Abstract
Flavoproteins catalyze a variety of reactions utilizing flavin mononucleotide or flavin adenine dinucleotide as cofactors. The oxidoreductase properties of flavoenzymes implicate them in redox homeostasis, oxidative stress, and various cellular processes, including programmed cell death. Here we explore three critical flavoproteins involved in apoptosis and redox signaling, ie, apoptosis-inducing factor (AIF), proline dehydrogenase, and NADPH oxidase. These proteins have diverse biochemical functions and influence apoptotic signaling by unique mechanisms. The role of AIF in apoptotic signaling is two-fold, with AIF changing intracellular location from the inner mitochondrial membrane space to the nucleus upon exposure of cells to apoptotic stimuli. In the mitochondria, AIF enhances mitochondrial bioenergetics and complex I activity/assembly to help maintain proper cellular redox homeostasis. After translocating to the nucleus, AIF forms a chromatin degrading complex with other proteins, such as cyclophilin A. AIF translocation from the mitochondria to the nucleus is triggered by oxidative stress, implicating AIF as a mitochondrial redox sensor. Proline dehydrogenase is a membrane-associated flavoenzyme in the mitochondrion that catalyzes the rate-limiting step of proline oxidation. Upregulation of proline dehydrogenase by the tumor suppressor, p53, leads to enhanced mitochondrial reactive oxygen species that induce the intrinsic apoptotic pathway. NADPH oxidases are a group of enzymes that generate reactive oxygen species for oxidative stress and signaling purposes. Upon activation, NADPH oxidase 2 generates a burst of superoxide in neutrophils that leads to killing of microbes during phagocytosis. NADPH oxidases also participate in redox signaling that involves hydrogen peroxide-mediated activation of different pathways regulating cell proliferation and cell death. Potential therapeutic strategies for each enzyme are also highlighted.
Keywords: apoptosis, flavoproteins, apoptosis-inducing factor, NADPH oxidase, proline dehydrogenase
Introduction
Apoptosis is a controlled, genetically regulated form of cell death. The term apoptosis (Greek, meaning “falling off”) was coined by Kerr et al in the early 1970s to describe the mechanism of programmed cell death in healthy human tissues.1 Programmed cell death is an evolutionarily conserved form of cell suicide and is crucial in plants and animals.2–5 This review will focus on the role of flavoproteins in programmed cell death and the interrelated extrinsic and intrinsic pathways of apoptosis. The mechanisms by which three different flavoproteins, ie, apoptosis-inducing factor (AIF), proline dehydrogenase, and NADPH oxidase, contribute to apoptosis and oxidative stress are described, along with therapeutic strategies for disease intervention.
Extrinsic and intrinsic pathway of apoptosis
Several comprehensive reviews are available for extrinsic and intrinsic apoptosis.6–8 Here we will only summarize some of the main features of extrinsic and intrinsic apoptosis. The extrinsic pathway of apoptosis is mediated at the cell surface by various death receptors such as Fas, tumor necrosis factor (TNF) receptor 1, death receptor (DR) 3, TNF-related apoptosis-inducing ligand (TRAIL) receptor 1, TRAIL R2, DR6, p75 nerve growth factor receptor, and ectodermal dysplasia receptor. Death receptors are activated by specific ligands, such as the Fas ligand, TNFα, and TRAIL.4,5,9,10 Ligand binding to death receptors transmits signals that result in recruitment of intracellular death domain-binding partners, such as Fas-associated death domain and TNF receptor-associated death domain. The Fas receptor-Fas-associated death domain complex leads to recruitment of specific adaptor proteins to form a larger complex, termed the death-inducing signaling complex, which subsequently activates procaspases 8 and 10.7,11 Activated caspases 8 and 10 then activate downstream caspases 3, 6, and 7.7,10,12 These activated caspases cleave nuclear DNA repair enzymes and other cellular proteins, such as poly(ADP-ribose) polymer (PARP), cytosolic phospholipase A2, actin, and lamin, leading to the formation of apoptotic bodies, chromatin condensation, and DNA fragmentation.5,6,13–15
The intrinsic or mitochondrial apoptotic pathway is primarily initiated in response to cellular oxidative stress or DNA damage. Ultraviolet radiation or DNA-damaging drugs induce the intrinsic pathway via p53-mediated transcriptional upregulation of proapoptotic factors of B cell lymphoma 2 (BCL-2) family members such as BAX, APAF-1, PUMA, and NOXA genes. These proapoptotic factors inhibit the action of antiapoptotic BCL-2 mediators like BCL-xL, BCL-w, and myeloid cell factor-1.10,11 Upregulation of the p53 modulator of apoptosis and other factors, such as dynamin-related protein-1 (a GTPase) and BCL-2 interacting mediator of cell death, promote the oligomerization of BAX (BCL-2-associated X protein)-BAK (BCL-2 homologous antagonist-killer) which leads to formation of pores in the outer mitochondrial membrane.16 These outer mitochondrial permeability transition pores alter the mitochondrial membrane potential and allow release of cytochrome c, AIF, and endonuclease G (a mitochondrial-specific nuclease) from the mitochondria.
The cytochrome c released binds to apoptotic protease activating factor-1 in the cytosol to form a complex called an apoptosome.7 The apoptosome provides a platform for activation of caspase 9, which subsequently activates downstream caspases 3, 6, and 7.17,18 AIF and endonuclease G contribute to apoptosis independently of caspase activity by translocating to the nucleus where they assist in chromatin condensation and DNA fragmentation.19–21 The extrinsic and intrinsic apoptotic pathways also involve interrelated pathways, which in certain cell types amplify the effect of apoptosis-inducing signals.21,22 The mitochondrial apoptotic pathway can be induced by the action of the death-inducing signaling complex which cleaves cytosolic BCL-2-interacting domain (BID) into truncated BID. Truncated BID then translocates into the mitochondria to facilitate BAX-BAK dimerization and mitochondrial permeability transition pore formation.23,24 In addition, calpain, a calcium-dependent cysteine protease, has been shown to cleave BID into truncated BID during induction of the extrinsic apoptotic pathway.25
Apoptosis-inducing factor
The role of AIF in mitochondrial processes and apoptosis has been an interesting area of research since the discovery of AIF in 1999.3 The initial identification of AIF as a flavoprotein suggested that it was a novel mitochondrial redox sensor. Subsequent studies have established that AIF is a proapoptotic bifunctional protein that exhibits distinct functions according to its compartmentalization in either the mitochondria or the cell nucleus. However, the physiological function of AIF in the mitochondria and the mechanisms by which it induces chromatinolysis and cell death are not yet fully known.
Newly synthesized AIF is targeted to the intermembrane space of the mitochondria where flavin adenine dinucleotide (FAD) is incorporated into the folded protein.26 A transmembrane peptide at the N-terminus of AIF anchors it to the inner mitochondrial membrane.27 The mature form of AIF appears to have a stabilizing influence on complex I activity of the respiratory chain.28 In response to apoptotic stimuli such as oxidative stress, AIF is proteolytically cleaved into a soluble form that translocates to the cell nucleus.29 Once in the nucleus, AIF facilitates chromatin condensation and DNA fragmentation, leading to apoptosis.30 Thus, similar to cytochrome c, AIF is a mitochondrial protein that is released into the cytosol and translocated to the nucleus, resulting in apoptosis. However, the mechanisms by which AIF participates in apoptosis are distinct from the intrinsic pathway involving cytochrome c and are caspase-independent. In the following sections, we review the structure and biochemical functions of AIF, translocation mechanisms, and the role of AIF in apoptosis and disease.
AIF structure
Human AIF is encoded as a 67 kDa polypeptide with 613 residues. The precursor protein contains a mitochondrial localization sequence (residues 1–55), a transmembrane region (residues 66–84), an FAD binding domain (residues 128–262 and 401–480), an NADH binding domain (residues 263–400), a nuclear localization sequence (residues 446–452), and a C-terminal domain (residues 481–613), which has a proline-rich motif (residues 543–554) and a proline/glutamate/serine/threonine sequence (residues 528–559).28,30 AIF does not have a canonical DNA binding domain; instead, DNA binding is mediated by positively charged residues from the FAD and C-terminal domains that form favorable electrostatic interactions between AIF and DNA.29 The mature form of AIF (residues 55–613, 57 kDa) is generated by removal of the mitochondrial localization signal peptide and is anchored to the inner mitochondrial membrane via the transmembrane binding peptide.
The structure of NADH-treated AIF with the FAD cofactor in the reduced form was reported, enabling comparison of the oxidized and reduced forms of AIF.28 Oxidized AIF is a monomer, whereas NADH reduction of the FAD cofactor generates an AIF dimer.28 The crystal structure indicates that changes on the surface of AIF occur upon reduction, promoting dimeric interactions. Also, the nuclear localization sequence is less solvent-accessible in the dimeric structure, suggesting translocation into the nucleus would be impeded for the AIF dimer relative to the monomeric form.28,31
Functions of AIF
AIF exhibits distinct functions depending on its subcellular localization in the cell. In the mitochondria, studies have focused on characterizing the enzymatic activity of AIF with NADPH and its contribution to mitochondrial function. The FAD cofactor in AIF can be reduced by both NADH and NADPH, but NADH is the preferred reducing substrate (100-fold preference).32,33 However, the rate of reduction of AIF by NADH is relatively slow in comparison with other NADPH-dependent flavoenzymes.33,34 Upon reduction by NADH, a charge transfer complex is formed between FADH− and bound NAD+. Interestingly, the FADH−-NAD+ charge transfer complex is very stable in air, with reduced AIF exhibiting a high affinity for NAD+ (Kd of 5–80 nM).33
AIF has been reported to generate superoxide, but the sluggish activity of reduced AIF with oxygen argues against AIF having meaningful NADH oxidase activity in vivo.32 Recently, Miseviciene et al tested the reactivity of mouse AIF with different quinones. AIF exhibited NADH:quinone oxidoreductase activity, but the rate was much lower (104-fold lower) than that of other quinone reductases, such as NADPH:quinone oxidoreductase.34 Thus, it was concluded that AIF does not contribute significantly to reduction of quinone drugs in vivo. Whether the quinone reductase activity of AIF is physiologically important is not clear. It was suggested that reactivity of AIF with quinones may interfere with NADH reduction of AIF and the overall redox homeostasis of mitochondrial AIF.34 Clearly, further studies are needed to identify other potential oxidizing substrates of AIF.
Despite not having a clear enzymatic role, it is well established that AIF has a significant influence on mitochondrial function. Studies have shown that AIF deficiency or low expression of AIF generally decreases oxidative phosphorylation, even though AIF is not part of complex I in the electron transport chain.35–38 Changes in AIF expression are strongly correlated with mitochondrial complex I activity, with loss of mitochondrial AIF generally leading to increased oxidative stress. Inactivation of AIF in the early mouse embryo impaired respiratory complex I activity and led to increased cell death.39 Thus, reduction of AIF function leads to lower mitochondrial respiration and altered energy metabolism.
Other results supporting a role for AIF in respiratory activity are from studies of AIF-deficient mice harboring a proviral insertion in the AIF gene, known as the Harlequin mutation.35 Knockout of AIF in mice is embryonically lethal, so Harlequin (Hq) mice are a widely used AIF-deficient animal model used to study the deficiency of AIF. In Hq mice, AIF expression is decreased by 80%–90% relative to wild-type mice, making these mice a useful model for evaluating the physiological role of AIF in complex I function and oxidative phosphorylation.35,40 The Hq mice are characterized by a 50% drop in complex I activity, and show evidence of elevated oxidative stress in the heart, brain, and cerebellum, with increased lipid peroxidation and mitochondrial DNA damage.35,41 In addition, T cells isolated from Hq mice are more sensitive to Fas ligand-induced cell death than are wild-type controls.26,41 Hq mice are also resistant to cell death induced by the PARP activator, which suggests a role for AIF in PARP activation.35 The observation of increased oxidative stress in Hq mice led to the proposal that AIF functions as an antioxidant enzyme.42,43 However, in vitro knockdown studies of AIF did not provide solid evidence for AIF having an antioxidant role.42 AIF-deficient cells were shown to have a dramatic loss of cell tumorigenicity in mice due to the loss of complex I activity.42,44 More studies are needed to understand the molecular details of how AIF influences redox homeostasis and protects complex I activity and mitochondrial bioenergetics.36,45
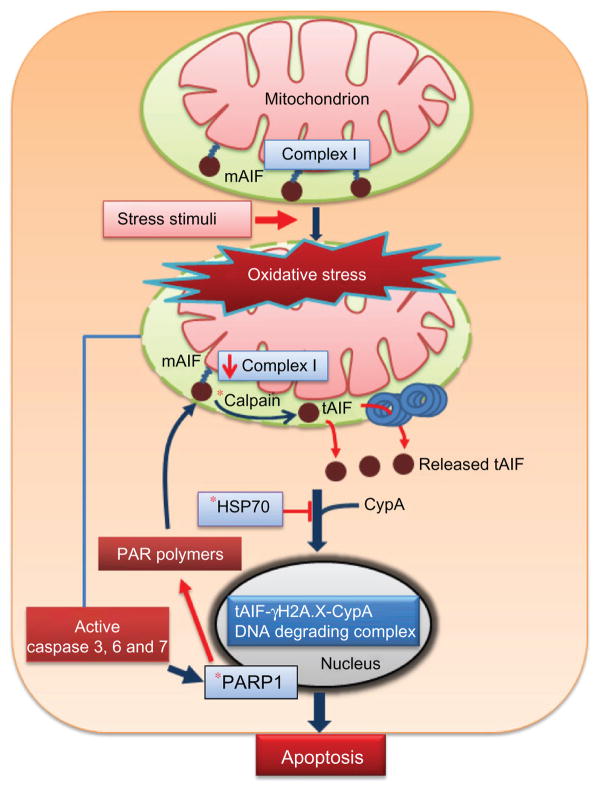
In the nucleus, the function of AIF is to promote chromatin condensation and DNA fragmentation. Thus, attention has focused on understanding the DNA binding properties of AIF and the mechanisms by which it initiates DNA fragmentation and apoptotic events. As mentioned above, AIF does not have a canonical DNA binding domain, but x-ray crystal structures of AIF have revealed positively charged surface areas that are complementary to the negatively charged phosphate backbone of the DNA.29,46 AIF has been shown to bind double-stranded and single-stranded DNA and RNA.47 AIF-DNA binding leads to condensation of the DNA, possibly through cooperative interactions of AIF proteins on the DNA.47 Thus, AIF-DNA interactions alone are sufficient to generate higher-ordered DNA structures, which presumably would facilitate chromatin condensation, a fundamental feature of apoptosis.47 The mechanisms by which AIF contribute to DNA fragmentation in the nucleus are less certain. It appears that AIF lacks nuclease activity, so AIF most likely helps to recruit other proteins to the DNA for chromatin degradation. One such protein is cyclophilin A, which normally catalyzes the cis-trans isomerization of peptidyl-prolyl bonds in target proteins, but has also been reported to have calcium-dependent nuclease activity.48 Although AIF shows no independent DNA nicking activity, the combination of AIF and cyclophilin A caused degradation of plasmid DNA in vitro, suggesting that AIF and cyclophilin A may work synergistically in the nucleus to degrade chromatin.49 A cyclophilin A interaction domain of about 30 amino acids (367–399) in AIF was identified, supporting the formation of an AIF-cyclophilin A complex.49 More recently, it was found that AIF interacts with histone H2AX, a member of the histone H2A family and part of the histone nucleosome core.50 Interactions between AIF and H2AX were suggested to involve the proline-rich motif of AIF.50 It was proposed that AIF, cyclophilin A, and H2AX form a complex in the nucleus that promotes chromatinolysis and programmed cell death. H2AX most likely provides AIF/cyclophilin A with access to the DNA to facilitate degradation of chromatin (see Figure 1).
Figure 1.
AIF and intrinsic pathway of apoptosis. Stress stimuli, such as oxidative stress, induce cleavage of mAIF in the mitochondria to tAIF by calpain. Newly formed tAIF escapes from the mitochondria and interacts with cyclophilin A in the cytosol to facilitate translocation into the nucleus. Alternatively, the chaperone, HSP70, can bind tAIF, resulting in sequestration of tAIF in the cytosol and inhibition of AIF-dependent apoptosis. In the nucleus, tAIF forms a DNA degrading complex with phosphorylated histone, γH2A.X, and cyclophilin A to induce DNA fragmentation and apoptosis. AIF release from the mitochondria is also increased by the action of poly (ADP-ribose) polymerase 1 (PARP-1) and PAR polymers.
Note: *Indicates potential target in the AIF pathway for therapeutic intervention.
Abbreviations: AIF, apoptosis-inducing factor; tAIF, truncated apoptosis-inducing factor; HSP, heat shock protein; mAIF, mature apoptosis-inducing factor.
AIF translocation and redox regulation
The function of AIF is determined by its localization in the cell. Thus, understanding the regulation of AIF localization is critical to understanding the mechanisms by which AIF induces apoptosis. Figure 1 summarizes the overall mechanism of AIF translocation to the nucleus and activation of apoptosis. The mitochondrial location of AIF and its relationship with complex I activity indicates a possible redox mechanism for AIF regulation. Evidence for redox regulation of AIF has been reported from analysis of the AIF oligomeric structure. In the presence of NADH, AIF is primarily a dimer whereas in the absence of NADH, AIF is mainly a monomer.28 Thus, AIF exists in dimer-monomer equilibrium, with NADH reduction of the bound FAD favoring the dimeric form and oxidation driving formation of the AIF monomer.28 AIF localization in the inner mitochondrial membrane is ideal for responding to intracellular redox changes, such as the NADH/NAD+ ratio. Decreased NADH levels associated with oxidative stress and apoptotic insults would be predicted to shift AIF toward the monomeric form.30 The significance of the redox regulation of AIF dimer-monomer equilibrium is that the AIF monomer appears to have increased susceptibility to proteolysis at the N-terminal membrane binding domain. Apoptotic AIF is generated by proteolytic processing of mature AIF upon apoptotic insult, oxidative stress, or mitochondrial membrane rupture. In particular, the Ca2+-dependent cysteine protease, μ-calpain (also known as calpain-I), is thought to have a major role in protease-dependent release of AIF from the mitochondrial inner membrane.3,8,45,51 Treatment of isolated mitochondria with a calpain inhibitor abolished AIF release.26 Thus, oxidative conditions which favor the monomeric form of AIF also lead to increased proteolytic susceptibility and processing.
Newly cleaved AIF breaks out of the mitochondria by traversing BAX/BAK pores in the outer mitochondrial membrane.27 AIF can also escape from the inner mitochondria membrane space via the mitochondrial permeability transition pores. This was shown by inhibition of AIF release by cyclosporin A, a permeability transition pore inhibitor.32 AIF release is also diminished by increased expression of BCL-2, an antiapoptotic protein which prevents formation of the mitochondrial membrane transition pore. Although liberation of AIF from the mitochondria is caspase-independent, it has been suggested that caspases increase mitochondrial permeabilization and thereby facilitate calpain-dependent release of AIF from the mitochondria.50,52 A consequence of AIF proteolysis and release from the inner membrane space is decreased complex I activity and oxidative phosphorylation. These effects eventually lead to increased generation of reactive oxygen species, mitochondrial swelling, and rupture of the outer membrane, resulting in accelerated release of AIF from the mitochondria.30
Upon leaving the mitochondria, AIF translocates to the cell nucleus where it assumes its apoptotic function. AIF has also been proposed to interact with a number of functionally diverse proteins in the cytosol that either inhibit or enhance its translocation to the cell nucleus. For example, heat shock protein (HSP) 70 has been shown to intercept AIF en route to the nucleus, forming an AIF-HSP70 complex and thereby retaining AIF in the cytosol and inhibiting apoptosis.53 Interactions between AIF and cyclophilin A seem to enhance translocation of AIF into the nucleus because AIF fails to induce apoptosis in cyclophilin A knockout cells.49 AIF has also been suggested to interact with phospholipid scramblase (an ATP-dependent phospholipid translocating enzyme), eukaryotic translation initiation factor 3, thioredoxin 2, T-cell ubiquitin ligand, and scythe.26 All of these proteins have been proposed to promote AIF translocation into the nucleus.30 Scythe (HLA-B-associated transcript 3) is a regulator of apoptosis in mammalian development. Inactivation of scythe in mice leads to embryonic or perinatal lethality due to development defects consistent with it having a role in facilitating the apoptotic function of AIF.30 Scythe-deficient cells are less sensitive to endoplasmic reticulum stress-induced apoptosis.54 Desmots et al demonstrated that scythe interacts with AIF in the cytosol in response to stress on the endoplasmic reticulum.54 It was proposed that AIF-scythe interaction functions as a regulatory complex to enhance apoptogenic activity of AIF.54
Another mechanism of AIF translocation relates to parthanatos cell death involving interactions with PARP.55 Activation of PARP-1, a nuclear DNA repair enzyme, leads to increased levels of poly(ADP-ribose) (PAR), which subsequently increase AIF translocation to the nucleus.27,56 PAR have been shown to bind to AIF in the mitochondria, forming an AIF-PAR complex that escapes from the mitochondria and translocates to the nucleus, resulting in chromatin condensation and DNA cleavage during parthanatos.55 Recently, Wang et al identified a PAR binding site in AIF which is distinct from its DNA binding site.57 The functional importance of the PAR binding site was demonstrated by characterization of the triple PAR binding mutant (Pbm)-AIF (R588A, K589A, K592A). Pbm-AIF defective in PAR binding was not translocated to the nucleus by activation of PARP-1 in response to cellular DNA damage.57 The small pool of AIF present in the outer mitochondrial membrane of the rat brain has also been proposed to translocate rapidly to the nucleus in response to PAR-induced parthanatos.58
Implications in disease
A deletion mutation in the AIF gene has been associated with the severe X-linked mitochondrial encephalomyopathy disorder characterized by axonal sensory and motor peripheral neuropathy, and severe muscular atrophy. A deletion of base pairs 601–603 in exon 5 of the human AIF gene results in the loss of the Arg201 codon.31 The AIF Arg201Δ mutant has been found in two patients.31 In the structure of human AIF, Arg201 (Arg 200 in the mouse) was shown to be part of the 191–203 β-hairpin which helps to form the FAD binding pocket and gives conformational stability to the flavoprotein. Arg201 was also observed to form a salt bridge with Glu531. The importance of Arg201 in FAD binding was further demonstrated by characterization of the AIF Arg201Δ mutant protein, which was found to contain less FAD relative to wild-type AIF.31 Thus, the deletion of the Arg201 codon has a clear functional consequence that leads to a less stable AIF protein.
Fibroblasts isolated from patients with AIF Arg201Δ showed partially reduced mitochondrial complex I, III, and IV activity and decreased oxidative phosphorylation.31 These fibroblasts upon exposure to staurosporine, a protein kinase C inhibitor that induces cell death through the mitochondria-mediated pathway, showed significantly higher levels of morphologically altered nuclei than control fibroblasts.31 Electron microscopy of fibroblasts from AIF Arg201Δ patients showed typical features of parthanatos, with high chromatinolysis but without any apoptotic bodies.31 These results suggested that the chromatinolysis activity of AIF is higher in the AIF Arg201Δ mutant relative to wild-type AIF. Treatment with riboflavin, an FAD precursor, partially improved mitochondrial complex I activities and staurosporine-induced cell death in AIF mutant fibroblasts.31 In addition, prolonged riboflavin supplementation was reported to ameliorate the clinical status of an AIF Arg201Δ patient.31 Further studies are needed to test and evaluate the prevalence of this mutation in other populations and the efficacy of riboflavin supplementation.
Another human disease related to AIF is familial autosomal dominant optic atrophy caused by mutations in the OPA1 gene which encodes optic atrophy 1 (OPA1), a dynamin GTPase. Dominant optic atrophy is characterized by blindness due to the loss of retinal ganglion cells, leading to optic neuropathy.59 Zanna et al showed OPA1 improved activity of mitochondrial complex enzymes by direct interactions with mitochondrial complex proteins, including AIF.59 Future studies are required to understand how AIF-OPA1 interactions influence mitochondrial complex 1 activity.
Recent evidence suggests an important apoptotic role for AIF in intestinal epithelial cells exposed to pathogenic bacteria. Infecting adenocarcinoma cells with Helicobacter pylori or human epithelial cells with Acinetobacter baumannii and Pseudomonas aeruginosa induces apoptosis and has been shown to involve AIF.60–63 AIF has also been implicated in intestinal epithelial apoptotic cell death upon exposure of cells to Citrobacter rodentium and enteropathogenic Escherichia coli.63,64 Knockdown of AIF in HeLa cells by siRNA blocked apoptosis induced by enteropathogenic E. coli, providing further evidence for an important apoptotic role of AIF in bacterial infections in the gut.63
Therapeutic outlook
Studies investigating the therapeutic potential of AIF have pursued strategies for either blocking or increasing AIF apoptotic activity. One approach for interfering with AIF apoptosis involves HSP70. As noted above, HSP70 is a cytosolic binding partner of AIF and acts to sequester AIF in the cytosol, blocking the apoptotic activity of AIF. Apoptosis and gastric lesions in the small intestine induced by the nonsteroidal anti-inflammatory drug, indomethacin, were significantly decreased in transgenic mice overexpressing HSP70.65 These transgenic mice were also protected against neonatal hypoxicischemic brain injury due to increased sequestering of AIF in the cytosol and therefore lower AIF-dependent apoptosis.66 Mice lacking HSP70 show increased apoptosis in the heart after ischemia-reperfusion, providing additional evidence for the regulation of AIF apoptosis by HSP70.67 An HSP70 gene polymorphism has been shown to be associated with schizophrenia,68 but whether the underlying mechanisms involved alter AIF apoptotic activity remains unknown. Studies of AIF binding to the HSP70 mutant would provide important insights into this question.
Strategies that prevent AIF release from the mitochondria have also been pursued. Calpain inhibitors, such as MDL28170, PDI150606, AK275, and AKT295, have been shown to block AIF release in rat models of ischemia, Parkinson’s disease, and retinitis pigmentosa, suggesting a new therapeutic approach.27 INO1-1001, a PARP-1 inhibitor, improves the function of the perfused heart in diabetic rat models. Inhibition of PARP-1 is predicted to decrease AIF release from the mitochondria and apoptosis. Thus, drugs that target PARP-1 block AIF apoptosis which would be a potential therapeutic benefit in the treatment of myocardial ischemia.27,69
Approaches to activate AIF apoptotic function have been of primary interest in cancer research. Overexpression of AIF in gastric cancer, hepatocellular carcinoma cells, and osteosarcoma cells resulted in increased drug-induced apoptosis.70 Thus, various therapeutic methods to increase AIF apoptotic activity have been explored. Drugs that target AIF release and apoptosis induction in cancer have recently been reviewed.27 Here, we will discuss just a few of the approaches.
One study sought to diminish the sequestering of AIF in the cytosol by disrupting HSP70-AIF interactions. In a colon cancer model, rats injected with the synthetic AIF peptide (150–228 residues) exhibited an improved response with cisplatin-induced apoptosis.55,71 Similar results were obtained in mouse melanoma cells treated with the AIF peptide, with no harmful effect on the normal cell population.70,71 These results show that targeting HSP-AIF interactions may be a promising method for sensitizing cancer cells to chemotherapy. Atiprimod (also known as azaspirane), a drug studied for treatment of multiple myeloma, was shown to trigger apoptosis in mantle cell lymphoma by a caspase-independent pathway involving AIF.72,73 Atriprimod activates c-Jun N-terminal kinase and upregulates Bax, Bak, and phosphorylated Bcl2, resulting in elevated AIF release from the mitochondria.73 Sulindac, a nonsteroidal anti-inflammatory drug widely used to treat colon cancer, has also been shown to act mainly by activating AIF-mediated cell death in colon cancer cells (HT29 cells) which have dysfunctional p53.70,74
Although there has been significant progress in our understanding of AIF in the short time since its discovery, significant gaps remain in our knowledge of AIF. Future studies need to explore physiological redox partners for AIF, determine the mechanisms by which AIF enhances complex I activity, and identify other accessory proteins involved in the translocation and apoptotic function of AIF. Tackling these challenging studies may lead to improved treatment of neurodegenerative diseases and cancer.
Proline dehydrogenase
Human proline dehydrogenase, also known as proline oxidase, is an inner mitochondrial membrane flavoenzyme encoded by the PRODH1 gene located on chromosome 22q11.2. Proline dehydrogenase catalyzes the first step in proline catabolism by oxidizing proline to Δ1-pyrroline 5-carboxylate (P5C) which is then hydrolyzed nonenzymatically to γ-glutamate semialdehyde.75,76 γ-Glutamate semialdehyde is next converted to glutamate in an NAD+-dependent step by the enzyme P5C dehydrogenase.75,77 P5C dehydrogenase is also localized in the mitochondria, thus, the conversion of proline to glutamate occurs exclusively in the mitochondrion.78
A kinetic study of purified human proline dehydrogenase has not yet been performed, but significant insights have been gained from the related proline utilization A (PutA) enzyme in Gram-negative bacteria and proline dehydrogenase from Thermus thermophilus (TtPRODH).79 PutA is a bifunctional enzyme which combines proline dehydrogenase and P5C dehydrogenase domains in the same polypeptide.79 The proline dehydrogenase domain in PutA from E. coli and TtPRODH share high sequence conservation in the active site with human proline dehydrogenase, thus providing important insights into the mechanism and structure of human proline dehydrogenase.80 The proline dehydrogenase domain of PutA (PutA/PRODH) and TtPRODH is a (βα)8 barrel core structure that noncovalently binds the FAD cofactor.79 The (βα)8 barrel core is predicted to be similar to the catalytic core of human proline dehydrogenase.81 Residues that are critical for proline binding in E. coli PutA, such as Arg555 and Arg556, are conserved in human proline dehydrogenase (Arg479 and Arg480).81 The proline dehydrogenase reaction can be separated into reductive and oxidative half-reactions. In the reductive half-reaction, proline dehydrogenase catalyzes the two- electron oxidation of proline, forming FADH2 and P5C.82 The reductive half-reaction has been proposed to occur by a hydride transfer mechanism from proline to the FAD bound in E. coli PutA/proline dehydrogenase.83 In the oxidative half-reaction, PutA/proline dehydrogenase catalyzes the electron transfer from reduced FAD to membrane-bound ubiquinone.82 The kinetic mechanism of the proline:ubiquinone oxidoreductase reaction was recently shown to occur for PutA/proline dehydrogenase via a ping-pong mechanism with proline and ubiquinone binding at two distinct sites.82 Human proline dehydrogenase may use a similar mechanism for transferring reducing equivalents from FADH2 to ubiquinone in the mitochondrial electron transport chain.77
Apoptosis and signaling
PRODH1 is a p53-inducible gene (PIG6) that has a crucial role in apoptosis.84 The mechanisms involving proline dehydrogenase and induction of apoptosis are illustrated in Figure 2. Overexpression or upregulation of proline dehydrogenase leads to increased proline oxidation in the mitochondria and superoxide production.85 In colon cancer cells expressing wild-type p53, proline dehydrogenase expression was induced by adriamycin, whereas in cells that lacked a functional p53, proline dehydrogenase expression was unresponsive to adriamycin.86–90 Thus, cytotoxic drugs such as adriamycin that cause DNA damage and apoptosis induce proline dehydrogenase expression via p53.
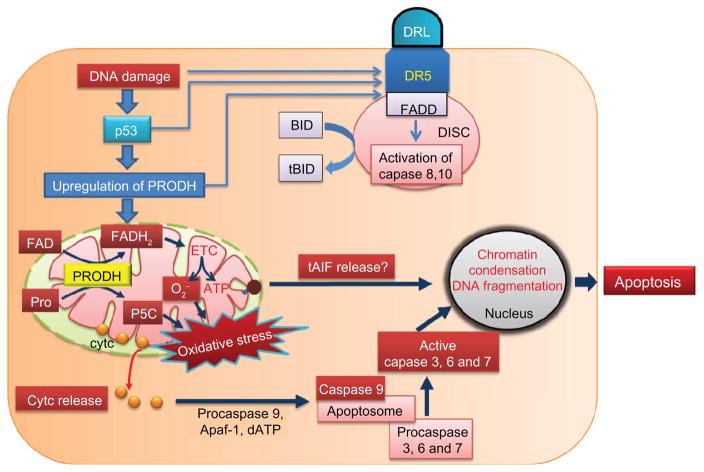
Figure 2.
Proline dehydrogenase and classical apoptotic pathway. DNA damage induces both the extrinsic and intrinsic pathway of apoptosis by increasing the expression of death receptors and p53, respectively. Proline dehydrogenase in the mitochondria is upregulated by p53. Oxidation of proline by proline dehydrogenase leads to production of pyrroline-5-carboxylate (P5C) and FADH2. Electrons from FADH2 are then transferred to the electron transport chain to produce ATP for survival or superoxide (O2−) and oxidative stress in the mitochondria. Oxidative stress generated by proline dehydrogenase induces the release of cytochrome c from the mitochondria, resulting in the formation of an apoptosome involving activated caspase 9, which then activates downstream caspases 3, 6, and 7 to induce apoptosis. It is not known whether proline dehydrogenase also induces mitochondrial release of AIF.
Abbreviations: PRODH, proline dehydrogenase; AIF, apoptosis inducing factor; cyt c, cytochrome c; FAD, flavin adenine dinucleotide; FADD, Fas-associated death domain; ETC, electron transport chain; BID, BCL-2 interacting domain; tBID, truncated BCL-2 interacting domain.
The mechanism by which proline dehydrogenase contributes to apoptosis has been carefully studied by Phang et al.75,77,85,87,91–95 Upregulation of proline dehydrogenase expression by p53 results in increased reactive oxygen species levels which initiate the intrinsic and extrinsic apoptotic cell death pathways.92 Intrinsic apoptosis is generally characterized by elevated levels of mitochondrial reactive oxygen species resulting in mitochondrial membrane damage, cytochrome c and AIF release, and activation of caspase 9.90 Consistent with intrinsic apoptosis, increased proline dehydrogenase expression causes cytochrome c release and caspase 9 activation.95 Addition of proline to proline dehydrogenase-overexpressing cell lines further augments reactive oxygen species-mediated cytochrome c release and caspase 9 activation.95 Proline dehydrogenase-induced apoptosis is inhibited in cells overexpressing manganese superoxide dismutase, consistent with an important role for reactive oxygen species in proline dehydrogenase-mediated apoptosis.96 In addition, P5C generated by proline dehydrogenase has been shown to promote apoptosis in bladder carcinoma cell lines and inhibit cell proliferation.86 Thus, the contribution of proline dehydrogenase to apoptosis may be two-fold, involving reactive oxygen species and the intrinsic apoptotic pathway, as well as P5C. Whether proline dehydrogenase overexpression causes release of AIF has yet to be determined.
In addition to the mitochondrial apoptotic pathway, evidence has been shown for proline dehydrogenase inducing the extrinsic cell death pathway. Liu et al showed proline dehydrogenase upregulates TRAIL and DR5 at the mRNA and protein levels.92 Also, inhibition of caspase 8 resulted in decreased apoptosis by proline dehydrogenase overexpression.92 The mechanism of TRAIL upregulation by proline dehydrogenase was shown to be mediated by nuclear factor activated T cells (NFAT).92 Increased cellular calcium levels are sensed by calmodulin, leading to activation of calcineurin, a Ser/Thr phosphatase known to activate NFAT by dephosphorylation. Activated NFAT then translocates from the cytosol to the nucleus to induce TRAIL expression and apoptosis.92 Increased reactive oxygen species are well known to affect intracellular calcium homeostasis. Proline dehydrogenase-generated reactive oxygen species were also suggested to mediate the Ca2+/calcineurin-NFAT cascade to activate apoptotic pathways.92 Peroxisome proliferator-activated receptor γ (PPARγ) ligand has been shown to induce the extrinsic apoptotic pathway in a variety of cancer cells.97,98 The mechanism by which the PPARγ ligand, troglitazone, induces apoptosis in colon cancer cell lines involves the upregulation of proline dehydrogenase and increased generation of reactive oxygen species.75,87 Triglitazone-induced proline dehydrogenase expression was absent in HCT cells lacking functional p53 and PPARγ, further indicating a role for PPARγ and p53 in the regulation of proline dehydrogenase expression.87
Proline dehydrogenase and stress
Proline oxidation plays an important role in cellular redox control that impacts cell survival and cancer cell growth.75 Earlier work by Phang et al showed that the enzyme actions of proline dehydrogenase and the cytosolic NADP+-dependent P5C reductase (P5CR) form a proline-P5C cycle that effectively shuttles reducing equivalents from the cytosol into the mitochondria.75,77 The proline-P5C cycle helps drive the pentose phosphate pathway by replenishing NADP+ levels during nutrient stress.87 Although the proline-P5C cycle seems to have a critical role in cellular redox homeostasis, it is not known yet whether mitochondria have a specific transporter for P5C in mammals.99
Proline has also been shown to serve as a microenvironmental stress substrate via proline dehydrogenase to generate ATP for cell survival under poor nutrient conditions (ie, low glucose).77,88 Proline dehydrogenase is also induced by AMP-activated protein kinase (AMPK), a central metabolic sensor kinase and an upstream inhibitor of the mammalian target of rapamycin complex 1 activity.85,88,100 During nutrient stress conditions, decreased cellular ATP levels and increased AMP levels act as an allosteric activator of AMPK by binding to one of the subunits of AMPK.88,101 Recently, it was shown that glucose deprivation results in increased activity of matrix metalloproteinases 2 and 9, leading to increased intracellular proline levels, higher proline dehydrogenase expression, and increased flux of the pentose phosphate pathway.88 This study also suggested that the proline oxidation pathway generates glutamate and α-ketoglutarate, which can have an anaplerotic role in the tricarboxylic acid cycle.88
We have previously shown that proline supplementation protects mammalian cells against various oxidative stress agents and carcinogen-induced apoptosis.102 In addition, over-expression of proline dehydrogenase decreases intracellular proline levels and cell survival upon exposure to oxidative stress induced by hydrogen peroxide.102 The mechanism by which proline protects against oxidative stress is unclear, but our unpublished data indicate that proline dehydrogenase is essential, consistent with the proline-P5C cycle having a significant impact on the intracellular redox state in mammalian cells. Thus, proline dehydrogenase expression levels appear to be pivotal in determining whether proline oxidation has a cell survival role or initiates apoptotic programmed cell death.
Therapeutic outlook
Targeting proline metabolism for potential therapeutic applications seems promising in cancers, schizophrenia, and certain gastrointestinal infections. Proline dehydrogenase expression is downregulated in many cancer tissues such as renal, colon, rectum, stomach, liver, and pancreas.93 Thus, proline dehydrogenase has been suggested to be a novel mitochondrial tumor suppressor and a potential target for cancer therapy. Recently, ectopic overexpression of miR-23b* in renal cancer cell lines was shown to decrease proline dehydrogenase expression.103 In miR23b* knockout cells, proline dehydrogenase expression was elevated relative to control cells. It was also confirmed that miR23b* binds to the 3′-untranslated region of proline dehydrogenase mRNA.103 Thus, it was suggested that inhibiting miR23b* could be a potential strategy for upregulating proline dehydrogenase in renal cancer cells.103 Additional studies addressing the upregulation of proline dehydrogenase as a potential cancer therapy are needed.
Inborn errors of proline dehydrogenase are manifested in type I hyperprolinemia and neurological disorders such as schizophrenia and increased incidence of seizures.104 The PRODH1 gene on chromosome 22q11 is located near the centromeric end of the region which is deleted in Velocardiofacial syndrome/DiGeorge syndrome. Jacquet et al identified a complete deletion of the PRODH1 gene locus in a subset of schizophrenic patients with hyperprolinemia.105,106 Another study showed microdeletion of PRODH1 is strongly associated with schizophrenia and 22q11 DiGeorge syndrome.107 Missense mutations in the PRODH1 gene found in patients with type I hyperprolinemia were characterized and shown to result in mild to severe reduction in proline dehydrogenase activity. Mutations associated with schizophrenia were found to have a severe reduction in proline dehydrogenase activity indicating that dysfunctional proline dehydrogenase may be a risk factor in schizophrenia.108 The mechanism by which aberrant proline dehydrogenase and proline metabolism possibly contributes to mental disorders is not known. Novel therapeutics aimed at maintaining normal proline levels in the central nervous system may be a worthwhile strategy for treating certain cases of schizophrenia.
Proline levels in the gastric juice of patients infected with H. pylori were reported to be 10-fold higher than in control subjects.109,110 Proline has been shown to be a preferred respiratory substrate of H. pylori in the gut.109 We and others have shown that knocking out the putA gene in Helicobacter significantly reduces infection rates and inflammation of the liver in mice relative to wild-type Helicobacter strains, indicating that PutA and proline metabolism is important for virulence.110,111 Thus, inhibiting PutA/proline dehydrogenase with small molecules such as tetrahydro-2-furoic acid, a competitive inhibitor of proline dehydrogenase, may be beneficial for treating infections of H. pylori, the causative agent of peptic ulcers.
NADPH oxidase
NADPH oxidase enzymes share a catalytic core comprising a FAD cofactor, two heme cofactors, and a transmembrane domain anchor.112 In the reaction catalyzed by NADPH oxidase, a hydride is transferred from NADPH to FAD followed by single electron transfer steps from the reduced FAD to the hemes, where molecular oxygen is reduced to superoxide. The overall electron transfer pathway involving NADPH (Em = −320 mV), FAD (Eox/sq = −256 mV, Esq/red = −304 mV),113 heme (Em = −245 mV),114 and the dioxygen/superoxide couple (Em = −160 mV) isthermodynamically favored. The transmembrane domain orients NADPH oxidase to enable electron flow across the membrane, with NADPH oxidation occurring on one side of the membrane and reduction of dioxygen occurring on the opposite side. Thus, electrons are effectively shuttled from cytosolic NADPH to molecular oxygen in intracellular or extracellular compartments. The physiological function of NADPH oxidase was originally considered only to be the phagocyte respiratory burst to kill invading microorganisms. With the discovery of several NADPH oxidase isoforms, additional cellular functions beyond host defense have now emerged.
NADPH oxidase subunits and regulation
The NADPH oxidase family of enzymes is comprised of seven isoforms (NADPH oxidase 1–5 and Duox1 and 2) which all share a redox catalytic core and a transmembrane binding domain. Previous reviews have provided excellent summaries of the structural features and regulation of the different NADPH oxidase isoforms.112,115–117 Here we will only provide a brief summary.
Nox2 (also known as gp91phox) is the isoform of NADPH oxidase expressed in white blood cells and is responsible for the respiratory burst associated with the phagocytic killing of microbes. Nox2 forms a heterodimer with p22phox, a membrane-associated subunit, to form a complex known as flavocytochrome b558.118 Regulatory subunits involved in activating Nox2/p22phox are p47phox, p67phox, p40phox, and Rac1/Rac2.118,119 Upon cell activation, these regulatory subunits assemble with Nox2/p22phox to form an active NADPH oxidase complex. The formation of an active Nox2 complex is GTP-dependent via Rac1/Rac2 and is upregulated by phosphorylation of cytosolic factors, p47phox and p40phox.116 Phosphorylated p47phox then binds the p67phox subunit to provide a binding site for GTP-Rac which then results in activation of Nox2 enzyme.112,120–122
Nox1 forms a heterodimer with p22phox and is activated by regulatory subunits Noxo1 (NADPH oxidase organizer 1, analogous to p47phox), Noxa1 (NADPH oxidase activator 1, homolog of p67phox), and GTP-Rac1.116 Nox1 and Nox2 function in host defense by protecting against invasive pathogenic bacteria in the colonocytes.123 Nox3 activity relies on p22phox and is primarily regulated by Noxa1. Nox4 also requires p22phox but appears to be active without any regulatory subunits. Thus, it has been suggested that Nox4/p22phox is constitutively active. Nox4 has been confirmed to have a role in mitochondrial dysfunction and cardiac damage during pressure overload.124 Recently, a role for Nox4 in DNA damage induced by conditional overexpression of oncogenic H-Ras in primary human thyrocytes was reported.125
Nox5 is distinguished by four EF-hand motifs in the N-terminus on the intracellular side of the membrane that allow regulation of NADPH oxidase activity by calcium. Duox 1 and 2 have two EF-hand motifs and an N-terminal extracellular peroxidase homology domain. Similar to Nox5, the EF-hand motifs of Duox 1 and 2 are on the intracellular membrane side, allowing for activation by calcium binding.112,116,126 The function of the peroxidase domain is not fully known because residues important for peroxidase activity are lacking. However, the peroxidase domain has been proposed to have superoxide dismutase activity which would facilitate the conversion of superoxide into hydrogen peroxide, the substrate for thyroperoxidase, which is essential for hormone synthesis.127
Apoptosis and signaling
NADPH oxidase isoforms have been shown to have beneficial and detrimental effects, depending on the type and amount of reactive oxygen species generated. NADPH oxidase enzymes can differentially modulate the intracellular redox state and impact cell signaling pathways that promote cell survival or apoptosis. Regulated reactive oxygen species generation is necessary for redox signaling, while prolonged reactive oxygen species generation favors apoptotic pathways.117 Figure 3 highlights how reactive oxygen species generated by the activation of NADPH oxidase lead to apoptosis. Several studies using different mammalian cell types have shown that NADPH oxidase activity inactivates protein tyrosine phosphatase.116,117,126 Downregulation of protein tyrosine phosphatase results in activation of the receptor tyrosine kinase and downstream pathways, such as extracellular signal-regulated kinase/mitogen activated protein kinase, c-Jun N-terminal kinase, and phosphatidylinositol 3-kinase/Akt (a serine/threonine protein kinase), which regulate cell survival and apoptosis in response to stress.116 Thus, NADPH oxidase-generated reactive oxygen species, presumably H2O2, can serve as a signaling molecule that mediates cell adaptation to stress and life/death outcomes.
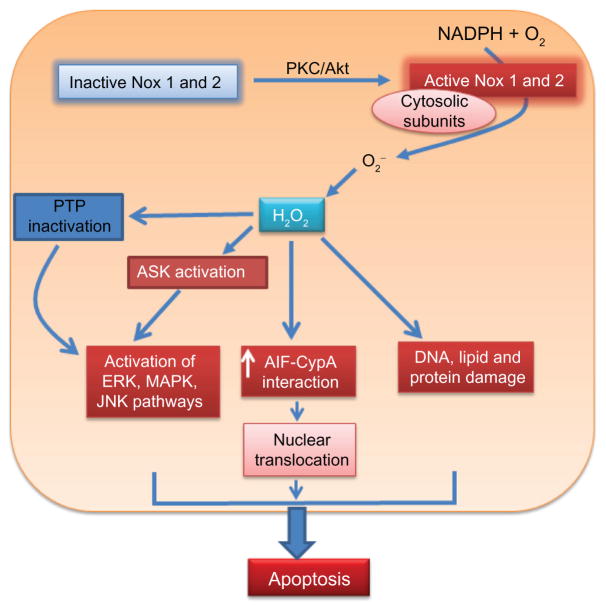
Figure 3.
NADPH oxidase and apoptotic signaling. Protein kinase C/Akt can activate Nox1 and Nox2 by phosphorylation of cytosolic subunits such as p47phox. Activated Nox1 and Nox2 produce superoxide radical (O2−) which dismutates to hydrogen peroxide (H2O2). NADPH oxidase-generated H2O2 acts as a signaling molecule that affects signaling pathways leading to apoptosis. H2O2/reactive oxygen species generated by NADPH oxidase inactivates protein tyrosine phosphatase, which indirectly activates the ERK, MAPK, and c-Jun N-terminal kinase pathways of apoptosis. NADPH oxidase-generated H2O2 can also activate apoptotic signal-regulating kinases, increase AIF-CypA interactions, and increase protein and DNA damage resulting in apoptosis.
Abbreviations: Nox, NADPH oxidase; AIF, apoptosis-inducing factor; PTP, protein tyrosine phosphatase; CypA, cyclophilin A; MAPK, mitogen-activated protein kinase; ERK, extracellular signal-regulated kinase; ASK, apoptosis signal-regulating kinase; JNK, c-Jun N-terminal kinase.
NADPH oxidase-generated reactive oxygen species have also been shown to stimulate the secretion of cyclophilin A (a main player in AIF-DNA binding complex) resulting in activation of extracellular signal-regulated kinase/mitogen activated protein kinase.128 NADPH oxidase-generated reactive oxygen species also upregulate AIF gene expression during cerulein-induced apoptosis in rat pancreatic acinar cells.129 Nox1 has been shown to facilitate formation of the TNFα-TNF receptor type 1 complex which includes associated death domain protein, receptor interacting protein, and Rac, resulting in c-Jun N-terminal kinase-mediated apoptosis.112,130 Nox4 has also been shown to be involved in TNF-α mediated apoptosis in cerebral vascular endothelial cells.131 Nox2 in macrophages and Nox4 in astrocytes result in reactive oxygen species which activates apoptosis by activating the apoptosis signal-regulating kinase 1-p38 mitogen activated protein kinas/c-Jun N-terminal kinase pathway leading to apoptosis.117,132 Recently, DR4 and DR5 have been shown to activate Nox1 directly, independent of death-inducing signaling complex-mediated apoptosis. Here, NADPH oxidase-generated superoxide was responsible for the sustained activation of c-Jun N-terminal kinase-dependent apoptosis.133,134
NADPH oxidase and disease
Involvement of NADPH oxidase in the pathogenesis of several diseases is well established. The most familiar case is Nox2 (gp91phox) deficiency, which causes chronic granulomatous disease, a primary immunodef iciency affecting phagocytosis of the innate immune system.135,136 Patients with chronic granulomatous disease are more susceptible to certain types of bacterial and fungal infections due to dysfunctional phagocytes that lack proper NADPH-dependent superoxide formation.136 Mice def icient in Nox2 develop symptoms of granulomatous disease as well, but also display a second phenotype of balance order dysfunction and defective otoconia formation.135 This suggests that Nox2 is involved in otoconium formation, a structure in the utricle of the inner ear, which helps in sensing gravity and linear acceleration.135
Evidence for NADPH oxidase-derived reactive oxygen species contributing to the progression of cardiovascular disease,137 pulmonary diseases, lung cancer,138 human atherosclerosis,139 myocardial angiogenesis in ischemic heart disease,140 and angiotensin II-initiated apoptosis of neonatal cardiomyocytes141 have been reported. In humans, atherosclerosis is associated with increased Nox1 and Nox2 expression.142 Atherogenic particles such as oxidized low-density lipoprotein have been shown to induce NADPH oxidase-dependent endothelial cell activation and injury which is the underlying cause of atherosclerotic plaque formation.143,144 Similar to oxidized low-density lipoprotein, advanced glycation end products can induce NADPH oxidase activity, leading to increased reactive oxygen species and induction of apoptosis in a caspase 3-dependent manner during diabetic retinopathy.133,134,145 In addition, the advanced glycation end product precursor, 3-deoxyglucosone, induces stress in the endoplasmic reticulum and apoptosis in dermal fibroblasts through activation of Nox4.146
Therapeutic outlook
Recent comprehensive reviews have discussed the inhibition of NADPH oxidase as a therapeutic target for cardiovascular disease, diabetes, fibrosis, hyperlipidemia, Alzheimer’s disease, and β-cell dysfunction.116,147–151 Here, we will provide some of the recent advances and perspectives.
Reactive oxygen species derived from Nox1 were shown to activate murine hepatic stellate cells.152,153 Activated hepatic stellate cells produce extracellular matrix proteins which lead to liver fibrosis.154 Recently, it was shown that Nox1 knockout mice are less susceptible to liver injury and fibrosis.155 The mechanism of liver injury and fibrosis associated with Nox1 was proposed to involve reactive oxygen species inactivation of phosphatase and tensin homolog (PTEN). Inactivation of PTEN leads to enhanced signaling of pathways involving the Akt/Fork head family of transcription factor Class O (Foxo4) to induce cell proliferation.155 NADPH oxidase enzyme activity has also been reported to lead to activation of pancreatic stellate cells and subsequent pancreatic fibrosis and inflammation in mice.156 Thus, targeting Nox1 may have therapeutic benefits for treatment of chronic liver and pancreatic fibrosis.
In contrast with apoptosis, Nox4 appears to have an important role in cancer cell growth and survival which can influence disease progression. Upregulation of Nox4 activity by overexpression of Akt in melanoma cells leads to increased cancer metastasis.157 Reactive oxygen species generated by Nox4 inhibits apoptosis of pancreatic cancer cells.158 Downregulation of Nox4 by AMPK was shown in a mouse model of type I diabetes, suggesting that low Nox4 activity may be a contributing factor in diabetes.159 The prosurvival role of NADPH oxidase has also been documented in prostate cancer cells160 and ovarian cancer cells.161 Thus, inhibition of the specific NADPH oxidase isoform would be a potential therapeutic strategy for the treatment of cancer.
Inhibition of NADPH oxidase in animal models was demonstrated with diphenyl iodonium, an inhibitor that disrupts the physiological oxidative half-reaction by acting as an alternative electron acceptor for reduced FAD. However, diphenyl iodonium is a nonspecific inhibitor of NADPH oxidase because it also inhibits several other flavoproteins, including mitochondrial complexes I and III, cytochrome P450 oxidoreductase, xanthine oxidase, and nitric oxide synthase.147,162,163 Another inhibitor of NADPH oxidase is the natural organic compound, apocynin, which can decrease superoxide production by Nox2 in phagocytes. Apocynin inhibits the assembly of activated Nox2 by interfering with interactions between the Nox2/p22phox heterodimer and the regulatory subunits, p47phox and p67phox.164 However, using apocynin as an NADPH oxidase inhibitor in nonphagocyte cell types is somewhat controversial. Apocynin has been reported to function as an antioxidant in vascular smooth muscle cells by forming an apocynin radical which then scavenges reactive oxygen species.165 In contrast, another group reported that apocynin helps generate reactive oxygen species in nonphagocytic cell types.166
One strategy that was pursued for specific inhibition of Nox1 and 2 was targeting p22phox interactions with the regulatory subunit p47phox which is essential for the activation of Nox1 and Nox2.118,121,167 Targeting the activation of Nox1 and 2 may be therapeutically useful for the diseases in which Nox1 and 2 activation have pathological consequences. Thus, synthetic peptides designed to interfere with p22phox–p47phox interactions have been tested. Synthetic peptides containing the p47phox interacting domain of Nox2 conjugated to the HIV transactivator transcription protein were shown to decrease reactive oxygen species generation significantly in vascular smooth muscle cells and attenuate systolic blood pressure in a mouse model of hypertension.168 Although peptide inhibitors of NADPH oxidase show promising effects in animal models, oral bioavailability and immune response over long-term use remains an open question.
New potent compounds developed by pharmaceutical companies such as pyrazolopyridines (GKT-136901 and GKT137891)169 and triazolopyrimidines (VAS2870 and VAS3947) have been reported to inhibit NADPH oxidase, with IC50 values below the μM range, and are bioavailable through oral administration.147,169 These compounds are proposed to be competitive inhibitors of NADPH oxidase with respect to NADPH. However, the above compounds appear to lack NADPH oxidase isoform specificity.151 Further studies are required to elucidate the clinical efficacy and safety of these NADPH oxidase inhibitors.
Conclusion
The three flavoproteins discussed here are involved in different pathways of apoptosis. Various approaches for manipulating AIF, proline dehydrogenase, and NADPH oxidase activity have been pursued in efforts to enhance or diminish apoptosis in the context of different diseases. Strategies for developing therapeutics involving AIF, proline dehydrogenase, and NADPH oxidase need to consider the multifaceted and complex roles of these enzymes in different cell types. Upregulation of AIF and proline dehydrogenase apoptotic mechanisms may be useful for developing novel adjuvant therapies in cancer treatment. Interestingly, blocking human AIF in combination with small molecule inhibitors of PutA/proline dehydrogenase may also be of therapeutic use in fighting intestinal pathogenic bacterial colonization/infections. The wide ranging physiological roles of NADPH oxidase provide unique opportunities for targeting specific NADPH oxidase isoforms in the treatment of several diseases, in particular cardiovascular disease, diabetes, and Alzheimer’s disease.
Acknowledgments
This work was supported in part by a grant from the National Institutes of Health (GM079393).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
This is an Open Access article which permits unrestricted noncommercial use, provided the original work is properly cited.
References
- 1.Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26(4):239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thompson CB. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267(5203):1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- 3.Susin SA, Lorenzo HK, Zamzami N, et al. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397(6718):441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- 4.Earnshaw WC, Martins LM, Kaufmann SH. Mammalian caspases: structure, activation, substrates, and functions during apoptosis. Annu Rev Biochem. 1999;68:383–424. doi: 10.1146/annurev.biochem.68.1.383. [DOI] [PubMed] [Google Scholar]
- 5.Ashkenazi A. Targeting death and decoy receptors of the tumournecrosis factor superfamily. Nat Rev Cancer. 2002;2(6):420–430. doi: 10.1038/nrc821. [DOI] [PubMed] [Google Scholar]
- 6.Nagata S. Fas ligand-induced apoptosis. Annu Rev Genet. 1999;33:29–55. doi: 10.1146/annurev.genet.33.1.29. [DOI] [PubMed] [Google Scholar]
- 7.Jiang X, Wang X. Cytochrome C-mediated apoptosis. Annu Rev Biochem. 2004;73:87–106. doi: 10.1146/annurev.biochem.73.011303.073706. [DOI] [PubMed] [Google Scholar]
- 8.Lorenzo HK, Susin SA. Therapeutic potential of AIF-mediated caspase-independent programmed cell death. Drug Resist Updat. 2007;10(6):235–255. doi: 10.1016/j.drup.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Solary E, Droin N, Bettaieb A, Corcos L, Dimanche-Boitrel M, Garrido C. Positive and negative regulation of apoptotic pathways by cytotoxic agents in hematological malignancies. Leukemia. 2000;14(10):1833–1849. doi: 10.1038/sj.leu.2401902. [DOI] [PubMed] [Google Scholar]
- 10.Zhang X, Vallabhaneni R, Loughran PA, et al. Changes in FADD levels, distribution, and phosphorylation in TNF alpha-induced apoptosis in hepatocytes is caspase-3, caspase-8 and BID dependent. Apoptosis. 2008;13(8):983–992. doi: 10.1007/s10495-008-0228-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas LR, Stillman DJ, Thorburn A. Regulation of Fas-associated death domain interactions by the death effector domain identified by a modified reverse two-hybrid screen. J Biol Chem. 2002;277(37):34343–34348. doi: 10.1074/jbc.M204169200. [DOI] [PubMed] [Google Scholar]
- 12.Wiezorek J, Holland P, Graves J. Death receptor agonists as a targeted therapy for cancer. Clin Cancer Res. 2010;16(6):1701–1708. doi: 10.1158/1078-0432.CCR-09-1692. [DOI] [PubMed] [Google Scholar]
- 13.Fan TJ, Han LH, Cong RS, Liang J. Caspase family proteases and apoptosis. Acta Biochim Biophys Sin (Shanghai) 2005;37(11):719–727. doi: 10.1111/j.1745-7270.2005.00108.x. [DOI] [PubMed] [Google Scholar]
- 14.Ramachandran A, Madesh M, Balasubramanian KA. Apoptosis in the intestinal epithelium: its relevance in normal and pathophysiological conditions. J Gastroenterol Hepatol. 2000;15(2):109–120. doi: 10.1046/j.1440-1746.2000.02059.x. [DOI] [PubMed] [Google Scholar]
- 15.Sakata S, Sakamaki K, Watanabe K, et al. Involvement of death receptor Fas in germ cell degeneration in gonads of Kit-deficient Wv/Wv mutant mice. Cell Death Differ. 2003;10(6):676–686. doi: 10.1038/sj.cdd.4401215. [DOI] [PubMed] [Google Scholar]
- 16.Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. The BCL-2 family reunion. Mol Cell. 2010;37(3):299–310. doi: 10.1016/j.molcel.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riedl SJ, Salvesen GS. The apoptosome: signalling platform of cell death. Nat Rev Mol Cell Biol. 2007;8(5):405–413. doi: 10.1038/nrm2153. [DOI] [PubMed] [Google Scholar]
- 18.Li J, Yuan J. Caspases in apoptosis and beyond. Oncogene. 2008;27(48):6194–6206. doi: 10.1038/onc.2008.297. [DOI] [PubMed] [Google Scholar]
- 19.Li LY, Luo X, Wang X. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature. 2001;412(6842):95–99. doi: 10.1038/35083620. [DOI] [PubMed] [Google Scholar]
- 20.Li H, Zhu H, Xu C, Yuan J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell. 1998;94(4):491–501. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- 21.Ott M, Norberg E, Zhivotovsky B, Orrenius S. Mitochondrial targeting of tBid/Bax: a role for the TOM complex? Cell Death Differ. 2009;16(8):1075–1082. doi: 10.1038/cdd.2009.61. [DOI] [PubMed] [Google Scholar]
- 22.Festjens N, van Gurp M, Van Loo G, Saelens X, Vandenabeele P. Bcl-2 family members as sentinels of cellular integrity and role of mitochondrial intermembrane space proteins in apoptotic cell death. Acta Haematol. 2004;111(1–2):7–27. doi: 10.1159/000074483. [DOI] [PubMed] [Google Scholar]
- 23.Yin XM. Bid, a BH3-only multifunctional molecule, is at the cross road of life and death. Gene. 2006;369(1):7–19. doi: 10.1016/j.gene.2005.10.038. [DOI] [PubMed] [Google Scholar]
- 24.Llambi F, Green DR. Apoptosis and oncogenesis: give and take in the BCL-2 family. Curr Opin Genet Dev. 2011;21(1):12–20. doi: 10.1016/j.gde.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen M, He H, Zhan S, Krajewski S, Reed JC, Gottlieb RA. Bid is cleaved by calpain to an active fragment in vitro and during myocardial ischemia/reperfusion. J Biol Chem. 2001;276(33):30724–30728. doi: 10.1074/jbc.M103701200. [DOI] [PubMed] [Google Scholar]
- 26.Joza N, Pospisilik JA, Hangen E, et al. AIF: not just an apoptosis-inducing factor. Ann N Y Acad Sci. 2009;1171:2–11. doi: 10.1111/j.1749-6632.2009.04681.x. [DOI] [PubMed] [Google Scholar]
- 27.Delavallée L, Cabon L, Galán Malo P, Lorenzo HK, Susin SA. AIF mediated caspase independent necroptosis: A new chance for targeted therapeutics. IUBMB Life. 2011;63(4):221–232. doi: 10.1002/iub.432. [DOI] [PubMed] [Google Scholar]
- 28.Sevrioukova IF. Redox-linked conformational dynamics in apoptosis-inducing factor. J Mol Biol. 2009;390(5):924–938. doi: 10.1016/j.jmb.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye H, Cande C, Stephanou NC, et al. DNA binding is required for the apoptogenic action of apoptosis inducing factor. Nat Struct Biol. 2002;9(9):680–684. doi: 10.1038/nsb836. [DOI] [PubMed] [Google Scholar]
- 30.Sevrioukova IF. Apoptosis-inducing factor: structure, function, and redox regulation. Antioxid Redox Signal. 2011;14(12):2545–2579. doi: 10.1089/ars.2010.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghezzi D, Sevrioukova I, Invernizzi F, et al. Severe X-linked mitochondrial encephalomyopathy associated with a mutation in apoptosis-inducing factor. Am J Hum Genet. 2010;86(4):639–649. doi: 10.1016/j.ajhg.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miramar MD, Costantini P, Ravagnan L, et al. NADH oxidase activity of mitochondrial apoptosis-inducing factor. J Biol Chem. 2001;276(19):16391–16398. doi: 10.1074/jbc.M010498200. [DOI] [PubMed] [Google Scholar]
- 33.Churbanova IY, Sevrioukova IF. Redox-dependent changes in molecular properties of mitochondrial apoptosis-inducing factor. J Biol Chem. 2008;283(9):5622–5631. doi: 10.1074/jbc.M709147200. [DOI] [PubMed] [Google Scholar]
- 34.Miseviciene L, Anusevicius Z, Sarlauskas J, Sevrioukova IF, Cenas N. Redox reactions of the FAD-containing apoptosis-inducing factor (AIF) with quinoidal xenobiotics: a mechanistic study. Arch Biochem Biophys. 2011;512(2):183–189. doi: 10.1016/j.abb.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klein JA, Longo-Guess CM, Rossmann MP, et al. The hHarlequin mouse mutation downregulates apoptosis-inducing factor. Nature. 2002;419(6905):367–374. doi: 10.1038/nature01034. [DOI] [PubMed] [Google Scholar]
- 36.Vahsen N, Cande C, Briere JJ, et al. AIF deficiency compromises oxidative phosphorylation. EMBO J. 2004;23(23):4679–4689. doi: 10.1038/sj.emboj.7600461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pospisilik JA, Knauf C, Joza N, et al. Targeted deletion of AIF decreases mitochondrial oxidative phosphorylation and protects from obesity and diabetes. Cell. 2007;131(3):476–491. doi: 10.1016/j.cell.2007.08.047. [DOI] [PubMed] [Google Scholar]
- 38.Koene S, Willems PH, Roestenberg P, Koopman WJ, Smeitink JA. Mouse models for nuclear DNA-encoded mitochondrial complex I deficiency. J Inherit Metab Dis. 2011;34(2):293–307. doi: 10.1007/s10545-009-9005-x. [DOI] [PubMed] [Google Scholar]
- 39.Brown D, Yu BD, Joza N, et al. Loss of AIF function causes cell death in the mouse embryo, but the temporal progression of patterning is normal. Proc Natl Acad Sci U S A. 2006;103(26):9918–9923. doi: 10.1073/pnas.0603950103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Culmsee C, Zhu C, Landshamer S, et al. Apoptosis-inducing factor triggered by poly(ADP-ribose) polymerase and Bid mediates neuronal cell death after oxygen-glucose deprivation and focal cerebral ischemia. J Neurosci. 2005;25(44):10262–10272. doi: 10.1523/JNEUROSCI.2818-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joza N, Susin SA, Daugas E, et al. Essential role of the mitochondrial apoptosis-inducing factor in programmed cell death. Nature. 2001;410(6828):549–554. doi: 10.1038/35069004. [DOI] [PubMed] [Google Scholar]
- 42.Apostolova N, Cervera AM, Victor VM, et al. Loss of apoptosis-inducing factor leads to an increase in reactive oxygen species, and an impairment of respiration that can be reversed by antioxidants. Cell Death Differ. 2006;13(2):354–357. doi: 10.1038/sj.cdd.4401776. [DOI] [PubMed] [Google Scholar]
- 43.Krantic S, Mechawar N, Reix S, Quirion R. Apoptosis-inducing factor: a matter of neuron life and death. Prog Neurobiol. 2007;81(3):179–196. doi: 10.1016/j.pneurobio.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 44.Urbano A, Lakshmanan U, Choo PH, et al. AIF suppresses chemical stress-induced apoptosis and maintains the transformed state of tumor cells. EMBO J. 2005;24(15):2815–2826. doi: 10.1038/sj.emboj.7600746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Modjtahedi N, Giordanetto F, Madeo F, Kroemer G. Apoptosis-inducing factor: vital and lethal. Trends Cell Biol. 2006;16(5):264–272. doi: 10.1016/j.tcb.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 46.Mate MJ, Ortiz-Lombardia M, Boitel B, et al. The crystal structure of the mouse apoptosis-inducing factor AIF. Nat Struct Biol. 2002;9(6):442–446. doi: 10.1038/nsb793. [DOI] [PubMed] [Google Scholar]
- 47.Vahsen N, Cande C, Dupaigne P, et al. Physical interaction of apoptosis-inducing factor with DNA and RNA. Oncogene. 2006;25(12):1763–1774. doi: 10.1038/sj.onc.1209206. [DOI] [PubMed] [Google Scholar]
- 48.Montague JW, Gaido ML, Frye C, Cidlowski JA. A calcium-dependent nuclease from apoptotic rat thymocytes is homologous with cyclophilin. Recombinant cyclophilins A, B, and C have nuclease activity. J Biol Chem. 1994;269(29):18877–18880. [PubMed] [Google Scholar]
- 49.Candé C, Vahsen N, Kouranti I, et al. AIF and cyclophilin A cooperate in apoptosis-associated chromatinolysis. Oncogene. 2004;23(8):1514–1521. doi: 10.1038/sj.onc.1207279. [DOI] [PubMed] [Google Scholar]
- 50.Artus C, Boujrad H, Bouharrour A, et al. AIF promotes chromatinolysis and caspase-independent programmed necrosis by interacting with histone H2AX. EMBO J. 2010;29(9):1585–1599. doi: 10.1038/emboj.2010.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu L, Xing D, Chen WR. Micro-calpain regulates caspase- dependent and apoptosis inducing factor-mediated caspase-independent apoptotic pathways in cisplatin-induced apoptosis. Int J Cancer. 2009;125(12):2757–2766. doi: 10.1002/ijc.24626. [DOI] [PubMed] [Google Scholar]
- 52.Yuste VJ, Moubarak RS, Delettre C, et al. Cysteine protease inhibition prevents mitochondrial apoptosis-inducing factor (AIF) release. Cell Death Differ. 2005;12(11):1445–1448. doi: 10.1038/sj.cdd.4401687. [DOI] [PubMed] [Google Scholar]
- 53.Ravagnan L, Gurbuxani S, Susin SA, et al. Heat-shock protein 70 antagonizes apoptosis-inducing factor. Nat Cell Biol. 2001;3(9):839–843. doi: 10.1038/ncb0901-839. [DOI] [PubMed] [Google Scholar]
- 54.Desmots F, Russell HR, Michel D, McKinnon PJ. Scythe regulates apoptosis-inducing factor stability during endoplasmic reticulum stress-induced apoptosis. J Biol Chem. 2008;283(6):3264–3271. doi: 10.1074/jbc.M706419200. [DOI] [PubMed] [Google Scholar]
- 55.Yu SW, Wang H, Poitras MF, et al. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297(5579):259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- 56.Wang Y, Dawson VL, Dawson TM. Poly(ADP-ribose) signals to mitochondrial AIF: a key event in parthanatos. Exp Neurol. 2009;218(2):193–202. doi: 10.1016/j.expneurol.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y, Kim NS, Haince JF, et al. Poly(ADP-ribose) (PAR) binding to apoptosis-inducing factor is critical for PAR polymerase-1-dependent cell death (parthanatos) Sci Signal. 2011;4(167):RA20. doi: 10.1126/scisignal.2000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu SW, Wang Y, Frydenlund DS, Ottersen OP, Dawson VL, Dawson TM. Outer mitochondrial membrane localization of apoptosis-inducing factor: mechanistic implications for release. ASN Neuro. 2009;1(5):e00021. doi: 10.1042/AN20090046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zanna C, Ghelli A, Porcelli AM, et al. OPA1 mutations associated with dominant optic atrophy impair oxidative phosphorylation and mitochondrial fusion. Brain. 2008;131(Pt 2):352–367. doi: 10.1093/brain/awm335. [DOI] [PubMed] [Google Scholar]
- 60.Jendrossek V, Fillon S, Belka C, Muller I, Puttkammer B, Lang F. Apoptotic response of Chang cells to infection with Pseudomonas aeruginosa strains PAK and PAO-I: molecular ordering of the apoptosis signaling cascade and role of type IV pili. Infect Immun. 2003;71(5):2665–2673. doi: 10.1128/IAI.71.5.2665-2673.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Choi CH, Lee EY, Lee YC, et al. Outer membrane protein 38 of Acinetobacter baumannii localizes to the mitochondria and induces apoptosis of epithelial cells. Cell Microbiol. 2005;7(8):1127–1138. doi: 10.1111/j.1462-5822.2005.00538.x. [DOI] [PubMed] [Google Scholar]
- 62.Ashktorab H, Dashwood RH, Dashwood MM, et al. H. pylori induced apoptosis in human gastric cancer cells mediated via the release of apoptosis inducing factor from mitochondria. Helicobacter. 2008;13(6):506–517. doi: 10.1111/j.1523-5378.2008.00646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Flynn AN, Wang A, McKay DM, Buret AG. Apoptosis-inducing factor contributes to epithelial cell apoptosis induced by enteropathogenic Escherichia coli. Can J Physiol Pharmacol. 2011;89(2):143–148. doi: 10.1139/y11-002. [DOI] [PubMed] [Google Scholar]
- 64.Flynn AN, Buret AG. Tight junctional disruption and apoptosis in an in vitro model of Citrobacter rodentium infection. Microb Pathog. 2008;45(2):98–104. doi: 10.1016/j.micpath.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 65.Suemasu S, Tanaka K, Namba T, et al. A role for HSP70 in protecting against indomethacin-induced gastric lesions. J Biol Chem. 2009;284(29):19705–19715. doi: 10.1074/jbc.M109.006817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Matsumori Y, Hong SM, Aoyama K, et al. Hsp70 overexpression sequesters AIF and reduces neonatal hypoxic/ischemic brain injury. J Cereb Blood Flow Metab. 2005;25(7):899–910. doi: 10.1038/sj.jcbfm.9600080. [DOI] [PubMed] [Google Scholar]
- 67.Choudhury S, Bae S, Ke Q, Lee JY, Kim J, Kang PM. Mitochondria to nucleus translocation of AIF in mice lacking Hsp70 during ischemia/reperfusion. Basic Res Cardiol. 2011;106(3):397–407. doi: 10.1007/s00395-011-0164-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pae CU, Kim TS, Kwon OJ, et al. Polymorphisms of heat shock protein 70 gene (HSPA1A, HSPA1B and HSPA1L) and schizophrenia. Neurosci Res. 2005;53(1):8–13. doi: 10.1016/j.neures.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 69.Xiao CY, Chen M, Zsengeller Z, Szabo C. Poly(ADP-ribose) polymerase contributes to the development of myocardial infarction in diabetic rats and regulates the nuclear translocation of apoptosis-inducing factor. J Pharmacol Exp Ther. 2004;310(2):498–504. doi: 10.1124/jpet.104.066803. [DOI] [PubMed] [Google Scholar]
- 70.Millan A, Huerta S. Apoptosis-inducing factor and colon cancer. J Surg Res. 2009;151(1):163–170. doi: 10.1016/j.jss.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 71.Schmitt E, Maingret L, Puig PE, et al. Heat shock protein 70 neutralization exerts potent antitumor effects in animal models of colon cancer and melanoma. Cancer Res. 2006;66(8):4191–4197. doi: 10.1158/0008-5472.CAN-05-3778. [DOI] [PubMed] [Google Scholar]
- 72.Hamasaki M, Hideshima T, Tassone P, et al. Azaspirane (NN-diethyl-8, 8-dipropyl-2-azaspiro [4.5] decane-2-propanamine) inhibits human multiple myeloma cell growth in the bone marrow milieu in vitro and in vivo. Blood. 2005;105(11):4470–4476. doi: 10.1182/blood-2004-09-3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang M, Zhang L, Han X, et al. Atiprimod inhibits the growth of mantle cell lymphoma in vitro and in vivo and induces apoptosis via activating the mitochondrial pathways. Blood. 2007;109(12):5455–5462. doi: 10.1182/blood-2006-12-063958. [DOI] [PubMed] [Google Scholar]
- 74.Park YC, Jeong JH, Park KJ, et al. Sulindac activates nuclear translocation of AIF, DFF40 and endonuclease G but not induces oligonucleosomal DNA fragmentation in HT-29 cells. Life Sci. 2005;77(16):2059–2070. doi: 10.1016/j.lfs.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 75.Phang JM, Donald SP, Pandhare J, Liu Y. The metabolism of proline, a stress substrate, modulates carcinogenic pathways. Amino Acids. 2008;35(4):681–690. doi: 10.1007/s00726-008-0063-4. [DOI] [PubMed] [Google Scholar]
- 76.Arentson BW, Sanyal N, Becker DF. Substrate channeling in proline metabolism. Front Biosci. 2012;17:375–388. doi: 10.2741/3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Phang JM, Liu W, Zabirnyk O. Proline metabolism and microenvironmental stress. Annu Rev Nutr. 2010;30:441–463. doi: 10.1146/annurev.nutr.012809.104638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Small WC, Jones ME. Pyrroline 5-carboxylate dehydrogenase of the mitochondrial matrix of rat liver. Purification, physical and kinetic characteristics. J Biol Chem. 1990;265(30):18668–18672. [PubMed] [Google Scholar]
- 79.Tanner JJ. Structural biology of proline catabolism. Amino Acids. 2008;35(4):719–730. doi: 10.1007/s00726-008-0062-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang M, White TA, Schuermann JP, Baban BA, Becker DF, Tanner JJ. Structures of the Escherichia coli PutA proline dehydrogenase domain in complex with competitive inhibitors. Biochemistry. 2004;43(39):12539–12548. doi: 10.1021/bi048737e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.White TA, Krishnan N, Becker DF, Tanner JJ. Structure and kinetics of monofunctional proline dehydrogenase from Thermus thermophilus. J Biol Chem. 2007;282(19):14316–14327. doi: 10.1074/jbc.M700912200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Moxley MA, Tanner JJ, Becker DF. Steady-state kinetic mechanism of the proline: ubiquinone oxidoreductase activity of proline utilization A (PutA) from Escherichia coli. Arch Biochem Biophys. 2011;516(2):113–120. doi: 10.1016/j.abb.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lee YH, Nadaraia S, Gu D, Becker DF, Tanner JJ. Structure of the proline dehydrogenase domain of the multifunctional PutA flavoprotein. Nat Struct Biol. 2003;10(2):109–114. doi: 10.1038/nsb885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389(6648):300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 85.Donald SP, Sun XY, Hu CA, et al. Proline oxidase, encoded by p53-induced gene-6, catalyzes the generation of proline-dependent reactive oxygen species. Cancer Res. 2001;61(5):1810–1815. [PubMed] [Google Scholar]
- 86.Maxwell SA, Davis GE. Differential gene expression in p53-mediated apoptosis-resistant vs apoptosis-sensitive tumor cell lines. Proc Natl Acad Sci U S A. 2000;97(24):13009–13014. doi: 10.1073/pnas.230445997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pandhare J, Cooper SK, Phang JM. Proline oxidase, a proapoptotic gene, is induced by troglitazone: evidence for both peroxisome proliferator-activated receptor gamma-dependent and -independent mechanisms. J Biol Chem. 2006;281(4):2044–2052. doi: 10.1074/jbc.M507867200. [DOI] [PubMed] [Google Scholar]
- 88.Pandhare J, Donald SP, Cooper SK, Phang JM. Regulation and function of proline oxidase under nutrient stress. J Cell Biochem. 2009;107(4):759–768. doi: 10.1002/jcb.22174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Maxwell SA, Kochevar GJ. Identification of a p53-response element in the promoter of the proline oxidase gene. Biochem Biophys Res Commun. 2008;369(2):308–313. doi: 10.1016/j.bbrc.2008.01.171. [DOI] [PubMed] [Google Scholar]
- 90.Cooper SK, Pandhare J, Donald SP, Phang JM. A novel function for hydroxyproline oxidase in apoptosis through generation of reactive oxygen species. J Biol Chem. 2008;283(16):10485–10492. doi: 10.1074/jbc.M702181200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Phang JM, Pandhare J, Zabirnyk O, Liu Y. PPARgamma and proline oxidase in cancer. PPAR Res. 2008;2008:542694. doi: 10.1155/2008/542694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu Y, Borchert GL, Surazynski A, Hu CA, Phang JM. Proline oxidase activates both intrinsic and extrinsic pathways for apoptosis: the role of ROS/superoxides, NFAT and MEK/ERK signaling. Oncogene. 2006;25(41):5640–5647. doi: 10.1038/sj.onc.1209564. [DOI] [PubMed] [Google Scholar]
- 93.Liu Y, Borchert G, Surazynski A, Phang J. Proline oxidase, a p53-induced gene, targets COX-2/PGE2 signaling to induce apoptosis and inhibit tumor growth in colorectal cancers. Oncogene. 2008;27(53):6729–6737. doi: 10.1038/onc.2008.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hu CA, Phang JM, Valle D. Proline metabolism in health and disease. Preface. Amino Acids. 2008;35(4):651–652. doi: 10.1007/s00726-008-0102-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hu CA, Donald SP, Yu J, et al. Overexpression of proline oxidase induces proline-dependent and mitochondria-mediated apoptosis. Mol Cell Biochem. 2007;295(1–2):85–92. doi: 10.1007/s11010-006-9276-6. [DOI] [PubMed] [Google Scholar]
- 96.Liu Y, Borchert GL, Donald SP, et al. MnSOD inhibits proline oxidase-induced apoptosis in colorectal cancer cells. Carcinogenesis. 2005;26(8):1335–1342. doi: 10.1093/carcin/bgi083. [DOI] [PubMed] [Google Scholar]
- 97.Zou W, Liu X, Yue P, Khuri FR, Sun SY. PPARgamma ligands enhance TRAIL-induced apoptosis through DR5 upregulation and c-FLIP downregulation in human lung cancer cells. Cancer Biol Ther. 2007;6(1):99–106. doi: 10.4161/cbt.6.1.3555. [DOI] [PubMed] [Google Scholar]
- 98.Elrod HA, Sun SY. PPARgamma and apoptosis in cancer. PPAR Res. 2008;2008:704165. doi: 10.1155/2008/704165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Miller G, Honig A, Stein H, Suzuki N, Mittler R, Zilberstein A. Unraveling delta1-pyrroline-5-carboxylate-proline cycle in plants by uncoupled expression of proline oxidation enzymes. J Biol Chem. 2009;284(39):26482–26492. doi: 10.1074/jbc.M109.009340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12(1):21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chiacchiera F, Simone C. The AMPK-FoxO3A axis as a target for cancer treatment. Cell Cycle. 2010;9(6):1091–1096. doi: 10.4161/cc.9.6.11035. [DOI] [PubMed] [Google Scholar]
- 102.Krishnan N, Becker DF, Dickman MB. Proline modulates the intracellular redox environment and protects mammalian cells against oxidative stress. Free Radical Biol Med. 2008;44(4):671–681. doi: 10.1016/j.freeradbiomed.2007.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu W, Zabirnyk O, Wang H, et al. miR-23b* targets proline oxidase, a novel tumor suppressor protein in renal cancer. Oncogene. 2010;29(35):4914–4924. doi: 10.1038/onc.2010.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Willis A, Bender HU, Steel G, Valle D. PRODH variants and risk for schizophrenia. Amino Acids. 2008;35(4):673–679. doi: 10.1007/s00726-008-0111-0. [DOI] [PubMed] [Google Scholar]
- 105.Jacquet H, Demily C, Houy E, et al. Hyperprolinemia is a risk factor for schizoaffective disorder. Mol Psychiatry. 2005;10(5):479–485. doi: 10.1038/sj.mp.4001597. [DOI] [PubMed] [Google Scholar]
- 106.Jacquet H, Raux G, Thibaut F, et al. PRODH mutations and hyperprolinemia in a subset of schizophrenic patients. Hum Mol Genet. 2002;11(19):2243–2249. doi: 10.1093/hmg/11.19.2243. [DOI] [PubMed] [Google Scholar]
- 107.Karayiorgou M, Simon TJ, Gogos JA. 22q11.2 microdeletions: linking DNA structural variation to brain dysfunction and schizophrenia. Nat Rev Neurosci. 2010;11(6):402–416. doi: 10.1038/nrn2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bender HU, Almashanu S, Steel G, et al. Functional consequences of PRODH missense mutations. Am J Hum Genet. 2005;76(3):409–420. doi: 10.1086/428142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Alderton WK, Cooper CE, Knowles RG. Nitric oxide synthases: structure, function and inhibition. Biochem J. 2001;357(Pt 3):593–615. doi: 10.1042/0264-6021:3570593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nakajima K, Inatsu S, Mizote T, et al. Possible involvement of put A gene in Helicobacter pylori colonization in the stomach and motility. Biomed Res. 2008;29(1):9–18. doi: 10.2220/biomedres.29.9. [DOI] [PubMed] [Google Scholar]
- 111.Krishnan N, Doster AR, Duhamel GE, Becker DF. Characterization of a Helicobacter hepaticus putA mutant strain in host colonization and oxidative stress. Infect Immun. 2008;76(7):3037–3044. doi: 10.1128/IAI.01737-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Brown DI, Griendling KK. Nox proteins in signal transduction. Free Radic Biol Med. 2009;47(9):1239–1253. doi: 10.1016/j.freeradbiomed.2009.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kakinuma K, Kaneda M, Chiba T, Ohnishi T. Electron spin resonance studies on a flavoprotein in neutrophil plasma membranes. Redox potentials of the flavin and its participation in NADPH oxidase. J Biol Chem. 1986;261(20):9426–9432. [PubMed] [Google Scholar]
- 114.Cross AR, Jones OTG, Harper AM, Segal AW. Oxidation-reduction properties of the cytochrome-B found in the plasma-membrane fraction of human-neutrophils – a possible oxidase in the respiratory burst. Biochem J. 1981;194(2):599–606. doi: 10.1042/bj1940599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fulton DJ. Nox5 and the regulation of cellular function. Antioxid Redox Signal. 2009;11(10):2443–2452. doi: 10.1089/ars.2009.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Drummond GR, Selemidis S, Griendling KK, Sobey CG. Combating oxidative stress in vascular disease: NADPH oxidases as therapeutic targets. Nat Rev Drug Discov. 2011;10(6):453–471. doi: 10.1038/nrd3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jiang F, Zhang Y, Dusting GJ. NADPH oxidase-mediated redox signaling: roles in cellular stress response, stress tolerance, and tissue repair. Pharmacol Rev. 2011;63(1):218–242. doi: 10.1124/pr.110.002980. [DOI] [PubMed] [Google Scholar]
- 118.Takeya R, Ueno N, Kami K, et al. Novel human homologues of p47phox and p67phox participate in activation of superoxide- producing NADPH oxidases. J Biol Chem. 2003;278(27):25234–25246. doi: 10.1074/jbc.M212856200. [DOI] [PubMed] [Google Scholar]
- 119.Ueyama T, Tatsuno T, Kawasaki T, et al. A regulated adaptor function of p40phox: distinct p67phox membrane targeting by p40phox and by p47phox. Mol Biol Cell. 2007;18(2):441–454. doi: 10.1091/mbc.E06-08-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Johnson JL, Park JW, Benna JE, Faust LRP, Inanami O, Babior BM. Activation of p47 phox, a cytosolic subunit of the leukocyte NADPH oxidase. J Biol Chem. 1998;273(52):35147–35152. doi: 10.1074/jbc.273.52.35147. [DOI] [PubMed] [Google Scholar]
- 121.Hoyal CR, Gutierrez A, Young BM, et al. Modulation of p47PHOX activity by site-specif ic phosphorylation: Akt-dependent activation of the NADPH oxidase. Proc Natl Acad Sci U S A. 2003;100(9):5130–5135. doi: 10.1073/pnas.1031526100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lopes LR, Dagher MC, Gutierrez A, et al. Phosphorylated p40PHOX as a negative regulator of NADPH oxidase. Biochemistry. 2004;43(12):3723–3730. doi: 10.1021/bi035636s. [DOI] [PubMed] [Google Scholar]
- 123.Kim KA, Kim JY, Lee YA, Song KJ, Min D, Shin MH. NOX1 participates in ROS-dependent cell death of colon epithelial Caco2 cells induced by Entamoeba histolytica. Microbes Infect. 2011;13(12–13):1052–1061. doi: 10.1016/j.micinf.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 124.Kuroda J, Ago T, Matsushima S, Zhai P, Schneider MD, Sadoshima J. NADPH oxidase 4 (Nox4) is a major source of oxidative stress in the failing heart. Proc Natl Acad Sci U S A. 2010;107(35):15565–15570. doi: 10.1073/pnas.1002178107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Weyemi U, Lagente-Chevallier O, Boufraqech M, et al. ROS-generating NADPH oxidase NOX4 is a critical mediator in oncogenic H-Ras-induced DNA damage and subsequent senescence. Oncogene. 2011 Aug 15; doi: 10.1038/onc.2011.327. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Babior BM. NADPH oxidase. Curr Opin Immunol. 2004;16(1):42–47. doi: 10.1016/j.coi.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 127.Ris-Stalpers C. Physiology and pathophysiology of the DUOXes. Antioxid Redox Signal. 2006;8(9–10):1563–1572. doi: 10.1089/ars.2006.8.1563. [DOI] [PubMed] [Google Scholar]
- 128.Pendyala S, Natarajan V. Redox regulation of Nox proteins. Respir Physiol Neurobiol. 2010;174(3):265–271. doi: 10.1016/j.resp.2010.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yu JH, Kim KH, Kim H. Role of NADPH oxidase and calcium in cerulein-induced apoptosis: involvement of apoptosis-inducing factor. Ann N Y Acad Sci. 2006;1090:292–297. doi: 10.1196/annals.1378.031. [DOI] [PubMed] [Google Scholar]
- 130.Matsuzawa A, Saegusa K, Noguchi T, et al. ROS-dependent activation of the TRAF6-ASK1-p38 pathway is selectively required for TLR4-mediated innate immunity. Nat Immunol. 2005;6(6):587–592. doi: 10.1038/ni1200. [DOI] [PubMed] [Google Scholar]
- 131.Basuroy S, Bhattacharya S, Leffler CW, Parfenova H. Nox4 NADPH oxidase mediates oxidative stress and apoptosis caused by TNF-alpha in cerebral vascular endothelial cells. Am J Physiol Cell Physiol. 2009;296(3):C422–C432. doi: 10.1152/ajpcell.00381.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Noguchi T, Ishii K, Fukutomi H, et al. Requirement of reactive oxygen species-dependent activation of ASK1-p38 MAPK pathway for extracellular ATP-induced apoptosis in macrophage. J Biol Chem. 2008;283(12):7657–7665. doi: 10.1074/jbc.M708402200. [DOI] [PubMed] [Google Scholar]
- 133.Park KJ, Lee CH, Kim A, Jeong KJ, Kim CH, Kim YS. Death receptors 4 and 5 activate Nox1 NADPH oxidase through riboflavin kinase to induce reactive oxygen species-mediated apoptotic cell death. J Biol Chem. 2011 Dec 9; doi: 10.1074/jbc.M111.309021. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mustapha NM, Tarr JM, Kohner EM, Chibber R. NADPH oxidase versus mitochondria-derived ROS in glucose-induced apoptosis of pericytes in early diabetic retinopathy. J Ophthalmol. 2010;2010:746978. doi: 10.1155/2010/746978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nakano Y, Longo-Guess CM, Bergstrom DE, Nauseef WM, Jones SM, Banfi B. Mutation of the Cyba gene encoding p22phox causes vestibular and immune defects in mice. J Clin Invest. 2008;118(3):1176–1185. doi: 10.1172/JCI33835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.de Oliveira-Junior EB, Bustamante J, Newburger PE, Condino-Neto A. The human NADPH oxidase: primary and secondary defects impairing the respiratory burst function and the microbicidal ability of phagocytes. Scand J Immunol. 2011;73(5):420–427. doi: 10.1111/j.1365-3083.2010.02501.x. [DOI] [PubMed] [Google Scholar]
- 137.Cave AC, Brewer AC, Narayanapanicker A, et al. NADPH oxidases in cardiovascular health and disease. Antioxid Redox Signal. 2006;8(5–6):691–728. doi: 10.1089/ars.2006.8.691. [DOI] [PubMed] [Google Scholar]
- 138.Griffith B, Pendyala S, Hecker L, Lee PJ, Natarajan V, Thannickal VJ. NOX enzymes and pulmonary disease. Antioxid Redox Signal. 2009;11(10):2505–2516. doi: 10.1089/ars.2009.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sorescu D, Weiss D, Lassegue B, et al. Superoxide production and expression of nox family proteins in human atherosclerosis. Circulation. 2002;105(12):1429–1435. doi: 10.1161/01.cir.0000012917.74432.66. [DOI] [PubMed] [Google Scholar]
- 140.Ushio-Fukai M, Nakamura Y. Reactive oxygen species and angiogenesis: NADPH oxidase as target for cancer therapy. Cancer Lett. 2008;266(1):37–52. doi: 10.1016/j.canlet.2008.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Grishko V, Pastukh V, Solodushko V, Gillespie M, Azuma J, Schaffer S. Apoptotic cascade initiated by angiotensin II in neonatal cardiomyocytes: role of DNA damage. Am J Physiol Heart Circ Physiol. 2003;285(6):H2364–H2372. doi: 10.1152/ajpheart.00408.2003. [DOI] [PubMed] [Google Scholar]
- 142.Sheehan AL, Carrell S, Johnson B, Stanic B, Banfi B, Miller FJ., Jr Role for Nox1 NADPH oxidase in atherosclerosis. Atherosclerosis. 2011;216(2):321–326. doi: 10.1016/j.atherosclerosis.2011.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Heinloth A, Heermeier K, Raff U, Wanner C, Galle J. Stimulation of NADPH oxidase by oxidized low-density lipoprotein induces proliferation of human vascular endothelial cells. J Am Soc Nephrol. 2000;11(10):1819–1825. doi: 10.1681/ASN.V11101819. [DOI] [PubMed] [Google Scholar]
- 144.Carmena R, Duriez P, Fruchart JC. Atherogenic lipoprotein particles in atherosclerosis. Circulation. 2004;109(23 Suppl 1):III2–III7. doi: 10.1161/01.CIR.0000131511.50734.44. [DOI] [PubMed] [Google Scholar]
- 145.Gotoh N, Niki E. Rates of interactions of superoxide with vitamin E, vitamin C and related compounds as measured by chemiluminescence. Biochim Biophys Acta. 1992;1115(3):201–207. doi: 10.1016/0304-4165(92)90054-x. [DOI] [PubMed] [Google Scholar]
- 146.Loughlin DT, Artlett CM. Precursor of advanced glycation end products mediates ER-stress-induced caspase-3 activation of human dermal fibroblasts through NAD(P)H oxidase 4. PLoS One. 2010;5(6):e11093. doi: 10.1371/journal.pone.0011093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jaquet V, Scapozza L, Clark RA, Krause KH, Lambeth JD. Small-molecule NOX inhibitors: ROS-generating NADPH oxidases as therapeutic targets. Antioxid Redox Signal. 2009;11(10):2535–2552. doi: 10.1089/ars.2009.2585. [DOI] [PubMed] [Google Scholar]
- 148.Guzik TJ, Harrison DG. Vascular NADPH oxidases as drug targets for novel antioxidant strategies. Drug Discov Today. 2006;11(12):524–533. doi: 10.1016/j.drudis.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 149.Guichard C, Moreau R, Pessayre D, Epperson TK, Krause KH. NOX family NADPH oxidases in liver and in pancreatic islets: a role in the metabolic syndrome and diabetes. Biochem Soc Trans. 2008;36(Pt 5):920–929. doi: 10.1042/BST0360920. [DOI] [PubMed] [Google Scholar]
- 150.Leto TL, Morand S, Hurt D, Ueyama T. Targeting and regulation of reactive oxygen species generation by Nox family NADPH oxidases. Antioxid Redox Signal. 2009;11(10):2607–2619. doi: 10.1089/ars.2009.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kim JA, Neupane GP, Lee ES, Jeong BS, Park BC, Thapa P. NADPH oxidase inhibitors: a patent review. Expert Opin Ther Pat. 2011;21(8):1147–1158. doi: 10.1517/13543776.2011.584870. [DOI] [PubMed] [Google Scholar]
- 152.Guimaraes EL, Empsen C, Geerts A, van Grunsven LA. Advanced glycation end products induce production of reactive oxygen species via the activation of NADPH oxidase in murine hepatic stellate cells. J Hepatol. 2010;52(3):389–397. doi: 10.1016/j.jhep.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 153.Ping J, Li JT, Liao ZX, Shang L, Wang H. Indole-3-carbinol inhibits hepatic stellate cells proliferation by blocking NADPH oxidase/reactive oxygen species/p38 MAPK pathway. Eur J Pharmacol. 2011;650(2–3):656–662. doi: 10.1016/j.ejphar.2010.10.057. [DOI] [PubMed] [Google Scholar]
- 154.Friedman SL. Liver fibrosis – from bench to bedside. J Hepatol. 2003;38(1):S38–S53. doi: 10.1016/s0168-8278(02)00429-4. [DOI] [PubMed] [Google Scholar]
- 155.Cui W, Matsuno K, Iwata K, et al. NOX1/nicotinamide adenine dinucleotide phosphate, reduced form (NADPH) oxidase promotes proliferation of stellate cells and aggravates liver fibrosis induced by bile duct ligation. Hepatology. 2011;53(3):948–958. doi: 10.1002/hep.24465. [DOI] [PubMed] [Google Scholar]
- 156.Masamune A, Watanabe T, Kikuta K, Satoh K, Shimosegawa T. NADPH oxidase plays a crucial role in the activation of pancreatic stellate cells. Am J Physiol Gastrointest Liver Physiol. 2008;294(1):G99–G108. doi: 10.1152/ajpgi.00272.2007. [DOI] [PubMed] [Google Scholar]
- 157.Govindarajan B, Sligh JE, Vincent BJ, et al. Overexpression of Akt converts radial growth melanoma to vertical growth melanoma. J Clin Invest. 2007;117(3):719–729. doi: 10.1172/JCI30102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Vaquero EC, Edderkaoui M, Pandol SJ, Gukovsky I, Gukovskaya AS. Reactive oxygen species produced by NAD(P)H oxidase inhibit apoptosis in pancreatic cancer cells. J Biol Chem. 2004;279(33):34643–34654. doi: 10.1074/jbc.M400078200. [DOI] [PubMed] [Google Scholar]
- 159.Eid AA, Ford BM, Block K, et al. AMP-activated protein kinase (AMPK) negatively regulates Nox4-dependent activation of p53 and epithelial cell apoptosis in diabetes. J Biol Chem. 2010;285(48):37503–37512. doi: 10.1074/jbc.M110.136796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Brar SS, Corbin Z, Kennedy TP, et al. NOX5 NAD(P)H oxidase regulates growth and apoptosis in DU 145 prostate cancer cells. Am J Physiol Cell Physiol. 2003;285(2):C353–C369. doi: 10.1152/ajpcell.00525.2002. [DOI] [PubMed] [Google Scholar]
- 161.Jiang Z, Fletcher NM, Ali-Fehmi R, et al. Modulation of redox signaling promotes apoptosis in epithelial ovarian cancer cells. Gynecol Oncol. 2011;122(2):418–423. doi: 10.1016/j.ygyno.2011.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Tew DG. Inhibition of cytochrome P450 reductase by the diphenyliodonium cation. Kinetic analysis and covalent modifications. Biochemistry. 1993;32(38):10209–10215. doi: 10.1021/bi00089a042. [DOI] [PubMed] [Google Scholar]
- 163.Bedard K, Krause KH. The NOX family of ROS-generating NADPH oxidases: physiology and pathophysiology. Physiol Rev. 2007;87(1):245–313. doi: 10.1152/physrev.00044.2005. [DOI] [PubMed] [Google Scholar]
- 164.Stolk J, Hiltermann TJ, Dijkman JH, Verhoeven AJ. Characteristics of the inhibition of NADPH oxidase activation in neutrophils by apocynin, a methoxy-substituted catechol. Am J Respir Cell Mol Biol. 1994;11(1):95–102. doi: 10.1165/ajrcmb.11.1.8018341. [DOI] [PubMed] [Google Scholar]
- 165.Heumüller S, Wind S, Barbosa-Sicard E, et al. Apocynin is not an inhibitor of vascular NADPH oxidases but an antioxidant. Hypertension. 2008;51(2):211–217. doi: 10.1161/HYPERTENSIONAHA.107.100214. [DOI] [PubMed] [Google Scholar]
- 166.Vejrazka M, Micek R, Stipek S. Apocynin inhibits NADPH oxidase in phagocytes but stimulates ROS production in non-phagocytic cells. Biochim Biophys Acta. 2005;1722(2):143–147. doi: 10.1016/j.bbagen.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 167.Yamamoto A, Kami K, Takeya R, Sumimoto H. Interaction between the SH3 domains and C-terminal proline-rich region in NADPH oxidase organizer 1 (Noxo1) Biochem Biophys Res Commun. 2007;352(2):560–565. doi: 10.1016/j.bbrc.2006.11.060. [DOI] [PubMed] [Google Scholar]
- 168.Rey FE, Cifuentes ME, Kiarash A, Quinn MT, Pagano PJ. Novel competitive inhibitor of NAD(P)H oxidase assembly attenuates vascular O(2)(−) and systolic blood pressure in mice. Circ Res. 2001;89(5):408–414. doi: 10.1161/hh1701.096037. [DOI] [PubMed] [Google Scholar]
- 169.Laleu B, Gaggini F, Orchard M, et al. First in class, potent, and orally bioavailable NADPH oxidase isoform 4 (Nox4) inhibitors for the treatment of idiopathic pulmonary fibrosis. J Med Chem. 2010;53(21):7715–7730. doi: 10.1021/jm100773e. [DOI] [PubMed] [Google Scholar]