Abstract
MDC1 is a key mediator of the DNA-damage response in mammals with several phosphorylation-dependent protein interaction domains. The function of its N-terminal forkhead-associated (FHA) domain remains elusive. Here, we show with structural, biochemical and cellular data that the FHA domain mediates phosphorylation-dependent dimerization of MDC1 in response to DNA damage. Crystal structures of the FHA domain reveal a face-to-face dimer with pseudo-dyad symmetry. We found that the FHA domain recognizes phosphothreonine 4 (pT4) at the N-terminus of MDC1 and determined its crystal structure in complex with a pT4 peptide. Biochemical analysis further revealed that in the dimer, the FHA domain binds in trans to pT4 from the other subunit, which greatly stabilizes the otherwise unstable dimer. We show that T4 is phosphorylated primarily by ATM upon DNA damage. MDC1 mutants with the FHA domain deleted or impaired in its ability to dimerize formed fewer foci at DNA-damage sites, but the localization defect was largely rescued by an artificial dimerization module, suggesting that dimerization is the primary function of the MDC1 FHA domain. Our results suggest a novel mechanism for the regulation of MDC1 function through T4 phosphorylation and FHA-mediated dimerization.
INTRODUCTION
Genomic DNA is constantly damaged by assaults from both exogenous factors (e.g. the ultraviolet components of sunlight) and endogenous factors (e.g. the free radicals generated from metabolic intermediates). The DNA-damage response (DDR) pathways detect and repair damaged DNA or induce apoptosis if the damage is too severe to be repaired. Defects in the DDR result in genome instability and disease (1).
DNA double-strand breaks (DSBs), the most severe form of DNA lesions, can be induced by genotoxic agents such as ionizing radiation (IR) and chemotherapeutic agents such as camptothecin (CPT) or can be generated during cellular processes. DSBs are repaired by two major mechanisms: homologous recombination (HR) and non-homologous end-joining (NHEJ) (2). DSBs are rapidly bound by the Ku70/Ku80 heterodimer, recruiting and activating the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs), a phosphoinositol-3 like kinase (PIKK), to initiate NHEJ (3). DSBs are also sensed by the Mre11-Rad50-NBS1 (MRN) complex, which recruits and activates another PIKK, ataxia-telangiectasia mutated (ATM), in preparation for HR-mediated repair (4). One key consequence of ATM activation is the rapid formation of the S139-phosphorylated histone variant H2AX (γ-H2AX) surrounding DNA-damage sites (5). γ-H2AX serves as a platform for the recruitment and enrichment of many DDR factors (6,7). Microscopy and chromatin immunoprecipitation (ChIP) experiments have revealed that many DDR factors, such as mediator of DNA-damage checkpoint 1 (MDC1, also known as nuclear factor with BRCT domains 1 or NFBD1), ATM and the MRN complex, co-localize with γ-H2AX foci that can extend up to 1–2 megabases on both sides of the DNA-damage sites (1,6-9).
MDC1 is the master organizer of assembly and maintenance of the γ-H2AX-decorated chromatin region (10-15). MDC1-knockout mice present growth retardation, male infertility, immune defects, chromosome instability, defects in DNA repair by HR, radiation sensitivity and cancer predisposition (16,17). A reduction or lack of MDC1 expression has been observed in a significant subset of breast cancers and lung cancers (18). MDC1 also participates in mitotic regulation. MDC1 is hyperphosphorylated upon nocodazole treatment, which activates the spindle assembly checkpoint (14). More recently, MDC1 was shown to regulate the metaphase-to-anaphase transition by modulating the Cdc20-dependent activation of anaphase-promoting complex through its BRCA1 C terminus domain (BRCT) domains (19,20).
MDC1 is a large protein (2089 residues) comprising an N-terminal forkhead-associated (FHA) domain, a C-terminal tandem-BRCT domain and a long linker without a known structural domain (Figure 1A) (21). The tandem-BRCT domain specifically binds the C-terminal phosphopeptide of γ-H2AX (22,23). The linker harbors several repetitive sequence motifs: the serine–aspartic acid–threonine (SDT) repeats, the TQXF repeats and the proline–serine–threonine (PST)-rich repeats. The SDT repeats are constitutively phosphorylated by casein kinase II at the serine and threonine residues, and the dual-phospho SDT motif recruits the MRN complex via both the FHA domain and BRCT domains of NBS1 (24-29). This interaction is important for MRN focus formation, DSB repair, the G2/M checkpoint and the intra-S-phase checkpoint. The TQXF repeats are phosphorylated by ATM in response to DNA damage, inducing a ubiquitination cascade that involves the E3 ubiquitin ligases RNF8, RNF168, HECR2 and BRCA1 (30-34). The PST repeats are implicated in HR and NHEJ repair of DSBs, but the underlying molecular mechanism remains unknown (35,36).
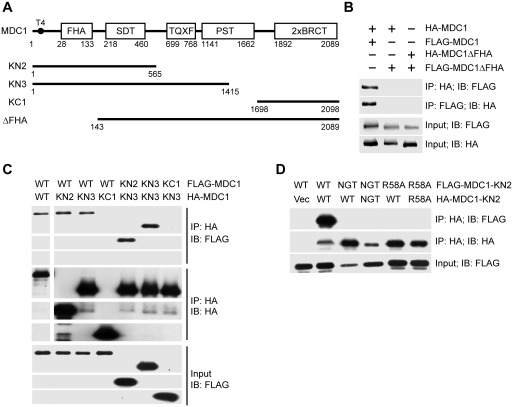
Figure 1.
MDC1 self-associates via its FHA domain. (A) Diagram of MDC1 domain organization. Boundaries are indicated for MDC1 truncation constructs. (B–D) HA-tagged MDC1 or mutants were co-transfected with FLAG-tagged MDC1 or mutants into Ad293 cells. After 48 h, the total cell lysates were immunoprecipitated with anti-FLAG and/or anti-HA antibodies and immunoblotted with anti-HA and anti-FLAG antibodies. The whole cell lysates were also blotted against anti-HA and -FLAG antibodies and are shown as the input. (B) Self-association of MDC1 and its FHA deletion mutant (MDC1ΔFHA) were analyzed by immunoprecipitation (IP) and immunoblotting (IB). (C) Self-association of wild-type MDC1 (WT) and its truncation mutants KN2, KN3 and KC1. (D) Self-association of KN2 and its mutants R58A and N96A/G97A/T98A (NGT).
FHA domains are a type of phosphothreonine (pT)-binding domain and mediate many phosphorylation-dependent events in DDR and cell cycle control (37,38). Deletion of the FHA domain in MDC1 results in defective G2/M checkpoint, inefficient DSB repair by sister chromatid recombination and reduced apoptosis in response to IR (11,16,36,39). Overexpression of the FHA and SDT domains impairs DNA damage-induced nuclear focus formation of several DDR factors (14). The FHA domain of MDC1 has been suggested to bind phosphopeptides of CHK2, ATM and the recombinase RAD51 (11,16,39); however, its function remains elusive (21).
Through structural, biochemical and cellular studies, we describe here a novel function of the FHA domain in mediating MDC1 dimerization. The FHA domain mediates dimerization by asymmetric FHA–FHA interaction and by intermolecular binding to phospho-T4 (pT4) in the N-terminal tail of MDC1. We determined the crystal structures of the FHA domain alone and in complex with a pT4 phosphopeptide. Additionally, we demonstrate that T4 is phosphorylated by ATM in response to DNA damage and that the trans FHA–pT4 interaction is critical for stable dimerization. Dimerization promotes the formation of MDC1 foci at DNA-damage sites, and forced dimerization by an unrelated domain has a similar effect. Our results support a novel mechanism by which MDC1 function is regulated through T4 phosphorylation and FHA-mediated dimerization.
MATERIALS AND METHODS
Reagents, antibodies and cell lines
The ATM inhibitor KU55993, the DNA-PKcs inhibitor NU7026 and the topoisomerase I inhibitor CPT were purchased from Sigma. The FK506 binding protein (FKBP) bivalent ligand AP20187 was a kind gift from Ariad Pharmaceuticals. All cancer cell lines were purchased from ATCC and cultured in high-glucose Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal bovine serum. The purchased mouse monoclonal antibodies included anti-hemagglutinin (HA) (Covance), anti-β-actin (AC15, Sigma), anti-FLAG (Sigma) and anti-γ-H2AX (Millipore). Rabbit polyclonal antibodies against MDC1 were described before (15). Rabbit polyclonal antibodies against HA and γ-H2AX were purchased from Bethyl and Cell Signaling Technology, respectively. The rabbit polyclonal anti-pT4-MDC1 antibody was raised against the phosphopeptide MED(pT)QAIDWDVEEE and affinity purified (Beijing CoWin Biotech).
Plasmids
The mammalian expression constructs pcDNA-MDC1 and its mutants KN3, KC1, KN2, KN2-R58A and KN2-NGT were previously described (14,15). HA-FKBP fusions of MDC1 were constructed in the pcDNAHA2FKBP vector (40). The domain-deletion mutants MDC1ΔFHA (deletion of residues 1–144) and MDC1ΔBRCT (deletion of residues 1698–2089) were prepared using two-step PCR mutagenesis (41). For bacterial expression, MDC1 fragments covering residues 27–138 and 2–133 were cloned into a modified pET28a(+) plasmid and were expressed as fusions to an N-terminal six-histidine tag and SMT3 domain (His-SMT3) (42). Point mutations were generated using QuikChange (Stratagene). All the expression constructs were verified by sequencing. The kinase domain of ATM (residues 2709–2964) was amplified by PCR from human ATM cDNA and subcloned into pGEX-4T, and the kinase-dead mutant D2870A was generated by QuikChange.
Immunoprecipitation and immunostaining experiments
Plasmid DNA was transfected into human cancer cells with FuGene 6 (Roche). Immunoprecipitation, immunoblotting and immunostaining were conducted as described previously (14,15).
Induced dimerization of MDC1
Induced dimerization assays were performed as previously described (40). Briefly, plasmids encoding HA-FKBP-MDC1 and its mutants were transfected into Ad293 cells. After 24 h, transfectants were selected with neomycin for 4 days. The cells were mock treated with 0.1% ethanol or treated with 10 nM AP20187 for 4 h to induce the dimerization of FKBP. Where indicated, transfectants were treated with 10 μM CPT for 1 h to induce DNA damage 3 h after adding AP20187. The cells were fixed and immunostained with anti-HA and anti-γ-H2AX antibodies.
ChIP assay
ChIP assays were performed as described previously (43,44). Immunoprecipitated DNA was amplified with the primers HRChIP.S (5′-TCTTCTTCAAGGACGACGGCAACT-3′) and HRChIP.R (5′-TTGTAGTTGTACTCCAGCTTGTGC-3′) by real-time PCR. The PCR signal intensity of each ChIP sample was divided by the signal of corresponding input sample that had the same I-SceI and AP20187 treatment but was not immunoprecipitated. The signal was further normalized to the signal of the ChIP sample without I-SceI and AP20187 treatment, yielding the fold of enrichment.
Protein expression and purification
MDC1 FHA proteins were expressed in Escherichia coli BL21(DE3) strain (Novagen). Protein expression was induced with 0.2 mM isopropyl β-d-1-thiogalactopyranoside for 8 h at 25°C. The harvested cells were resuspended in buffer P300 (50 mM phosphate pH 7.6 and 300 mM KCl) and disrupted by sonication. After the cell lysates were clarified, the supernatant was passed through a 0.45-μm membrane filter before loading onto a 5-ml HisTrap column (GE healthcare). The protein was washed by 20 mM imidazole in P300 and eluted by 500 mM imidazole in P300. The His-SMT3 tag was cleaved by ULP1 protease for 30 min at room temperature and removed by passage through a HisTrap column. The flow-through containing the cleaved target protein was concentrated and further purified with a HiLoad 16/60 Superdex 75 column (GE healthcare) equilibrated in 5 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid/KOH (HEPES-K) (pH 7.6) and 100 mM KCl.
The T98A MDC12–133 mutant protein was expressed at 16°C and purified with the His-SMT3 tag uncleaved through HisTrap, Q and size exclusion chromatography. The size exclusion step was conducted in buffer P250 (20 mM sodium phosphate pH 7.6 and 250 mM NaCl).
The purified proteins were concentrated using Amicon Ultra-15 centrifugal filter units and stored at −80°C. The molar concentrations of MDC127–138 and MDC12–133 referred to the monomeric form and were calculated with extinction coefficients at 280 nm of 6990 and 12490 M−1 cm−1, respectively. The phosphopeptide MED(pT)QAID (pT4-8 P) was chemically synthesized, purified by HPLC to >99% purity and lyophilized (Bootech, Shanghai). The amount of peptide was estimated by weighting. The GST fusions of ATM (2709–2964) and its kinase-dead mutant D2870A were purified with glutathione-Sepharose beads.
In vitro phosphorylation of MDC1 FHA proteins
The purified MDC1 FHA proteins (4 μg) were incubated in a 20 μl reaction containing 0.2 μl of 17 mg/ml nuclear extracts of HeLa cells (NE), 25 mM HEPES–Na (pH 7.6), 50 mM KCl, 5 mM MgCl2, 0.2 mM EGTA, 0.1 mM ethylenediaminetetraacetic acid (EDTA), 1 mM dithiothreitol (DTT), 1.0 mM adenosine-5′-triphosphate (ATP) and 1 μCi 32P-γ-ATP for 30 min at 30°C. Large-scale phosphorylation reactions were conducted in a 30-ml volume containing the same buffer as above, 50–100 mg MDC12–133 protein, 500 μl NE and 1.5 mM unlabeled ATP for 1 h. The phosphorylated protein was further purified with a HiLoad 16/60 Superdex 75 pg column in 5 mM HEPES–Na (pH 7.6) and 250 mM NaCl.
Crystallization and structure determination
MDC127–138 crystals were grown using the hanging drop vapor diffusion method at 20°C by mixing 1 μl 20 mg/ml protein in 5 mM HEPES–K (pH 7.6) and 100 mM KCl and 1 μl of the well solution containing 0.1 M HEPES (pH 7.5) and 20% PEG-3350. Two types of crystals with different shapes belonging to space groups P212121 and P21 grew from the same drops in 2–3 days. The P212121 crystals were derivatized in 1 mM KAu(CN)2 in the well solution for 5 h. The crystals were cryoprotected in the well solution plus 15% glycerol and flash frozen in liquid nitrogen. The native data sets were collected at the Shanghai Synchrotron Research Facility (SSRF) beamline BL17U and processed by HKL2000 (45). The gold derivative data were collected at a wavelength of 1.5418 Å using a Rigaku machine and processed by Denzo and Scalepack (45).
The structure of the MDC1 FHA domain was first solved for the P212121 crystal form by the single isomorphous replacement with anomalous scattering (SIRAS) method. Heavy atoms were located by SHELXD (46), and experimental phase calculation and density modification were performed in SHARP (47). The structure model was automatically built with the Autobuild module of PHENIX (48), followed by several rounds of manual adjustment in COOT (49) and refinement in Refmac (50). The asymmetric unit (ASU) of the P212121 crystal contains two FHA molecules arranged as an asymmetric dimer. The P212121 structure was refined to 1.65 Å resolution and contains residues 29–133 in both protomers and 170 water molecules.
The P21 structure was solved by molecular replacement (MR) in PHASER using a single FHA subunit of the P212121 structure as a search model (51). The ASU of the P21 crystal contains one FHA dimer that has virtually identical protomer structure and dimer arrangement as the P212121 dimer except for slight conformational variations at several loop regions. Only the structure of P212121 space group is illustrated in our figures. The P21 structure was refined to 1.8 Å and contains residues 29–101, 105–109 and 111–133 in one protomer, residues 29–134 in the other protomer and 84 water molecules.
Phosphopeptide pT4-8P was dissolved in DMSO and mixed at a molar ratio of 2:1 with 0.5 mg/ml MDC127–138. The complex was concentrated to 19 mg/ml in buffer containing 5 mM HEPES–Na (pH 7.6) and 250 mM NaCl. For crystallization, 1 μl of the protein solution was mixed with 1 μl of the reservoir solution containing 0.1 M HEPES–Na (pH 6.6), 0.2 M ammonium acetate and 57% 2-methyl-2,4-pentanediol in sitting drops. Crystals appeared in 1–2 days at 20°C and were flash frozen in liquid nitrogen without further treatment. The crystal belongs to the P3121 space group and contains one molecule of the MDC127–138 and pT4-8P complex in the ASU. The structure was solved by MR in PHASER with one FHA monomer structure as search model. The final model was refined to 1.7 Å and contains MDC1 residues 27–133, pT4-8P residues 1–8 and 71 water molecules. The data collection and refinement statistics are listed in Supplementary Table S1. The structural figures were created in PyMOL (52).
Isothermal titration calorimetry
MDC127–138 and its mutants R58A and L127R were exchanged into buffer P150 (20 mM sodium phosphate pH 7.6 and 150 mM NaCl) with a Superdex-75 10/300 GL column and supplemented with 1% DMSO before titration. Phosphopeptide pT4-8P was dissolved in 100% DMSO and diluted in P150 to a final concentration of 1% DMSO. For experiments with SMT3-MDC12–133-T98A and related controls, buffer P150 was replaced by P250. Isothermal titration calorimetry (ITC) experiments were performed with an iTC200 microcalorimeter (Microcal Inc.) at 25°C. The sample cell contained 200 μl of 150 μM MDC1 FHA or 140 μM SMT3-fused MDC1 FHA. Approximately 2 mM of the peptide was injected via a syringe, with 0.4 μl for the first injection and 2 μl for the next 19 injections. The dilution effect was measured by injecting the peptide into the respective buffer plus 1% DMSO. The integrated heat data after subtracting the dilution effect were analyzed using a one-set-of-sites model in Origin according to the manufacturer's instructions. The first data point was excluded in the analysis. The binding parameters ΔH (reaction enthalpy change in cal mol−1), K (binding constant in M−1) and n (bound peptide per FHA) were floating in the fit. The binding free energy ΔG and reaction entropy ΔS were calculated using the relationships ΔG = −RTlnK (R = 1.9872 cal mol−1 K−1, T = 298 K) and ΔG = ΔH−TΔS. The dissociation constant Kd was calculated as 1/K.
Size exclusion chromatography
Size exclusion chromatography was carried out with AKTA Purifier-10 (GE Healthcare). Approximately 0.5 mg of each protein (400 μM) was loaded in 100 μl to a Superdex 200 10/300 GL column running in buffer HEPES–Na pH 7.6 and 200 mM NaCl or P250 for the T98A mutant. The columns were calibrated with thyroglobulin (669 kD), ferritin (440 kD), catalase (232 kD), aldolase (158 kD), bovine serum albumin (67 kD), ovalbumin (43 kD), chymotrypsinogen (25 kD), cytochrome C (12.4 kD) and aprotinin (6.5 kD). In subunit exchange assays, 50 nmol of MDC12–133 or its variants were mixed with 50 nmol of MDC127–138 in 120 μl P150 and incubated for 1 h at 37 °C before loading onto a Superdex 75 10/300 GL column running in P150.
Analytical ultracentrifugation
The experiments were conducted using a Beckman-Coulter XL-I analytical ultracentrifuge equipped with a 6-channel centerpiece and a Beckman An-60 Ti rotor. MDC12–133 and pT4-MDC12–133 were exchanged into buffer P150 using a Superdex 75 10/300 GL column. MDC12–133 was mixed in a 1:2 molar ratio with phosphopeptide pT4-8P dissolved in DMSO. For each protein, three samples in 80, 40 and 8 μM concentrations were centrifuged at 25 000 rpm at 25°C for 24 h to establish sedimentation equilibrium. Concentration profiles were measured at 280 nm. The protein partial specific volume was calculated to be 0.7349 cm3 g−1 for MDC12–133 from its amino acid composition. The solvent density was estimated to be 1.00596 g/ml by SEDNTERP (J. Philo, http://www.jphilo.mailway.com/default.htm). Global fitting of the absorbance profiles at three protein concentrations was performed in SEDPHAT (53), yielding the dimerization constant Ka. The MDC12–133 and pT4-MDC12–133 data were analyzed by a monomer–dimer equilibrium model. The MDC12–133 in the presence of pT4-8P data were analyzed by a model that accounts for MDC12–133 dimerization and the association between MDC12–133 and pT4-8P. The dissociation constant of the MDC1 pT4-8P complex was shown to be 20 μM after the first round of fitting. This value was consistent with the ITC value, so it was fixed in the following rounds of fitting. The bottom of the cell, protein concentration and Ka were floating in the fitting, whereas the extinction coefficient and meniscus were fixed. The covariance matrix approach was employed to estimate the errors in log(Ka). The log(Ka) value is 5.036 ± 0.144 for MDC12–133, 7.456 ± 0.793 for pT4-MDC12–133 and 4.789 ± 0.107 for the MDC12–133 and pT4-8P complex.
Accession numbers
The atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 3UMZ for the MDC127–138 structure in the P212121 space group, 3UNM for the MDC127–138 structure in the P21 space group and 3UNN for the MDC127–138 and pT4-8P complex structure.
RESULTS
MDC1 self-associates via its FHA domain
We found that FLAG-tagged MDC1 and HA-tagged MDC1 co-immunoprecipitated (Figure 1B), suggesting that MDC1 self-associates in vivo. Domain mapping experiments further indicate that the tandem-BRCT domain is neither required for MDC1 self-association (KN3) nor involved in self-association (KC1), whereas an FHA-deletion mutant cannot complex with itself or with wild-type MDC1 (Figure 1A-C). Moreover, a fragment KN2 containing the FHA and SDT domain was sufficient for self-association (Figure 1C-D). These results point to a key role for the FHA domain in MDC1 self-association.
As FHA domains are often involved in binding pT, we asked whether any FHA–pT recognition event is involved in MDC1 self-association. To this end, we made two mutants of KN2, one with a single alanine mutation at residue R58 (R58A) and the other with three alanine mutations, at residues N96, G97 and T98 (NGT). These residues are critical for pT-binding as illustrated in known FHA–pT complex structures (54), and both of the mutant proteins failed to self-associate (Figure 1D). We can conclude that the FHA domain and its pT-binding ability are essential for MDC1 self-association.
The FHA domain forms a dimer
To reveal how the FHA domain mediates self-association, we determined the crystal structure of an MDC1 fragment containing residues 27–138 (MDC127–138) in space group P212121 at 1.65 Å resolution and in space group P21 at 1.8 Å resolution (Supplementary Table S1). Despite the different crystal packing environments, the FHA domain adopts a nearly identical dimeric structure in both crystal forms (Supplementary Figure S1). The FHA domain contains eleven strands named β1 through β11, which are assembled into a twisted β-sandwich with two β-sheets (Figure 2A). One β-sheet that mediates dimerization is composed of six antiparallel strands arranged in the order β2–β1–β11–β10–β7–β8. The other β-sheet is formed by five strands in the order β4–β3–β5–β6–β9, which are antiparallel to each other except for the peripheral strand β4, which is parallel to strand β3. The MDC1 FHA structure is highly similar to other FHA structures (37). For example, alignment with the CHK2 FHA structure yields a root mean square deviation (rmsd) of 1.2 Å over 65 Cα pairs (55).
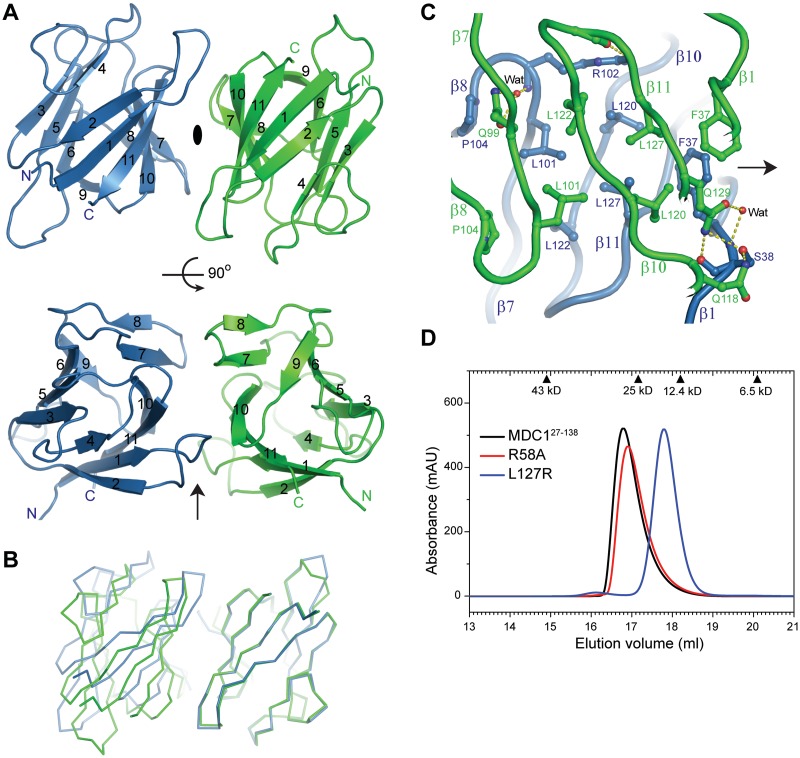
Figure 2.
Dimeric structure of the MDC1 FHA domain. (A) Ribbon representation of the FHA dimer structure in the P212121 space group. Two orthogonal views are displayed. The pseudo-dyad axis is shown as an ellipse when perpendicular to the paper or as an arrow when in the paper. The two subunits are colored green and blue. The 11 strands are labeled by numbers, and the N- and C-termini are marked. (B) Alignment of two dimers by the non-equivalent protomers to the right. The other protomers to the left do not overlap following the alignment, indicating asymmetry in the dimer arrangement. (C) Interactions at the dimer interface. Protein backbones are shown as tubes, interacting residues as sticks and hydrogen bonds as dashed lines. Carbon atoms are green in one subunit and blue in the other subunit, oxygen atoms are red and nitrogen atoms are blue. (D) MDC127–138 and its mutants R58A and L127R were analyzed in a Superdex 200 10/300 GL column running in HEPES–Na (pH 7.6) and 200 mM NaCl. Each protein (400 μM) was loaded in a 100 μl volume. The elution volume of calibration standards are indicated on the top.
In the FHA dimer, the six-stranded β-sheet of one subunit packs against its mate face-to-face. The dyad axis is roughly perpendicular to the direction of strands, resulting in the two subunits opposite to each other along the long dimension of the β-sandwich (Figure 2A). Interestingly, the dyad symmetry is not perfect, leading to many non-reciprocal interactions at the dimer interface. The two subunits are related by a 174° rotation along the pseudo-dyad axis, rather than by a 180° rotation in a dimer with perfect dyad symmetry. Consequently, when one subunit of a dimer is aligned to the non-equivalent subunit of a second dimer, the other subunits would end up unaligned with an average Cα displacement of as large as 5.5 Å (Figure 2B).
The two subunits dimerize at three adjacent strands β10, β11 and β7 with minor contributions from strand β1 and the β7–β8 loop (Figure 2C), burying a moderate 490 Å2 solvent accessible area per subunit. The hydrophobic residues F37, L101, L120, L122, L127 and P104 from both subunits mediate major intermolecular contacts at the center of the dimer interface. There are also hydrogen bonding and water-mediated interactions on the periphery of the dimer interface. The side chains of Q129 and Q118 of one subunit are within hydrogen bonding distance of the side-chain hydroxyl and backbone carbonyl of S38 of the other subunit but not vice versa due to asymmetry in the dimer. R102 of one subunit non-reciprocally contacts the D125 backbone carbonyl of the other subunit. The asymmetric dimer interface is identical in two different crystal lattices, confirming it is not a crystal packing artifact (Supplementary Figure S1).
MDC127–138 also forms a dimer in solution, according to its elution peak in size exclusion column (Figure 2D). Replacement of L127, a hydrophobic residue at the center of the dimer interface, to a charged arginine apparently disassociates the FHA dimer into a monomer (Figure 2D). This confirms that the observed interface in crystal is responsible for dimerization in solution.
The FHA domain recognizes pT4 at the N-terminus of MDC1
The above co-immunoprecipitation experiments with the R58A and N96A/G97A/T98A mutants implicate an involvement of the FHA–pT interaction in MDC1 dimerization (Figure 1D). It has been documented that the FHA domain of the checkpoint kinase CHK2 makes an intermolecular interaction with phosphothreonine 68 (pT68) in its N-terminal tail, mediating CHK2 dimerization and activation (40,56). By analogy, we wondered whether the FHA domain of MDC1 interacts with a pT from the other molecule in dimer. We noticed that the extreme N-terminal sequences of MDC1 (1-MEDTQAID-8) harbor a TQ motif at positions 4–5, in which T4 is a potential phosphorylation site for the PIKK kinases ATM, ATR and DNA-PKcs. Moreover, the sequences surrounding T4 conform to the in vitro selected binding target for MDC1 FHA domain, particularly at the −3 (M), +1 (Q) and +3 (I) positions (54). Therefore, we tested whether the MDC1 FHA domain binds to an N-terminal eight-residue phosphopeptide MED(pT)QAID (called pT4-8P). ITC measurements yielded an apparent dissociation constant Kd of 40 ± 3 μM, suggesting that the phosphorylated T4 is a binding target of MDC1 FHA (Figure 3A).
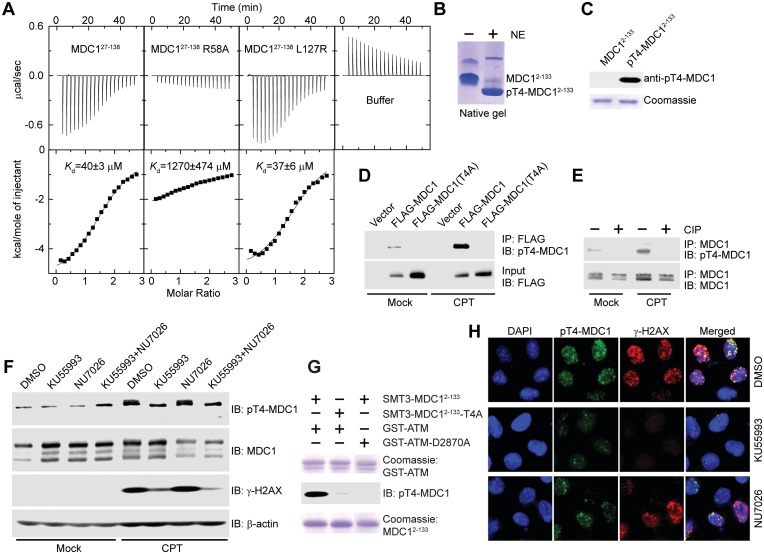
Figure 3.
MDC1 T4 is phosphorylated by ATM in response to DNA damage, and pT4 is recognized by the FHA domain. (A) Microcalorimetric titration of phosphopeptide pT4-8P into MDC127–138, its mutants R58A and L127R, or 20 mM sodium phosphate (pH 7.6) and 150 mM NaCl buffer. The heat peaks were integrated, corrected for the ligand dilution effect and fit to a one-set-of-sites binding model. The dissociation constant, Kd, values are indicated. (B) MDC12–133 was phosphorylated by nuclear extracts (NE) of HeLa cells and resolved in a native gel. (C) Affinity purified rabbit polyclonal anti-pT4-MDC1 antibody specifically recognizes phosphorylated MDC12–133 but not unphosphorylated MDC12–133. (D) pT4 in transiently expressed MDC1. Empty vector or vectors expressing FLAG-MDC1 or FLAG-MDC1(T4A) were transfected into U2OS cells. The cells were mock treated or treated with CPT for 1 h, followed by immunoprecipitation with anti-FLAG antibody and immunoblotting with anti-FLAG and anti-pT4-MDC1 antibodies. (E) pT4 in endogenous MDC1. U2OS cells were mock treated or treated with CPT for 1 h. The total cell lysates were immunoprecipitated with anti-MDC1 antibody, treated with or without calf intestine phosphatase (CIP), and blotted with anti-pT4-MDC1 and anti-MDC1 antibodies. (F) T4 is primarily phosphorylated by ATM. U2OS cells were treated with DMSO, ATM inhibitor KU55993 (10 μM), DNA-PKcs inhibitor NU7026 (10 μM) or both KU55993 and NU7026 for 2 h before treatment of 0 or 10 μM CPT for 1 h. The total cell lysates were resolved in SDS-PAGE and blotted with anti-pT4-MDC1, anti-MDC1, anti-γ-H2AX and anti-β-actin antibodies. Endogenous MDC1 presents multiple isoforms when blotted with anti-MDC1 antibody; only the slowest migrating one was recognized by anti-pT4-MDC1 antibody. (G) SMT3-MDC12–133, but not its T4A mutant, can be phosphorylated in vitro by a recombinant GST-fused kinase domain of ATM (GST-ATM, residues 2709–2964). GST-ATM-D2870A is a kinase-dead mutant. (H) U2OS cells were treated as in (F) and were immunostained with anti-pT4-MDC1 (green) and γ-H2AX (red). DNA was stained by DAPI (blue).
T4 is phosphorylated in response to DNA damage
Encouraged by the finding that the MDC1 FHA domain binds to pT4 in vitro, we asked if T4 is indeed phosphorylated in cells. We prepared a recombinant MDC1 fragment containing residues 2–133 (MDC12–133) and phosphorylated MDC12–133 with nuclear extracts of HeLa cells. The phosphorylated MDC12–133 protein appeared as a major single species that migrated faster than the unphosphorylated protein in native gel (Figure 3B). With mass spectrometry analysis, we detected T4 as the only site phosphorylated (data not shown). Moreover, alanine substitution of T4 largely blocked phosphorylation of MDC12–133 (see below Figure 5A). These results indicate that MDC12–133 is phosphorylated by nuclear extracts primarily at the T4 site.
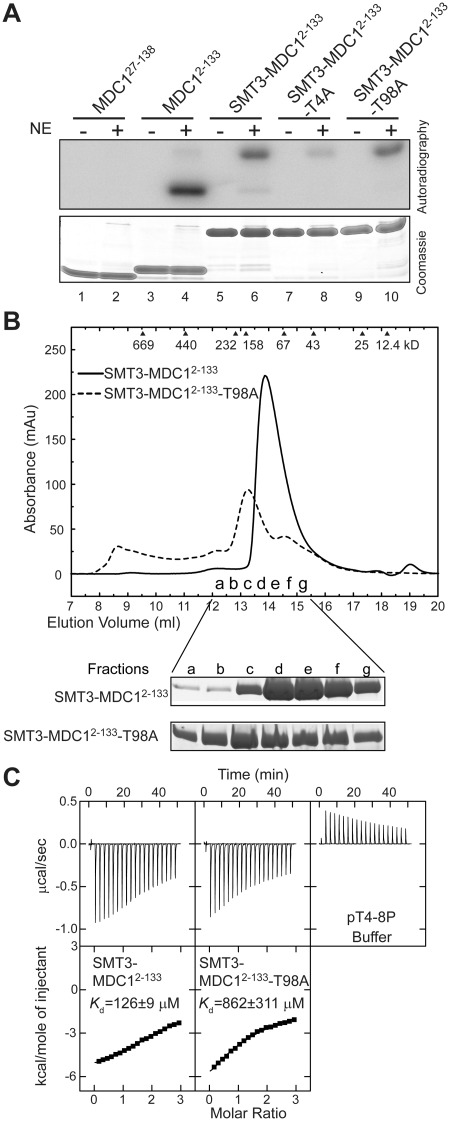
Figure 5.
T98 is not a phosphorylation site and the T98A mutation disturbs the folding and pT-binding of the FHA domain. (A) Phosphorylation of MDC1 FHA proteins by HeLa cell nuclear extracts (NE) in the presence of 32P-γ-ATP. (B) SMT3-MDC12–133 and its T98A mutant were eluted in a Superdex 200 10/300 GL column and in buffer 20 mM sodium phosphate (pH 7.6) and 250 mM NaCl. SDS-PAGE gels of the fractions are shown at the bottom. (C) ITC analysis of SMT3-MDC12–133 and its T98A mutant with phosphopeptide pT4-8P in 20 mM sodium phosphate (pH 7.6) and 250 mM NaCl.
We raised a rabbit polyclonal antibody against a pT4 phosphopeptide containing the N-terminal 14 amino acids of MDC1. The antibody was specific to pT4-MDC1, as it recognized phosphorylated, but not unphosphorylated MDC12–133 protein (Figure 3C). The specificity of the anti-pT4-MDC1 antibody was further demonstrated by its recognition of transiently expressed and endogenous MDC1 but not the T4A mutant of MDC1 and the dephosphorylated protein (Figure 3D and E).
T4 was phosphorylated at a background level in transiently expressed MDC1 (Figure 3D) and endogenous MDC1 (Figure 3E). The level of pT4 significantly increased after CPT treatment in both cases, indicating that T4 is phosphorylated in response to DNA damage.
T4 is phosphorylated primarily by ATM
The epitope sequence of T4 and its DNA damage-induced phosphorylation suggest that PIKKs are responsible for T4 phosphorylation. The ATM inhibitor KU55993 greatly inhibited the CPT-induced phosphorylation of MDC1 T4 as well as H2AX, whereas the DNA-PKcs inhibitor NU7026 had a minor effect (Figure 3F). This finding suggests that ATM is the primary kinase targeting MDC1 T4. Moreover, a recombinant ATM kinase domain was able to phosphorylate MDC12–133 but not its T4A mutant in vitro (Figure 3G). The phosphorylation was abolished if a dead ATM kinase domain was used, indicating that the activity is specifically mediated by ATM rather by other contaminating kinases.
We next examined whether pT4-MDC1 is enriched at DNA-damage sites. Immunofluorescence staining with anti-pT4-MDC1 antibody revealed that pT4-MDC1 formed foci that co-localized with γ-H2AX foci, after induction of DSBs by CPT, bleomycin, adriamycin or mitomycin C treatment (Figure 3H and data not shown). Moreover, the ATM inhibitor KU55993 abolished the formation of CPT-induced γ-H2AX and pT4-MDC1 foci, whereas the DNA-PKcs inhibitor NU7026 had a minor effect. These results suggest that pT4-MDC1 is enriched at DNA-damage sites in an ATM-dependent manner.
Structures of the FHA–pT4 complex
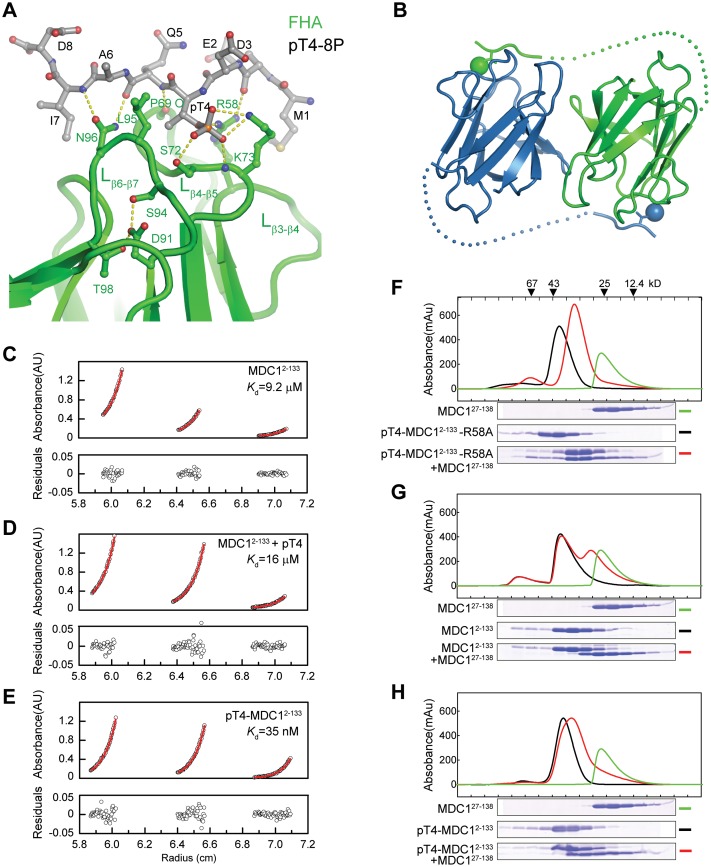
To understand the mechanism of the MDC1 FHA–pT4 interaction, we determined a structure of MDC127–138 in complex with phosphopeptide pT4-8P to 1.7 Å resolution (Figure 4A and Supplementary Figure S2). The MDC1 FHA domain binds the pT4 phosphopeptide at a pocket composed of loops β3–β4, β4–β5 and β6–β7. The phosphate group of pT4 is recognized by multiple hydrogen bonds from the side chains of S72, K73 and R58 and the amide nitrogen of K73. In addition, the backbone atoms of the phosphopeptide form multiple hydrogen bonds to the side chains of N96 and R58 and the P69 carbonyl oxygen. These interactions with the phosphate and peptide backbone would make highly specific recognition of pT at position 4. However, the sequences surrounding pT4 appear to be only weakly recognized. Some specificity may be directed to the two acidic residues E2 and D3 preceding pT4, which are within electrostatic interaction distance of K73 and R58, and to I7, which makes a van der Waals contact with the side chain of N96. The pT4 peptide binding mode is quite similar to previously observed binding modes in other structures of FHA–pT peptide complex (Supplementary Figure S3) (54,55). The phosphopeptide association induces little conformational change in the FHA domain, including the pT-binding pocket (Supplementary Figure S4).
Figure 4.
MDC FHA domain binds to pT4 in trans. (A) The monomeric structure of MDC127–138 in complex with phosphopeptide pT4-8P. The phosphopeptide and important ligand-binding residues are shown as sticks and balls and colored red for oxygen, blue for nitrogen, gray for carbon in the phosphopeptide and green for carbon in the FHA domain. (B) A structural model depicting the trans FHA–pT4 interaction in the FHA dimer. Dots denote putative positions of residues 9–26. Phosphate groups are shown as spheres. (C–E) Analytic ultracentrifugation sedimentation equilibrium analysis of MDC12–133(C), MDC12–133 in the presence of pT4-8P (D) and pT4-MDC12–133(E). The dimer dissociation constants Kd are indicated. (F–H) Subunit exchange between MDC12–133 and MDC127–138 homodimers. pT4-MDC12–133-R58A (F), MDC12–133 (G) and pT4-MDC12–133 (H) were analyzed by a Superdex 75 10/300 GL column in the absence and presence of MDC127–138. The fractions were resolved with SDS-PAGE, and the gels are shown below the elution profiles. The same elution profile and gel image of MDC127–138 are displayed in each panel for comparison.
Puzzlingly, pT4-bound MDC127–138 did not form a dimer in crystal like free MDC127–138 or present an alternative and reasonable dimer interface. The dimer interface, which is rather weak by itself (see below), was likely disrupted because of crystal packing interaction (Supplementary Figure S5).
The dimeric FHA domain binds pT4 in trans
Our structural evidence so far cannot distinguish whether the MDC1 FHA domain binds pT4 from the same (cis binding) or a different subunit (trans binding) in the dimer. If the FHA domain binds pT4 in trans, the subunit association would be stabilized by such intermolecular interactions (Figure 4B). To distinguish the two FHA–pT4 interaction modes, we measured the dimer dissociation constant Kd via analytic ultracentrifugation sedimentation equilibrium assay (Figure 4C–E). The T4-phosphorylated MDC12–133 dimer (Kd = 35 nM) was 260-fold more stable against subunit dissociation than the unphosphorylated dimer (Kd = 9.2 μM). The increased stability was not due to pT association per se, as binding of an isolated pT4-8P phosphopeptide did not change the dimer stability (Kd = 16 μM). These results suggest that each subunit of MDC1 FHA dimer binds to pT4 from the other subunit, significantly stabilizing the dimer formation.
To further investigate the mode of FHA–pT4 interaction, we examined subunit exchange between the pT4-MDC12–133 homodimer with mutation R58A, which is defective in pT-binding (Figure 3A), and the MDC127–138 homodimer, which contains functional pT-binding pockets but no pT4. If the FHA–pT4 interaction occurs in trans, then subunit dissociation and re-association would lead to the formation of a more stable heterodimer in which pT4 in the pT4-MDC12–133-R58A subunit binds to the pT-binding pocket of the MDC127–138 subunit. However, if the FHA–pT4 interaction occurs in cis, then the heterodimer would have a similar stability as the two homodimers and not be more energetically favored. Indeed, mixing of pT4-MDC12–133-R58A and MDC127–138 yielded predominantly the heterodimer, which migrated between the two homodimers in size exclusion chromatography (Figure 4F). The symmetric shape of the elution peak also suggests that the heterodimer is more stable than the two homodimers that have trailing peaks. The heterodimer formation depends critically on the presence of pT4 as unphosphorylated MDC12–133 cannot form a stable heterodimer with MDC127–138 (Figure 4G). Finally, wild-type pT4-MDC12–133 and MDC127–138 form only a small amount of heterodimer (Figure 4H). This is reasonable given that the pT4-MDC12–133 homodimer that contains a pair of trans FHA–pT4 interactions is expected to be more stable than the pT4-MDC12–133/MDC127–138 heterodimer that contains only one trans FHA–pT4 interaction. These subunit exchange results further support that MDC1 FHA domain recognizes pT4 in trans.
The MDC1 FHA domain without pT4 interaction will still form a dimer if the protein concentration is much higher than the intrinsic dissociation constant of FHA domain (Kd = 9.2 μM). For example, MDC127–138 is dimeric in the size exclusion chromatography experiments and in crystal because the protein concentration in these cases is in the submillimolar to millimolar range (Figure 2). In contrast, endogenous MDC1, which is expected to have a low concentration in cells, would adopt a monomeric structure if T4 is not phosphorylated. Notably, T4 is phosphorylated at a basal level in untreated cells (Figure 3D–F), which may account for the detection of dimerization of overexpressed MDC1 without induction of DNA damage (Figure 1).
T98 is inaccessible for phosphorylation
A very recent study reported that T98 in the FHA domain can be phosphorylated by ATM and that phospho-T98 (pT98) is bound by the FHA domain, inducing oligomerization of MDC1 (57). However, according to our crystal structure, the side chain of T98 is totally buried and should be inaccessible for phosphorylation in a folded FHA domain (Figure 4A). Indeed, MDC127–138 that lacks the N-terminal tail could not be phosphorylated by nuclear extracts of HeLa cells (Figure 5A), indicating that no phosphorylation site is located within the FHA domain. In the context of SMT3-fused MDC12–133, where the SMT3 tag was used to increase the solubility of the T98A mutant protein, the phosphorylation level was reduced ∼7-fold by the T4A mutation but was unchanged by the T98A mutation (Figure 5A). These findings further support that T4 is the primary phosphorylation site in MDC12–133.
The T98A mutation disturbs the FHA structure and the pT-binding pocket
The T98A mutation was shown to abolish MDC1 oligomerization (57). If T98 is not a phosphorylation site, then how could the T98A mutation affect MDC1 dimerization? Our structure suggests that T98 is important for the structural stability of FHA domain and the conformation of loop β6–β7, which is involved in phosphopeptide interaction (Figure 4A). Specifically, the methyl group of T98 is packed inside the hydrophobic core of FHA domain, and its hydroxyl group is involved in a hydrogen bond network that connects the side chains of T98, D91 and S94.
We found that recombinant MDC12–133 with the T98A mutation was poorly soluble and needed to be purified with a solubility-enhancing SMT3 tag. SMT3-MDC12–133-T98A was also more heterogeneous and larger in size than wild-type SMT3-MDC12–133 in size exclusion chromatography (Figure 5B), suggesting that the T98A mutation affects the folding and solubility of the FHA domain. Moreover, ITC measurements indicated that the T98A mutation reduced the binding affinity of phosphopeptide pT4-8P by 7-fold (Figure 5C). To increase the solubility of the mutant T98A protein, the ITC experiments were conducted in 250 mM NaCl, which also reduced the binding affinity of the wild-type protein compared to that measured in 150 mM NaCl (Figure 3A). In summary, our data suggest that the T98A mutation disrupts MDC1 dimerization by affecting the folding and pT binding of the FHA domain, rather than by eliminating a pT site.
Dimerization promotes localization of MDC1 at DNA-damage sites
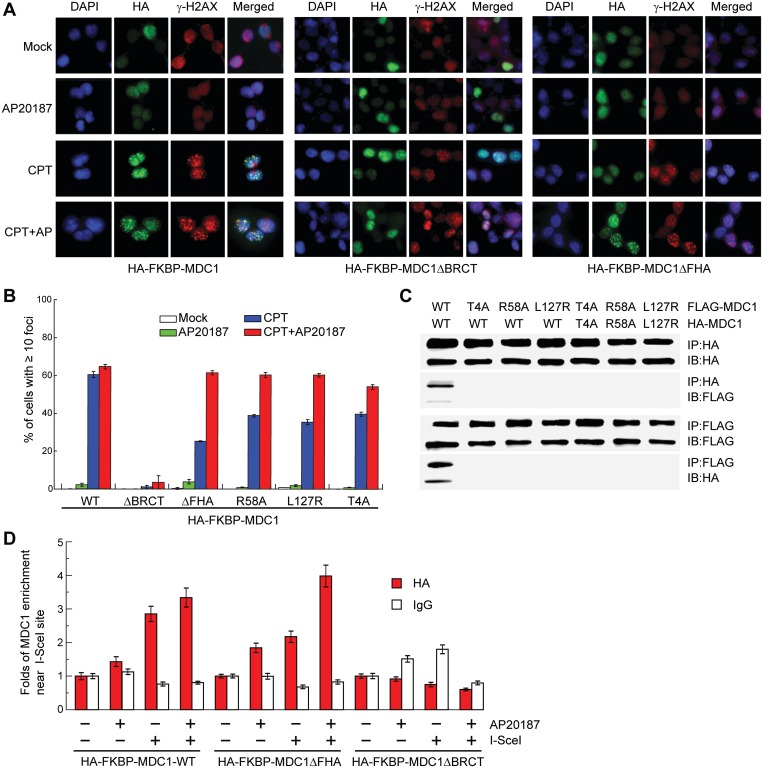
MDC1 is quickly recruited to DSB sites and forms discrete foci that co-localize with γ-H2AX (10,11,13,15). We asked whether FHA-mediated dimerization is important for the formation of MDC1 foci at DSB sites and whether the dimerization or the FHA domain itself is important. To modulate dimerization, MDC1 was fused to the C-terminus of a tandem FKBP tag, which is a monomer but can be induced to dimerize by the bivalent ligand AP20187 (40,58,59).
Upon CPT induction of DNA damage, HA-tagged FKBP-MDC1 efficiently formed foci irrespective of AP21087, suggesting that both the FKBP tag and AP21087 did not interfere with the behavior of MDC1 (Figure 6A and B). Deleting the tandem-BRCT domain completely abolished focus formation, which is consistent with the essential role of this domain in recognizing the DSB marker γ-H2AX (22). The removal of the FHA domain decreased the fraction of cells with 10 or more CPT-induced MDC1 foci by approximately half. This result suggests the FHA domain plays a role in promoting MDC1 focus formation. Interestingly, the focus formation of the FHA deletion mutant could be largely restored by forced dimerization of FKBP with AP20187. This finding suggests that the primary role of the FHA domain is to mediate MDC1 dimerization. Moreover, when the dimerization was disrupted by three single-site mutations that interfere with either the intermolecular FHA–pT4 interaction (T4A and R58A) or FHA–FHA association (L127R) (Figures 2D, 3A and 6C), the MDC1 foci formation was also reduced (Figure 6B). These mutants were better than the FHA deletion mutant in focus formation, probably because they could partially dimerize with endogenous wild-type MDC1. The defective focus formation of these dimerization mutants could also be compensated for by the forced dimerization of FKBP.
Figure 6.
Dimerization promotes enrichment of MDC1 at DNA-damage sites. (A) HA-FKBP-MDC1, HA-FKBP-MDC1ΔBRCT or HA-FKBP-MDC1ΔFHA was stably expressed in Ad293 cells. The cells were mock treated or treated with 10 μM CPT for 1 h, with 10 nM AP20187 for 4 h to induce dimerization of FKBP, or with 10 nM AP20187 for 4 h and 10 μM CPT for 1 h. Cells were fixed and immuno-stained with anti-HA (green) and anti-γ-H2AX (red). DNA was stained with DAPI (blue). (B) Percentage of cells with 10 or more HA-foci in (A). Average of three independent experiments ± SD. In each treatment, 100–200 cells were counted. (C) The T4A, R58A or L127R mutations disrupt MDC1 dimerization. HA- and FLAG-tagged MDC1 or the indicated mutants were transiently expressed in Ad293 cells for 48 h. The total cell lysates were immunoprecipitated with anti-HA and anti-FLAG antibodies and immunoblotted with anti-HA and anti-FLAG antibodies. (D) ChIP analysis of MDC1 localization in defined DSBs. HA-FKBP-MDC1, HA-FKBP-MDC1ΔBRCT or HA-FKBP-MDC1ΔFHA was stably expressed in DFGFP U2OS cells. The cells were transfected with or without an I-SceI expression construct for 24 h, treated with 0 or 10 nM AP20187, and harvested for ChIP assays with anti-HA antibody or a control IgG. The input sample prior to immunoprecipitation and the ChIP elutant were amplified by real-time PCR using a pair of primers covering a region 190–335 bp from the I-SceI cutting site. The signal intensity of the HA or IgG ChIP samples was first divided by the signal intensity of the input samples of same treatment and then normalized to the signal intensity of the ChIP samples without AP20187 and I-SceI treatment. Average of three measures ± SD.
We also analyzed the recruitment of MDC1 to a defined DSB by ChIP assays in U2OS cells (Figure 6D). A DRGFP gene is integrated into the genome of U2OS cells and contains an I-SceI restriction site that will generate a single DNA break in the presence of endonuclease I-SceI (60). HA-FKBP-MDC1, but not its BRCT deletion mutant, was enriched near the I-SceI-induced DSB sites independent of AP20187 treatment. The FHA deletion mutant showed reduced enrichment near the I-SceI site, which could be restored by AP20187 treatment. The ChIP results agree with the MDC1 immunofluorescence results and further suggest the importance of FHA-mediated dimerization for MDC1 function.
DISCUSSION
In this study, we found a novel function of the MDC1 FHA domain in mediating phosphorylation-dependent dimerization. The dimerization is mediated by both the FHA–FHA association and the trans interaction between the FHA domain of one MDC1 molecule and the N-terminal pT4 from the other MDC1 molecule in the dimer. Our results suggest that ATM phosphorylates the T4 residue of MDC1 in response to DNA damage, thereby inducing dimerization of the FHA domain and facilitating localization of MDC1 to DSBs. We present several lines of evidence to support this new regulation mechanism of MDC1 function. First, our biochemical and crystallographic data show that the MDC1 FHA domain recognizes the N-terminal pT4 phosphopeptide. Second, the FHA domain is critically dependent on the trans pT4 interaction for stable dimerization. Third, T4 is primarily phosphorylated by ATM in response to DNA damage. Finally, the FHA domain and its dimerization function are important for optimal MDC1 focus formation at DNA-damage sites.
Luo and colleagues recently reported that T98 of MDC1 is phosphorylated by ATM and interacts with the FHA domain, inducing MDC1 self-association (57). This result is contradictory to our finding about the role of pT4 in mediating MDC1 dimerization. We demonstrate that T98 is totally buried in the structure and is inaccessible to phosphorylation. The T98A mutation appears to disrupt MDC1 dimerization by disturbing the folding and pT binding of the FHA domain, rather than by blocking a phosphorylation site. The impairment of FHA folding and solubility may complicate the interpretation of mutational effects of T98A. Analyses in MDC1-knockout mouse embryonic fibroblasts have shown that the MDC1 T98A mutant was efficiently recruited to DNA-damage sites immediately after damage but was defective in forming full-sized foci in the late stage (57). Moreover, the T98A mutant was shown to partially compromise the intra-S-phase and G2/M checkpoint response after DNA damage, DNA repair efficiency and ATM signaling (57). We also found that MDC1 dimerization mutants form fewer foci at DNA-damage sites. FHA-mediated dimerization appears to play a role in the maintenance and development of MDC1 foci, but the exact molecular mechanism remains speculative.
MDC1 is a key mediator that recruits many DDR factors through its multiple protein–protein interaction modules (21). Dimeric MDC1 may be more efficient at recruiting downstream DDR factors and at amplifying and spreading the DNA-damage signal along the chromatin. In particular, the two tandem-BRCT domains in dimeric MDC1 are separated by approximately 3500 residues and could bridge two γ-H2AX molecules located in distant nucleosomes. Recently, the tandem-BRCT domain of MDC1 was shown to recruit MMSET, a methyltransferase for histone H4 lysine 20, to DSBs (62). The dimeric structure of MDC1 is well suited to serve as a bivalent linker connecting γ-H2AX and MMSET.
We propose that dimerization is the primary function of the MDC1 FHA domain because its role in MDC1 focus formation can largely be replaced by an artificial dimerization module. Interestingly, the FHA-mediated dimerization appears to be evolutionarily conserved in MDC1 proteins. The FHA domain of Xenopus tropicalis MDC1 is dimeric in size exclusion chromatography (S. Luo and K. Ye, unpublished data). Moreover, the sequences of Xenopus and mammalian MDC1 are highly conserved in the FHA domain and the N-terminal residues, including the T4 site, suggesting that FHA- and phosphorylation-dependent dimerization are conserved in Xenopus MDC1. Mutator 2 is the putative ortholog of MDC1 in Drosophila melanogaster (63). We recently found that the Mutator 2 FHA domain forms a stable dimer but lacks a functional pT-binding pocket, indicating that dimerization, but not pT-binding, is conserved between Mutator 2 and MDC1 FHA domains (S. Luo and K. Ye, manuscript in preparation).
In summary, we have discovered a novel function of the MDC1 FHA domain as a phosphorylation-dependent dimerization module that facilitates MDC1 localization to sites of DNA damage. Phosphorylation of MDC1 T4 and induced MDC1 dimerization could constitute a new layer of regulation of the DDR.
ACCESSION NUMBERS
PDB, 3UMZ, 3UNM, 3UNN.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–5.
FUNDING
Ministry of Science and Technology of China (863 project 2008AA022310 and 973 project 2010CB835402 to K.Y. and 973 project 2010CB911904 to X.X.); Beijing Municipal Government (to K.Y.); Capital Normal University (a startup grant to X.X.); National Natural Science Foundation of China (30570371, 90608014, 31071190 and 31130017 to X.X.); Funding Project for Academic Human Resources Development in Institutions of Higher Learning Under the Jurisdiction of Beijing Municipality (PHR20110508 to X.X.); National Institutes of Health (R01CA082257 to D.F.S. and R01CA133093, R01ES016354 and R21NS061748 to B.X.). Funding for open access charge: Ministry of Science and Technology of China (2010CB835402).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully acknowledge the staff of the Shanghai Synchrotron Radiation Facility beamline BL17U for help with data collection, Xiaoxia Yu at Institute of Biophysics, Chinese Academy of Sciences, for analytical ultracentrifugation experiments, Dr Bing Zhu for providing HeLa cell nuclear extracts and Shanshan Wang for technical assistance. We thank Drs Li-Lin Du and Zhiyuan Shen for critically reading the article and comments. We thank other members in the Ye laboratory and the Xu laboratory for help. Author contributions: X.X. initiated the MDC1 dimerization study when he worked at the D.F.S. laboratory. J.L., H.Z., J.L., J.L. and X.X. conducted in vivo biochemical and cellular experiments (Figures 1, 3C–H and 6). S.L. performed X-ray crystallographic and in vitro biochemical experiments (Figures 2, 3A and B, 4 and 5). X.X. and K.Y. conceived and supervised the project. C.Y. and B.X. provided the GST-ATM expression constructs. K.Y. and X.X. wrote the article.
REFERENCES
- 1.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohnishi T, Mori E, Takahashi A. DNA double-strand breaks: their production, recognition, and repair in eukaryotes. Mutat. Res. 2009;669:8–12. doi: 10.1016/j.mrfmmm.2009.06.010. [DOI] [PubMed] [Google Scholar]
- 3.Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem. J. 2009;417:639–650. doi: 10.1042/BJ20080413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem. Cell Biol. 2007;85:509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- 5.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 6.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 7.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iacovoni JS, Caron P, Lassadi I, Nicolas E, Massip L, Trouche D, Legube G. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 2010;29:1446–1457. doi: 10.1038/emboj.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Savic V, Yin B, Maas NL, Bredemeyer AL, Carpenter AC, Helmink BA, Yang-Iott KS, Sleckman BP, Bassing CH. Formation of dynamic gamma-H2AX domains along broken DNA strands is distinctly regulated by ATM and MDC1 and dependent upon H2AX densities in chromatin. Mol. Cell. 2009;34:298–310. doi: 10.1016/j.molcel.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldberg M, Stucki M, Falck J, D'Amours D, Rahman D, Pappin D, Bartek J, Jackson SP. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature. 2003;421:952–956. doi: 10.1038/nature01445. [DOI] [PubMed] [Google Scholar]
- 11.Lou Z, Minter-Dykhouse K, Wu X, Chen J. MDC1 is coupled to activated CHK2 in mammalian DNA damage response pathways. Nature. 2003;421:957–961. doi: 10.1038/nature01447. [DOI] [PubMed] [Google Scholar]
- 12.Shang YL, Bodero AJ, Chen PL. NFBD1, a novel nuclear protein with signature motifs of FHA and BRCT, and an internal 41-amino acid repeat sequence, is an early participant in DNA damage response. J. Biol. Chem. 2003;278:6323–6329. doi: 10.1074/jbc.M210749200. [DOI] [PubMed] [Google Scholar]
- 13.Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421:961–966. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- 14.Xu X, Stern DF. NFBD1/MDC1 regulates ionizing radiation-induced focus formation by DNA checkpoint signaling and repair factors. FASEB J. 2003;17:1842–1848. doi: 10.1096/fj.03-0310com. [DOI] [PubMed] [Google Scholar]
- 15.Xu X, Stern DF. NFBD1/KIAA0170 is a chromatin-associated protein involved in DNA damage signaling pathways. J. Biol. Chem. 2003;278:8795–8803. doi: 10.1074/jbc.M211392200. [DOI] [PubMed] [Google Scholar]
- 16.Lou Z, Minter-Dykhouse K, Franco S, Gostissa M, Rivera MA, Celeste A, Manis JP, van Deursen J, Nussenzweig A, Paull TT, et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol. Cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 17.Minter-Dykhouse K, Ward I, Huen MS, Chen J, Lou Z. Distinct versus overlapping functions of MDC1 and 53BP1 in DNA damage response and tumorigenesis. J. Cell. Biol. 2008;181:727–735. doi: 10.1083/jcb.200801083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bartkova J, Horejsi Z, Sehested M, Nesland JM, Rajpert-De Meyts E, Skakkebaek NE, Stucki M, Jackson S, Lukas J, Bartek J. DNA damage response mediators MDC1 and 53BP1: constitutive activation and aberrant loss in breast and lung cancer, but not in testicular germ cell tumours. Oncogene. 2007;26:7414–7422. doi: 10.1038/sj.onc.1210553. [DOI] [PubMed] [Google Scholar]
- 19.Townsend K, Mason H, Blackford AN, Miller ES, Chapman JR, Sedgwick GG, Barone G, Turnell AS, Stewart GS. Mediator of DNA damage checkpoint 1 (MDC1) regulates mitotic progression. J. Biol. Chem. 2009;284:33939–33948. doi: 10.1074/jbc.M109.009191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coster G, Hayouka Z, Argaman L, Strauss C, Friedler A, Brandeis M, Goldberg M. The DNA damage response mediator MDC1 directly interacts with the anaphase-promoting complex/cyclosome. J. Biol. Chem. 2007;282:32053–32064. doi: 10.1074/jbc.M705890200. [DOI] [PubMed] [Google Scholar]
- 21.Jungmichel S, Stucki M. MDC1: The art of keeping things in focus. Chromosoma. 2010;119:337–349. doi: 10.1007/s00412-010-0266-9. [DOI] [PubMed] [Google Scholar]
- 22.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 23.Lee MS, Edwards RA, Thede GL, Glover JN. Structure of the BRCT repeat domain of MDC1 and its specificity for the free COOH-terminal end of the gamma-H2AX histone tail. J. Biol. Chem. 2005;280:32053–32056. doi: 10.1074/jbc.C500273200. [DOI] [PubMed] [Google Scholar]
- 24.Chapman JR, Jackson SP. Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep. 2008;9:795–801. doi: 10.1038/embor.2008.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melander F, Bekker-Jensen S, Falck J, Bartek J, Mailand N, Lukas J. Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. J. Cell. Biol. 2008;181:213–226. doi: 10.1083/jcb.200708210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spycher C, Miller ES, Townsend K, Pavic L, Morrice NA, Janscak P, Stewart GS, Stucki M. Constitutive phosphorylation of MDC1 physically links the MRE11-RAD50-NBS1 complex to damaged chromatin. J. Cell. Biol. 2008;181:227–240. doi: 10.1083/jcb.200709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu L, Luo K, Lou Z, Chen J. MDC1 regulates intra-S-phase checkpoint by targeting NBS1 to DNA double-strand breaks. Proc. Natl. Acad. Sci. U S A. 2008;105:11200–11205. doi: 10.1073/pnas.0802885105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lloyd J, Chapman JR, Clapperton JA, Haire LF, Hartsuiker E, Li J, Carr AM, Jackson SP, Smerdon SJ. A supramodular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA damage. Cell. 2009;139:100–111. doi: 10.1016/j.cell.2009.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williams RS, Dodson GE, Limbo O, Yamada Y, Williams JS, Guenther G, Classen S, Glover JN, Iwasaki H, Russell P, et al. Nbs1 flexibly tethers Ctp1 and Mre11-Rad50 to coordinate DNA double-strand break processing and repair. Cell. 2009;139:87–99. doi: 10.1016/j.cell.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 31.Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 34.Wang B, Elledge SJ. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc. Natl. Acad. Sci. U S A. 2007;104:20759–20763. doi: 10.1073/pnas.0710061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lou Z, Chen BP, Asaithamby A, Minter-Dykhouse K, Chen DJ, Chen J. MDC1 regulates DNA-PK autophosphorylation in response to DNA damage. J. Biol. Chem. 2004;279:46359–46362. doi: 10.1074/jbc.C400375200. [DOI] [PubMed] [Google Scholar]
- 36.Xie A, Hartlerode A, Stucki M, Odate S, Puget N, Kwok A, Nagaraju G, Yan C, Alt FW, Chen J, et al. Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol. Cell. 2007;28:1045–1057. doi: 10.1016/j.molcel.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahajan A, Yuan C, Lee H, Chen ES, Wu PY, Tsai MD. Structure and function of the phosphothreonine-specific FHA domain. Sci. Signal. 2008;1:re12. doi: 10.1126/scisignal.151re12. [DOI] [PubMed] [Google Scholar]
- 38.Mohammad DH, Yaffe MB. 14-3-3 proteins, FHA domains and BRCT domains in the DNA damage response. DNA Repair (Amst) 2009;8:1009–1017. doi: 10.1016/j.dnarep.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J, Ma Z, Treszezamsky A, Powell SN. MDC1 interacts with Rad51 and facilitates homologous recombination. Nat. Struct. Mol. Biol. 2005;12:902–909. doi: 10.1038/nsmb991. [DOI] [PubMed] [Google Scholar]
- 40.Xu X, Tsvetkov LM, Stern DF. Chk2 activation and phosphorylation-dependent oligomerization. Mol. Cell Biol. 2002;22:4419–4432. doi: 10.1128/MCB.22.12.4419-4432.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu X, Liao J, Creek KE, Pirisi L. Human keratinocytes and tumor-derived cell lines express alternatively spliced forms of transforming growth factor-alpha mRNA, encoding precursors lacking carboxyl-terminal valine residues. Oncogene. 1999;18:5554–5562. doi: 10.1038/sj.onc.1203091. [DOI] [PubMed] [Google Scholar]
- 42.Mossessova E, Lima CD. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- 43.Rauch T, Zhong X, Pfeifer GP, Xu X. 53BP1 is a positive regulator of the BRCA1 promoter. Cell Cycle. 2005;4:1078–1083. [PubMed] [Google Scholar]
- 44.Wang H, Zhao A, Chen L, Zhong X, Liao J, Gao M, Cai M, Lee DH, Li J, Chowdhury D, et al. Human RIF1 encodes an anti-apoptotic factor required for DNA repair. Carcinogenesis. 2009;30:1314–1319. doi: 10.1093/carcin/bgp136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 46.Sheldrick GM. A short history of SHELX. Acta. Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 47.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol. Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 48.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta. Crystallogr D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 50.Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr D Biol. Crystallogr. 1999;55:247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- 51.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DeLano WL. The PyMOL user's manual. San Carlos, CA, USA: Delano Scientific; 2002. [Google Scholar]
- 53.Schuck P. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal. Biochem. 2003;320:104–124. doi: 10.1016/s0003-2697(03)00289-6. [DOI] [PubMed] [Google Scholar]
- 54.Durocher D, Taylor IA, Sarbassova D, Haire LF, Westcott SL, Jackson SP, Smerdon SJ, Yaffe MB. The molecular basis of FHA domain:phosphopeptide binding specificity and implications for phospho-dependent signaling mechanisms. Mol. Cell. 2000;6:1169–1182. doi: 10.1016/s1097-2765(00)00114-3. [DOI] [PubMed] [Google Scholar]
- 55.Li J, Williams BL, Haire LF, Goldberg M, Wilker E, Durocher D, Yaffe MB, Jackson SP, Smerdon SJ. Structural and functional versatility of the FHA domain in DNA-damage signaling by the tumor suppressor kinase Chk2. Mol. Cell. 2002;9:1045–1054. doi: 10.1016/s1097-2765(02)00527-0. [DOI] [PubMed] [Google Scholar]
- 56.Ahn JY, Li X, Davis HL, Canman CE. Phosphorylation of threonine 68 promotes oligomerization and autophosphorylation of the Chk2 protein kinase via the forkhead-associated domain. J. Biol. Chem. 2002;277:19389–19395. doi: 10.1074/jbc.M200822200. [DOI] [PubMed] [Google Scholar]
- 57.Luo K, Yuan J, Lou Z. Oligomerization of MDC1 protein is important for proper DNA damage response. J. Biol. Chem. 2011;286:28192–28199. doi: 10.1074/jbc.M111.258087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Clackson T, Yang W, Rozamus LW, Hatada M, Amara JF, Rollins CT, Stevenson LF, Magari SR, Wood SA, Courage NL, et al. Redesigning an FKBP-ligand interface to generate chemical dimerizers with novel specificity. Proc. Natl. Acad. Sci. U S A. 1998;95:10437–10442. doi: 10.1073/pnas.95.18.10437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Spencer DM, Wandless TJ, Schreiber SL, Crabtree GR. Controlling signal transduction with synthetic ligands. Science. 1993;262:1019–1024. doi: 10.1126/science.7694365. [DOI] [PubMed] [Google Scholar]
- 60.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cai Z, Chehab NH, Pavletich NP. Structure and activation mechanism of the CHK2 DNA damage checkpoint kinase. Mol. Cell. 2009;35:818–829. doi: 10.1016/j.molcel.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 62.Pei H, Zhang L, Luo K, Qin Y, Chesi M, Fei F, Bergsagel PL, Wang L, You Z, Lou Z. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature. 2011;470:124–128. doi: 10.1038/nature09658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dronamraju R, Mason JM. Recognition of double strand breaks by a mutator protein (MU2) in Drosophila melanogaster. PLoS Genet. 2009;5:e1000473. doi: 10.1371/journal.pgen.1000473. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.