Abstract
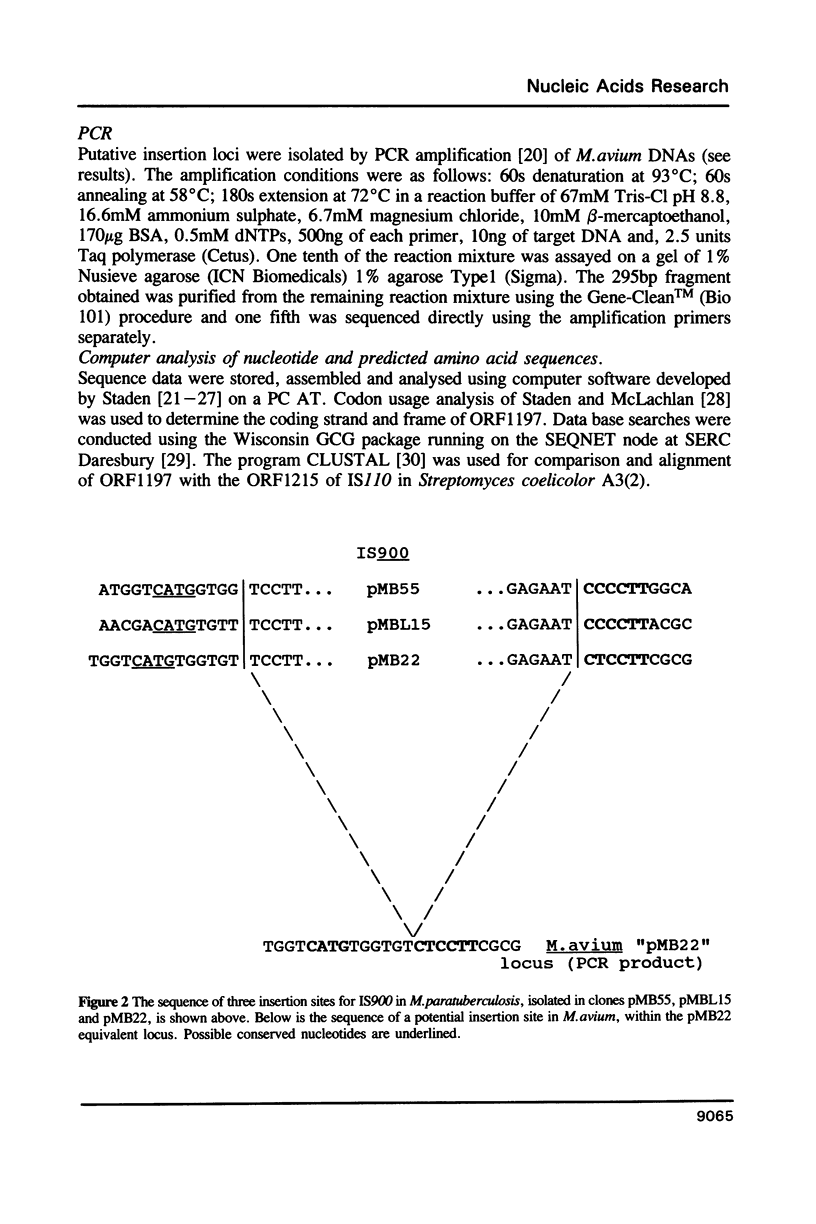
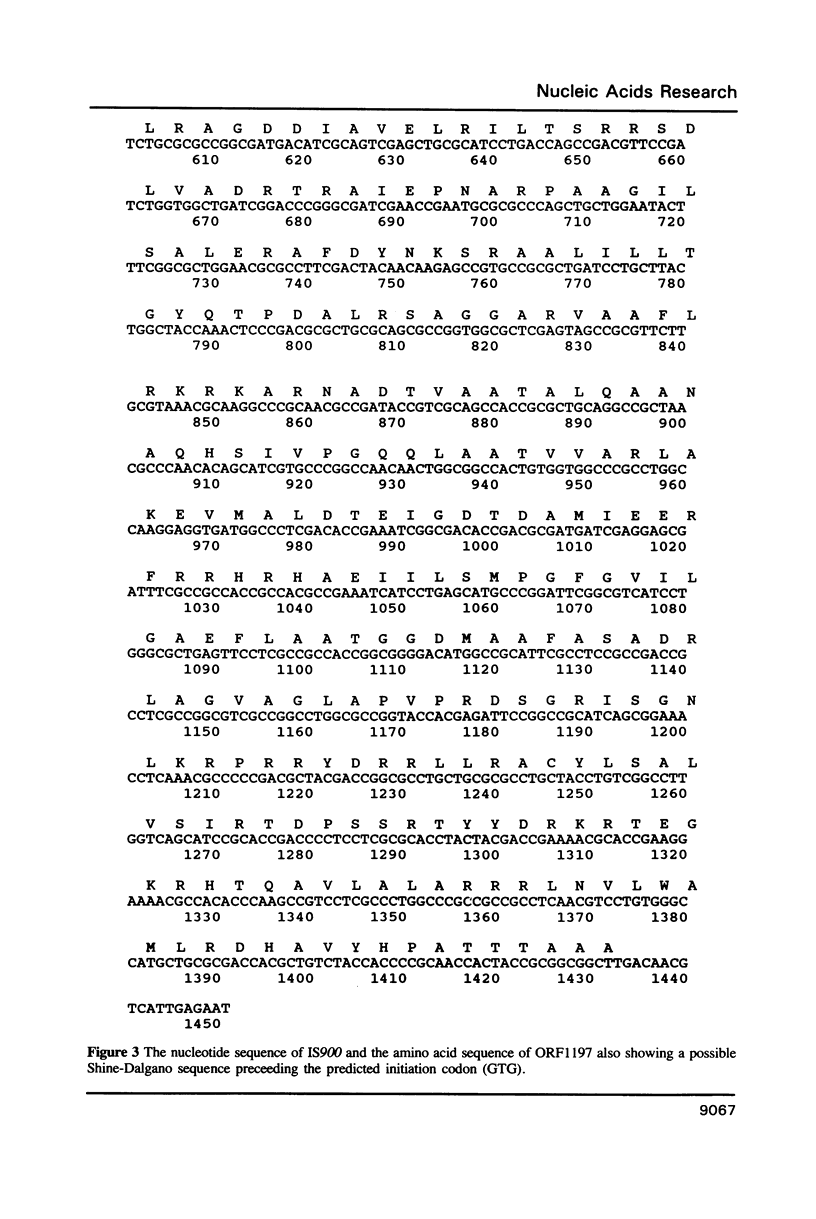
The complete sequence of an insertion element IS900 in Mycobacterium paratuberculosis is reported. This is the first characterised example of a mycobacterial insertion element. IS900 consists of 1451bp of which 66% is G + C. It lacks terminal inverted and direct repeats, characteristic of Escherichia coli insertion elements but shows a degree of target sequence specificity. A single open reading frame (ORF 1197) coding for 399 amino acids is predicted. This amino acid sequence, and to a lesser extent the nucleotide sequence, show significant homologies to IS110, an insertion element of Streptomyces coelicolor A3(2). It is proposed that IS900, IS110, and similar insertion elements recently identified in disease isolates of Mycobacterium avium are members of a phylogenetically related family. IS900 will provide highly specific markers for the precise identification of Mycobacterium paratuberculosis, useful in defining its relationship to animal and human diseases.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bartlett D. H., Silverman M. Nucleotide sequence of IS492, a novel insertion sequence causing variation in extracellular polysaccharide production in the marine bacterium Pseudomonas atlantica. J Bacteriol. 1989 Mar;171(3):1763–1766. doi: 10.1128/jb.171.3.1763-1766.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg D. E. Structural requirement for IS50-mediated gene transposition. Proc Natl Acad Sci U S A. 1983 Feb;80(3):792–796. doi: 10.1073/pnas.80.3.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruton C. J., Chater K. F. Nucleotide sequence of IS110, an insertion sequence of Streptomyces coelicolor A3(2). Nucleic Acids Res. 1987 Sep 11;15(17):7053–7065. doi: 10.1093/nar/15.17.7053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell A., Starlinger P., Berg D. E., Botstein D., Lederberg E. M., Novick R. P., Szybalski W. Nomenclature of transposable elements in prokaryotes. Plasmid. 1979 Jul;2(3):466–473. doi: 10.1016/0147-619x(79)90030-1. [DOI] [PubMed] [Google Scholar]
- Chater K. F., Bruton C. J., Foster S. G., Tobek I. Physical and genetic analysis of IS110, a transposable element of Streptomyces coelicolor A3(2). Mol Gen Genet. 1985;200(2):235–239. doi: 10.1007/BF00425429. [DOI] [PubMed] [Google Scholar]
- Chiodini R. J. Crohn's disease and the mycobacterioses: a review and comparison of two disease entities. Clin Microbiol Rev. 1989 Jan;2(1):90–117. doi: 10.1128/cmr.2.1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Curtiss J. E., Docherty M. A. A species-specific repetitive sequence in Mycobacterium leprae DNA. J Infect Dis. 1989 Jan;159(1):7–15. doi: 10.1093/infdis/159.1.7. [DOI] [PubMed] [Google Scholar]
- DasSarma S., RajBhandary U. L., Khorana H. G. High-frequency spontaneous mutation in the bacterio-opsin gene in Halobacterium halobium is mediated by transposable elements. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2201–2205. doi: 10.1073/pnas.80.8.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenach K. D., Crawford J. T., Bates J. H. Repetitive DNA sequences as probes for Mycobacterium tuberculosis. J Clin Microbiol. 1988 Nov;26(11):2240–2245. doi: 10.1128/jcm.26.11.2240-2245.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green P. M., Bentley D. R., Mibashan R. S., Nilsson I. M., Giannelli F. Molecular pathology of haemophilia B. EMBO J. 1989 Apr;8(4):1067–1072. doi: 10.1002/j.1460-2075.1989.tb03474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley N. D., Sherratt D. J. Sequence analysis at IS1 insertion sites: models for transposition. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1257–1261. doi: 10.1101/sqb.1979.043.01.142. [DOI] [PubMed] [Google Scholar]
- Higgins D. G., Sharp P. M. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988 Dec 15;73(1):237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- Machida Y., Sakurai M., Kiyokawa S., Ubasawa A., Suzuki Y., Ikeda J. E. Nucleotide sequence of the insertion sequence found in the T-DNA region of mutant Ti plasmid pTiA66 and distribution of its homologues in octopine Ti plasmid. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7495–7499. doi: 10.1073/pnas.81.23.7495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy M. H. Frameshift mutations in the lactose operon of E. coli. Cold Spring Harb Symp Quant Biol. 1966;31:189–201. doi: 10.1101/sqb.1966.031.01.027. [DOI] [PubMed] [Google Scholar]
- McFadden J. J., Butcher P. D., Chiodini R., Hermon-Taylor J. Crohn's disease-isolated mycobacteria are identical to Mycobacterium paratuberculosis, as determined by DNA probes that distinguish between mycobacterial species. J Clin Microbiol. 1987 May;25(5):796–801. doi: 10.1128/jcm.25.5.796-801.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden J. J., Butcher P. D., Thompson J., Chiodini R., Hermon-Taylor J. The use of DNA probes identifying restriction-fragment-length polymorphisms to examine the Mycobacterium avium complex. Mol Microbiol. 1987 Nov;1(3):283–291. doi: 10.1111/j.1365-2958.1987.tb01934.x. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Rappuoli R., Perugini M., Ratti G. DNA element of Corynebacterium diphtheriae with properties of an insertion sequence and usefulness for epidemiological studies. J Bacteriol. 1987 Jan;169(1):308–312. doi: 10.1128/jb.169.1.308-312.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saedler H., Starlinger P. 0 degree mutations in the galactose operon in E. coli. I. Genetic characterization. Mol Gen Genet. 1967;100(2):178–189. doi: 10.1007/BF00333604. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. A. Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1933–1937. doi: 10.1073/pnas.76.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Automation of the computer handling of gel reading data produced by the shotgun method of DNA sequencing. Nucleic Acids Res. 1982 Aug 11;10(15):4731–4751. doi: 10.1093/nar/10.15.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Computer methods to locate signals in nucleic acid sequences. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):505–519. doi: 10.1093/nar/12.1part2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Graphic methods to determine the function of nucleic acid sequences. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):521–538. doi: 10.1093/nar/12.1part2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R., McLachlan A. D. Codon preference and its use in identifying protein coding regions in long DNA sequences. Nucleic Acids Res. 1982 Jan 11;10(1):141–156. doi: 10.1093/nar/10.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Measurements of the effects that coding for a protein has on a DNA sequence and their use for finding genes. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):551–567. doi: 10.1093/nar/12.1part2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. The current status and portability of our sequence handling software. Nucleic Acids Res. 1986 Jan 10;14(1):217–231. doi: 10.1093/nar/14.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]



