Abstract
MicroRNAs (miRNAs) have emerged as key regulators of gene expression. Intragenic miRNAs account for ∼50% of mammalian miRNAs. Classic studies reported that they are usually coexpressed with host genes. Here, using genome-wide miRNA and gene expression profiles from five sample sets, we show that evolutionarily conserved (‘old’) intragenic miRNAs tend to be coexpressed with host genes, but non-conserved (‘young’) ones rarely do so. This result is robust: in all sample sets, the coexpression rate of young miRNAs is significantly lower than that of conserved ones even after controlling for abundance. As a result, although young miRNAs dominate in human genome, the majority of intragenic miRNAs that show coexpression with host genes are phylogenetically old ones. For younger miRNAs, extrapolation of their expression profiles from those of their host genes should be treated with caution. We propose a model to explain this phenomenon in which the majority of young miRNAs are unlikely to be coexpressed with host genes; however, for some fraction of young miRNAs coexpression with their host genes, initially imbued by chromatin level effects, is advantageous and these are the ones likely to embed into the system and evolve ever higher levels of coexpression, possibly by evolving piggybacking mechanisms.
INTRODUCTION
Evidence is emerging that microRNAs (miRNAs), an abundant class of small (∼22nt) non-coding RNAs, are key regulators of gene expression in both health and disease (1–9). Around 50% of the mammalian miRNAs are located within introns or exons of protein-coding genes and on the same strand (10–13). Most of these same-strand intragenic miRNAs are located within introns of host genes and are referred to as intronic miRNAs, whereas the remaining small portion are overlapping with exons of their host genes and are thus called exonic miRNAs. MiRNAs on the opposite strand are also known. Unless stated otherwise, our analysis considers only sense strand intronic and exonic miRNAs.
Early evidence suggested that intragenic miRNAs and host mRNAs might be processed from the same RNA substrate (14). In analyses of expressed sequence tag (EST) libraries, a population of chimeric miRNA precursor-mRNA transcripts were observed in normal human and mouse tissues (15), and some EST fragments that are located either immediately upstream or downstream of miRNAs were partially spliced, with either 5′- or 3′-ends matching the putative Drosha cleavage sites (16). In addition, some studies indicated that the biogenesis of intronic miRNAs were enhanced in the presence of flanking exons (17) and that some miRNA microprocessor complex-associated proteins were identified also as splicing factors or involved in pre-mRNA processing (14,18–20). More importantly, several previous studies showed that intragenic miRNAs are often coexpressed with their host genes (10,21,22). Thus, a commonly considered model was proposed suggesting that mammalian intragenic miRNAs are derived from the same primary transcripts as, and thereby are coexpressed with their host genes (14–16,21,23–28).
If this coexpression model is correct, it is important for at least three reasons. First, it suggests that miRNAs commonly piggy-back on the transcription of the host gene rather than have their own independent promoters. Consequently, disruption to the expression of the host gene is likely to result in disruption to the expression of the miRNA making the phenotypic impact of such disruption both more acute and more complex. Second, based on this model, expression profiles of host genes are used as surrogates for those of the intragenic miRNAs to predict miRNA target genes (29–32). If not true, however, this shortcut cannot be defended and all prior results derived from this assumption would require reappraisal. Third, the model suggests a potentially important mode of regulation in which upregulation of one gene (containing the miRNA) by necessity leads to downregulation of others (the targets of the embedded miRNA). Thus, we might expect the protein-coding genes containing miRNAs that are upregulated to have opposite functions to those downregulated by the miRNAs. Thus, there might be a logic as to which miRNAs are in which genes. However, if miRNA and host gene are not coexpressed as frequently as observed in the previous studies, then such coupling is not expected.
Previous results, however, were derived from expression studies of only a small number (<35) of intragenic miRNAs. In fact, using recent versions of miRBase and human genome annotation, we identified over 600 possible human intragenic miRNAs, although many are of uncertain validity. Here, then we re-evaluate the correlation of expression between intragenic miRNAs and their host genes. Our analysis is based on genome-wide miRNA and gene array data from five human sample sets that we experimentally derived or we collected from publically available databases. Our results suggest that evolutionarily conserved intragenic miRNAs do tend to be coexpressed with their host genes, but, in contrast, poorly conserved ones rarely do so. We do not wish to assert that these young ones are or are not coexpressed with their host genes as the answer to this would be sensitive to, among other details, which tissues are analyzed. Rather, we wish to point out a difference between young and old miRNAs in the likelihood that their expression is closely coupled with that of their host. Further to this finding, we then propose an evolutionary model in which miRNAs can undergo an embedding process, part of which can be increase in coordination in expression between the miRNA and the host gene.
MATERIALS AND METHODS
Annotation of intragenic miRNAs and host genes
We focused on intragenic miRNAs that are located within genes with reference sequences (RefSeqs), as those without Refseq might not be well annotated. Furthermore, according to a common criterion (23), only the miRNAs that have the same orientation with their host genes were counted as intragenic miRNAs. The University of California Santa Cruz (UCSC) Genome Browser (human genome hg19), RefSeq mRNA annotation and miRBase (Release 17) miRNA annotation were used to identify intronic and exonic miRNAs.
miRNA and mRNA expression profiling assays and data normalization
An in-house sample set including 81 human primary acute myeloid leukemia patient samples was used in this study. Exiqon miRCURY LNA™ Arrays (v11.0; covering over 1000 human) and Affymetrix Human Exon 1.0 ST Arrays were used to produce miRNA and mRNA expression profiling, respectively. See Supplementary Information for details about the data normalization. The data set has been deposited at GEO database (GSE27370; www.ncbi.nlm.nih.gov/geo/).
In addition, we obtained four data sets from GEO database (accession numbers GSE17306, GSE20161, GSE19783, GSE21032), which include miRNA and mRNA expression profiles of human myeloma (n = 52; primary patient samples), EBV transformed lymphoblastoid cell (n = 90; cell lines), breast cancer (n = 101; primary patient samples) and prostate (n = 139; human primary and metastatic prostate cancer samples and control normal adjacent benign prostate) samples. Agilent Human miRNA arrays, Illumina Human miRNA beadchip, Agilent Human Genome Microarray (probe name version), Affymetrix Human Genome U133 Plus 2.0 Array, Affymetrix Human Exon 1.0 ST arrays and Illumina Human expression beadchip, respectively, were used to derive the miRNA and gene expression profiles (33–36). For data set GSE21032, the data normalization were performed as described above. For data sets GSE17306, GSE20161 and GSE19783, we used their own normalized intensity.
Data analyses and statistics
Pearson's product-moment correlation was used to assess expression correlation between intragenic miRNAs and host genes (the sample number of each tissue was described above as well as in Tables 2–4). P < 0.05 was considered as significant. Evolutionary conservation information of miRNAs was downloaded from TargetScan (37) (http://www.targetscan.org/). Partek Genomics Suite (Partek Inc, St Louis, MI, USA), WinSTAT (R. Fitch Software; Bad Krozingen, Germany) and Bioconductor R packages were used for the data and statistics analyses.
Table 2.
Intragenic miRNAs that exhibit a significantly positive (P < 0.05) correlation of expression with their host genes
| Intragenic miRNAs | Leukemia sample set (n = 81), n/N (%) |
Myeloma sample set (n = 52), n/N (%) |
Lymphoblastoid cell sample set (n = 90), n/N (%) |
Breast cancer sample set (n = 101), n/N (%) |
Prostate sample set (n = 139), n/N (%) |
On average (%) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rawa | Bonferroni correctionb | Raw | Bonferroni correction | Raw | Bonferroni correction | Raw | Bonferroni correction | Raw | Bonferroni correction | Raw | Bonferroni correction | |
| Intragenic miRNAs | ||||||||||||
| Whole | 58/240 (24) | 21/240 (9) | 25/161 (16) | 6/161 (4) | 36/155 (23) | 18/155 (12) | 63/105 (60) | 48/105 (46) | 55/92 (60) | 32/92 (35) | 37 | 21 |
| Conserved | 40/78 (51) | 18/78 (23) | 20/73 (27) | 5/73 (7) | 21/67 (31) | 10/67 (15) | 47/61 (77) | 36/61 (59) | 47/58 (81) | 28/58 (48) | 53 | 30 |
| Non-Conserved | 18/162 (11) | 3/162 (2) | 5/88 (6) | 1/88 (1) | 15/88 (17) | 8/88 (9) | 16/44 (36) | 12/44 (27) | 8/34 (24) | 4/34 (12) | 19 | 10 |
| Intronic miRNAs | ||||||||||||
| Whole | 51/206 (25) | 19/206 (9) | 22/143 (15) | 5/143 (3) | 35/143 (24) | 17/143 (12) | 55/92 (60) | 43/92 (47) | 51/80 (64) | 29/80 (36) | 38 | 21 |
| Conserved | 37/71 (52) | 16/71 (23) | 17/65 (26) | 4/65 (6) | 20/64 (31) | 9/64 (14) | 42/55 (76) | 33/55 (60) | 44/52 (85) | 25/52 (48) | 54 | 30 |
| Non-conserved | 14/135 (10) | 3/135 (2) | 5/78 (6) | 1/78 (1) | 15/79 (19) | 8/79 (10) | 13/37 (35) | 10/37 (27) | 7/28 (25) | 4/28 (14) | 19 | 11 |
| Exonic miRNAs | ||||||||||||
| Whole | 7/34 (21) | 2/34 (6) | 3/18 (17) | 1/18 (6) | 1/12 (8) | 1/12 (8) | 8/13 (62) | 5/13 (38) | 4/12 (33) | 3/12 (25) | 28 | 17 |
| Conserved | 3/7 (43) | 2/7 (29) | 3/8 (38) | 1/8 (13) | 1/3 (33) | 1/3 (33) | 5/6 (83) | 3/6 (50) | 3/6 (50) | 3/6 (50) | 49 | 35 |
| Non-Conserved | 4/27 (15) | 0/27 (0) | 0/10 (0) | 0/10 (0) | 0/9 (0) | 0/9 (0) | 3/7 (43) | 2/7 (29) | 1/6 (17) | 0/6 (0) | 15 | 6 |
aThe original P-value from Pearson's correlation test was used to determine the significance.
bAdjusted P-value from Bonferroni correction was used to determine the significance.
Table 3.
Comparison of rates of coexpression with host genes between selected groups of conserved and non-conserved intragenic miRNAs
| Intragenic miRNAs | Leukemia sample set (n = 81) | Myeloma sample set (n = 52) | Lymphoblastoid cell sample set (n = 90) | Breast cancer sample set (n = 101) | Prostate sample set (n = 139) |
|---|---|---|---|---|---|
| Conserved | |||||
| Number of miRNAs | 53 | 72 | 42 | 25 | 21 |
| Expression range | 0.1–1.09 | 0.02–8.96 | 2.65–12.17 | 1.12–6.77 | 2.04–7.23 |
| Average expression level | 0.36 | 2.88 | 6.38 | 3.89 | 5.92 |
| Rate of coexpression with host genes, n/N (%) | 27/53 (51) | 20/72 (28) | 13/42 (31) | 20/25 (80) | 17/21 (81) |
| Non-conserved | |||||
| Number of miRNAs | 61 | 35 | 88 | 44 | 26 |
| Expression range | 0.1–0.79 | 0.42–8.49 | 2.68–15.4 | 0.78–10.92 | 4.00–10.35 |
| Average expression level | 0.34 | 2.87 | 6.12 | 3.95 | 5.93 |
| Rate of coexpression with host genes, n/N (%) | 3/61 (5) | 4/35 (11) | 15/88 (17) | 16/44 (36) | 7/26 (27) |
Table 4.
Antisense intragenic miRNAs that exhibit a significantly positive (P < 0.05) correlation of expression with their sense genes
| Antisense intragenic miRNAs | Leukemia sample set (n = 81), n/N (%) |
Myeloma sample set (n = 52), n/N (%) |
Lymphoblastoid cell sample set (n = 90), n/N (%) |
Breast cancer sample set (n = 101), n/N (%) |
Prostate sample set (n = 139), n/N (%) |
On average (%) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rawa | Bonferroni correctionb | Raw | Bonferroni correction | Raw | Bonferroni correction | Raw | Bonferroni correction | Raw | Bonferroni correction | Raw | Bonferroni correction | |
| Antisense intragenic miRNAs | ||||||||||||
| Whole | 5/35 (14) | 1/35 (3) | 1/23 (4) | 0/23 (0) | 3/27 (11) | 1/27 (4) | 2/13 (15) | 0/13 (0) | 3/13 (23) | 2/13 (15) | 13 | 4 |
| Conserved | 5/21 (24) | 1/21 (5) | 1/16 (6) | 0/16 (0) | 2/19 (11) | 1/19 (5) | 2/10 (2) | 0/10 (0) | 2/10 (20) | 2/10 (20) | 13 | 6 |
| Non-conserved | 0/14 (0) | 0/14 (0) | 0/7 (0) | 0/7 (0) | 1/8 (13) | 0/8 (0) | 0/3 (0) | 0/3 (0) | 1/3 (33) | 0/3 (0) | 9 | 0 |
| Antisense intronic miRNAs | ||||||||||||
| Whole | 4/26 (15) | 1/26 (4) | 1/22 (5) | 0/22 (0) | 3/21 (14) | 1/21 (5) | 2/11 (18) | 0/11 (0) | 3/10 (30) | 2/10 (20) | 16 | 6 |
| Conserved | 4/17 (24) | 1/17 (6) | 1/15 (7) | 0/15 (0) | 2/14 (14) | 1/14 (7) | 2/9 (22) | 0/9 (0) | 2/8 (25) | 2/8 (25) | 18 | 8 |
| Non-conserved | 0/9 (0) | 0/9 (0) | 0/7 (0) | 0/7 (0) | 1/7 (14) | 0/7 (0) | 0/2 (0) | 0/2 (0) | 1/2 (50) | 0/2 (0) | 13 | 0 |
| Antisense exonic miRNAs | ||||||||||||
| Whole | 1/9 (11) | 0/9 (0) | 0/1 (0) | 0/1 (0) | 0/6 (0) | 0/6 (0) | 0/2 (0) | 0/2 (0) | 0/3 (0) | 0/3 (0) | 2 | 0 |
| Conserved | 1/4 (25) | 0/4 (0) | 0/1 (0) | 0/1 (0) | 0/5 (0) | 0/5 (0) | 0/1 (0) | 0/1 (0) | 0/2 (0) | 0/2 (0) | 5 | 0 |
| Non-conserved | 0/5 (0) | 0/5 (0) | 0/1 (0) | 0/1 (0) | 0/1 (0) | 0/1 (0) | 0/1 (0) | 0/1 (0) | 0/1 (0) | 0/1 (0) | 0 | 0 |
aThe original P-value from Pearson's correlation test was used to determine the significance.
bAdjusted P-value from Bonferroni correction was used to determine the significance.
Database set up
To facilitate examination of the genomic loci of the intragenic miRNAs and their position relationship with corresponding host genes, as well as the positions of the probes of the host genes included in the Affymetrix exon arrays, we also developed a web-based database (http://chenlab.uchicago.edu/database/intragenic_mir.php).
RESULTS
Identification of intragenic miRNAs and host genes from the human genome
Based on RefSeq mRNA annotation (human genome hg19) in UCSC Genome Browser and miRNA annotation in miRBase (Release 17), we identified 657 human intragenic miRNAs that are embedded within genomic loci of 594 human RefSeq genes, respectively (Table 1 and Supplementary Table S1). Of them, 552 (84%) human intragenic miRNAs are intronic miRNAs, which are located in the introns of their host genes, whereas 105 (16%) human intragenic miRNAs are overlapping with exons of their host genes and thereby are referred to as exonic miRNAs (Table 1 and Supplementary Table S1).
Table 1.
Statistics of human intragenic miRNAs
| All (number of unique host genes) | 657 (594) |
|---|---|
| Genomic location, n (%) | |
| Intronic | 552 (84) |
| Exonic | 105 (16) |
| Evolutionary conservationa | |
| Conserved | 98 (32%) |
| Intronic | 89 |
| Exonic | 9 |
| Non-conserved | 209 (68%) |
| Intronic | 175 |
| Exonic | 34 |
aThe conservational subtypes were classed according to the categories of TargetScan (37) (Release 5.1; http://www.targetscan.org/). The ‘Highly conserved’ and ‘Conserved’ miRNAs were referred as ‘Conserved’ ones, that are conserved across most mammals. The ‘poorly conserved’ miRNAs were referred as ‘Non-Conserved’ ones. A total of 317 intragenic miRNAs have conservation information in TargetScan. The remaining 340 intragenic miRNAs were not included in TargetScan (37), and were excluded from our further analyses.
In contrast to prior reports that exonic miRNAs solely overlapping with exons of non-coding genes (10,38), we find that the host genes of exonic miRNAs are mainly protein-coding genes. As shown in Supplementary Table S2, 82% (86/105) of the unique host genes of the exonic miRNAs are protein-coding genes, whereas only 18% (19/105) are non-coding RNAs.
According to the criteria of TargetScan (37) (http://www.targetscan.org/), we classified intragenic miRNAs into two subgroups based on their evolution conservation degrees, including ‘conserved’ (i.e. those that are conserved across most mammals, including both ‘highly conserved’ and ‘conserved’ ones annotated in Targetscan) and ‘non-conserved’ ones (i.e. those that are identified as ‘poorly conserved’ in TargetScan). Among the 657 human intragenic miRNAs, 317 have conservation information in TargetScan (37) (http://www.targetscan.org/) and are thought to be highly reliable miRNAs. The remaining 340 miRNAs were not included in TargetScan. Among the 317 intragenic miRNAs, we found that the majority of intragenic miRNAs are ‘non-conserved’ ones: 209 (68%) are ‘non-conserved’, whereas only 98 (32%) are ‘conserved’ ones (Table 1). One may expect that most of the remaining 340 miRNAs might also be ‘non-conserved’ ones. Thus, these results accord with the notion that miRNAs are both readily gained and readily lost, resulting in an excess of young, non-conserved miRNAs (39,40).
Only a small fraction of intragenic miRNAs are likely coexpressed with their host genes in human leukemia samples
The concept that intragenic miRNAs are always coexpressed with their host genes was largely derived from several previous studies that focused on only a small number of intragenic miRNAs (10,21,22). For example, Rodriguez et al. (10) and Ronchetti et al. (22) experimentally assessed expression correlation of five (i.e. miR-9-3, miR-22, miR-137, miR-153 and miR-219-2) and three (miR-335, miR-342-3p and miR-561) intragenic miRNAs with their host genes, respectively. The largest set previously reported, the study of Baskerville and Bartel (21), focused on 34 intragenic miRNAs. In addition, since in early studies evolutionary conservation was used as a common criterion in identification of miRNAs (41,42), all miRNAs (including intragenic miRNAs) identified early on are almost exclusively evolutionarily conserved ones. Indeed, all the five miRNAs studied by Rodriguez et al. (10) and two of the three miRNAs studied by Ronchetti et al. (22) are ‘conserved’ ones. Of the 34 miRNAs studied by Baskerville and Bartel (21), 31 (91%) are ‘conserved’ and only 3 (9%) are ‘non-conserved’ ones.
As hundreds of intragenic miRNAs have been identified in mammalian genomes now and the majority of them are poorly conserved ones (Table 1), it is important to determine whether the current conception that most of the intragenic miRNAs are coexpressed with host genes is still true when we assess expression correlation of a much larger number of intragenic miRNAs with their host genes, and whether poorly conserved (i.e. ‘new’) intragenic miRNAs exhibit a similar trend in coexpression with their host genes as the evolutionarily conserved ones.
To this end, we conducted both miRNA and mRNA expression profiling assays of 81 human primary leukemia samples by use of Exiqon miRNA arrays and Affymetrix exon arrays, respectively (‘Materials and Methods’ section). We obtained reliable expression profiles for 240 intragenic miRNAs and their host genes. Surprisingly, we found that only 24% (58) of the 240 intragenic miRNAs exhibited a significantly positive correlation (P < 0.05, Pearson Correlation) of expression with their host genes (Table 2). After controlling for the probability of false positives (type I errors), 9% (21/240) of the intragenic miRNAs are coexpressed (adjusted P < 0.05, Bonferroni correction) with their host genes.
The discrepancy between our observation and the previous reports is likely attributed to the difference in proportions of conserved intragenic miRNAs
There was no P-value available in the Baskerville and Bartel set (21), but if we define a coexpressed pair as a pair of intragenic miRNA and host gene with a correlation coefficient of expression (i.e. r) > 0.3, 26 out of the 34 (i.e. 76%) intragenic miRNAs are coexpressed with their host genes in the Baskerville and Bartel set, which is significantly higher (P < 0.0001, chi-square test) than that (24%) we observed in our human leukemia set.
Tissue type and expression profiling technology differences might be able to explain part of the large discrepancy. However, ∼90% (31/34) of intragenic miRNAs studied by Baskerville and Bartel are conserved ones (21), and this ratio is significantly (P < 0.0001, chi-square test) greater than that (∼33%; 78/240) in our leukemia set (note: all the 240 intragenic miRNAs in the leukemia set have conservation information from TargetScan). Remarkably, we found that non-conserved intragenic miRNAs have a much lower possibility to be coexpressed with their host genes than conserved ones. As shown in Table 2, only 11% (18/162) of the poorly conserved intragenic miRNAs in our leukemia set are likely coexpressed with their host genes, over four times lower (P < 0.0001, chi-square test) than the coexpression rate (51%; 40/78) of the conserved ones. If only considering intragenic miRNAs included in the Baskerville and Bartel set (21) in our leukemia set, the coexpression rate would be dramatically increased from 24% (58/240) to 70% (19/27), close to that (76%) reported by Baskerville and Bartel (21). Thus, our data indicate that evolutionarily conserved intragenic miRNAs have a significantly (P < 0.0001, chi-square test) higher possibility than non-conserved ones in coexpression with host genes, and that the discrepancy between our observation and the previous reports is largely attributed to the difference in proportions of conserved intragenic miRNAs.
Evolutionarily conserved intragenic miRNAs exhibit a significantly higher rate of coexpression with host genes than non-conserved ones within and across individual sets
To examine the robustness and generality of our above finding from human leukemia sample set, we obtained miRNA and mRNA expression profiles of four additional human sample sets from a public database (GEO; ‘Materials and Methods’ section); in each set, we have more than 50 individual samples and over 90 intragenic miRNA–host gene pairs (Table 2). We then correlated expression of each intragenic miRNA with that of the host gene in each sample set.
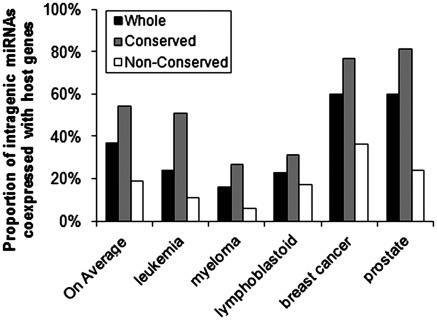
Although the rate of potential coexpression between intragenic miRNAs and host genes varies greatly between different data sets (e.g. only 16–24% in the human leukemia, myeloma or lymphoblastoid cell samples, but as high as 60% in both human breast cancer and prostate samples; Pearson correlation; based on raw P-values), we observed that conserved intragenic miRNAs exhibited a significantly (P < 0.01, chi-square test) higher rate of coexpression with their host genes than non-conserved ones in each data set (Table 2 and Figure 1; all the intragenic miRNAs with reliable expression values in each data set have conservation information from TargetScan). On average, 53% (based on raw P-values) or 30% (Bonferroni correction) of ‘conserved’, whereas only 19% (based on raw P-values) or 10% (Bonferroni correction) of ‘non-conserved’ intragenic miRNAs are likely coexpressed with their host genes across the five sample sets (P < 0.05, Wilcoxon rank test—based on either raw or adjusted P-values; Figure 1 and Table 2). The difference of coexpression rate between ‘conserved’ and ‘non-conserved’ subgroups was also significant (P < 0.05, Wilcoxon rank test) in both intronic and exonic miRNAs (Table 2).
Figure 1.
Proportion of intragenic miRNAs that are likely coexpressed with their host genes in each individual sample set and the average proportions across the five sample sets. Based on the raw P-values.
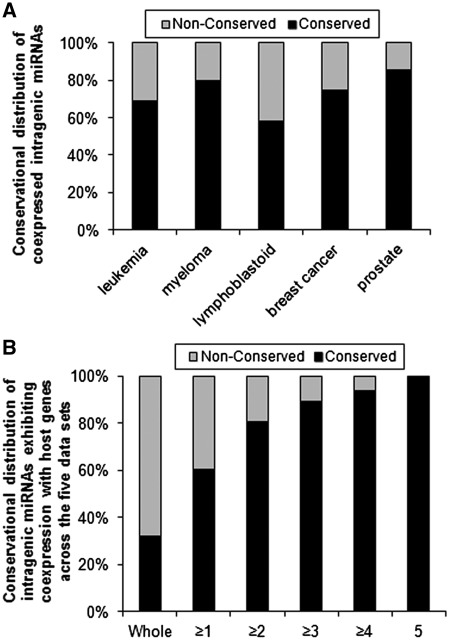
As a result, the majority of the intragenic miRNAs that are likely coexpressed with their host genes are evolutionarily conserved ones in each data set (Table 2 and Figure 2A). The fact that we can recover high coexpression rates for conserved genes supports the view that our coexpression scoring method is adequately powerful to detect coexpression if it is present.
Figure 2.
The majority or most of the coexpressed intragenic miRNAs are conserved ones, which also exhibit coexpression with host genes in more data sets than non-conserved ones. (A) Conservational distribution of coexpressed intragenic miRNAs in individual sample sets based on the raw P-values. (B) Conservational distribution of the intragenic miRNAs that exhibit coexpression (based on raw P-values) with their host genes in more than one, more than two, more than three, more than four and all the five sample sets, respectively.
We also analyzed the coexpression probability of each intragenic miRNA with its host gene across the five sample sets. There are a total of 90 conserved and 189 non-conserved unique intragenic miRNAs that have reliable expression profiles with their host genes in at least one of the five sample sets (Supplementary Table S3). As expected, conserved intragenic miRNAs have a much higher possibility to be coexpressed with their host genes across the five sample sets (Figure 2B and Supplementary Table S3). For example, 74% (67/90), 60% (54/90) and 37% (33/90) of the conserved intragenic miRNAs are likely coexpressed with their host genes in at least one, two and three sample sets, respectively; in contrast, the ratios for non-conserved intragenic miRNAs are 23% (44/189), 7% (13/189) and 2% (4/189), respectively (Supplementary Table S3 and Figure 2B). The average number of sample sets in which a given conserved intragenic miRNA exhibits a significant (raw P < 0.05) coexpression with its host gene is ∼2, significantly (P < 0.05, Mann–Whitney U-test) greater than that (0.3) of non-conserved intragenic miRNAs.
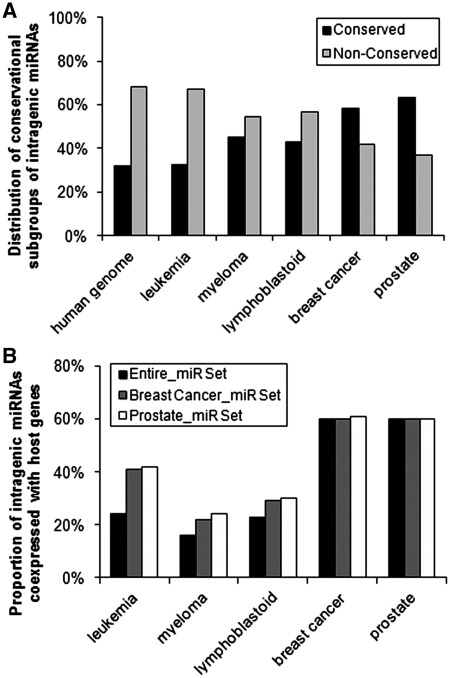
The higher coexpression rate in breast cancer or prostate sample set is likely related to a higher proportion of conserved intragenic miRNAs and some other factors
The higher coexpression rate in breast cancer or prostate sample set (60%) than in other three data sets (16–24%) is likely related with a higher proportion of conserved intragenic miRNAs (Figure 3A). If only considering miRNAs in breast cancer or prostate cancer sample set, the coexpression rate was also increased in the other three sample sets compared to that of entire miRNA set (Figure 3B).
Figure 3.
High rates of coexpression of intragenic miRNAs with host genes in the breast cancer and prostate cancer sample sets are, at least in part, related to the predominance of ‘conserved’ intragenic miRNAs. (A) Proportions of ‘conserved’ and ‘non-conserved’ intragenic miRNAs in the five sample sets. (B) Proportion of intragenic miRNAs that coexpressed with their host genes when using intragenic miRNAs from the breast cancer set and prostate cancer sample set, respectively. Based on raw P-values.
Nevertheless, even if considering similar sets of intragenic miRNAs, the coexpression rates are not the same between different sample sets (Figure 3B), suggesting that some other factors may also have influence on coexpression probability of intragenic miRNAs with their host genes. For example, sample size (e.g. the breast cancer and prostate sample sets have a greater number of individual samples than the other three sets), variability of sample types in each set (e.g. the prostate set contains both tumor and normal control samples, whereas the other sets have more unique sample types), and tissue specificity (e.g. intragenic miRNAs may tend to be coexpressed with host genes in some types of tissues rather than in other types) may also have some influence.
The difference of coexpression rates between conserved and non-conserved intragenic miRNAs is not due to potential differences in abundance
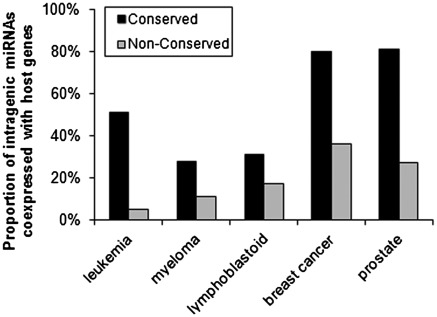
One may argue that many poorly conserved miRNAs are expressed at very low levels and are not detected reliably by microarrays so that non-conserved intragenic miRNAs exhibited a lower rate of coexpression with host genes as detected by microarrays. In order to eliminate the potential effects of different expression abundance between conserved and non-conserved intragenic miRNAs in microarray experiments, we selected conserved and non-conserved miRNA groups that have comparable mean expression levels. As shown in Table 3, the range and average expression abundance of the selected group of non-conserved intragenic miRNAs are similar to those of the selected group of conserved intragenic miRNAs in each sample set.
Notably, we still observed significantly (P < 0.01, chi-square test) higher coexpression rates between intragenic miRNAs and their host genes in ‘conserved’ than in ‘non-conserved’ ones in each sample set: 51% (27/53) of ‘conserved’ and 5% (3/61) of ‘non-conserved’ in leukemia sample set; 28% (20/72) of ‘conserved’ and 11% (4/35) of ‘non-conserved’ in myeloma sample set; 31% (13/42) of ‘conserved’ and 17% (15/88) of ‘non-conserved’ in lymphoblastoid cell line sample set; 80% (20/25) of ‘conserved’ and 36% (16/44) of ‘non-conserved’ in breast cancer sample set; 81% (17/21) of ‘conserved’ and 27% (7/26) of ‘non-conserved’ in prostate sample set, respectively (Table 3 and Figure 4). On average, 54% of the ‘conserved’ intragenic miRNAs are likely coexpressed with their host genes across the five sample sets, significantly (P < 0.05, Wilcoxon rank test) more frequent than the coexpression rate (19%) of ‘non-conserved’ ones.
Figure 4.
Comparison of rates of coexpression with host genes between selected groups of conserved and non-conserved intragenic miRNAs that have similar expression abundance in each sample set (Table 3). Based on raw P-values.
To avoid the problem of arbitrarily classifying a given miRNA as either coexpressed with its host gene or not, we also consider for each of the five samples, an ANCOVA in which we ask whether conserved miRNAs have higher raw coexpression values than non-conserved ones, controlling for abundance. For each of the five samples, we started by testing for an interaction effect (which would preclude the use of ANCOVA). In no case did we observe a significant interaction term (input gene set the same as in Table 3). In all five samples, we observe that conserved miRNAs have higher r values than non-conserved ones, controlling for abundance (Supplementary Figures S1–S5). From Fisher's method, we can combine these results to one chi-squared test which is very highly significant (Fisher's method, χ2 = 80.4, df = 10, P < 0.0001). The same is found if we consider all genes, not just those matched for expression level (Fisher's method, χ2 = 82.8, df = 10, P < 0.0001). We conclude that conserved miRNAs are consistently more highly coexpressed than younger ones controlling for any difference in abundance.
Sense strand intragenic miRNAs are more likely to be coexpressed with their host genes than are antisense miRNAs
Previous studies (43–47) suggest that genomically neighboring genes tend to be coexpressed across tissues or through time. For example, the coexpression of some neighboring genes might be attributable to the open chromatin domain of that entire region (48–50), as supported by transgene analysis (51). Similarly, coexpression of some intragenic miRNAs and host genes might also be attributable to chromatin-level regulation, rather than sharing primary transcripts. To evaluate the effect of chromatin-level regulation on the rate of coexpression between intragenic miRNAs and host genes, we assessed the correlation of expression between antisense miRNAs, i.e. those that reside on the opposite strand to protein-coding sense strand, and the sense genes. By definition, the antisense miRNAs cannot be driven off the same promoter nor be splice products of the sense strand mRNA. As the analysis is also using miRNAs, this approach also controls for methodological noise.
We identified a total of 167 antisense miRNAs in the human genome (Supplementary Tables S4 and S5). Among them, 35, 23, 27, 13 and 13 antisense miRNAs have expression values available in the human leukemia, myeloma, lymphoblastoid cell, breast cancer and prostate samples, respectively, along with expression values of their corresponding sense genes. On average, only 13% (based on raw P-values) or 4% (based on the adjusted P-values via Bonferroni correction) of the antisense miRNAs are likely coexpressed with the sense host genes (Table 4), which is significantly less frequent than do (sense) intragenic miRNAs and host genes (on average, 53% or 30% based on the raw or adjusted P-values; Table 2) in each sample set (P < 0.01, Fisher's exact test) or across the five sample sets (P < 0.05, Wilcoxon rank test). The difference of coexpression probability with host genes between antisense and sense intragenic miRNAs is significant (on average, 13% versus 53%; P < 0.05, Wilcoxon rank test) in the conserved population, but not significant (on average, 9% versus 19%; P = 0.2, Wilcoxon rank test) in the non-conserved population.
Thus, our data suggest that the high frequency (on average, 53%; Table 2) of coexpression between conserved intragenic miRNAs and their host genes is likely largely due to the sharing of promoters or primary transcripts, whereas only a small fraction (13% out of the 53%; derived from the coexpression rate between conserved antisense intragenic miRNAs and sense host genes; Table 4) of the signal of coexpression can be accounted for in terms of shared chromosomal location. The low frequency (on average, 19%; Table 2) of coexpression between non-conserved intragenic miRNAs and their host genes is likely partially (9% out of the 19%; derived from the coexpression rate between non-conserved antisense intragenic miRNAs and sense host genes; Table 4) due to the shared chromosomal location, and the remaining part might be also owing to the shared promoters or primary transcripts, which might occur by chance.
DISCUSSION
In contrast to the prior conception, that intragenic miRNAs are always coexpressed with their host genes due to sharing the same promoters or primary transcripts (10,21,22), we showed here that not all types of intragenic miRNAs have the same coexpression trend. We observed that the probability of coexpression of intragenic miRNA with host genes is strongly related to their evolutionary conservation degree. Evolutionarily conserved intragenic miRNAs have a much higher rate of coexpression with host genes than poorly conserved ones (Table 2).
The covariance with age is important for several reasons. First, the result runs contrary to the expectations of prior analyses (52), which suppose that coexpression between miRNA and host gene is the ancestral state and necessary for the emergence of an miRNA. Second, the age effect argues against a methodological problem. One might suppose that as miRNA and mRNA are on separate decay paths that we might never expect a correlation between miRNA and mRNA levels even if piggybacking was occurring. If this is a bona fide technical problem, however, we should not expect to see any systematic patterns of coexpression associated with miRNA age. Furthermore, as shown in Tables 2 and 4, intragenic miRNAs (sense) exhibit a much (53% versus 13%; >4-fold; P < 0.05, Fisher's exact test) higher rate of coexpression with host genes than the antisense intragenic miRNAs do with their sense genes. Such data suggest that besides the shared chromatin regulation effect (as reflected by the coexpression probability of antisense miRNAs with sense genes), a certain number of intragenic miRNAs, particularly the evolutionarily conserved ones, may really share promoters/primary transcripts with their host genes.
Why then do the old (i.e. conserved) intronic miRNAs tend to be cotranscribed with their host genes more than the new (i.e. non-conserved) ones? We suggest that most new miRNAs are expressed serendipitously and weakly but not necessarily always from the same transcripts as the host genes. Some are coexpressed owing to shared chromatin dynamics, just as some antisense miRNAs are coexpressed with the corresponding sense genes. Let us then conjecture that for some of these their coexpression is beneficial and selection favors them to be what we might call ‘embedded’, meaning they evolve stronger coexpression, which in turn favors conservation of miRNA target sites on 3′-UTRs of desirable target genes (and possibly selection for undesirable targets to evolve away from being targets). For instance, some intronic miRNAs may functionally cooperate with their host genes. Indeed, a ‘conserved’ miRNA, miR-338 has been reported to be cotranscribed with its host gene, apoptosis-associated tyrosine kinase (AATK), and more importantly, exhibits functional cooperation with its host gene by repressing genes that are functionally antagonistic to the host gene (27). The strong coexpression is perhaps best achieved by sharing promoters/transcripts. Eventually, then we see old intragenic miRNAs exhibiting a greater possibility of being coexpressed with host genes than the newer less embedded ones do. Antisense miRNA cannot easily become embedded as they do not have the option of coopting the sense transcripts nor the sense promoter. This is by no means the only possible model to explain our results. It could be that for a few miRNAs sharing of the promoter and strong coexpression with the host gene was the ancestral condition. It is not, however, clear why these should necessarily also be more ancient. In such an instance, we might also expect that among the set of genes with young miRNAs we should see a bimodal distribution of coexpression r values, such that some (transcript sharing ones) would be centered around a high coexpression score and others around a much lower value. However, when we test young miRNAs for deviation from unimodality of the coexpression values for each of the five samples, we see no evidence such deviation [Hartigan and Hartigan dip test (53), P > 0.05 in all instances].
Assuming our results are as robust as they appear to be, how then can we explain the two pre-eminent observations that were the stimulus to the notion that young miRNAs must piggy-back on their host genes (52)? First, that so many miRNAs are intragenic (and young ones tend to more commonly be intragenic). Second, that when they are reported intragenically, there are approximately four times more in the sense strand than antisense (657 versus 167 in humans, Table 1 and Supplementary Table S4). As most of the genome is not protein coding, randomly inserted/generated miRNA one would also expect to be in intergene spacer. Similarly, the simplest null would predict an equal number sense and antisense. For both observations, we can rule out ascertainment bias as most of the miRNAs are identified from small RNA libraries without pre-selection in recent years.
Both facts can, at least in part, be explained by the idea that miRNAs are more likely to persist if they can have their expression piggybacking on that of a host gene. Nonetheless, as the majority of both sense and antisense intragenic miRNAs are poorly conserved ones, the above model cannot explain the two facts thoroughly. Perhaps, we might also suppose that new sense and antisense intragenic miRNAs both have expression enabled largely by being in open chromatin associated with transcriptional activity of host genes; however, frequent RNA polymerase collisions might select against intragenic antisense transcripts, much as RNA polymerase DNA polymerase collisions in bacteria is thought to affect strand bias (54). As a result, even among poorly conserved intragenic miRNAs, most of them are located in sense with the host genes (Table 1 and Supplementary Table S4).
Meanwhile, that miRNAs are commonly intragenic accords with the observation that miRNAs are enriched in transposon-free regions that tend also to be domains of high protein-coding density (55). There exist multiple mutational/insertion bias models alongside selectionist models to explain both observations. Perhaps, newly created miRNAs are more likely to become expressed if within a gene owing to being in open chromatin at some point in time (whenever the host gene is in open chromatin), expression of the miRNA being a minimal criterion for its persistence? Alternatively, there might exist insertion biases for miRNAs produced via retroposition-like events, much as retroviruses often prefer to insert near transcriptionally active genes (56). Alternatively, if introns are under selection to have particular stem–loop structures, perhaps to aid exon junction recognition, they might be genomic sweet spots for the emergence of miRNA-like structures (57). A better understanding of the fate of very young miRNAs, where they come from, how they become expressed should shed light on these issues.
Our data indicate that a fraction of conserved and the majority of non-conserved intragenic miRNAs are unlikely to be strongly coexpressed with their host genes, which is accord with recent studies that have suggested that over one-third of intronic miRNAs have their own promoters (Polymerase II or III), whose expression occurs independently from host gene transcription (11,12,58–61). For examples, Fisher and colleagues (11) conducted nucleosome positioning analyses (62) and chromatin immunoprecipitation (ChIP)-chip assays (63) in two human melanoma (UACC62 and MALME) lines and one breast cancer (MCF7) cell line and observed that one-third of intragenic miRNAs may have their own promoters. Corcoran et al. (60) performed Polymerase II ChIP-chip assays in A549 human lung epithelial cells and showed that over 26% of the intragenic miRNAs may be transcribed from their own Pol II promoters. Similarly, Monteys et al. (12) observed that ∼35% of intragenic miRNAs have upstream regulatory elements consistent with Pol II (30%) or Pol III (5%) promoter function in human genome. More importantly, they further cloned intronic regions composed of miRNAs and their upstream Pol II (for miR-107, miR-126, miR-208b, miR-548f-2, miR-569 and miR-590) or Pol III (for miR-566 and miR-128-2) sequences into a promoterless plasmid, and confirmed that miRNA expression occurs independent of host gene transcription (12). In Caenorhabditis. elegans, Martinez et al. (59) conducted genome-wide assays of promoters of 89 miRNAs (66% of all predicted miRNAs) using transgenic promoter–reporter constructs and their results indicated that intronic miRNAs are likely controlled by their own, rather than the promoters of host genes. Similarly, Isik et al. (61) reported that over one-third of intronic miRNAs in C. elegans have their own promoters and could be transcribed independently from host genes. Given the fact that on average over 50% of intragenic miRNAs might not be coexpressed with host genes (Table 2), we expect that probably more than one-third of intragenic miRNAs may have their own promoters or relevant transcriptional regulatory elements. It was reported that miRNAs can be transcribed from promoters located several kilobases away (60), and thus it is possible that promoters of many intragenic miRNAs have not been identified yet.
Given the above findings, caution should be paid to those analyses in which expression profiles of host genes were used as a proxy for the expression of the corresponding intragenic miRNAs, such as in the cases where expression profiles of host genes were used to predict miRNA target genes (29–32). If the intragenic miRNA is not an evolutionarily conserved one, cases where the upregulation of one gene in turn causes, by miRNA-mediated effects, the downregulation of others should probably be considered the exception not the rule.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5, Supplementary Tables 1–5, Supplementary Methods and Supplementary References [64–66].
FUNDING
G. Harold and Leila Y. Mathers Charitable Foundation (to J.C.); the National Institutes of Health (NIH) (R01 CA127277) (to J.C.); Gabrielle's Angel Foundation (to J.C., Z.L. and H.H.) and Leukemia & Lymphoma Society Special Fellow (to Z.L.). LDH is a Royal Society Wolfson Research Merit Award Holder. Funding for open access charge: G. Harold and Leila Y. Mathers Charitable Foundation (to J.C.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank H.S. Xiao and Y. Qu in Shanghai Microarray Company for suggestion on exon array analysis.
REFERENCES
- 1.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 4.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu W, Sun M, Zou GM, Chen J. MicroRNA and cancer: Current status and prospective. Int. J. Cancer. 2007;120:953–960. doi: 10.1002/ijc.22454. [DOI] [PubMed] [Google Scholar]
- 6.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu. Rev. Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 7.Ritchie W, Rajasekhar M, Flamant S, Rasko JE. Conserved expression patterns predict microRNA targets. PLoS Comput. Biol. 2009;5:e1000513. doi: 10.1371/journal.pcbi.1000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ritchie W, Flamant S, Rasko JE. mimiRNA: a microRNA expression profiler and classification resource designed to identify functional correlations between microRNAs and their targets. Bioinformatics. 2010;26:223–227. doi: 10.1093/bioinformatics/btp649. [DOI] [PubMed] [Google Scholar]
- 9.Chen J, Odenike O, Rowley JD. Leukaemogenesis: more than mutant genes. Nat. Rev. Cancer. 2010;10:23–36. doi: 10.1038/nrc2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ozsolak F, Poling LL, Wang Z, Liu H, Liu XS, Roeder RG, Zhang X, Song JS, Fisher DE. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monteys AM, Spengler RM, Wan J, Tecedor L, Lennox KA, Xing Y, Davidson BL. Structure and activity of putative intronic miRNA promoters. RNA. 2010;16:495–505. doi: 10.1261/rna.1731910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saini HK, Enright AJ, Griffiths-Jones S. Annotation of mammalian primary microRNAs. BMC Genomics. 2008;9:564. doi: 10.1186/1471-2164-9-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shomron N, Levy C. MicroRNA-biogenesis and Pre-mRNA splicing crosstalk. J. Biomed. Biotechnol. 2009;2009:594678. doi: 10.1155/2009/594678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smalheiser NR. EST analyses predict the existence of a population of chimeric microRNA precursor-mRNA transcripts expressed in normal human and mouse tissues. Genome Biol. 2003;4:403. doi: 10.1186/gb-2003-4-7-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pawlicki JM, Steitz JA. Primary microRNA transcript retention at sites of transcription leads to enhanced microRNA production. J. Cell Biol. 2008;182:61–76. doi: 10.1083/jcb.200803111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shiohama A, Sasaki T, Noda S, Minoshima S, Shimizu N. Nucleolar localization of DGCR8 and identification of eleven DGCR8-associated proteins. Exp. Cell Res. 2007;313:4196–4207. doi: 10.1016/j.yexcr.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 19.Wen X, Tannukit S, Paine ML. TFIP11 interacts with mDEAH9, an RNA helicase involved in spliceosome disassembly. Int. J. Mol. Sci. 2008;9:2105–2113. doi: 10.3390/ijms9112105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 21.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ronchetti D, Lionetti M, Mosca L, Agnelli L, Andronache A, Fabris S, Deliliers GL, Neri A. An integrative genomic approach reveals coordinated expression of intronic miR-335, miR-342, and miR-561 with deregulated host genes in multiple myeloma. BMC Med. Genomics. 2008;1:37. doi: 10.1186/1755-8794-1-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ying SY, Lin SL. Current perspectives in intronic micro RNAs (miRNAs) J. Biomed. Sci. 2006;13:5–15. doi: 10.1007/s11373-005-9036-8. [DOI] [PubMed] [Google Scholar]
- 24.Ying SY, Chang DC, Lin SL. The microRNA (miRNA): overview of the RNA genes that modulate gene function. Mol. Biotechnol. 2008;38:257–268. doi: 10.1007/s12033-007-9013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saini HK, Griffiths-Jones S, Enright AJ. Genomic analysis of human microRNA transcripts. Proc. Natl Acad. Sci. USA. 2007;104:17719–17724. doi: 10.1073/pnas.0703890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin SL, Miller JD, Ying SY. Intronic microRNA (miRNA) J. Biomed. Biotechnol. 2006;2006:26818. doi: 10.1155/JBB/2006/26818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barik S. An intronic microRNA silences genes that are functionally antagonistic to its host gene. Nucleic Acids Res. 2008;36:5232–5241. doi: 10.1093/nar/gkn513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morlando M, Ballarino M, Gromak N, Pagano F, Bozzoni I, Proudfoot NJ. Primary microRNA transcripts are processed co-transcriptionally. Nat. Struct. Mol. Biol. 2008;15:902–909. doi: 10.1038/nsmb.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gennarino VA, Sardiello M, Avellino R, Meola N, Maselli V, Anand S, Cutillo L, Ballabio A, Banfi S. MicroRNA target prediction by expression analysis of host genes. Genome Res. 2009;19:481–490. doi: 10.1101/gr.084129.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Radfar M, Wong W, Morris QD. Predicting the target genes of intronic microRNAs using large-scale gene expression data. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2010;1:791–794. doi: 10.1109/IEMBS.2010.5626505. [DOI] [PubMed] [Google Scholar]
- 31.Radfar MH, Wong W, Morris Q. Computational prediction of intronic microRNA targets using host gene expression reveals novel regulatory mechanisms. PLoS One. 2011;6:e19312. doi: 10.1371/journal.pone.0019312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gennarino VA, Sardiello M, Mutarelli M, Dharmalingam G, Maselli V, Lago G, Banfi S. HOCTAR database: a unique resource for microRNA target prediction. Gene. 2011;480:51–58. doi: 10.1016/j.gene.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Enerly E, Steinfeld I, Kleivi K, Leivonen SK, Aure MR, Russnes HG, Ronneberg JA, Johnsen H, Navon R, Rodland E, et al. miRNA-mRNA integrated analysis reveals roles for miRNAs in primary breast tumors. PLoS One. 2011;6:e16915. doi: 10.1371/journal.pone.0016915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L, Tang H, Thayanithy V, Subramanian S, Oberg AL, Cunningham JM, Cerhan JR, Steer CJ, Thibodeau SN. Gene networks and microRNAs implicated in aggressive prostate cancer. Cancer Res. 2009;69:9490–9497. doi: 10.1158/0008-5472.CAN-09-2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou Y, Chen L, Barlogie B, Stephens O, Wu X, Williams DR, Cartron MA, van Rhee F, Nair B, Waheed S, et al. High-risk myeloma is associated with global elevation of miRNAs and overexpression of EIF2C2/AGO2. Proc. Natl Acad. Sci. USA. 2010;107:7904–7909. doi: 10.1073/pnas.0908441107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 38.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 39.Marco A, Hui JH, Ronshaugen M, Griffiths-Jones S. Functional shifts in insect microRNA evolution. Genome Biol. Evol. 2010;2:686–696. doi: 10.1093/gbe/evq053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu J, Shen Y, Wu Q, Kumar S, He B, Shi S, Carthew RW, Wang SM, Wu CI. The birth and death of microRNA genes in Drosophila. Nat. Genet. 2008;40:351–355. doi: 10.1038/ng.73. [DOI] [PubMed] [Google Scholar]
- 41.Berezikov E, Cuppen E, Plasterk RH. Approaches to microRNA discovery. Nat. Genet. 2006;38(Suppl.):S2–S7. doi: 10.1038/ng1794. [DOI] [PubMed] [Google Scholar]
- 42.Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 43.Lercher MJ, Blumenthal T, Hurst LD. Coexpression of neighboring genes in Caenorhabditis elegans is mostly due to operons and duplicate genes. Genome Res. 2003;13:238–243. doi: 10.1101/gr.553803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lercher MJ, Urrutia AO, Hurst LD. Clustering of housekeeping genes provides a unified model of gene order in the human genome. Nat. Genet. 2002;31:180–183. doi: 10.1038/ng887. [DOI] [PubMed] [Google Scholar]
- 45.Deng Y, Dai X, Xiang Q, Dai Z, He C, Wang J, Feng J. Genome-wide analysis of the effect of histone modifications on the coexpression of neighboring genes in Saccharomyces cerevisiae. BMC Genomics. 2010;11:550. doi: 10.1186/1471-2164-11-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Purmann A, Toedling J, Schueler M, Carninci P, Lehrach H, Hayashizaki Y, Huber W, Sperling S. Genomic organization of transcriptomes in mammals: Coregulation and cofunctionality. Genomics. 2007;89:580–587. doi: 10.1016/j.ygeno.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 47.Woo YH, Walker M, Churchill GA. Coordinated expression domains in mammalian genomes. PLoS One. 5:e12158. doi: 10.1371/journal.pone.0012158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hurst LD, Pal C, Lercher MJ. The evolutionary dynamics of eukaryotic gene order. Nat. Rev. Genet. 2004;5:299–310. doi: 10.1038/nrg1319. [DOI] [PubMed] [Google Scholar]
- 49.Singer GA, Lloyd AT, Huminiecki LB, Wolfe KH. Clusters of coexpressed genes in mammalian genomes are conserved by natural selection. Mol. Biol. Evol. 2005;22:767–775. doi: 10.1093/molbev/msi062. [DOI] [PubMed] [Google Scholar]
- 50.Batada NN, Urrutia AO, Hurst LD. Chromatin remodelling is a major source of coexpression of linked genes in yeast. Trends Genet. 2007;23:480–484. doi: 10.1016/j.tig.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 51.Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Campo-Paysaa F, Semon M, Cameron RA, Peterson KJ, Schubert M. microRNA complements in deuterostomes: origin and evolution of microRNAs. Evol. Dev. 2011;13:15–27. doi: 10.1111/j.1525-142X.2010.00452.x. [DOI] [PubMed] [Google Scholar]
- 53.Hartigan JA, Hartigan PM. The Dip Test of Unimodality. Ann. Stat. 1985;13:70–84. [Google Scholar]
- 54.Rocha EP. The organization of the bacterial genome. Annu. Rev. Genet. 2008;42:211–233. doi: 10.1146/annurev.genet.42.110807.091653. [DOI] [PubMed] [Google Scholar]
- 55.Simons C, Pheasant M, Makunin IV, Mattick JS. Transposon-free regions in mammalian genomes. Genome Res. 2006;16:164–172. doi: 10.1101/gr.4624306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang GP, Ciuffi A, Leipzig J, Berry CC, Bushman FD. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007;17:1186–1194. doi: 10.1101/gr.6286907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berezikov E. Evolution of microRNA diversity and regulation in animals. Nat. Rev. Genet. 2011;12:846–860. doi: 10.1038/nrg3079. [DOI] [PubMed] [Google Scholar]
- 58.Aboobaker AA, Tomancak P, Patel N, Rubin GM, Lai EC. Drosophila microRNAs exhibit diverse spatial expression patterns during embryonic development. Proc. Natl Acad. Sci. USA. 2005;102:18017–18022. doi: 10.1073/pnas.0508823102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martinez NJ, Ow MC, Reece-Hoyes JS, Barrasa MI, Ambros VR, Walhout AJ. Genome-scale spatiotemporal analysis of Caenorhabditis elegans microRNA promoter activity. Genome Res. 2008;18:2005–2015. doi: 10.1101/gr.083055.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Corcoran DL, Pandit KV, Gordon B, Bhattacharjee A, Kaminski N, Benos PV. Features of mammalian microRNA promoters emerge from polymerase II chromatin immunoprecipitation data. PLoS One. 2009;4:e5279. doi: 10.1371/journal.pone.0005279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Isik M, Korswagen HC, Berezikov E. Expression patterns of intronic microRNAs in Caenorhabditis elegans. Silence. 2010;1:5. doi: 10.1186/1758-907X-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ozsolak F, Song JS, Liu XS, Fisher DE. High-throughput mapping of the chromatin structure of human promoters. Nat. Biotechnol. 2007;25:244–248. doi: 10.1038/nbt1279. [DOI] [PubMed] [Google Scholar]
- 63.Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 64.Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, Holloway A, Smyth GK. A comparison of background correction methods for two-colour microarrays. Bioinformatics. 2007;23:2700–2707. doi: 10.1093/bioinformatics/btm412. [DOI] [PubMed] [Google Scholar]
- 65.Berger JA, Hautaniemi S, Jarvinen AK, Edgren H, Mitra SK, Astola J. Optimized LOWESS normalization parameter selection for DNA microarray data. BMC Bioinformatics. 2004;5:194. doi: 10.1186/1471-2105-5-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.