracellular and intercellular network underlies regulation of cytoplasmic mRNA decay. Our current appreciation of this network is attributable to the development of methods to purify and characterize the constituents of mRNPs. For example, after immunoprecipitating a particular mRNA-binding protein, the associated proteins and their post-translational modifications can now be identified by using mass spectrometry and the associated transcripts can be identified by using microarray analyses or, most recently, deep sequencing. The localization of each protein and ncRNA to a particular mRNA sequence and/or mRNP sub-complex together with experiments that assess the functional consequence of mRNA binding under different conditions provide information essential to understanding mRNA metabolism, including the control of mRNA half-life.
Currently, studies of the conditional regulation of mRNA decay represent only a small fraction of what is undoubtedly a vast and interconnected cellular repertoire. This review highlights our current understanding of how the cytoplasmic decay of mammalian-cell mRNAs is regulated, thereby establishing a base for future work. We begin with the regulation of nonsense-mediated mRNA decay (NMD), which unlike most mRNA decay pathways appears to be restricted to newly synthesized transcripts. We then discuss: the regulation of histone mRNA decay, to exemplify a cell cycle-regulated process; AU-rich element (ARE)-mediated mRNA decay, which involves 3′-untranslated region (UTR) destabilizing elements; and mRNA decay in response to nuclear receptors. In addition we examine how endonucleolytic mRNA decay activities are controlled. This review illustrates ways in which mRNA decay endows cells with effective responses to changing milieus, and provides some examples of how disregulation of mRNA decay can impact disease-associated processes.
Nonsense-mediated mRNA decay
Since mRNAs function primarily as templates for protein synthesis, it reasons that cells have evolved translation-dependent quality-control (QC) mechanisms to dispose of defective mRNAs that synthesize abnormal proteins. Nonsense-mediated mRNA decay (NMD), which occurs in all eukaryotes that have been studied, eliminates mRNAs that prematurely terminate translation as a means to dampen the potentially toxic effects of defective transcripts that are routinely generated during gene expression5. Downregulating the essential NMD factor up-frameshift 1 (UPF1) using siRNA upregulates ~5% of properly functioning transcripts6 in HeLa cells. A similar percentage (~4%) is upregulated using siRNA to the UPF2 NMD factor7. Thus, a significant number of physiologic cellular transcripts are likely to be either direct NMD targets or downregulated by a direct NMD target, suggesting that NMD functions in cellular processes in addition to QC. The recent proposal that NMD targets are the primary source of antigenic peptides for the major histocompatibility class I pathway, which depletes immunoreactive T cells that recognize endogenous proteins8, provides another emerging role for NMD in humans and, most likely, all mammals.
Besides NMD, other mammalian-cell QC mechanisms consist of no-go mRNA decay and nonstop mRNA decay9. NMD is the best characterized of the three and the only one studied in any detail from a regulatory perspective. Thus, we will restrict our QC comments to NMD (see references5,10 for recent detailed reviews).
Targets of NMD
NMD is one of a number of mammalian-cell mRNA decay pathways that is coupled to the translation of its target. Therefore, regulators of translation must be considered regulators of NMD. While not everyone agrees, data indicate that mammalian-cell NMD largely if not exclusively degrades newly synthesized mRNAs that are bound at their 5′ caps by the cap-binding protein (CBP) complex CBP80-CBP20 (CBC), at their poly(A) tails by poly(A)-binding protein N1 (PABPN1) and PABPC1 and, if they derived from pre-mRNA splicing, an exon-junction complex (EJC) at exon-exon junctions during what has been termed the pioneer round of translation5. Notably, this “round” can be associated with more than one ribosome, depending on the translation initiation efficiency and length of the reading frame. Mammalian-cell NMD does not appear to target older mRNAs, which are more abundant and direct the bulk of cellular protein synthesis. Older mRNAs are bound at their caps and poly(A) tails by, respectively, eukaryotic translation initiation factor 4E (eIF4E) and PABPC1, and they lack detectable EJCs even if they derived from pre-mRNA splicing. For simplicity, here we refer to the new mRNAs as CBC-bound and the older mRNAs as eIF4E-bound. It should be noted that NMD does not appear to be restricted to newly synthesized mRNAs in non-mammalian cells. In mammals, cap-bound CBP80 transiently and/or weakly interacts with a key NMD factor, UPF1, prior to the pioneer round of translation (Fig. 1). This interaction promotes NMD should the mRNA harbor a translation termination codon, often a premature termination codon (PTC), more than ~50–55-nucleotides upstream of an EJC-bound exon-exon junction11 (Fig. 1). The absence of CBP80 and EJCs on eIF4E-bound mRNAs at least in part explains the immunity of these mRNAs to NMD.
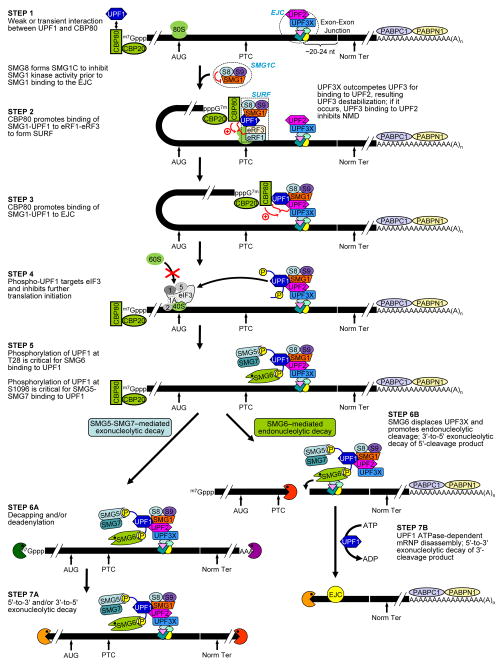
Figure 1. NMD factors as regulators of NMD.
NMD in mammalian cells occurs during the translation of mRNA that is bound at its 5′ m7Gppp cap by CBP80 and CBP20. STEP1 involves a weak and/or transient interaction of UPF1 with cap-bound CBP80 – an interaction that typifies all newly synthesized mRNAs11. In STEP2, the suppressor of morphogenetic effect on genitalia (SMG)1, which is a PIK-related serine-threonine protein kinase that phosphorylates multiple residues in the C-terminal region of UPF125, binds together with UPF1 to the two translation termination factors, eukaryotic release factor (eRF)1 and eRF3, to form SURF at a PTC. During the formation of the SURF complex SMG1 kinase activity is suppressed by SMG8 via the so-called SMG1 complex (SMG1C)114. In STEP3 SMG1-UPF1 is joined to an EJC that resides downstream of the PTC11. This is dependent upon UPF3X out-competing its functionally less-effective paralog UPF3 for binding at the exon-exon junction complex27. The SMG1-mediated phosphorylation of UPF1 that occurs in STEP 4 occurs as a consequence of SMG1-UPF1 binding directly to the UPF2 EJC constituent. Phosphorylated UPF1 binds to eIF3, should a pre-initiation complex form at the AUG translation initiation codon inhibiting additional rounds of translation initiation23. In STEP5, SMG6 binds UPF1 dependent on the phosphorylation of UPF1 at threonine 28, and SMG5 and SMG7 bind UPF1 dependent on UPF1 phosphorylation at serine 109624. In STEP 6A, the SMG5- and SMG7-mediated decay of the NMD target occurs by decapping and/or deadenylation followed in STEP 7A by degradation of the body of the transcript in the 5′-to-3′ direction from the decapped 5′-end and/or 3′-to-5′direction from the 3′-deadenylated end115. A parallel pathway in STEP 6B involves the SMG6-mediated endonucleolytic cleavage of the NMD target and the accompanying rapid decay of the resulting 5′-cleavage product and 3′-cleavage product in STEP 7B116,117. Decay of the 3′-cleavage product requires mRNP disassembly that is mediated by UPF1118.
Inhibitors and activators of translation-dependent mRNA decay
NMD is inhibited by negative regulators of the pioneer round of translation, which sometimes also negatively regulate the translation of older mRNAs. For example, some stresses – such as amino acid starvation, viral infection, heat shock, oncogenic stress created by c-myc overexpression, and early stages of hypoxia – inhibit the translation of both CBC-bound mRNAs and eIF4E-bound mRNAs. These stresses trigger a signaling cascade that results in phosphorylation of the translation initiation factor eIF2alpha and/or the proteolysis of translation initiation factors that are required for both types of translation (Fig. 2; see12,13). In the case of early stages of hypoxia, inhibition of both forms of translation involves inhibition of the mammalian target of rapamycin complex 1 (mTORC1)14, which activates both CBC-bound and eIF4E-bound mRNA translation (Fig. 2). Inhibition of translation of CBC-bound mRNAs (and hence NMD) only occurs during early stages of hypoxia whereas inhibition of translation of eIF4E-bound mRNAs persists through to late stages of hypoxia. This may be beneficial because several mRNAs that contribute to the cellular response to stress are NMD targets15. For example, activating transcription factor (ATF)4 mRNA is an NMD target, and ATF4 triggers transcription of some proteins that are toxic in unstressed cells.
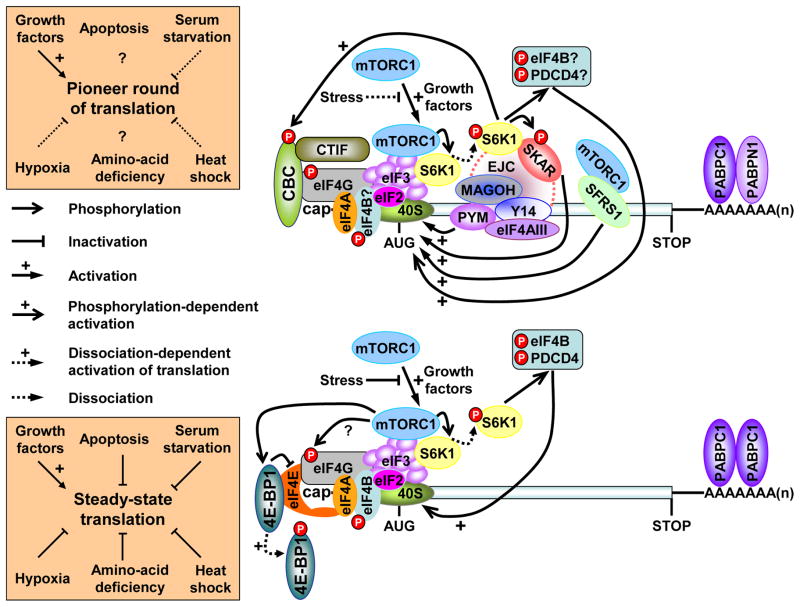
Figure 2. Signaling pathways that regulate the pioneer round of translation and NMD.
The pioneer-round of translation utilizes newly synthesized mRNA bound by the cap-binding protein heterodimer CBP80-CBP20 (CBC) and, provided the mRNA derived from splicing, at least one exon-junction complex (EJC, which consists of but is not limited to MAGOH, Y14, PYM and eIF4AIII) situated ~ 20–24-nucleotides upstream of such a junction. This round of translation (and, thus, NMD for those mRNAs that are NMD targets) is promoted upon activation of the mTORC signaling pathway, i.e, in the absence of stress and the presence of growth factors, by multiple pathways. mTOR-mediated translational activation is best understood for eIF4E-bound mRNAs119. Translational activation, and thus the activation of any associated mRNA decay pathways, involves mTORC1 binding to eIF3, which results in the phosphorylation of the 4E-BP1 translational repressor and consequently the dissociation of inactivated 4E-BP1 from mRNA-bound eIF4E. eIF4G can then interact with and stabilize the binding of eIF4E, eIF4A and PABPC1 to mRNA to ensure efficient translation. mTORC1 binding to eIF3 also results in the phosphorylation of the S6 kinase 1 (S6K1) translational activator and consequently the dissociation of activated S6K1 from mRNA-bound eIF3. Dissociated S6K1 phosphorylates eIF4B and tumor suppressor programmed cell death 4 (PDCD4), which augment scanning of the 43S preinitiation complex to the translation initiation codon. Taking cues from how mTOR activates eIF4E-bound mRNA translation, the mTOR-mediated translational activation of CBC-bound mRNA, and thus the activation of NMD, may also involve mTORC1 binding to eIF3 and the dissociation of activated S6K1 from CBC-bound mRNA. Activated S6K1 is then recruited by the EJC constituent SKAR, which promotes the pioneer round of translation by enhancing the phosphorylation of SKAR and other downstream effectors that may include eIF4B and PDCD4, which associate with not only eIF4E-bound mRNA but also CBC-bound mRNA120. Occurring in parallel, S6K1 phosphorylation by the Cdc42 kinase may augment binding of CBC to mRNA caps, the splicing factor SFRS1 may initiate S6K1 signaling via mTORC1, and the interaction of PYM with the EJC-core proteins Y14 and MAGOH and the 40S ribosomal subunit all appear to promote the pioneer round of translation and, when appropriate, NMD. (After Figure 2 in5.)
NMD is activated by positive regulators of the pioneer round of translation. CBC-bound mRNA translation, like eIF4E-bound mRNA translation, is activated via the mTOR pathway, although by a mechanism that involves the EJC constituent S6K1 ALY-REF-like target (SKAR), which is detected on CBC-bound but not eIF4E-bound mRNA (Fig. 2). Additional proteins that promote the translation of CBC-bound mRNA and are absent from eIF4E-bound mRNA include other EJC constituents16, such as PYM17, and the splicing factor serine-arginine-rich 1 (SFSR1, formerly called SF2 or ASF). SFSR1 recruits mTOR and another translational activator, TAP18,19 (Fig. 2). The finding that SKAR augments cell size and thus presumably global protein synthesis20 is reminiscent of data indicating that PYM or SFSR1 somehow likewise increase mRNA translation beyond the pioneer round of translation.
NMD factors as regulators of NMD
NMD can be regulated at the level of individual NMD factors or in feedback loops that coordinately control the abundance of one or more different NMD factors. The steps of the mammalian NMD pathway are shown in Figure 1, (however, there is an alternative, less well-understood pathway of NMD as well21,22). Phosphorylation of NMD factors is an important mode of regulation of their activity, and thus regulation of NMD and some key examples are noted here. The phosphorylation of UPF1 by a phosphatidylinositol kinase (PIK)-related serine-threonine protein kinase called SMG1 is triggered when both UPF1 and SMG1 interact with a downstream EJC11(STEP 3, Fig. 1). The phosphorylated form of UPF1 prevents additional rounds of translation initiation on the NMD target23 (STEP 4, Fig. 1). SMG5-SMG7 or SMG6 at some point bind to phosphorylated UPF124 (STEP 5, Fig. 1) resulting in exonucleolytic degradation (STEP 6A and STEP 7B, Fig. 1) or endonucleolytic degradation (STEP 6B and 7B, Fig. 1), respectively. SMG5-SMG7- and/or SMG6 also promote dephosphorylation of UPF1 by protein phosphatase 2A (PP2A), which enables UPF1 recycling25,26. It is uncertain whether this occurs on the NMD target and in a way that is essential for the decay of that target or if it occurs after UPF1 is released from the NMD target23,24,26.
The abundance of NMD factors can also be regulated. Arguably, the best characterized example of this is the interplay between UPF3 (also called UPF3A) and UPF3X (also called UPF3B). UPF3X has a higher affinity for the EJC that does its paralog UPF3, and the failure of UPF3 to compete effectively with UPF3X for EJC binding results in UPF3 destabilization27 (Fig. 1).
Feedback loops also coordinately regulate the level of NMD factors28,29. A multipronged negative feedback regulatory network acts on the seven NMD factors – UPF1, UPF3X, UPF2, SMG1, SMG5, SMG6 and SMG7 – that are rate-limiting to NMD in HeLa cells; this feedback network buffers the cells from perturbations and maintains homeostasis29. The HeLa-cell mRNAs that encode these factors are regulated by different UPF1-dependent branches of NMD: UPF1 and SMG5 mRNAs are the only of the seven NMD-factor mRNAs upregulated upon depletion of the eIF4AIII EJC constituent; SMG1 mRNA is selectively upregulated upon depletion of the EJC constituent RNPS1; and UPF1 and SMG7 mRNAs are selectively upregulated upon downregulation of either or both UPF3 or UPF3X29. This intricate network is not limited to HeLa cells (see below) or even mammalian cells, and it appears to minimize side-effects when NMD is perturbed by a change in the level of one or more NMD factors.
The efficiency of NMD is also affected by microRNAs (miRNAs). For example, the efficiency of NMD for mRNAs that bind the miRNA-induced silencing complex appears to be muted due to miRNA-mediated translational repression of CBC-bound mRNA30. Of course, miRNAs that target one or more NMD factors would also influence the efficiency of NMD (see below).
NMD cell- and tissue-specific differences
Variations in the efficiency of NMD are evident when comparing different cell lines or different tissues31. For example, the efficiency of NMD decreases during the differentiation of mouse C2C12 myoblasts to myotubes. During differentiation, the abundance of the NMD factor UPF2 decreases more than the abundance of Staufen1 (STAU1), which functions during STAU1-mediated mRNA decay (SMD) (Box 2). STAU1 thus competes more effectively than UPF2 for binding to UPF132. The cause of the changes in UPF1 and STAU1 abundance during myogenesis has yet to be determined. As another example, the efficiency of NMD in brain is decreased by miRNA-128, which targets UPF1 and the EJC core constituent metastatic lymph node gene 51 (MLN51; also known as CASC3 or barentz) protein, and miR-128 levels increase during brain development and in differentiating neuronal cells33. Intrinsic differences among cell-types in the specific negative regulatory feedback mechanisms that are active also exist depending on which NMD factor(s) happen to be rate-limiting29. For example, in contrast to UPF3X-deficient HeLa cells that expressed elevated levels of UPF1 and SMG7 mRNAs, lymphoblastoid cells from patients with UPF3X deficiency had upregulated levels of not only UPF1 and SMG7 mRNAs but also UPF2 mRNA relative to control lymphoblastoid cells.
BOX 2. Staufen1-mediated mRNA decay.
Staufen (STAU)1-mediated mRNA decay (SMD) targets mRNAs that contain a 3′UTR STAU1-binding site (SBS) provided translation terminates sufficiently upstream of the SBS102. After translation termination, usually at the normal termination codon, the direct interaction of SBS-bound STAU1 with the key NMD factor UPF1 results in UPF1 phosphorylation and SMD. This series of events appears to be analogous to what occurs during NMD: when translation terminates sufficiently upstream of an EJC, usually at a premature termination codon, the direct interaction of the EJC with UPF1 triggers UPF1 phosphorylation and NMD. It seems reasonable to think that the SMD and NMD pathways converge following UPF1 phosphorylation. However unlike NMD, SMD occurs independently of CBP80 and EJCs and targets newly synthesized CBC-bound mRNAs and older eIF4E-bound mRNAs103. Cell cycle-regulated histone mRNA decay also likely also occurs during both pioneer and subsequent rounds of translation since SLBP is required for histone mRNA 3′-end formation within nuclei104 and thus probably binds not only eIF4E-bound but also CBC-bound histone mRNA.
Differences in the efficiency of NMD also occur in a developmentally controlled fashion, the molecular basis of which may reflect the relative abundance of NMD factors or the ability of a particular newly synthesized mRNA to undergo the pioneer round of translation34. In the case of the latter, mRNAs may be sequestered into translationally inactive particles until properly localized in time and in space.
Histone mRNA decay
The decay of histone mRNAs in metazoans provides a particular example of cell cycle-restricted expression mediated by the regulation of mRNA degradation. The half-life of such histone mRNAs decreases from between 45 and 60 minutes during S phase to 10 min at the end of S phase. This decrease plays a major role in coordinating histone protein synthesis with DNA replication. Although histones are required to package newly replicated DNA during S phase, histone mRNA transcription continues beyond S phase, albeit at a reduced rate. Cell cycle-regulated histone mRNAs are exceptional because they lack a poly(A) tail and instead end in a conserved stem-loop structure that binds stem-loop-binding protein (SLBP). SLBP, which is present only during S phase, is critical for efficient histone mRNA 3′-end formation in the nucleus and efficient histone mRNA translation in the cytoplasm35. At the end of S phase, when histone mRNA terminates translation SLBP bound to the histone mRNA stem-loop recruits the NMD factor UPF136 (presumably analogously to how STAU1 bound to an STAU1-binding site) recruits UPF1 when translation terminates normally on a target of SMD) so as to trigger mRNA decay. While UPF2 and UPF3 do not function in either histone mRNA decay or SMD, SMG1 does, presumably by phosphorylating UPF1 (W.F. Marzluff, pers. comm.; L.E.M, unpub. data).
Regulated histone mRNA decay also requires the PIK-related protein kinase ataxia telangiectasia-mutated and Rad 3-related (ATR)36, which is a checkpoint kinase that is active during normal S-phase progression and properly schedules the initiation of DNA replication. ATR mediates the decay of histone mRNA in a manner independent of its role in DNA synthesis. However, the ATR target(s) that function in degrading histone mRNA remain unknown. Regulated histone mRNA decay also requires cyclin A2/CDK1 and casein kinase 2 (CK2), which phosphorylate SLBP at the end of S phase, thus triggering SLBP degradation by an undefined ubiquitin ligase37,38.
Another example of cell-cycle regulated mRNA degradation may be found in the degradation of mRNA for the cell-cycle regulator cyclin B by an RNase called mitochondrial RNA processing (MRP). Mutated variants of MRP in patients with cartilage hair hypoplasia (reviewed in39) result in an increase in the level of cyclin B mRNA. In yeast, downregulation of cyclin B is known to be regulated by MRP.
ARE-mediated mRNA decay
AU-rich elements
AU-rich elements (AREs) are the largest and most studied group of cis-acting mRNA instability determinants. The canonical AREs generally have one or more copies of the AUUUA pentamer usually embedded in a U-rich context. If classified by sequence alone, canonical ARE-containing mRNAs constitute up to 9% of cellular mRNAs40. AREs have been grouped into three broad categories based on the number and context of the AUUUA repeats41. The decay of ARE-containing mRNAs begins with either synchronous or distributive poly(A) shortening42, with the subsequent steps targeting the body of the mRNA from both ends (Box 1, panel A) via the actions of the 5′-3′ and 3′-5′ exonucleases43. There are other cis-acting instability elements, the best studied of which are the GU-rich elements (GREs)44. GREs function in post-transcriptional regulons involving pre-mRNA splicing and mRNA decay that are coordinated by binding of the CUG-BP Elav-like family (CELF) proteins, the best known of which is CELF1 (also called CUG-BP1)45,46. Less is known about the regulation of GRE-mediated decay, and the reader is directed to a recent review47.
AREs are found in the tightly-regulated mRNAs that encode protooncogene proteins (e.g., c-fos) and inflammatory mediators (e.g., TNFα, IL-1, IL-2, IL-3, GM-CSF). Together with their associated binding proteins (ARE-BPs), AREs have significant physiological and pathological functions, not all of which are the result of rapid mRNA turnover48–52. The inflammatory response provides a good example of the physiological and pathological ramifications of ARE-mediated mRNA decay. The cytokines and chemokines that are elaborated by macrophages and monocytes after injury neutralize the damaging agent and repair damaged tissue; however, these proteins can also harm healthy tissue, and conditions that interfere with the destabilization of the mRNAs encoding proinflammatory proteins contribute to atherosclerosis, rheumatoid arthritis and asthma53.
The complexity of the ARE mRNA transcriptome is seen in the changing steady-state levels of ARE-containing mRNAs subsequent to treatment of resting human T cells with antibodies that bind T-cell receptors and stimulate proliferation and changes in gene expression54. Out of 900 ARE-containing mRNAs assayed by using microarrays, the steady-state levels of 250 declined within 30 minutes of stimulation. mRNAs in this group encode regulators of quiescence, such as phosphatase and tensin homolog (PTEN), inositol-trisphosphate 3-kinase B (ITPKB) and calcineurin A, suggesting that the decay of these ARE-containing mRNAs plays a role in the signal transduction-mediated transition from quiescence to proliferation. It is not clear from this or many other studies why, under different conditions, some mRNAs with one or more AREs undergo accelerated decay and others do not. This may result from constraints imposed by sequence context, secondary structure, and binding of proteins and/or ncRNAs at adjacent or overlapping sites (see below). Furthermore, not all ARE-binding proteins (ARE-BPs) confer mRNA instability (see below). As noted (BOX 3), widespread alternative 3′-end processing55 complicates the challenge of identifying mRNAs that contain functional AREs.
BOX 3. Regulation of mRNA decay by alternative 3′-end formation.
Cleavage and polyadenylation sites exist to form the 3′ ends of pre-mRNAs and, thus, the mRNAs that derive from them. Almost all human pre-mRNAs possess multiple cleavage and polyadenylation signals in their 3′ UTRs, and the analysis of expressed sequence tags indicates that about half of pre-mRNAs use alternative cleavage and polyadenylation (APA) sites105,106. Since most cis-acting elements that regulate mRNA half-life are situated within 3′UTRs, mRNAs containing shorter 3′UTRs have often lost one or more regulatory pathways dictated by, e.g., microRNA binding sites, and have greater stability55. APA can be regulated by changes in the abundance of one of the ~90 proteins involved in 3′-end maturation and/or a number of signal transduction pathways. For example, in day-18 embryonic rats, the use of upstream APA sites is activated for ~10% of transcripts that are upregulated by the transcriptional activator myocyte enhancer factor-2 (MEF-2) in response to extracellular signaling that increases synaptic activity in hippocampal neurons107. As another example, in response to oxidative stress, CKIalpha, which is sensitive to the lipid messenger PI4,5P2, and CKIepsilon, phosphorylates and thereby upregulates the activity of the non-canonical nuclear poly(A) polymerase (PAP) called Speckel targeted phosphatidylinositol-4-phosphate 5-kinase Ialpha-regulated (STAR)-PAP so as to enhance STAR-PAP cleavage and polyadenylation on select target pre-mRNAs108.
ARE-BPs
ARE-BPs can be grouped by their stabilizing or destabilizing effects on the mRNA, type of RNA-binding motif, and the proteins that modify their action, and some of the better studied ARE-BPs are listed in Box 4, panel A. ARE-BPs can be regulated by kinases, phosphatases and at least in one case by an arginine methyltransferase. As a general rule, ARE-BPs function as multimers and can be phosphorylated at the same site by different kinases, or at different sites by different kinases.
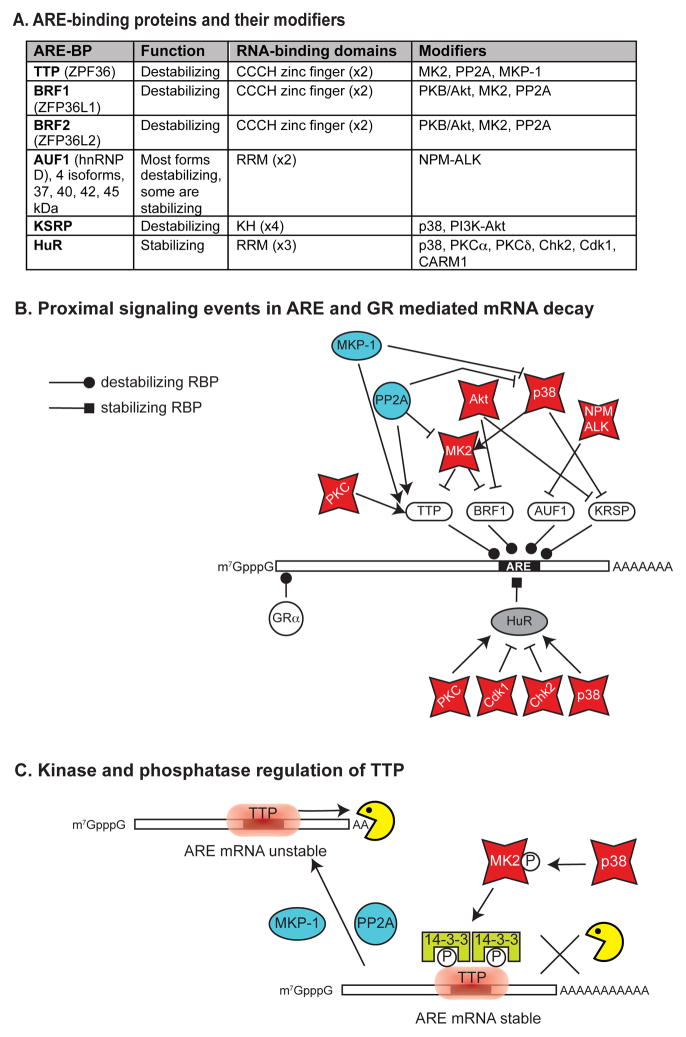
BOX 4. Signaling pathways that impact directly on mRNA decay.
AREs have different functions, RNA-binding motifs, and upstream modifiers (panel a). A representative mRNA is shown with a 3′UTR ARE and a 5′UTR glucocorticoid receptor-binding site indicated in their respective locations (see text). The ARE-binding proteins shown in white ovals (panel b) exert a destabilizing effect on ARE-containing target mRNAs by recruit exonucleases (Box 1) to the mRNA, and stabilization results from interference with this process. ARE-containing mRNAs are stabilized by MK2 phosphorylation of TTP and BRF158,63, by p38 MAPK phosphorylation of KSRP66, by AKT phosphorylation of BRF1 and KSRP64, and by NPM-ALK phosphorylation of AUF169. In contrast, the destabilizing activity of TTP is increased following phosphorylation by PKC. Each of these kinases is activated by upstream signaling processes, such as the p38 MAPK – MK2 pathway shown here. The phosphatases MKP-1 and PP2A exert stabilizing effects on ARE-containing mRNAs by dephosphorylating the upstream kinases (eg. p38 MAPK, MK2)71,or by dephosphorylating TTP70 (see panel b). HuR (grey) has an overall stabilizing effect on ARE-containing mRNA, and its binding is regulated both positively (eg. by PKC and p38 MAPK) and negatively (eg. by Cdk1 and Chk2) by phosphorylation (reviewed in48). The destabilizing activity of TTP has been studied in detail (panel c, reviewed in109). Non-phosphorylated TTP bound to the ARE recruits deadenylases to activate the decay process. This is inhibited by the phosphorylation of TTP at multiple sites by MK2, which functions downstream of activated p38 MAPK57,58. mRNA stabilization results from the binding of 14-3-3 proteins to phosphorylated TTP59,60, which blocks the interaction of TTP with the deadenylases61,62. This process is reversed by removal of the inactivating phosphates by MKP-1 or PP2A70, rendering the ARE-containing mRNA unstable.
Box 4.
Phosphorylation regulates cytoplasmic ARE-BP function
For simplicity, the rapid degradation of ARE-containing mRNAs can be thought of as a default process, with the destabilizing proteins TTP, BRF1. BRF2, KSRP and some forms of AUF1 functioning to recruit one or more of the degradative enzymes shown in Box 1 to mRNPs56. When considered this way, stabilization results from the dissociation of a destabilizing ARE-BP, interference of the interaction between a destabilizing ARE-BP and one or more ribonucleases, and/or replacement of a destabilizing ARE-BP by a stabilizing ARE-BP.
Phosphorylation regulates the binding and function of ARE-BPs, and the p38 mitogen-activated protein kinase (MAPK), MAPK-activated protein kinase 2 (MK2) and the non-specific serine-threonine protein Akt kinase pathways each play major roles in regulating the binding and activity of ARE-BPs (Box 4, panel B). Of these proteins, TTP is the best studied with regard to upstream signaling processes, and the interplay of kinases and phosphatases in the regulation of its destabilizing activity is shown in Box 4, panel C. In mammals p38 MAPK is activated by stress or by inflammatory stimuli, for example by treating macrophages with bacterial lipopolysaccharide (LPS). p38 MAPK then phosphorylates MK2, which activates MK2 phosphorylation of TTP57,58. TTP is phosphorylated at multiple sites, and the subsequent binding of one or more of the 14-3-3 adapter proteins59,60 interferes with the TTP-mediated recruitment of deadenylases that are required to initiate mRNA decay61,62. Similar results were reported for BRF163, which is phosphorylated by Akt in response to activation of an upstream PI-3-kinase pathway64,65. Note that Akt can also be activated by MK2, and constitutively active Akt is a common feature of many cancers.
KSRP, which acts like TTP and BRF1 by recruiting one or more degradative enzymes to ARE-containing mRNPs, is also regulated by phosphorylation, albeit directly by p38 MAPK66. However, phosphorylation reduces KRSP binding to RNA without altering its interaction with the exosome or the poly(A) ribonuclease PARN67. While AUF1 can be phosphorylated in vitro by protein kinase A (PKA) and glycogen synthase kinase 3β (GSK3β)68, only the NPM-ALK tyrosine kinase has been shown in vivo to stabilize ARE-containing mRNAs that are targeted by AUF169. NPM-ALK is a constitutively active kinase that is expressed in anaplastic large-cell lymphoma as a result of the fusion between the genes for nucleophosmin (NPM) and the anaplastic-lymphoma kinase (ALK) due to a chromosomal translocation. Although as yet unproven, the stabilization of ARE-containing mRNAs by the NPM-ALK-mediated phosphorylation of AUF1 may play a role in promoting malignant transformation.
The ARE-BP HuR counteracts the destabilizing effects of TTP, BRF1, KSRP and AUF1 (Box 4, panel B). Although HuR is primarily regulated at the level of nucleocytoplasmic shuttling (see below), its ARE-binding affinity is also both positively and negatively regulated by phosphorylation. An example of this is its stress-induced phosphorylation (e.g., after ionizing radiation) by the checkpoint homolog-kinase 2 (Chk2). HuR is phosphorylated at the same sites by PKCgamma and, as described above, by p38 MAPK (see48 for a detailed review).
The role of phosphatases in regulating ARE-mediated mRNA decay is less well studied. The destabilizing activity of TTP is increased by PP2A acting directly to remove the inactivating phosphates from the protein70, but the signaling process and mechanism of this targeting are currently unknown. The phosphorylation state of TTP is further reduced by PP2A acting upstream to dephosphorylate and thus reduce the activity of the p38 MAPK and MK2 kinases. p38 MAPK and MK2 are also targets of MAP kinase phosphatase 1 (MKP-1), whose activity is stimulated by CK267,71.
Regulation of cytoplasmic ARE-BP function by nuclear export
The nucleocytoplasmic distribution of ARE-BPs is a major regulatory nexus, with phosphorylation having different effects on TTP, AUF1 and HuR. For example, the export of TTP from the nucleus to the cytoplasm depends on its phosphorylation in the nucleus by MK272, and the binding of different 14-3-3 proteins distinguishes the export process from phosphorylation-dependent changes in activity. The converse is true for AUF1, whose dephosphorylation by MKP-1 promotes its redistribution to the cytoplasm73.
Phosphorylation at different sites on HuR controls the subcellular distribution of HuR, and its distribution between the nucleus and cytoplasm varies with the stage of the cell cycle. The cyclin mRNAs are among those whose stability is regulated by HuR, and HuR is retained in the nucleus at late G2 by Cdk1phosphorylation of residues within the HuR nucleocytoplasmic shuttling (HNS) motif74. Phosphorylation at other sites can have the converse effect, and the amount of cytoplasmic HuR is increased following phosphorylation by PKCalpha in response to activation of the G protein-coupled purinergic P2Y2 receptor75, by PKCgamma in response to angiotensin binding to its G protein coupled receptor76, and by p38 MAPK77. The cytoplasmic accumulation of HuR can also be increased by methylation within the HNS, for example by coactivator-associated arginine methyltransferase 1 (CARM1) in LPS-treated macrophages78.
Targets of HuR
A recent study using photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) to identify HuR-binding sites within the human embryonic kidney-cell transcriptome identified 137,305 clusters, 106,840 of which mapped to protein-coding genes79. The degree of HuR binding was directly related to the degree of mRNA stabilization, and mRNA stabilization was greater for transcripts with both intronic and 3′UTR HuR-binding sites compared to transcripts with either site alone. This finding suggests some as yet unknown link between nuclear pre-mRNA processing and stability of these mRNAs in the cytoplasm. The proximity of HuR- and microRNA-binding sites adds a layer of regulatory complexity to ARE-mediated mRNA decay, and may explain how the stress-induced redistribution of HuR overcomes a specific case of miRNA-mediated silencing (miR-122 targeting CAT1 mRNA)80. Since the PAR-CLIP results derive from normally cycling cells, they provide a general picture of the scope of HuR binding. The influences of stress, hormones, or neurotransmitter signaling that change the intracellular distribution of HuR and its affinity for its target mRNAs will undoubtedly further enrich these regulatory processes.
Nuclear receptors can be ligand-dependent activators of mRNA decay
Nuclear receptors are ligand-dependent transcription factors that are responsible for activating or repressing gene expression in response a range of molecules, including vitamin D and steroid or thyroid hormones among others. Some of these are always found in the nucleus whereas for others the unliganded form is in equilibrium between the cytoplasm and the nucleus. Once bound to their respective ligands these proteins undergo conformational changes and concentrate in the nucleus where they interact with transcriptional coactivators or corepressors.
Nuclear receptors also impact post-transcriptional processes, and the glucocorticoid receptor (GR) is one of the few that have been studied in detail. Corticosteroids are used clinically to treat inflammation, and some of their anti-inflammatory effects result from changes in gene expression that indirectly impact ARE-mediated mRNA decay. For example, corticosteroids acting through GR suppress the transcription of the p38 MAPK gene81 and induce the transcription of the MKP-182 and TTP genes82,83. However, GR can also binds directly to some mRNAs and activate their decay84,85. This was initially observed in airway epithelial cells and arterial smooth muscle cells for the mRNAs encoding the inflammatory chemokines CCL2 and CCL784,85. RNA-binding protein immunoprecipitation (RIP-ChIP) identified an additional ~500 mRNAs that are bound by GR in vivo84. A consensus binding motif was identified but it remains to be determined how GR binding occurs and activates mRNA decay. Furthermore, it is not known if ligand-dependent mRNA binding and destabilization is unique to GR or is also a property of other nuclear receptors.
Regulation of mRNA degradative activities
Remarkably little is known about how ribonucleases (Box 1) are themselves regulated. Here we discuss endonucleases because to date there are no examples of direct regulation of any of the other mRNA degradative enzymes. It is not clear whether this is because no one has looked for exonuclease regulation or whether most exonucleases are regulated through the proteins with which they interact.
Signaling processes activate PMR1-mediated mRNA decay
Polysomal ribonuclease 1 (PMR1) is one of five endonucleases that have been definitively shown to function in mRNA decay. PMR1-mediated mRNA decay depends on the ability of the PMR1 to associate with polysome-bound substrate mRNA86. Binding of PMR1 to polysomes requires phosphorylation by c-Src of the PMR1 C-terminal domain87,88 to generate a binding site for a SH2-domain protein on the translating mRNP. In the one case studied c-Src is stimulated by epidermal-growth factor (EGF)88, but it is likely that PMR1 phosphorylation is also regulated by other upstream activators of c-Src. To date, the activation of PMR1 by c-Src and of AUF1 by NPM-ALK (see above) provide the only examples of mRNA decay being regulated by oncogenic tyrosine kinases. The activity of polysome-bound PMR1 is further increased when estrogen-responsive cells are exposed to estrogen, but the mechanism for this is not known. PMR1 also binds to the cytoskeleton regulatory proteins mammalian homologue of Drosophila enabled (Mena) and vasodilator-stimulated phosphoprotein (VASP), and these associations localize PMR1 to the leading edge of motile cells where it stimulates cell motility89. This effect is likely the result of localized mRNA decay.
IRE1 autophosphorylation and the unfolded-protein response
The unfolded-protein response (UPR) is activated by the accumulation of misfolded or unfolded proteins in the endoplasmic reticulum (ER). Its major function is to restore balance within the ER, although it induces apoptosis under conditions of prolonged or chronic stress. Inositol-requiring enzyme 1 (IRE1) functions as the principal sensor of unfolded proteins and is the primary effector of the UPR. Its oligomerization during the UPR results in autophosphorylation and activation of the cytoplasmic IRE1 endonuclease domain90. The N-terminal sensor domain of IRE1, which is situated within the ER lumen, is normally bound by the HSP70 chaperone binding immunoglobulin protein (BiP), and its displacement by unfolded proteins is one model for the subsequent oligomerization and activation of IRE1. The endonuclease activity participates in the non-canonical splicing of mRNA encoding the transcription factor X box-binding protein-1 (XBP-1), which increases BiP levels. IRE1 also cleaves and destabilizes mRNAs encoding some of the proteins that transit through the ER (i.e., membrane-bound and secreted proteins)91,92 in a process termed rapid IRE1-dependent decay (RIDD). Together with increased BiP levels, RIDD restores balance within the ER by reducing the amount of protein entering the secretory pathway. Because IRE1 substrate selectivity is determined by an N-terminal signal sequence that causes mRNAs associated with nascent peptides bearing this signal to be localized to the ER, RIDD is likely to be regulated by signaling that influences translation initiation.
The selective loss of ER-associated mRNAs by RIDD contrasts with generalized stabilization of the mRNA transcriptome that occurs during oxidative stress or amino acid starvation 93. Stress-induced stabilization is particularly evident for ARE-containing mRNAs, where the accumulation of HuR in stress granules and P bodies is proposed to protect these transcripts by displacing destabilizing ARE-BPs94. It remains to be determined if IRE1 acts only on ER-associated mRNAs, or if RIDD can be activated independently of the UPR.
Induction of Zc3h12a by Toll-like receptor signaling
Zc3h12a (MCPIP1) is the only example so far of an mRNA endonuclease that is transcriptionally induced95,96. There is normally little Zc3h12a in cells. Zc3h12a synthesis is induced as part of the response by macrophages and monocytes to the binding of microbial cell-wall components to Toll-like receptors. Zc3h12a-/- mice treated with bacterial LPS overexpress IL-1, IL-6, TNFalpha and other cytokines as a consequence of failing to degrade the corresponding lymphokine mRNAs, resulting in a runaway inflammatory response. Unlike PMR1 and IRE1, which do not bind directly to their substrate mRNAs, Zc3h12a binds to its targets through a CCCH zinc finger that is similar to that of TTP and BRF-196. Although most Zc3h12a targets contain a 3′UTR ARE, sensitivity to Zc3h12a is imparted by a stem-loop structure that lies upstream of the ARE95,97. Zc3h12a cleaves within this loop to initiate decay.
Zc3h12a is also able to cleave within the loop structures of pre-miRNAs during microRNA processing, where it acts in opposition to Dicer98. This has potential clinical relevance since elevated Zc3h12a expression is linked to the reduced expression of some microRNAs in lung cancer98. There is also emerging evidence that much of the mRNA transcriptome is subject to endonucleolytic cleavage99, thus extending the action of endonucleases beyond the initiation of mRNA decay. Given that the activity and/or quantity of each of the major endonucleases is regulated in some fashion, challenges for the future are to identify the remaining endonucleases, determine how they are regulated at the cellular and organismal level, and identify their targets.
Conclusions
It is clear that the field of mRNA decay is approaching the end of its beginning. Tremendous progress has been made toward understanding mRNA sequences that regulate mRNA stability and the enzymatic activities that mediate mRNA decay. The few examples provided in this review illustrate the complexity that will arise when all of the mRNA decay pathways and their targets are considered collectively. Indeed, future advances will depend on elucidating the complicated, multifactorial webs of regulatory events that coordinate the half-lives of cellular mRNAs depending on the stage of organismal development, the type of tissue, and the environmental conditions under which individual cells exist.
Genetic and biochemical approaches have identified the deadenylases, decapping enzymes, and exonucleases that catalyze mRNA decay. Within the next several years, we should have catalogued and characterized most if not all of the endonucleases. Sequence comparison has not been very successful in identifying endonucleases that act on mRNAs, thus their identification will require careful application of biochemical and molecular biological approaches. Identification of the targets and cleavage sites of these enzymes will be possible though the application of RNA-Seq. Together with improvements in computational approaches to RNA structure determinations, the knowledge gained from such studies will play a crucial role in defining the molecular code that directs particular transcripts to regulated decay processes, and should also provide insights into the mechanisms responsible for generating many of the poorly characterized small RNAs.
Given the advent of high-throughput proteomic and sequencing technologies, it is also becoming possible to identify the constituents of individual mRNPs: RIP-SEQ can be used to deep sequence mRNAs that co-immunoprecipitate with a particular protein as well as ncRNAs that could regulate at least some of the mRNAs; mass spectrometry can be used to identify proteins in these complexes and their post-translational modifications. Once established, it then becomes important to uncover how the sequences and structures of mRNAs, including their post-transcriptional modifications (consider, e.g., the terminal-U transferases that promote exosome-mediated mRNA decay), and their associated proteins and ncRNAs interact dynamically in response to signal transduction pathways to determine mRNA half-lives.
Integrating the many mRNA-specific regulatory pathways with those pathways that modulate the half-lives of multiple mRNAs will be a daunting task requiring sophisticated mathematical modeling. The complexity of such a task is compounded by the direct participation of nuclear receptors, indicating that mRNA half-lives could be regulated by cell environment, age and type. After understanding how mRNA decay is regulated by intracellular signal transduction at the single-cell level, it becomes important to evaluate intracellular signal transduction-mediated regulation at the organ and, ultimately, organismal levels, the latter of which will undoubtedly require considerations of age, gender and diet. As an added caveat, while mRNAs largely function to produce proteins, growing support for the idea that they can also serve as sinks for regulatory proteins and antisense ncRNAs such as microRNAs by functioning as “competing cellular RNAs, or ceRNAs100, indicates that the regulation of mRNA decay may cast a very broad net and affect as yet unappreciated cellular processes.
In the end, it is hoped that new insights into how mRNA decay is regulated will uncover new therapies to treat inherited and acquired diseases (BOX 5). Developing such therapies is at a forefront of a number of biotechnology companies and academic laboratories.
BOX 5. mRNA degradation and disease.
Many diseases are due to the misregulation of mRNA decay (for a review see53). For example, the level of the UPF3X NMD factor is downregulated in patients with syndromic or non-syndromic intellectual disability who harbor mutations in the UPF3X gene, which as its name implies is X-linked; as a consequence, the elevated level of the UPF3 NMD factor appears to compensate for the lack of UPF3X and lessens the disability110,111.
Another recent example involves the von Hippel-Lindau (VHL) tumor suppressor that is inactivated in patients with VHL disease, an autosomally dominant cancer, or clear-cell renal carcinoma. VHL was found to regulate the stability of the ARE-containing mRNA that encodes vascular endothelial growth factor (and undoubtedly other ARE-containing mRNAs) by targeting at least some isoforms of AUF1 for ubiquitination and proteasome-mediated destruction112.
It follows from this and many other studies that the targeted decay of particular mRNAs using, e.g., small interfering (si)RNA is at the forefront of developing therapies. siRNA-based disease treatments are challenging considering difficulties in delivering siRNAs to tissues and the inherent instability of siRNAs once delivered. In a recent approach holding promise, the arginine-glycine-aspartate tripeptide that mimics cell adhesion proteins and binds integrins has been used to “functionalize” siRNA nanoparticles that target mRNA encoding the signal transducer and activator of transcription (STAT)1, which upregulates genes in response to signals by type I, type II or type III inteferons113. The nanoparticles were found to be resistant to serum-mediated degradation, effectively taken up by tissues, and effectively delivered to the joints of rheumatoid-arthritic mice so as to alleviate disease113.
Acknowledgments
We thank Myriam Gorospe for her helpful comments and Chenguang Gong for assistance constructing figures. We also apologize to colleagues whose work we could not cite because of page and/or reference limitations. Research in the Schoenberg and Maquat labs is supported by grants from the U.S. National Institute of General Medical Sciences.
Glossary
- 14-3-3 proteins
A group of seven ubiquitously expressed phosphoserine/phosphothreonine binding proteins. They can assemble into homo- or heterodimers, mediate protein:protein interactions, and function in many cellular processes
- APA
Alternative cleavage and polyadenylation, which provides a means to vary mRNA 3′-end formation and, thus, the regulatory sequences often present within 3′UTR sequences
- EJC
Exon-junction complex. A protein complex that is deposited ~20–24-nucleotides upstream of splicing-generated exon-exon junctions. It includes the NMD factors UPF3X and UPF3 among many others
- MAPK
Generic term for mitogen-activated protein kinases.. These proteins function in signal transduction by amplifying and integrating signals from different receptors followed by delivering each signal to one or more endpoint effector proteins
- mTORC1
Mammalian target of rapamycin complex 1. This complex consists of the phosphatidylinositol 3-kinase (PIK)-related serine-threonine protein kinase mTOR, raptor and LST8. mTORC1 is inhibited by low nutrient levels, growth factor deprivation and reductive stress so that cellular protein synthesis is concomitantly inhibited
- No-go mRNA decay
A pathway that degrades faulty mRNAs associated with stalled ribosomes. Decay is initiated by endonucleolytic cleavage near the stall site to release sequestered ribosomes and associated translation factors for the translation
- Nonstop mRNA decay
A pathway that degrades mRNAs that lack a stop codon and direct translation either to the poly(A) tail, due to, e.g., premature polyadenylation, or to an mRNA breakpoint. It facilitates the recycling of ribosomes and translation factors
- NMD
Nonsense-mediated mRNA decay. In mammalian cells, this pathway targets newly synthesized mRNAs that are undergoing a pioneer round of translation. It generally eliminates spliced mRNAs that prematurely terminate translation but also has some physiologic targets. It competes with Staufen-mediated decay
- P bodies
Processing bodies. Small cytoplasmic RNA granules that are enriched for decapping proteins, activators of decapping, XRN1 and non-translating mRNAs. P bodies function as sites for mRNA storage and possibly decay
- PAR-CLIP
, Photoactivatable-Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation. A method for profiling RNA bound to a specific protein. Cells are grown in medium containing 4-thiouridine or 6-thioguanosine, which when incorporated into RNA allow for efficient UV crosslinking to RNA-binding proteins. The immunoprecipitated protein:RNA complex is then used to generate libraries for deep sequencing
- PTC
Premature termination codon. A stop codon positioned 5′ to the normal termination codon. Usually activates NMD when situated >50 nucleotides upstream of a splicing-generated exon-exon junction
- RNA-binding protein immunoprecipitation
A method for recovering RNAs by virtue of their binding by a particular protein. This uses either an antibody specific to a particular RNA-binding protein, or antibody to an epitope tag on a recombinant protein expressed in target cells
- SMD
STAU1-mediated mRNA decay. A pathway that degrades mRNAs that harbor a STAU1-binding site within their 3′ UTR in a mechanism that depends on translation and the NMD factor UPF1
- Stress Granules
Large cytoplasmic foci containing non-translating mRNAs bound by the 40S ribosomal subunit. Accumulate in stressed cells, commonly as a result of translation inhibition secondary to the phosphorylation of eIF2alpha
- SURF
Complex of SMG1, UPF1, eRF1 and eRF3 that recognizes a premature termination codon
- Toll-like receptor
Single-chain membrane-bound receptors that function in the innate immune response. Binding of bacterial cell wall components, such as lipopolysaccharides or lipomannins, activates binding of adapter proteins that leads to the activation of NF-kB and associated changes in transcription
References
- 1.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Chen CY, Shyu AB. Mechanisms of deadenylation3dependent decay. Wiley Interdisc Rev : RNA. 2010;2:167–183. doi: 10.1002/wrna.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Y, Kiledjian M. Regulation of mRNA decapping. Wiley Interdiscip Rev RNA. 2010;1:253–265. doi: 10.1002/wrna.15. [DOI] [PubMed] [Google Scholar]
- 4.Schoenberg DR. Mechanisms of endonuclease-mediated mRNA decay. Wiley Interdisc Rev : RNA. 2011;2:582–600. doi: 10.1002/wrna.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maquat LE, Tarn WY, Isken O. The pioneer round of translation: features and functions. Cell. 2010;142:368–374. doi: 10.1016/j.cell.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 7.Wittmann J, Hol EM, Jack HM. hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol Cell Biol. 2006;26:1272–1287. doi: 10.1128/MCB.26.4.1272-1287.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Apcher S, et al. Major source of antigenic peptides for the MHC class I pathway is produced during the pioneer round of mRNA translation. Proc Natl Acad Sci USA. 2011;108:11572–11577. doi: 10.1073/pnas.1104104108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghosh S, Jacobson A. RNA decay modulates gene expression and controls its fidelity. Wiley Interdisc Rev : RNA. 2010;1:351–361. doi: 10.1002/wrna.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicholson P, Muhlemann O. Cutting the nonsense: the degradation of PTC-containing mRNAs. Biochem Soc Trans. 2010;38:1615–1620. doi: 10.1042/BST0381615. [DOI] [PubMed] [Google Scholar]
- 11.Hwang J, Sato H, Tang Y, Matsuda D, Maquat LE. UPF1 association with the cap-binding protein, CBP80, promotes nonsense-mediated mRNA decay at two distinct steps. Mol Cell. 2010;39:396–409. doi: 10.1016/j.molcel.2010.07.004. Provides mechanistic rationale for why NMD in mammalian cells appears to be restricted to CBC-bound mRNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang D, Wengrod J, Gardner LB. Overexpression of the c-myc oncogene inhibits nonsense-mediated RNA decay in B lymphocytes. J Biol Chem. 2011;286:40038–40043. doi: 10.1074/jbc.M111.266361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang D, et al. Inhibition of nonsense-mediated RNA decay by the tumor microenvironment promotes tumorigenesis. Mol Cell Biol. 2011;31:3670–3680. doi: 10.1128/MCB.05704-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cam H, Easton JB, High A, Houghton PJ. mTORC1 signaling under hypoxic conditions is controlled by ATM-dependent phosphorylation of HIF-1alpha. Mol Cell. 2010;40:509–520. doi: 10.1016/j.molcel.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gardner LB. Hypoxic inhibition of nonsense-mediated RNA decay regulates gene expression and the integrated stress response. Mol Cell Biol. 2008;28:3729–3741. doi: 10.1128/MCB.02284-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee HC, Choe J, Chi SG, Kim YK. Exon junction complex enhances translation of spliced mRNAs at multiple steps. Biochem Biophys Res Commun. 2009;384:334–340. doi: 10.1016/j.bbrc.2009.04.123. [DOI] [PubMed] [Google Scholar]
- 17.Diem MD, Chan CC, Younis I, Dreyfuss G. PYM binds the cytoplasmic exon-junction complex and ribosomes to enhance translation of spliced mRNAs. Nat Struct Mol Biol. 2007;14:1173–1179. doi: 10.1038/nsmb1321. [DOI] [PubMed] [Google Scholar]
- 18.Sato H, Hosoda N, Maquat LE. Efficiency of the pioneer round of translation affects the cellular site of nonsense-mediated mRNA decay. Mol Cell. 2008;29:255–262. doi: 10.1016/j.molcel.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 19.Michlewski G, Sanford JR, Caceres JF. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol Cell. 2008;30:179–189. doi: 10.1016/j.molcel.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 20.Richardson CJ, et al. SKAR is a specific target of S6 kinase 1 in cell growth control. Curr Biol. 2004;14:1540–1549. doi: 10.1016/j.cub.2004.08.061. [DOI] [PubMed] [Google Scholar]
- 21.Matsuda D, Hosoda N, Kim YK, Maquat LE. Failsafe nonsense-mediated mRNA decay does not detectably target eIF4E-bound mRNA. Nat Struct Mol Biol. 2007;14:974–979. doi: 10.1038/nsmb1297. [DOI] [PubMed] [Google Scholar]
- 22.Eberle AB, Stalder L, Mathys H, Orozco RZ, Muhlemann O. Posttranscriptional gene regulation by spatial rearrangement of the 3′ untranslated region. PLoS Biol. 2008;6:e92. doi: 10.1371/journal.pbio.0060092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isken O, et al. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell. 2008;133:314–327. doi: 10.1016/j.cell.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okada-Katsuhata Y, et al. N- and C-terminal Upf1 phosphorylations create binding platforms for SMG-6 and SMG-5:SMG-7 during NMD. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kashima I, et al. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohnishi T, et al. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol Cell. 2003;12:1187–1200. doi: 10.1016/s1097-2765(03)00443-x. [DOI] [PubMed] [Google Scholar]
- 27.Chan WK, et al. A UPF3-mediated regulatory switch that maintains RNA surveillance. Nat Struct Mol Biol. 2009;16:747–753. doi: 10.1038/nsmb.1612. [DOI] [PubMed] [Google Scholar]
- 28.Yepiskoposyan H, Aeschimann F, Nilsson D, Okoniewski M, Muhlemann O. Autoregulation of the nonsense-mediated mRNA decay pathway in human cells. RNA. 2011;17:2108–2118. doi: 10.1261/rna.030247.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang L, et al. RNA homeostasis governed by cell type-specific and branched feedback loops acting on NMD. Mol Cell. 2011;43:950–961. doi: 10.1016/j.molcel.2011.06.031. Provides first evidence for the feedback regulation of NMD via rate-limiting NMD factors as means to maintain cellular homeostasis considering the role of NMD in normal cellular physiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choe J, Cho H, Lee HC, Kim YK. microRNA/Argonaute 2 regulates nonsense-mediated messenger RNA decay. EMBO Rep. 2010;11:380–386. doi: 10.1038/embor.2010.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Linde L, Boelz S, Neu-Yilik G, Kulozik AE, Kerem B. The efficiency of nonsense-mediated mRNA decay is an inherent character and varies among different cells. Eur J Hum Genet. 2007;15:1156–1162. doi: 10.1038/sj.ejhg.5201889. [DOI] [PubMed] [Google Scholar]
- 32.Gong C, Kim YK, Woeller CF, Tang Y, Maquat LE. SMD and NMD are competitive pathways that contribute to myogenesis: effects on PAX3 and myogenin mRNAs. Genes Dev. 2009;23:54–66. doi: 10.1101/gad.1717309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bruno IG, et al. Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay. Mol Cell. 2011;42:500–510. doi: 10.1016/j.molcel.2011.04.018. Presents evidence for a mechanistic link between the brain-specific miR-128-mediated pathway that targets at least two NMD factors and NMD. This link appears to be conserved among diverse species as a means to control neuron development and activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hwang J, Maquat LE. Nonsense-mediated mRNA decay (NMD) in animal embryogenesis: to die or not to die, that is the question. Curr Opin Genet Dev. 2011;21:422–430. doi: 10.1016/j.gde.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaygun H, Marzluff WF. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat Struct Mol Biol. 2005;12:794–800. doi: 10.1038/nsmb972. [DOI] [PubMed] [Google Scholar]
- 37.Zheng L, et al. Phosphorylation of stem-loop binding protein (SLBP) on two threonines triggers degradation of SLBP, the sole cell cycle-regulated factor required for regulation of histone mRNA processing, at the end of S phase. Mol Cell Biol. 2003;23:1590–1601. doi: 10.1128/MCB.23.5.1590-1601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koseoglu MM, Dong J, Marzluff WF. Coordinate regulation of histone mRNA metabolism and DNA replication: cyclin A/cdk1 is involved in inactivation of histone mRNA metabolism and DNA replication at the end of S phase. Cell Cycle. 2010;9:3857–3863. doi: 10.4161/cc.9.19.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin AN, Li Y. RNase MRP RNA and human genetic diseases. Cell Res. 2007;17:219–226. doi: 10.1038/sj.cr.7310120. [DOI] [PubMed] [Google Scholar]
- 40.Bakheet T, Williams BR, Khabar KS. ARED 3.0: the large and diverse AU- rich transcriptome. Nucleic Acids Res. 2006;34:D111–D114. doi: 10.1093/nar/gkj052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 42.Yamashita A, et al. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol. 2005;12:1054–1063. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- 43.Murray EL, Schoenberg DR. A+U-rich instability elements differentially activate 5′-3′ and 3′-5′ mRNA decay. Mol Cell Biol. 2007;27:2791–2799. doi: 10.1128/MCB.01445-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vlasova-St Louis I, Bohjanen PR. Coordinate regulation of mRNA decay networks by GU-rich elements and CELF1. Curr Opin Genet Dev. 2011 doi: 10.1016/j.gde.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beisang D, Rattenbacher B, Vlasova-St Louis IA, Bohjanen PR. Regulation of CUG-binding protein 1 (CUGBP1) binding to target transcripts upon T cell activation. J Biol Chem. 2012;287:950–960. doi: 10.1074/jbc.M111.291658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang L, Lee JE, Wilusz J, Wilusz CJ. The RNA-binding protein CUGBP1 regulates stability of tumor necrosis factor mRNA in muscle cells: implications for myotonic dystrophy. J Biol Chem. 2008;283:22457–22463. doi: 10.1074/jbc.M802803200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dasgupta T, Ladd AN. The importance of CELF control: molecular and biologcial roles of the CUG-BP, Elav-like family of RNA-binding proteins. Wiley Interdisc Rev : RNA. 2012;3:104–121. doi: 10.1002/wrna.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abdelmohsen K, Gorospe M. Posttranscriptional regulation of cancer traits by HuR. Wiley Interdisc Rev : RNA. 2010;1:214–229. doi: 10.1002/wrna.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.von Roretz C, Di Marco S, Mazroui R, Gallouzi IE. Turnover of AU-rich-containing mRNAs during stress: a matter of survival. Wiley Interdiscip Rev RNA. 2011;2:336–347. doi: 10.1002/wrna.55. [DOI] [PubMed] [Google Scholar]
- 50.Gratacos FM, Brewer G. The role of AUF1 in regulated mRNA decay. Wiley Interdisc Rev : RNA. 2010;1:457–473. doi: 10.1002/wrna.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanduja S, Blanco FF, Dixon DA. The roles of TTP and BRF proteins in regulated mRNA decay. Wiley Interdisc Rev : RNA. 2011;2:42–57. doi: 10.1002/wrna.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schott J, Stoecklin G. Networks controlling mRNA decay in the immune system. Wiley Interdisc Rev : RNA. 2010;1:432–456. doi: 10.1002/wrna.13. [DOI] [PubMed] [Google Scholar]
- 53.Eberhardt W, Doller A, Akool e-S, Pfeilschifter J. Modulation of mRNA stability as a novel therapeutic approach. Pharmacol Ther. 2007;114:56–73. doi: 10.1016/j.pharmthera.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 54.Raghavan A, et al. Patterns of coordinate down-regulation of ARE-containing transcripts following immune cell activation. Genomics. 2004;84:1002–1013. doi: 10.1016/j.ygeno.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 55.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lykke-Andersen J, Wagner E. Recruitment and activation of mRNA decay enzymes by two ARE-mediated decay activation domains in the proteins TTP and BRF-1. Genes Dev. 2005;19:351–361. doi: 10.1101/gad.1282305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mahtani KR, et al. Mitogen-activated protein kinase p38 controls the expression and posttranslational modification of tristetraprolin, a regulator of tumor necrosis factor alpha mRNA stability. Mol Cell Biol. 2001;21:6461–6469. doi: 10.1128/MCB.21.9.6461-6469.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chrestensen CA, et al. MAPKAP kinase 2 phosphorylates tristetraprolin on in vivo sites including Ser178, a site required for 14–3–3 binding. J Biol Chem. 2004;279:10176–10184. doi: 10.1074/jbc.M310486200. [DOI] [PubMed] [Google Scholar]
- 59.Stoecklin G, et al. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 2004;23:1313–1324. doi: 10.1038/sj.emboj.7600163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hitti E, et al. Mitogen-activated protein kinase-activated protein kinase 2 regulates tumor necrosis factor mRNA stability and translation mainly by altering tristetraprolin expression, stability, and binding to adenine/uridine-rich element. Mol Cell Biol. 2006;26:2399–2407. doi: 10.1128/MCB.26.6.2399-2407.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marchese FP, et al. MAPKAP kinase 2 blocks tristetraprolin-directed mRNA decay by inhibiting CAF1 deadenylase recruitment. J Biol Chem. 2010;285:27590–27600. doi: 10.1074/jbc.M110.136473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Clement SL, Scheckel C, Stoecklin G, Lykke-Andersen J. Phosphorylation of tristetraprolin by MK2 impairs AU-rich element mRNA decay by preventing deadenylase recruitment. Mol Cell Biol. 2011;31:256–266. doi: 10.1128/MCB.00717-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maitra S, et al. The AU-rich element mRNA decay-promoting activity of BRF1 is regulated by mitogen-activated protein kinase-activated protein kinase 2. RNA. 2008;14:950–959. doi: 10.1261/rna.983708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmidlin M, et al. The ARE-dependent mRNA-destabilizing activity of BRF1 is regulated by protein kinase B. EMBO J. 2004;23:4760–4769. doi: 10.1038/sj.emboj.7600477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Graham JR, Hendershott MC, Terragni J, Cooper GM. mRNA degradation plays a significant role in the program of gene expression regulated by phosphatidylinositol 3-kinase signaling. Mol Cell Biol. 2010;30:5295–5305. doi: 10.1128/MCB.00303-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Briata P, et al. p38-dependent phosphorylation of the mRNA decay-promoting factor KSRP controls the stability of select myogenic transcripts. Mol Cell. 2005;20:891–903. doi: 10.1016/j.molcel.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 67.Winzen R, et al. Functional analysis of KSRP interaction with the AU-rich element of interleukin-8 and identification of inflammatory mRNA targets. Mol Cell Biol. 2007;27:8388–8400. doi: 10.1128/MCB.01493-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wilson GM, et al. Regulation of A + U-rich element-directed mRNA turnover involving reversible phosphorylation of AUF1. J Biol Chem. 2003;278:33029–33038. doi: 10.1074/jbc.M305772200. [DOI] [PubMed] [Google Scholar]
- 69.Fawal M, et al. A “liaison dangereuse” between AUF1/hnRNPD and the oncogenic tyrosine kinase NPM-ALK. Blood. 2006;108:2780–2788. doi: 10.1182/blood-2006-04-014902. [DOI] [PubMed] [Google Scholar]
- 70.Sun L, et al. Tristetraprolin (TTP)-14-3-3 complex formation protects TTP from dephosphorylation by protein phosphatase 2a and stabilizes tumor necrosis factor-alpha mRNA. J Biol Chem. 2007;282:3766–3777. doi: 10.1074/jbc.M607347200. [DOI] [PubMed] [Google Scholar]
- 71.Lee WH, et al. Casein kinase 2 regulates the mRNA-destabilizing activity of tristetraprolin. J Biol Chem. 2011;286:21577–21587. doi: 10.1074/jbc.M110.201137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brook M, et al. Posttranslational regulation of tristetraprolin subcellular localization and protein stability by p38 mitogen-activated protein kinase and extracellular signal-regulated kinase pathways. Mol Cell Biol. 2006;26:2408–2418. doi: 10.1128/MCB.26.6.2408-2418.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yu H, Sun Y, Haycraft C, Palanisamy V, Kirkwood KL. MKP-1 regulates cytokine mRNA stability through selectively modulation subcellular translocation of AUF1. Cytokine. 2011;56:245–255. doi: 10.1016/j.cyto.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim HH, et al. Nuclear HuR accumulation through phosphorylation by Cdk1. Genes Dev. 2008;22:1804–1815. doi: 10.1101/gad.1645808. A detailed analysis of nucleocytoplasmic partitioning of HuR as a function of site-specific phosphorylation during the cell cycle. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Doller A, et al. Protein kinase C alpha-dependent phosphorylation of the mRNA-stabilizing factor HuR: implications for posttranscriptional regulation of cyclooxygenase-2. Mol Biol Cell. 2007;18:2137–2148. doi: 10.1091/mbc.E06-09-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Doller A, et al. Posttranslational modification of the AU-rich element binding protein HuR by protein kinase Cdelta elicits angiotensin II-induced stabilization and nuclear export of cyclooxygenase 2 mRNA. Mol Cell Biol. 2008;28:2608–2625. doi: 10.1128/MCB.01530-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lafarga V, et al. p38 Mitogen-activated protein kinase- and HuR-dependent stabilization of p21(Cip1) mRNA mediates the G(1)/S checkpoint. Mol Cell Biol. 2009;29:4341–4351. doi: 10.1128/MCB.00210-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li H, et al. Lipopolysaccharide-induced methylation of HuR, an mRNA-stabilizing protein, by CARM1. Coactivator-associated arginine methyltransferase. J Biol Chem. 2002;277:44623–44630. doi: 10.1074/jbc.M206187200. [DOI] [PubMed] [Google Scholar]
- 79.Mukherjee N, et al. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell. 2011;43:327–339. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 81.Lasa M, Brook M, Saklatvala J, Clark AR. Dexamethasone destabilizes cyclooxygenase 2 mRNA by inhibiting mitogen-activated protein kinase p38. Mol Cell Biol. 2001;21:771–780. doi: 10.1128/MCB.21.3.771-780.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Smoak K, Cidlowski JA. Glucocorticoids regulate tristetraprolin synthesis and posttranscriptionally regulate tumor necrosis factor alpha inflammatory signaling. Mol Cell Biol. 2006;26:9126–9135. doi: 10.1128/MCB.00679-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ishmael FT, et al. Role of the RNA-binding protein tristetraprolin in glucocorticoid-mediated gene regulation. J Immunol. 2008;180:8342–8353. doi: 10.4049/jimmunol.180.12.8342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ishmael FT, et al. The human glucocorticoid receptor as an RNA-binding protein: global analysis of glucocorticoid receptor-associated transcripts and identification of a target RNA motif. J Immunol. 2011;186:1189–1198. doi: 10.4049/jimmunol.1001794. First demonstration of a direct and functional interaction between a nuclear receptor and mRNAs that are regulated by the hormone ligand. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dhawan L, Liu B, Blaxall BC, Taubman MB. A novel role for the glucocorticoid receptor in the regulation of monocyte chemoattractant protein-1 mRNA stability. J Biol Chem. 2007;282:10146–10152. doi: 10.1074/jbc.M605925200. [DOI] [PubMed] [Google Scholar]
- 86.Yang F, Schoenberg DR. Endonuclease-mediated mRNA decay involves the selective targeting of PMR1 to polyribosome-bound substrate mRNA. Mol Cell. 2004;14:435–445. doi: 10.1016/j.molcel.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 87.Yang F, Peng Y, Schoenberg DR. Endonuclease-mediated mRNA decay requires tyrosine phosphorylation of polysomal ribonuclease 1 (PMR1) for the targeting and degradation of polyribosome-bound substrate mRNA. J Biol Chem. 2004;279:48993–49002. doi: 10.1074/jbc.M409776200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Peng Y, Schoenberg DR. c-Src activates endonuclease-mediated mRNA decay. Mol Cell. 2007;25:779–787. doi: 10.1016/j.molcel.2007.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Peng Y, Murray EL, Sarkar M, Liu X, Schoenberg DR. The cytoskeleton-associated Ena/VASP proteins are unanticipated partners of the PMR1 mRNA endonuclease. RNA. 2009;15:576–587. doi: 10.1261/rna.1206209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tirasophon W, Lee K, Callaghan B, Welihinda A, Kaufman RJ. The endoribonuclease activity of mammalian IRE1 autoregulates its mRNA and is required for the unfolded protein response. Genes Dev. 2000;14:2725–2736. doi: 10.1101/gad.839400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Han D, et al. IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 2009;138:562–575. doi: 10.1016/j.cell.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hollien J, et al. Regulated IRE1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol. 2009;186:323–331. doi: 10.1083/jcb.200903014. Han et al (ref. 91) and Hollien et al. (ref. 92) are complementary papers that describe the endonuclease decay of ER-associated mRNAs catalyzed by IRE1 in response to ER stress during the unfolded protein response. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10:430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- 94.von Roretz C, Di Marco S, Mazroui R, Gallouzi IE. Turnover of AU-rich-containing mRNAs during stress: a matter of survival. Wiley Interdisc Rev : RNA. 2011;2:336–347. doi: 10.1002/wrna.55. [DOI] [PubMed] [Google Scholar]
- 95.Matsushita K, et al. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature. 2009;458:1185–1190. doi: 10.1038/nature07924. [DOI] [PubMed] [Google Scholar]
- 96.Mizgalska D, et al. Interleukin-1-inducible MCPIP protein has structural and functional properties of RNase and participates in degradation of IL-1beta mRNA. FEBS J. 2009;276:7386–7399. doi: 10.1111/j.1742-4658.2009.07452.x. [DOI] [PubMed] [Google Scholar]
- 97.Paschoud S, et al. Destabilization of interleukin-6 mRNA requires a putative RNA stem-loop structure, an AU-rich element, and the RNA-binding protein AUF1. Mol Cell Biol. 2006;26:8228–8241. doi: 10.1128/MCB.01155-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Suzuki HI, et al. MCPIP1 ribonuclease antagonizes Dicer and terminates microRNA biogenesis through precursor microRNA degradation. Mol Cell. 2011;44:424–436. doi: 10.1016/j.molcel.2011.09.012. Describes a new mechanism whereby endonucleolytic cleavage in the unpaired loop of a pre-microRNA prior to its processing by Dicer regulates the abundance of mature microRNA. This has broad implications for the action of other endonucleases in regulating the biogenesis of regulatory RNAs. [DOI] [PubMed] [Google Scholar]
- 99.Karginov FV, et al. Diverse endonucleolytic cleavage sits in the mammalian transcriptome depend on microRNAs, Drosha and additional nucleases. Mol Cell. 2010;38:781–788. doi: 10.1016/j.molcel.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lykke-Andersen S, Tomecki R, Jensen TH, Dziembowski A. The eukaryotic RNA exosome: same scaffold but variable catalytic subunits. RNA Biol. 2011;8:61–66. doi: 10.4161/rna.8.1.14237. [DOI] [PubMed] [Google Scholar]
- 102.Kim YK, Furic L, Desgroseillers L, Maquat LE. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 103.Hosoda N, Kim YK, Lejeune F, Maquat LE. CBP80 promotes interaction of Upf1 with Upf2 during nonsense-mediated mRNA decay in mammalian cells. Nat Struct Mol Biol. 2005;12:893–901. doi: 10.1038/nsmb995. [DOI] [PubMed] [Google Scholar]
- 104.Dominski Z, Marzluff WF. Formation of the 3′ end of histone mRNA: getting closer to the end. Gene. 2007;396:373–390. doi: 10.1016/j.gene.2007.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yan J, Marr TG. Computational analysis of 3′-ends of ESTs shows four classes of alternative polyadenylation in human, mouse, and rat. Genome Res. 2005;15:369–375. doi: 10.1101/gr.3109605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Flavell SW, et al. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Laishram RS, Barlow CA, Anderson RA. CKI isoforms {alpha} and {varepsilon} regulate Star-PAP target messages by controlling Star-PAP poly(A) polymerase activity and phosphoinositide stimulation. Nucleic Acids Res. 2011;39:7961–7973. doi: 10.1093/nar/gkr549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sandler H, Stoecklin G. Control of mRNA decay by phosphorylation of tristetraprolin. Biochem Soc Trans. 2008;36:491–496. doi: 10.1042/BST0360491. [DOI] [PubMed] [Google Scholar]
- 110.Tarpey PS, et al. Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat Genet. 2007;39:1127–1133. doi: 10.1038/ng2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nguyen LS, et al. Transcriptome profiling of UPF3B/NMD-deficient lymphoblastoid cells from patients with various forms of intellectual disability. Mol Psychiatry. 2011 doi: 10.1038/mp.2011.163. Demonstrates the importance of UPF3B (UPF3X) in regulating the neuronal-cell transcriptome via, e.g., the ARGHGAP24 GTPase-activating protein, which derives from an NMD target and promotes axon and dendrite outgrowth and branching. Additionally, through studies of UPF3B-deficient patients together with Chan et al., 2009, correlates disease-severity with the compensatory level of UPF3A (UPF3), which is destabilized by UPF3B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xin H, et al. Association of the von Hippel-Lindau protein with AUF1 and post- transcriptional regulation of Vascular Endothelial Growth Factor A mRNA. Mol Cancer Res. 2011 doi: 10.1158/1541-7786.MCR-11-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Scheinman RI, Trivedi R, Vermillion S, Kompella UB. Functionalized STAT1 siRNA nanoparticles regress rheumatoid arthritis in a mouse model. Nanomedicine (Lond) 2011;6:1699–1682. doi: 10.2217/nnm.11.90. [DOI] [PubMed] [Google Scholar]
- 114.Yamashita A, et al. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev. 2009;23:1091–1105. doi: 10.1101/gad.1767209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Unterholzner L, Izaurralde E. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol Cell. 2004;16:587–596. doi: 10.1016/j.molcel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 116.Huntzinger E, Kashima I, Fauser M, Sauliere J, Izaurralde E. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA. 2008;14:2609–2617. doi: 10.1261/rna.1386208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol. 2009;16:49–55. doi: 10.1038/nsmb.1530. [DOI] [PubMed] [Google Scholar]
- 118.Franks TM, Singh G, Lykke-Andersen J. Upf1 ATPase-dependent mRNP disassembly Is required for completion of nonsense- mediated mRNA decay. Cell. 2010;143:938–950. doi: 10.1016/j.cell.2010.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 120.Ma XM, Yoon SO, Richardson CJ, Julich K, Blenis J. SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell. 2008;133:303–313. doi: 10.1016/j.cell.2008.02.031. [DOI] [PubMed] [Google Scholar]