Abstract
Human Ebola virus causes severe hemorrhagic fever disease with high mortality and there is no vaccine or treatment. Antibodies in survivors occur early, are sustained, and can delay infection when transferred into nonhuman primates. Monoclonal antibodies (mAbs) from survivors exhibit potent neutralizing activity in vitro and are protective in rodents. To better understand targets and mechanisms of neutralization, we investigated a panel of mAbs shown previously to react with the envelope glycoprotein (GP). While one non-neutralizing mAb recognized a GP epitope in the nonessential mucin-like domain, the rest were specific for GP1, were neutralizing, and could be further distinguished by reactivity with secreted GP. We show that survivor antibodies, human KZ52 and monkey JP3K11, were specific for conformation-dependent epitopes comprising residues in GP1 and GP2 and that neutralization occurred by two distinct mechanisms; KZ52 inhibited cathepsin cleavage of GP whereas JP3K11 recognized the cleaved, fusion-active form of GP.
Keywords: Virus, Ebola, Immunity, Neutralization, Antibody, Human, Nonhuman primate, Rodent
Introduction
Ebola viruses (EBOV) are enveloped, nonsegmented, negative-strand RNA viruses belonging to the family Filoviridae (Sanchez et al., 2001). Infection by four of the five identified species, including Zaire (ZEBOV), Sudan (SEBOV), Ivory Coast (CIEBOV) and the recently discovered Bundibugyo (Towner et al., 2008), causes acute, severe viral hemorrhagic fever disease with high mortality in humans. While an animal reservoir for the virus has yet to be determined, it is likely that fruit bats play a role in the natural cycle of EBOV (Leroy et al., 2005, Leroy et al., 2009). The Centers for Disease Control and Prevention has classified EBOV as a potential biological threat and Category A Select Agent (Rotz et al., 2002) due in part to its high fatality rate, potential for aerosol transmission, and the lack of a vaccine or therapeutic treatment for infection.
Adaptive immunity contributes to protection against EBOV and has been demonstrated using vaccines in nonhuman primates, where symptoms and mortality rates resemble those observed during human infection (Bradfute et al., 2008, Jones et al., 2005, Sullivan et al., 2000, Sullivan et al., 2003, Sullivan et al., 2009, Warfield et al., 2007). Immune protection in animal models is associated with the development of both cellular and humoral immunity (Baize et al., 1999, Gupta et al., 2001, Parren et al., 2002, Takada et al., 2003b, Takada et al., 2007, Wilson et al., 2000). In human survivors, recovery is associated with early and vigorous antibody responses that are long lasting (Wauquier et al., 2009), whereas defective humoral responses are observed in lethal cases (Baize et al., 1999). This may be a consequence of impaired adaptive immunity due to EBOV replication in antigen-presenting cells (APCs) (Bosio et al., 2004, Mahanty et al., 2003, Warfield et al., 2004) resulting in a delayed antibody response (Baize et al., 1999), or a B-cell frequency too low to mediate virus clearance (Sanchez et al., 2001). Alternatively, antibody specificities or binding properties may be suboptimal for efficient virus clearance (Takada et al., 2001, Takada et al., 2003a). Since administration of mAbs confers protection in rodent models of lethal EBOV (Parren et al., 2002, Takada et al., 2003b, Takada et al., 2007, Wilson et al., 2000), identification of neutralizing antibodies (NAbs) and their mechanisms of activity may be important for developing vaccines and immunotherapies against EBOV (Sullivan et al., 2009).
A central target for NAbs is the EBOV structural envelope glycoprotein since it is accessible on the virion surface and essential for virus entry (Chan et al., 2001, Simmons et al., 2003, Takada et al., 2004, Wool-Lewis and Bates, 1998, Wool-Lewis and Bates, 1999). GP is synthesized as a polyprotein that is post-translationally modified into two subunits, GP1 and membrane-bound GP2, which covalently interact to form a monomer of the trimeric GP complex on virions. A key functional domain that is a potential target for NAbs is the putative receptor binding domain (RBD) in GP1 (Brindley et al., 2007, Kuhn et al., 2006, Manicassamy et al., 2005). However, access to this domain may be obscured by the heavily glycosylated mucin-like domain (MUC) in GP1 that serves as a major target for the humoral immune response (Wilson et al., 2000) and is a pathogenic determinant during EBOV infection (Dowling et al., 2006, Francica et al., 2009, Jeffers et al., 2002, Yang et al., 2000). Unlike the N-terminal RBD, MUC is nonessential (Simmons et al., 2002, Takada et al., 2004) and its removal by endosomal proteolysis is required for virus entry (Chandran et al., 2005, Kaletsky et al., 2007, Schornberg et al., 2006).
Several forms of GP have been identified in natural infection and may serve as targets for humoral immunity. Viral polymerase-driven expression from the EBOV GP gene yields a secreted form of GP, sGP, which is the most abundant GP protein synthesized during infection and constitutes greater than 80% of total GP (Volchkov et al., 1998). Its main role in viral pathogenesis is unknown but it is detected at high concentrations in the blood (Sanchez et al., 2001) and is hypothesized to act as an immune decoy (Maruyama et al., 1999) by serving as a target for virus specific antibodies (Wilson et al., 2000). The synthesis of full length virion-bound GP is directed only when the polymerase inserts a non-templated adenosine during transcription. Such tight control of GP expression could be necessary due to cytopathic effects exerted when the protein is expressed at high levels (Yang et al., 2000).
Cleavage of virion-associated GP by host cell cathepsins yields another functionally distinct form of GP (GPCatL) that is critical for EBOV entry (Chandran et al., 2005, Schornberg et al., 2006) and may expose critical Ab determinants within conserved domains of the RBD. EBOV is thought to enter hosts cells via receptor-mediated endocytosis in clathrin-coated pits and caveolae (Sanchez, 2007) and virus entry is affected by changes in endosomal pH (Chan et al., 2000). Although, acidification is not likely responsible for direct triggering of the envelope protein as in classic pH-dependent viruses like influenza, it is a requirement for the activity of host cell cathepsins responsible for protein modification of EBOV and other viruses including Marburg virus (Sanchez, 2007), reovirus (Ebert et al., 2002), severe acute respiratory syndrome (SARS) coronavirus (Huang et al., 2006, Simmons et al., 2005), mouse hepatitis virus 2 (Qiu et al., 2006), Hendra virus (Pager and Dutch, 2005) and Nipah virus (Pager et al., 2006). Unlike SARS coronavirus, where endosomal processing by cathepsins is necessary for entry only after receptor binding and would otherwise render particles noninfectious (Simmons et al., 2005), EBOV pretreatment with cathepsins renders a stable viral intermediate that is fully infectious (Chandran et al., 2005, Kaletsky et al., 2007, Schornberg et al., 2006). The function of cathepsins in EBOV entry may be analogous to CD4 binding for HIV; modification of the glycoprotein exposes key determinants of infection shielded by variable sequences that may otherwise serve as targets for antibody-mediated neutralization. Therefore, inhibition of cathepsin cleavage or antibody binding to GPCatL may provide additional targets for neutralization of EBOV. Herein we focused primarily on two NAbs from survivors of EBOV infection to explore functional determinants in GP as targets for neutralization.
Results
Neutralizing antibodies bind representative GP targets of natural infection
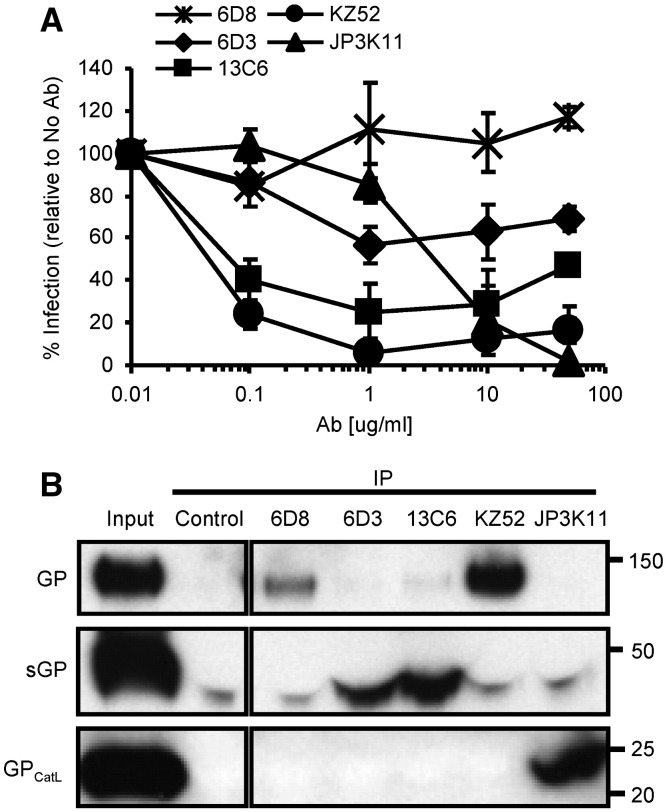
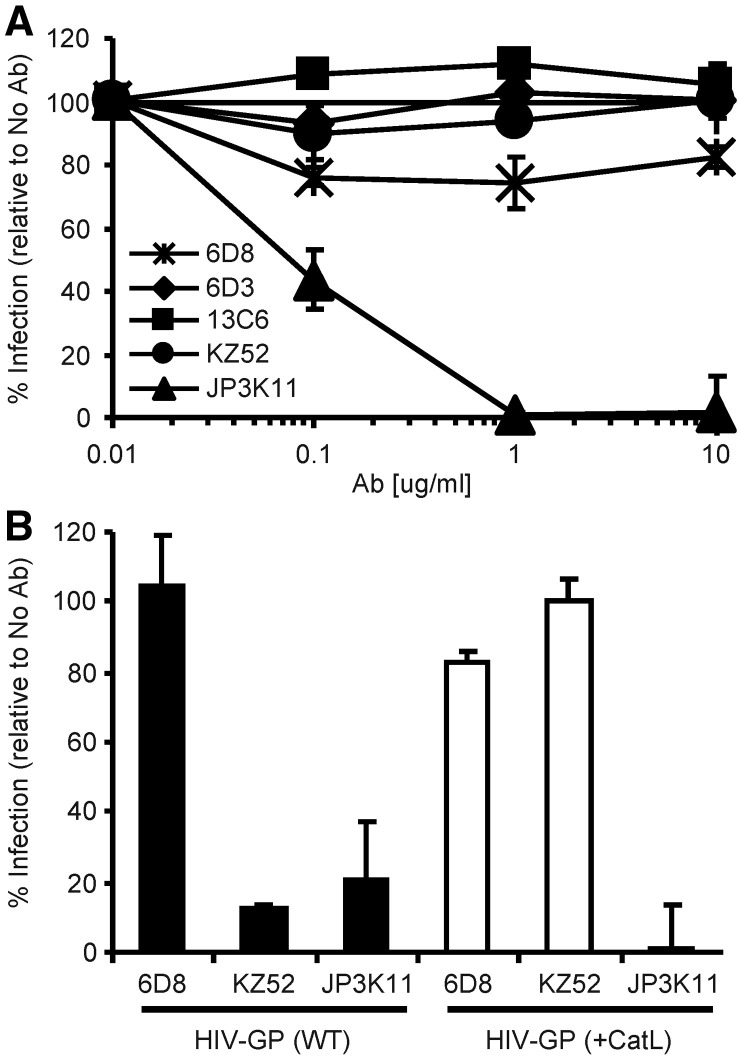
To gain a better understanding of the viral targets and mechanisms of neutralization by humoral immunity, we studied a set of select EBOV-specific antibodies; three mouse mAbs 6D8, 6D3 and 13C6 that conferred protection against a mouse-adapted strain of EBOV (Wilson et al., 2000), one human mAb, KZ52, (Lee et al., 2008) that protected guinea pigs (Parren et al., 2002) but not monkeys (Oswald et al., 2007), and one monkey mAb, JP3K11, previously shown by in vitro analysis to react with EBOV (Meissner et al., 2002). Neutralization studies were performed using EBOV GP-pseudotyped retroviral particles containing the luciferase reporter gene to assess the inhibitory capacity of the antibodies (Fig. 1A). EBOV GP viral pseudotypes were incubated with various concentrations of mAbs for 1 h prior to infection of HUVEC cells, a primary cell type infectable by EBOV (Wahl-Jensen et al., 2005, Yang et al., 1998). Luciferase activity resulting from pseudovirus entry was analyzed 72 h post infection and graphed as a percentage of infection in control samples with no antibody added. Results for mAbs 6D3, 13C6, and KZ52 extend previous data in that they were all inhibitory (Wilson et al., 2000), the former two being only moderately so; approximately 40% neutralizing for 6D2 and 65% for 13C6. MAb 6D8, specific for amino acids present in the GP MUC, did not interfere with the infection of target cells. JP3K11 was also inhibitory, but only at mAb concentrations above 1 µg/ml. Thus, results confirm the neutralization properties for all mAbs but one, 6D8, for an EBOV GP-bearing target pseudovirus. Since the capacity for Ab-mediated neutralization is affected by the presence, location and/or exposure of the respective binding epitope in EBOV GP, we next explored recognition by the antibodies for EBOV GP targets mimicking those present during natural infection. The virion-bound GP trimer, the secreted and nonessential sGP, and the fusion-active conformation of GP that results from proteolytic cleavage events were assessed for relative mAb recognition in a competitive immunoprecipitation (IP) assay (Fig. 1B; GP, sGP, and GPCatL were approximately 130, 40, and 22 kD, respectively). EBOV GP viral pseudotypes were pelleted, incubated for 30 min at 37 °C in the presence or absence of CatL enzyme, and reactions were terminated by the addition of a cathepsin inhibitor, E-64, and boiling. CatL-cleaved virion preparations served as the source material for GPCatL while untreated virions were used for to assess recognition of native GP. GP, GPCatL, and free sGP which was harvested from the supernatant of sGP(Z) plasmid-transfected cells, were combined in IP reactions at equivalent target protein concentrations as determined by immunoblotting IP reaction input material (Input), and little background was observed for the unlabeled bead control (Control). Antibody 6D8, known to bind a linear epitope in MUC (Wilson et al., 2000) and classified as non-neutralizing in Fig. 1, exhibited strong and exclusive reactivity with GP, but not with sGP or GPCatL, the latter species of which completely lacks amino acids present in MUC. While all five mAbs tested immunoprecipitated GP as confirmed by overexposure of the film (data not shown), neutralizing mAbs demonstrated various binding preferences for the different GP forms. Neutralizing mAbs, 6D3 and 13C6, recognize conformational epitopes in GP and sGP (Wilson et al., 2000) and were found here to exhibit a greater affinity for sGP than for virion GP under competitive conditions. Preference for sGP-binding by these mAbs is likely due to increased exposure of the epitope on sGP compared to GP, possibly owing to partial obstruction on the native trimer of N-terminal (sGP-homologous) residues by MUC or slightly different conformations between the sGP dimer and GP trimer (Yang et al., 1998). Consistent with previous findings, KZ52 bound exclusively to full length GP under competitive conditions as expected since the epitope is known to comprise amino acids present in both GP1 and GP2 (Lee et al., 2008, Maruyama et al., 1999). In contrast to KZ52, JP3K11 exhibited preferential recognition of GPCatL and was the sole mAb to bind GP in its Cat-cleaved conformation; increased exposure times confirmed that no mAb in this panel other than JP3K11 reacted with cathepsin-processed GP under competitive conditions (data not shown). These data demonstrate that the JP3K11 epitope does not comprise residues in MUC since they are not present in GPCatL. Altogether, competitive immunoprecipitation for three separate GP products of natural EBOV infection allowed for qualitative assessments of binding preference for epitopes presented on different forms of GP, and showed notable differences between antibodies.
Fig. 1.
EBOV neutralizing antibodies bind natural GP products of infection. (A) EBOV GP viral pseudotypes were incubated with various concentrations of mouse 6D8, 6D3 and 13C6, human KZ52, and monkey JP3K11 for 1 h prior to infection of HUVEC cells. Luciferase activity was read 72 h post infection and graphed as % infection relative to infection in the absence of antibody. Error bars represent ± SEM. (B) Competitive IP for natural GP products of infection: WT or CatL-processed (GPCatL) GP-bearing pseudovirus and sGP. IP material control (Input) and IP samples were analyzed by SDS-PAGE and Western immunoblotting using JCB antibody reagent for detection. Experiments were performed independently at least three times with similar results.
Neutralization epitopes require N-terminal GP1 residues
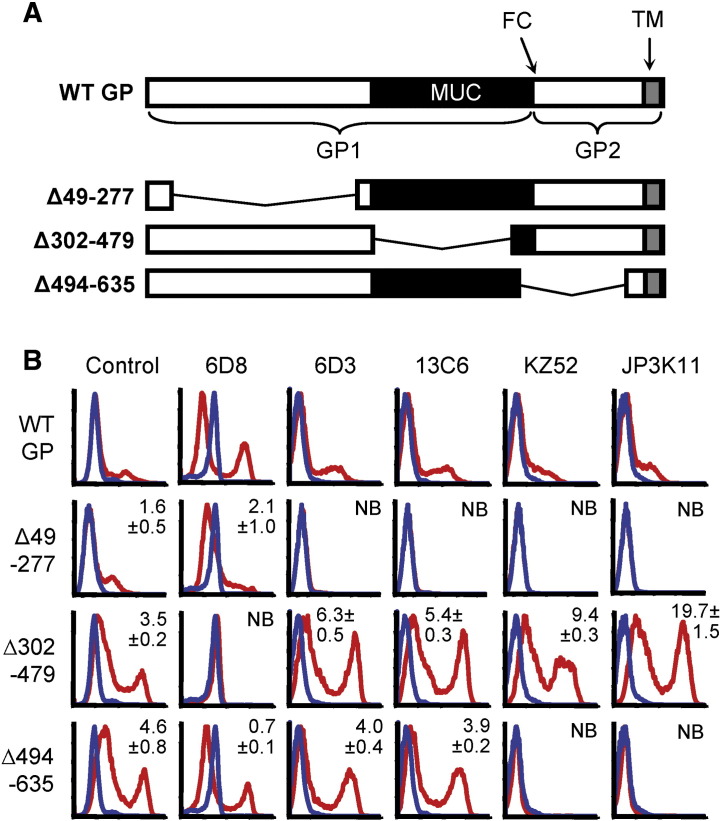
We have shown that the epitopes for EBOV-specific NAbs are expressed differentially in GP products representative of those during natural infection. To explore the molecular basis for these differences, a more detailed molecular characterization of NAb epitopes was undertaken. To this end, gross epitope-mapping was performed to determine the contribution of amino acids from N-terminal GP1, MUC, and GP2 to mAb binding. Antibody binding analysis was performed on GP mutants with broad deletions in the N-terminal domain (Δ49–277), MUC (Δ302–479), or GP2 region (Δ494–635) (Fig. 2A). FACS staining of each of the GP deletion mutants or empty vector-transfected when expressed in 293 cells was performed using control serum (sGP/GP), 6D3, 13C6, KZ52, or JP3K11. Significant binding ratios (MFI of staining for mutant over WT GP) were observed for Control (sGP/GP), 6D3, and 13C6 binding to Δ302–479 or Δ 494–635 (p < 0.001); 6D8, KZ52, and JP3K11 binding to Δ302–479 (p < 0.05). As expected for non-neutralizing mAb 6D8, which is known to be specific for a linear epitope in the MUC (Wilson et al., 2000), binding was ablated when the MUC domain was absent. For the neutralizing mAbs 6D3 and 13C6, which were shown above to be strongly specific for epitopes also present in sGP, deletion of the N-terminal GP1 region eliminated antibody binding (Fig. 2B). Similarly, binding for KZ52 and JP3K11 was lost upon deletion of the N-terminus, as expected for KZ52 (Lee et al., 2008). Deletion mutations may ablate binding by these NAbs either by removing critical binding residues within the epitope or by disrupting a conformation required for epitope formation. The latter explanation is likely true for KZ52 since its contact residues identified in co-crystals are present in the mutant GP (Lee et al., 2008). Removal of the highly glycosylated MUC, on the contrary, did not affect antibody recognition of GP by these four NAbs. Indeed, binding to this form of GP appeared to be even stronger than to WT GP for this subset of antibodies, owing either to increased cell surface expression levels of this protein, increased accessibility of the epitope due to removal of a MUC shielding effect, or both. Finally, the removal of amino acids present in GP2 revealed a notable difference in GP binding among the NAbs; recognition by 6D3 and 13C6 was independent of GP2 residues while KZ52 and JP3K11 binding was dependent, consistent with reported KZ52 contact residues between GP 505–514 and 549–556 when the antibody was co-crystalized with a modified form of GP (Lee et al., 2008). These data delineate a difference among N-terminal GP1-binding NAbs based on the requirement for GP2 residues, on which recognition by KZ52 and JP3K11 is critically dependent.
Fig. 2.
N-terminal GP1 contains potent neutralization epitopes. (A) Cartoon of WT GP and deletion mutants Δ49–277, Δ302–479, and Δ494–635; GP1 and GP2 subunits, furin cleavage site (FC), MUC (black) and transmembrane (TM; gray) are indicated. (B) FACS staining of GP deletion mutants expressed in 293 cells; GP expressing- (red lines) or empty vector-transfected (blue lines) cells were stained with control serum (sGP/GP), 6D8, 6D3, 13C6, KZ52, or JP3K11. Experiments were performed at least four times with similar results and numbers represent binding ratios (MFI relative to WT) ± SEM.
KZ52 blocks CatL proteolysis of GP
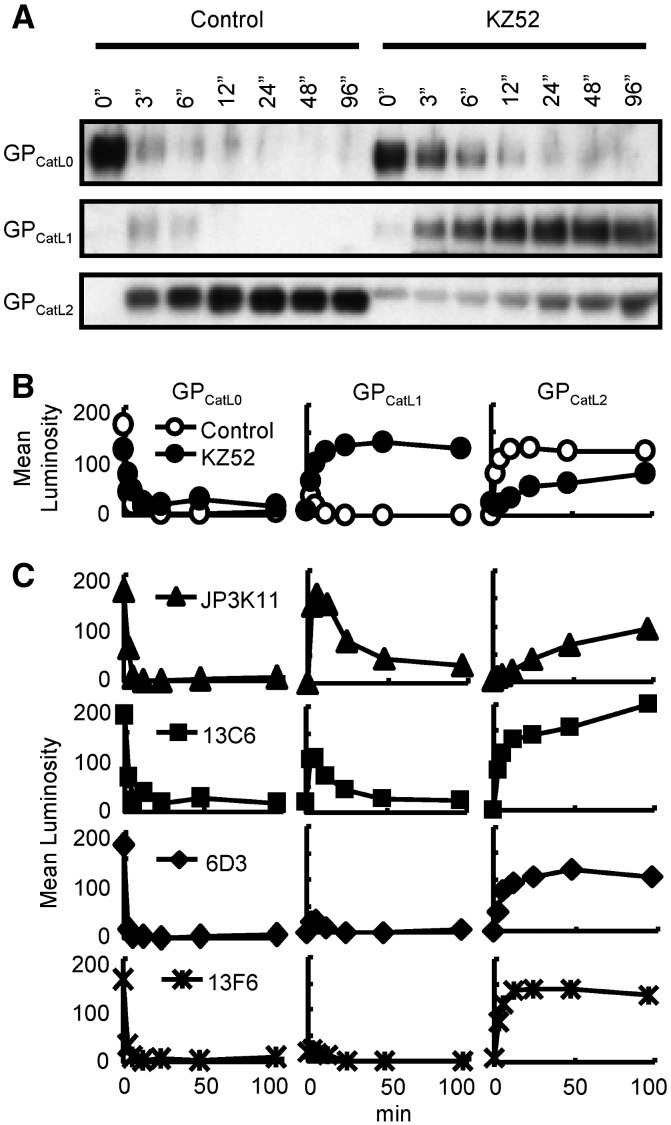
The observed dependence of NAb binding on N-terminal EBOV GP1 determinants suggests that these antibodies may block infection by interfering with essential functions that are associated with this portion of GP, receptor binding and CatL cleavage. Since KZ52 preferentially recognizes full length GP prior to cathepsin cleavage, we asked whether it might inhibit virus entry by interfering with this critical GP modification. To address this question, GP-pseudovirus was treated with CatL in the presence of 50 μg of KZ52 and the reaction was terminated at various times by the addition of a cathepsin inhibitor, E-64, and boiling. As negative controls, KZ52 inactivated by heating to 100 °C and a MUC-binding mAb 13F6 that is not neutralizing (data not shown) were processed similarly and all reaction products were evaluated by SDS-PAGE (Fig. 3 ). Uncleaved GP was observed to migrate as a 148-kD fragment (GPCatL0) at time zero, and was efficiently processed by the CatL enzyme (Figs. 3A and B) as reported previously (Chandran et al., 2005). Two dominant proteolytic products were observed as a result of CatL cleavage; an intermediate 56-kD fragment (indicated as GPCatL1) observed by others (Kaletsky et al., 2007, Schornberg et al., 2006), and the final 22-kD product (GPCatL2) that is capable of mediating membrane fusion. In the control reactions using either heated KZ52 or 13F6, intermediate fragment GPCatL1 constituted a minor transient cleavage product that was processed completely to fusogenic GPCatL2 (Figs. 3B and C). Processing to the fusion-active GPCatL2 occurred efficiently within the first 3 min and was complete within 20 min of incubation with CatL. In contrast, the presence of intact KZ52 delayed this reaction significantly and complete proteolysis was not achieved even after 96 min of incubation. These results show that KZ52 binding to the EBOV glycoprotein inhibits a critical CatL cleavage event required for the formation of fusion-active GP.
Fig. 3.
KZ52 inhibits cathepsin L-activation of fusogenic GP. (A) CatL cleavage of virion-associated GP in the presence of boiled (Control) or native KZ52 as assessed over time by immunoblotting using control JCB antibody reagent. Indicated are full length GP1 (GPCatL0), CatL-cleaved intermediate fragment (GPCatL1), and CatL-cleaved fusogenic GP (GPCatL2). (B) Mean luminosity of immunoblot data for boiled KZ52 (Control; open circles) and native KZ52 (closed circles). (C) Densitometry of immunoblot data for CatL cleavage in the presence of J3PK11 (triangles), 13C6 (squares), 6D3 (diamonds), and control, non-neutralizing, MUC-binding mAb 13F6 (stars) over time and expressed as mean luminosity. Data are from the same gel and experiments were performed at least twice with similar results.
Additional biochemical studies were performed using the remaining NAbs examined in this report since it may be possible that, similar to KZ52, their binding epitopes are comprised of residues either within or overlapping the putative CatL cleavage site. As noted above, CatL cleavage was unaffected by the presence of a MUC-binding control NAb, 13F6, and efficient processing of GP1 was observed. Likewise, addition of the NAb 6D3 had no effect on GP1 proteolysis. In contrast, 13C6 and JP3K11 demonstrated interference in the proteolysis of intermediate GPCatL1 to GPCatL2, but the block in GPCatL1 processing by JP3K11 and 13C6 was transient and the reaction eventually resulted in the complete proteolysis to GPCatL2. These data demonstrate that JP3K11, and to a much lesser extent 13C6, delay CatL cleavage, suggesting that this capacity may contribute in part to their neutralization activity.
JP3K11 binds a neutralization determinant in CatL-processed GP
We showed above that JP3K11 recognizes an epitope determinant that is dependent on the presence of both GP1 and GP2 residues. This was also confirmed for KZ52 (Lee et al., 2008). The binding properties of these two NAbs can be differentiated by the preference for binding to CatL-processed GP, which was exhibited exclusively by JP3K11. Therefore, we examined next the capacity of the antibody panel to block entry mediated by a CatL-cleaved EBOV GP-pseudovirus, which is fully infectious (Chandran et al., 2005, Kaletsky et al., 2007). CatL-treated pseudovirions were incubated for 1 h in the presence of mAbs at various concentrations, ranging from 0.01 to 10 μg/ml, for 1 h prior to infection of HUVEC cells, and luciferase activity was measured 72 h later (Fig. 4 ). While mAbs 6D3, 13C6, KZ52, and JP3K11, but not 6D8, neutralized GP-pseudovirus (Fig. 1A), only JP3K11 was capable of efficiently blocking transduction by CatL-GP pseudoviruses. The lack of neutralization by KZ52 is consistent with the preferential reactivity of this antibody with native, pre-cleaved GP shown in Fig. 1B, and lack of CatL-GP recognition. Thus, although both KZ52 and JP3K11 demonstrate dependence for binding on similar determinants within GP, only JP3K11 is able to bind and neutralize CatL-processed GP-bearing pseudoviruses. These data suggest that JP3K11 and KZ52 may neutralize pseudovirus at distinct stages of the entry process, which may help to explain the reduced potency observed for JP3K11-mediated neutralization of virions bearing native GP shown in Fig. 1.
Fig. 4.
Recognition of fusogenic GP by JP3K11 blocks EBOV entry. (A) CatL-treated (CatL-GP) viral pseudotypes were incubated with various concentrations of 6D8, 6D3, 13C6, KZ52, and JP3K11 for 1 h prior to infection of HUVEC cells and luciferase activity was measured 72 h later. Data is displayed as % infection relative to infection in the absence of antibody. (B) Neutralization of (black bars) WT (Native GP) and (white bars) CatL-GP pseudoviruses by KZ52, JP3K11, and a Control, 6D8. The experiments were performed three times independently and error bars represent ± SEM.
Discussion
In order to gain a better understanding of the targets and mechanisms of EBOV NAbs we studied a panel of known EBOV-specific antibodies; three mouse mAbs that conferred protection in mice against a mouse-adapted EBOV (Wilson et al., 2000), one human mAb (Lee et al., 2008) that protected guinea pigs (Parren et al., 2002) but not monkeys (Oswald et al., 2007), and one monkey survivor mAb specific for GP (Meissner et al., 2002). The N-terminus of GP1 contains the putative RBD that is required for receptor binding and virus fusion (Brindley et al., 2007, Kuhn et al., 2006, Manicassamy et al., 2005) and therefore may serve as a potent target for NAbs. The neutralizing mAbs examined herein had epitopes comprising N-terminal GP1 amino acids since deletion of the N-terminal GP1 region ablated Ab-binding. These NAbs could be further segregated based on their dependence on residues within GP2; only KZ52 and JP3K11 required the presence of GP2 for recognition. 6D3 and 13C6 were capable of binding free sGP, which does not contain GP2 residues. Thus, it would be reasonable to hypothesize that binding by these N-terminal specific Abs may interfere with a number of events required for virus entry including cell attachment, receptor binding, and/or membrane fusion. It is noteworthy that the binding determinants for 6D3 and 13C6 are more accessible in sGP than virion GP (Fig. 1), suggesting that a weaker binding affinity for native GP may explain their reduced potency for neutralization when compared with KZ52. Additionally, it should be noted that the efficacy of NAbs that recognize free sGP, like 6D3 and 13C6, may be diminished in vivo by the fact that their ligand of greatest affinity, sGP is present at higher frequencies than virus-associated GP during natural infection and may form complexes (and deplete) antibodies with this specificity. On the contrary, while recognition of GP by KZ52 and JP3K11 was also dependent on amino acids present in N-terminal GP1, binding was dependent on the presence of GP2 residues which are not present in sGP. Binding results for KZ52 are consistent with KZ52-GP contacts identified in co-crystals that reveal specific residues in GP2 that are situated at a distant location from the putative RBD in GP1 (Lee et al., 2008). While KZ52 was previously the only known mAb to bridge both attachment (GP1) and fusion (GP2) subunits of any viral GP (Lee et al., 2008), the present findings identify another antibody, JP3K11, with similar properties.
Neutralization studies herein revealed that all mAbs examined but one, 6D8, were capable of neutralizing an EBOV GP-bearing pseudovirus in vitro. The lack of neutralization by 6D8 was not predicted since studies in mice show protection by passive transfer (Wilson et al., 2000). However, it is quite possible that the protection observed in this model may be influenced by the use of a serially-passaged virus adapted for infection of non-natural host species (adult mice are naturally resistant to EBOV infection), in which important biological differences in viral pathology and GP amino acid sequences between the mouse-adapted and the naïve viruses may affect the recapitulation of clinical severity (Bray et al., 1998). Also, such protection could be mediated by an alternative mechanism distinct from interference with virus entry, such as antibody-dependent cell-mediated cytotoxicity (ADCC). The lack of neutralization by MUC-directed 6D8 in this report is consistent with a model whereby the EBOV MUC expresses a protective role against antibody binding not unlike the “glycan shield” of HIV which protects the receptor binding domain from NAbs (Wei et al., 2003), since the putative RBD of EBOV is recessed beneath a glycan cap (Lee et al., 2008). This domain is completely dispensable for infection (Simmons et al., 2002, Takada et al., 2004) and is immunodominant for humoral responses during non-lethal EBOV infection since the majority of EBOV-specific Abs reported to date recognize continuous epitopes in this region (Wilson et al., 2000), likely the result of MUC comprising the most exposed elements of virus-bound GP (Vanderzanden et al., 1998). Thus, the value of such a domain that protects critical functional regions from humoral immunity seems evident.
Although KZ52 and JP3K11 both bound epitopes dependent on amino acids in GP1 and GP2, only JP3K11 was able to bind and neutralize cathepsin-processed GP-pseudoviruses. These results demonstrate that JP3K11 recognizes a critical neutralizing determinant that is differentially accessible in native and CatL-cleaved GP. The mechanism of JP3K11 neutralization is consistent with a model whereby JP3K11 binding, occurring either before or after endosomal compartmentalization, blocks triggering of a final, activated form of GP, which may require the participation of a cellular reductase (Schornberg et al., 2006) or another cofactor(s), the fusion event itself, or downstream events required for virus entry. Moreover, data demonstrate that J3PK11 may also exhibit a capacity for CatL inhibition by virtue of its proximity to the GPCatL1–2 cleavage site, in addition to its primary mechanism of neutralization as described herein. However, since the mechanism of EBOV entry has yet to be fully characterized it is difficult to predict at what point during virus entry the JP3K11 neutralizing determinant is accessible to this antibody. To date, JP3K11 is the first mAb described to preferentially bind the CatL-cleaved form of EBOV GP. Importantly, this characteristic was associated with a greater potency for neutralization, albeit at higher mAb concentrations, than other antibodies studied. Since KZ52 was not as potent as JP3K11 at higher concentrations in neutralizing virus in vitro, antibody-mediated interference of CatL cleavage alone may not be as effective in neutralizing virus as a blocking mechanism acting on later events required for fusion, such as that observed by JP3K11. Additionally, CatL cleavage eventually progresses, despite KZ52 binding, following longer incubation periods, raising the possibility that “leakiness” by KZ52 could explain the observation of virus escape after passive transfer into macaques. While these data do not directly explain why passively transferred KZ52 alone failed to protect monkeys from lethal virus challenge (Oswald et al., 2007), they do suggest a mechanism for KZ52 that may be suboptimal for potent in vivo neutralization when compared to the multiple neutralization activities exhibited by JP3K11.
Since KZ52 did not bind CatL-processed GP nor inhibit CatL-cleaved viral pseudotypes, we hypothesize that proteolytic cleavage, which is critically required for viral entry (Chandran et al., 2005), may act to disrupt the binding epitope. Ablation of KZ52 binding may possibly occur by the removal of contact residues or by disrupting GP conformation. Indeed, the presence of KZ52 was sufficient to inhibit formation of the critical 22-kD fragment required for viral entry, and a larger intermediary fragment containing the RBD was observed (56-kD fragment or intermediate GPCatL1). Intermediates of cathepsin proteolysis of GP1 have been observed (Kaletsky et al., 2007, Schornberg et al., 2006), and data herein show that EBOV GP contains at least two separate CatL cleavage sites; the GPCatL1–2 cleavage site occurs before the MUC yielding the 22-kD fragment containing the RBD and the second CatL cleavage site, GPCatL0–1, likely occurs somewhere in the MUC. Furthermore, due to the proximity of the KZ52 binding site to the presumptive RBD, the contribution of direct receptor antagonism by the Ab should not be discounted entirely. Thus, these data are consistent with a model of Ab-mediated neutralization whereby KZ52 interferes with cathepsin cleavage of GP by blocking enzyme attachment or activity, and hinders cathepsin-activation of the fusogenic form of GP containing the RBD or receptor binding directly.
In conclusion, we show here for the first time that two neutralizing Abs, KZ52 and JP3K11, inhibit viral transduction by two fundamentally different mechanisms. Neutralization by KZ52 exploits the EBOV requirement for endosomal proteolysis required for virus entry. On the contrary, JP3K11 expresses a classic mechanism of Ab-mediated neutralization by inhibiting triggering of fusogenic GP, fusion events, and/or receptor binding. In addition, this NAb also exhibits a moderate capacity for inhibiting CatL proteolysis of EBOV GP. Since there is currently no vaccine or treatment for EBOV disease, future vaccine strategies may be more effective in the provision of antibodies like JP3K11 that recognize post-cleavage GP. While it remains to be determined whether JP3K11 can provide protection in vivo, its evaluation in future studies will be informative.
Materials and methods
Pseudovirus construction and neutralization assay
ZEBOV GP-pseudotyped lentiviruses were produced as previously described (Yang et al., 2000). Briefly, human embryonal kidney 293 cells were transfected with pCMVΔR8.2, pHR′CMV-Luc, and pVR1012 plasmid vectors using ProFection Mammalian Transfection System with calcium phosphate (Promega). Mammalian pVR1012 vectors encoded GP from ZEBOV-Mayinga 1976 (GenBank accession #U23187), are under control of the CMV enhancer promoter, and have been described (Hartikka et al., 1996, Sullivan et al., 2000). Supernatants were harvested 40–56 h post-transfection, cleared by low-speed centrifugation and 0.45 µm filtration, and then stored at − 80 °C. Infectivity into human umbilical vein endothelial cells (HUVECs; Cambrex), which are highly infectable by EBOV (Wahl-Jensen et al., 2005, Yang et al., 1998), in the presence of NAbs was measured as a function of luciferase reporter activity as previously described (Sullivan et al., 2006, Yang et al., 1998, Yang et al., 2000). Briefly, culture media from cells plated in 96-well plates 1 day prior to infection was replaced with pseudotyped virus that was first incubated at 37 °C for 1 h in the presence or absence of antibodies. 72 h post infection cells were lysed and assayed by Luciferase Assay System (Promega) and enzyme activity was determined using a Veritas Microplate Luminometer (Turner Biosystems). Cathepsin-treated virions used in the neutralization assay were produced under conditions as described below and normalized by p24 ELISA.
In vitro proteolysis of virion GP
CatL proteolysis of GP-pseudovirus was performed as previously described (Chandran et al., 2005). Briefly, for the competitive immunoprecipitation assay (Fig. 1), CatL kinetics experiment (Fig. 3) and the entry assay (Fig. 4), pelleted pseudovirus preparations (2.5 to 3.5 μg protein) were incubated for 30 min at 37 °C with CatL (0.03 to 0.15 μg; specific activity, 60,000 mU per mg; Calbiochem) in acetate buffer at pH 5.5 (100 mM sodium acetate [pH 5.5] and 1 mM EDTA). Enzymatic reactions were terminated for the competitive immunoprecipitation assay and the CatL kinetics experiment by the addition of 90 µM of the cathepsin inhibitor, E-64 (Sigma), or boiling, respectively. Mock treatments were performed in identical buffers in the absence of enzyme. Cleaved preparations were analyzed by SDS-PAGE using NuPAGE® Gel System (Invitrogen) and immunoblotting using RBD-specific JCB reagent, or they were used to infect HUVECs. For virus entry assays, GP-pseudovirus following incubation in the presence or absence of CatL was pelleted and then resuspended in EGM-2 (Clonetics® Endothelial Cell Growth Medium; Lonza, Walkersville, MD) prior to infection of HUVECs.
Antibody reagents
ZEBOV-specific Abs were the focus of this study; mouse mAbs IgG2a 13F6, and IgG2a 6D8 are specific for defined linear epitopes in the MUC of GP1; IgG1 6D3 and IgG2a 13C6 recognize discontinuous epitopes in GP1 and sGP, and all five antibodies are protective in mice against a mouse-adapted strain of ZEBOV (Wilson et al., 2000); human recombinant IgG1 KZ52 is specific for GP1 and is protective by passive transfer in guinea pigs (Maruyama et al., 1999) but not monkeys (Oswald et al., 2007); monkey IgG JP3K11 is GP-specific (Meissner et al., 2002). GP-specific rabbit control serum was generated by immunization with ZEBOV GP. JCB reagent is a polyclonal rabbit serum, monospecific for a defined linear epitope in N-terminal ZEBOV GP1 between amino acids 83 to 97.
Immunoprecipitation and immunoblotting
ZEBOV material, or GP target mixture, including WT GP-pseudovirus (as described above), CatL-cleaved GP-pseudovirus (as described above) and free sGP (harvested from supernatant from sGP(Z)-transfected cells), was prepared for usage in the competitive immunoprecipitation assay. Ab (10 μg) was incubated with 100 µl of immobilized protein G plus (Pierce) in NP40 lysis buffer containing protease inhibitors for 1 h at room temperature. The Ab-bead mixture was then blocked with 5% normal calf serum and 5% normal goat serum for an additional 1 h. Beads were washed three times in NP40 lysis buffer before resuspending in protein G-cleared, GP target mixture in sodium acetate buffer containing wild type (WT) and CatL-treated GP-pseudovirus (2.5 to 3.5 μg protein each) and free sGP (0.5 to 1.0 μg protein). IP reactions were at least 4 h at 4 °C in the presence of protease inhibitors and E-64 (Sigma), beads were washed four times with NP40 lysis buffer, and protein was eluted at 100 °C in 2× PAGE loading dye buffer for 5 min. Samples were analyzed by SDS-PAGE and Western immunoblotting using Invitrolon™ PVDF membranes of 0.45 µm pore size (Invitrogen). Unconjugated JCB reagent was used at a dilution of 1:6000 to detect WT GP, sGP and CatL-processed GP, and horseradish peroxidase-conjugated goat anti-rabbit IgG antibodies (Santa Cruz Biotechnology) were used at a dilution of 1:12,000 for secondary detection. Densitometry values were the mean luminosity of protein bands on inverted, gray-scale gel images using Adobe® Photoshop 7.0 (Adobe, San Jose, CA).
Vector construction and transfections
Plasmid vectors pVR1012, WT GP(Z), sGP(Z), Δ302–479, and Δ494–635 have been described (57, 58). PCR-directed mutagenesis was used for GP(Z) vectors Δ49–277, Δ302–479, and Δ494–635. ProFection Mammalian Transfection System with calcium phosphate was used per manufacturer's instructions (Promega) for transfection of human embryonal kidney 293 cells in Dulbecco's Modified Eagle Medium (Invitrogen, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS; Invitrogen) and antibiotics. Cells were harvested 20 to 22 h post-transfection using ice-cold PBS (Invitrogen) containing 0.3 mM EDTA. GP mutants expressed on cells were used as binding targets for NAbs, while pVR1012-transfected cells served as negative controls, and Ab-binding was assessed by FACS. Surface expression of WT GP and Δ49–277 protein is low since the WT protein is extremely toxic and the Δ49–277 protein is mainly secreted since the mutation affects covalent association between GP1 and GP2 (Jeffers et al., 2002).
Flow cytometry
Immunostaining procedures were described previously (Sullivan et al., 2006). Transfected cells were stained with unlabeled control GP-specific rabbit serum or mAbs, 6D8, 6D3, 13C6, KZ52 or JP3K11. Secondary detection antibodies included PE-conjugated donkey anti-human IgG (for detection of KZ52 and JP3K11; Jackson ImmunoResearch Laboratories), sheep anti-mouse IgG and goat anti-rabbit IgG (Sigma-Aldrich). Expression of the empty plasmid vector, pVR1012, in 293 cells was used as a control for non-specific Ab-reactivity. Live 293 cells were acquired on a Becton Dickenson LSR II (BD Immunocytometry Systems), and analyzed using FlowJo software (Tree Star, Ashland, OR).
Statistical analysis
All values are reported as the mean ± SEM. Comparison of MFI of mutant to WT (Fig. 2B) was completed by ANOVA with a post hoc Dunnett's test to adjust for multiple comparisons to one control group (WT GP). All statistical analysis was carried out using the Statistical Package for the Social Sciences (SPSS, Chicago, IL).
Acknowledgments
We thank A. Tislerics, B. Hartman and D. Jeffers for manuscript assistance, M. Cichanowski for graphics, G.J. Nabel, M. Roederer and R. Wyatt for helpful discussions, and G.J.N. for the generous gift of mouse-derived antibodies; A. Sanchez for generously providing GP-specific rabbit serum; J.R. Mascola for critical reading of the manuscript.
Role of the funding source
This research was supported in part by the Intramural Research Program of the NIH, Vaccine Research Center, NIAID and NIH grant #AI048053. The funding source had no role in study design; collection, analysis, or interpretation of data; writing of the report; or the decision to submit the paper for publication.
References
- Baize S., Leroy E.M., Georges-Courbot M.C., Capron M., Lansoud-Soukate J., Debre P., Fisher-Hoch S.P., McCormick J.B., Georges A.J. Defective humoral responses and extensive intravascular apoptosis are associated with fatal outcome in Ebola virus-infected patients. Nat. Med. 1999;5:423–426. doi: 10.1038/7422. [DOI] [PubMed] [Google Scholar]
- Bosio C.M., Moore B.D., Warfield K.L., Ruthel G., Mohamadzadeh M., Aman M.J., Bavari S. Ebola and Marburg virus-like particles activate human myeloid dendritic cells. Virology. 2004;326:280–287. doi: 10.1016/j.virol.2004.05.025. [DOI] [PubMed] [Google Scholar]
- Bradfute S.B., Warfield K.L., Bavari S. Functional CD8+ T cell responses in lethal Ebola virus infection. J. Immunol. 2008;180:4058–4066. doi: 10.4049/jimmunol.180.6.4058. [DOI] [PubMed] [Google Scholar]
- Bray M., Davis K., Geisbert T., Schmaljohn C., Huggins J. A mouse model for evaluation of prophylaxis and therapy of Ebola hemorrhagic fever. J. Infect. Dis. 1998;178:651–661. doi: 10.1086/515386. [DOI] [PubMed] [Google Scholar]
- Brindley M.A., Hughes L., Ruiz A., McCray P.B., Jr., Sanchez A., Sanders D.A., Maury W. Ebola virus glycoprotein 1: identification of residues important for binding and post binding events. J. Virol. 2007 doi: 10.1128/JVI.02433-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S.Y., Speck R.F., Ma M.C., Goldsmith M.A. Distinct mechanisms of entry by envelope glycoproteins of Marburg and Ebola (Zaire) viruses. J. Virol. 2000;74:4933–4937. doi: 10.1128/jvi.74.10.4933-4937.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S.Y., Empig C.J., Welte F.J., Speck R.F., Schmaljohn A., Kreisberg J.F., Goldsmith M.A. Folate receptor-alpha is a cofactor for cellular entry by Marburg and Ebola viruses. Cell. 2001;106:117–126. doi: 10.1016/s0092-8674(01)00418-4. [DOI] [PubMed] [Google Scholar]
- Chandran K., Sullivan N.J., Felbor U., Whelan S.P., Cunningham J.M. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling W., Thompson E., Badger C., Mellquist J.L., Garrison A.R., Smith J.M., Paragas J., Hogan R.J., Schmaljohn C. The influences of glycosylation on the antigenicity, immunogenicity, and protective efficacy of Ebola virus GP DNA vaccines. J. Virol. 2006;81:1821–1837. doi: 10.1128/JVI.02098-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert D.H., Deussing J., Peters C., Dermody T.S. Cathepsin L and cathepsin B mediate reovirus disassembly in murine fibroblast cells. J. Biol. Chem. 2002;277:24609–24617. doi: 10.1074/jbc.M201107200. [DOI] [PubMed] [Google Scholar]
- Francica J.R., Matukonis M.K., Bates P. Requirements for cell rounding and surface protein down-regulation by Ebola virus glycoprotein. Virology. 2009;383:237–247. doi: 10.1016/j.virol.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M., Mahanty S., Bray M., Ahmed R., Rollin P.E. Passive transfer of antibodies protects immunocompetent and imunodeficient mice against lethal Ebola virus infection without complete inhibition of viral replication. J. Virol. 2001;75:4649–4654. doi: 10.1128/JVI.75.10.4649-4654.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartikka J., Sawdey M., Cornefert-Jensen F., Margalith M., Barnhart K., Nolasco M., Vahlsing H.L., Meek J., Marquet M., Hobart P., Norman J., Manthorpe M. An improved plasmid DNA expression vector for direct injection into skeletal muscle. Hum. Gene Ther. 1996;7:1205–1217. doi: 10.1089/hum.1996.7.10-1205. [DOI] [PubMed] [Google Scholar]
- Huang I.C., Bosch B.J., Li F., Li W., Lee K.H., Ghiran S., Vasilieva N., Dermody T.S., Harrison S.C., Dormitzer P.R., Farzan M., Rottier P.J., Choe H. SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells. J. Biol. Chem. 2006;281:3198–3203. doi: 10.1074/jbc.M508381200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffers S.A., Sanders D.A., Sanchez A. Covalent modifications of the Ebola virus glycoprotein. J. Virol. 2002;76:12463–12472. doi: 10.1128/JVI.76.24.12463-12472.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S.M., Feldmann H., Stroher U., Geisbert J.B., Fernando L., Grolla A., Klenk H.D., Sullivan N.J., Volchkov V.E., Fritz E.A., Daddario K.M., Hensley L.E., Jahrling P.B., Geisbert T.W. Live attenuated recombinant vaccine protects nonhuman primates against Ebola and Marburg viruses. Nat. Med. 2005;11:786–790. doi: 10.1038/nm1258. [DOI] [PubMed] [Google Scholar]
- Kaletsky R.L., Simmons G., Bates P. Proteolysis of the Ebola virus glycoproteins enhances virus binding and infectivity. J. Virol. 2007;81:13378–13384. doi: 10.1128/JVI.01170-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn J.H., Radoshitzky S.R., Guth A.C., Warfield K.L., Li W., Vincent M.J., Towner J.S., Nichol S.T., Bavari S., Choe H., Aman M.J., Farzan M. Conserved receptor-binding domains of Lake Victoria Marburgvirus and Zaire Ebolavirus bind a common receptor. J. Biol. Chem. 2006;281:15951–15958. doi: 10.1074/jbc.M601796200. [DOI] [PubMed] [Google Scholar]
- Lee J.E., Fusco M.L., Hessell A.J., Oswald W.B., Burton D.R., Saphire E.O. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leroy E.M., Kumulungui B., Pourrut X., Rouquet P., Hassanin A., Yaba P., Delicat A., Paweska J.T., Gonzalez J.P., Swanepoel R. Fruit bats as reservoirs of Ebola virus. Nature. 2005;438:575–576. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- Leroy E.M., Epelboin A., Mondonge V., Pourrut X., Gonzalez J.P., Muyembe-Tamfum J.J., Formenty P. Human Ebola outbreak resulting from direct exposure to fruit bats in Luebo, Democratic Republic of Congo, 2007. Vector Borne Zoonotic Dis. 2009;9:723–728. doi: 10.1089/vbz.2008.0167. [DOI] [PubMed] [Google Scholar]
- Mahanty S., Hutchinson K., Agarwal S., McRae M., Rollin P.E., Pulendran B. Cutting edge: impairment of dendritic cells and adaptive immunity by Ebola and Lassa viruses. J. Immunol. 2003;170:2797–2801. doi: 10.4049/jimmunol.170.6.2797. [DOI] [PubMed] [Google Scholar]
- Manicassamy B., Wang J., Jiang H., Rong L. Comprehensive analysis of Ebola virus GP1 in viral entry. J. Virol. 2005;79:4793–4805. doi: 10.1128/JVI.79.8.4793-4805.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama T., Rodriguez L.L., Jahrling P.B., Sanchez A., Khan A.S., Nichol S.T., Peters C.J., Parren P.W., Burton D.R. Ebola virus can be effectively neutralized by antibody produced in natural human infection. J. Virol. 1999;73:6024–6030. doi: 10.1128/jvi.73.7.6024-6030.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner F., Maruyama T., Frentsch M., Hessell A.J., Rodriguez L.L., Geisbert T.W., Jahrling P.B., Burton D.R., Parren P.W. Detection of antibodies against the four subtypes of Ebola virus in sera from any species using a novel antibody-phage indicator assay. Virology. 2002;300:236–243. doi: 10.1006/viro.2002.1533. [DOI] [PubMed] [Google Scholar]
- Oswald W.B., Geisbert T.W., Davis K.J., Geisbert J.B., Sullivan N.J., Jahrling P.B., Parren P.W., Burton D.R. Neutralizing antibody fails to impact the course of Ebola virus infection in monkeys. PLoS Pathog. 2007;3:e9. doi: 10.1371/journal.ppat.0030009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pager C.T., Dutch R.E. Cathepsin L is involved in proteolytic processing of the Hendra virus fusion protein. J. Virol. 2005;79:12714–12720. doi: 10.1128/JVI.79.20.12714-12720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pager C.T., Craft W.W., Jr., Patch J., Dutch R.E. A mature and fusogenic form of the Nipah virus fusion protein requires proteolytic processing by cathepsin L. Virology. 2006;346:251–257. doi: 10.1016/j.virol.2006.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parren P.W., Geisbert T.W., Maruyama T., Jahrling P.B., Burton D.R. Pre- and postexposure prophylaxis of Ebola virus infection in an animal model by passive transfer of a neutralizing human antibody. J. Virol. 2002;76:6408–6412. doi: 10.1128/JVI.76.12.6408-6412.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Z., Hingley S.T., Simmons G., Yu C., Das Sarma J., Bates P., Weiss S.R. Endosomal proteolysis by cathepsins is necessary for murine coronavirus mouse hepatitis virus type 2 spike-mediated entry. J. Virol. 2006;80:5768–5776. doi: 10.1128/JVI.00442-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotz L.D., Khan A.S., Lillibridge S.R., Ostroff S.M., Hughes J.M. Public health assessment of potential biological terrorism agents. Emerg. Infect. Dis. 2002;8:225–230. doi: 10.3201/eid0802.010164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez A. Analysis of filovirus entry into vero e6 cells, using inhibitors of endocytosis, endosomal acidification, structural integrity, and cathepsin (B and L) activity. J. Infect. Dis. 2007;196(Suppl 2):S251–S258. doi: 10.1086/520597. [DOI] [PubMed] [Google Scholar]
- Sanchez A., Khan A.S., Zaki S.R., Nabel G.J., Ksiazek T.G., Peters C.J. In: Fields Virology. Four ed. Knipe D.M., Howley P.M., editors. vol. One. Lippincott Williams & Wilkins; Philadelphia: 2001. Filoviridae: Marburg and Ebola viruses; pp. 1279–1304. (Two vols.). [Google Scholar]
- Schornberg K., Matsuyama S., Kabsch K., Delos S., Bouton A., White J. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J. Virol. 2006;80:4174–4178. doi: 10.1128/JVI.80.8.4174-4178.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Wool-Lewis R.J., Baribaud F., Netter R.C., Bates P. Ebola virus glycoproteins induce global surface protein down-modulation and loss of cell adherence. J. Virol. 2002;76:2518–2528. doi: 10.1128/jvi.76.5.2518-2528.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Rennekamp A.J., Chai N., Vandenberghe L.H., Riley J.L., Bates P. Folate receptor alpha and caveolae are not required for Ebola virus glycoprotein-mediated viral infection. J. Virol. 2003;77:13433–13438. doi: 10.1128/JVI.77.24.13433-13438.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. U. S. A. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan N.J., Sanchez A., Rollin P.E., Yang Z.Y., Nabel G.J. Development of a preventive vaccine for Ebola virus infection in primates. Nature. 2000;408:605–609. doi: 10.1038/35046108. [DOI] [PubMed] [Google Scholar]
- Sullivan N.J., Geisbert T.W., Geisbert J.B., Xu L., Yang Z.Y., Roederer M., Koup R.A., Jahrling P.B., Nabel G.J. Accelerated vaccination for Ebola virus haemorrhagic fever in non-human primates. Nature. 2003;424:681–684. doi: 10.1038/nature01876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan N.J., Geisbert T.W., Geisbert J.B., Shedlock D.J., Xu L., Lamoreaux L., Custers J.H., Popernack P.M., Yang Z.Y., Pau M.G., Roederer M., Koup R.A., Goudsmit J., Jahrling P.B., Nabel G.J. Immune protection of nonhuman primates against Ebola virus with single low-dose adenovirus vectors encoding modified GPs. PLoS Med. 2006;3:e177. doi: 10.1371/journal.pmed.0030177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan N.J., Martin J.E., Graham B.S., Nabel G.J. Correlates of protective immunity for Ebola vaccines: implications for regulatory approval by the animal rule. Nat. Rev. Microbiol. 2009;7:393–400. doi: 10.1038/nrmicro2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada A., Watanabe S., Okazaki K., Kida H., Kawaoka Y. Infectivity-enhancing antibodies to Ebola virus glycoprotein. J. Virol. 2001;75:2324–2330. doi: 10.1128/JVI.75.5.2324-2330.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada A., Feldmann H., Ksiazek T.G., Kawaoka Y. Antibody-dependent enhancement of Ebola virus infection. J. Virol. 2003;77:7539–7544. doi: 10.1128/JVI.77.13.7539-7544.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada A., Feldmann H., Stroeher U., Bray M., Watanabe S., Ito H., McGregor M., Kawaoka Y. Identification of protective epitopes on Ebola virus glycoprotein at the single amino acid level by using recombinant vesicular stomatitis viruses. J. Virol. 2003;77:1069–1074. doi: 10.1128/JVI.77.2.1069-1074.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada A., Fujioka K., Tsuiji M., Morikawa A., Higashi N., Ebihara H., Kobasa D., Feldmann H., Irimura T., Kawaoka Y. Human macrophage C-type lectin specific for galactose and N-acetylgalactosamine promotes Filovirus entry. J. Virol. 2004;78:2943–2947. doi: 10.1128/JVI.78.6.2943-2947.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada A., Ebihara H., Jones S., Feldmann H., Kawaoka Y. Protective efficacy of neutralizing antibodies against Ebola virus infection. Vaccine. 2007;25:993–999. doi: 10.1016/j.vaccine.2006.09.076. [DOI] [PubMed] [Google Scholar]
- Towner J.S., Sealy T.K., Khristova M.L., Albarino C.G., Conlan S., Reeder S.A., Quan P.L., Lipkin W.I., Downing R., Tappero J.W., Okware S., Lutwama J., Bakamutumaho B., Kayiwa J., Comer J.A., Rollin P.E., Ksiazek T.G., Nichol S.T. Newly discovered Ebola virus associated with hemorrhagic fever outbreak in Uganda. PLoS Pathog. 2008;4:e1000212. doi: 10.1371/journal.ppat.1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderzanden L., Bray M., Fuller D., Roberts T., Custer D., Spik K., Jahrling P., Huggins J., Schmaljohn A., Schmaljohn C. DNA vaccines expressing either the GP or NP genes of Ebola virus protect mice from lethal challenge. Virology. 1998;246:134–144. doi: 10.1006/viro.1998.9176. [DOI] [PubMed] [Google Scholar]
- Volchkov V.E., Volchkova V.A., Slenczka W., Klenk H.D., Feldmann H. Release of viral glycoproteins during Ebola virus infection. Virology. 1998;245:110–119. doi: 10.1006/viro.1998.9143. [DOI] [PubMed] [Google Scholar]
- Wahl-Jensen V.M., Afanasieva T.A., Seebach J., Stroher U., Feldmann H., Schnittler H.J. Effects of Ebola virus glycoproteins on endothelial cell activation and barrier function. J. Virol. 2005;79:10442–10450. doi: 10.1128/JVI.79.16.10442-10450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warfield K.L., Perkins J.G., Swenson D.L., Deal E.M., Bosio C.M., Aman M.J., Yokoyama W.M., Young H.A., Bavari S. Role of natural killer cells in innate protection against lethal Ebola virus infection. J. Exp. Med. 2004;200:169–179. doi: 10.1084/jem.20032141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warfield K.L., Swenson D.L., Olinger G.G., Kalina W.V., Aman M.J., Bavari S. Ebola virus-like particle-based vaccine protects nonhuman primates against lethal Ebola virus challenge. J. Infect. Dis. 2007;196(Suppl 2):S430–S437. doi: 10.1086/520583. [DOI] [PubMed] [Google Scholar]
- Wauquier N., Becquart P., Gasquet C., Leroy E.M. Immunoglobulin g in Ebola outbreak survivors, Gabon. Emerg. Infect. Dis. 2009;15:1136–1137. doi: 10.3201/eid1507.090402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., Decker J.M., Wang S., Hui H., Kappes J.C., Wu X., Salazar-Gonzalez J.F., Salazar M.G., Kilby J.M., Saag M.S., Komarova N.L., Nowak M.A., Hahn B.H., Kwong P.D., Shaw G.M. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- Wilson J.A., Hevey M., Bakken R., Guest S., Bray M., Schmaljohn A.L., Hart M.K. Epitopes involved in antibody-mediated protection from Ebola virus. Science. 2000;287:1664–1666. doi: 10.1126/science.287.5458.1664. [DOI] [PubMed] [Google Scholar]
- Wool-Lewis R.J., Bates P. Characterization of Ebola virus entry by using pseudotyped viruses: identification of receptor-deficient cell lines. J. Virol. 1998;72:3155–3160. doi: 10.1128/jvi.72.4.3155-3160.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wool-Lewis R.J., Bates P. Endoproteolytic processing of the Ebola virus envelope glycoprotein: cleavage is not required for function. J. Virol. 1999;73:1419–1426. doi: 10.1128/jvi.73.2.1419-1426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Delgado R., Xu L., Todd R.F., Nabel E.G., Sanchez A., Nabel G.J. Distinct cellular interactions of secreted and transmembrane Ebola virus glycoproteins. Science. 1998;279:1034–1037. doi: 10.1126/science.279.5353.1034. [DOI] [PubMed] [Google Scholar]
- Yang Z.Y., Duckers H.J., Sullivan N.J., Sanchez A., Nabel E.G., Nabel G.J. Identification of the Ebola virus glycoprotein as the main viral determinant of vascular cell cytotoxicity and injury. Nat. Med. 2000;6:886–889. doi: 10.1038/78645. [DOI] [PubMed] [Google Scholar]