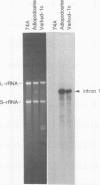
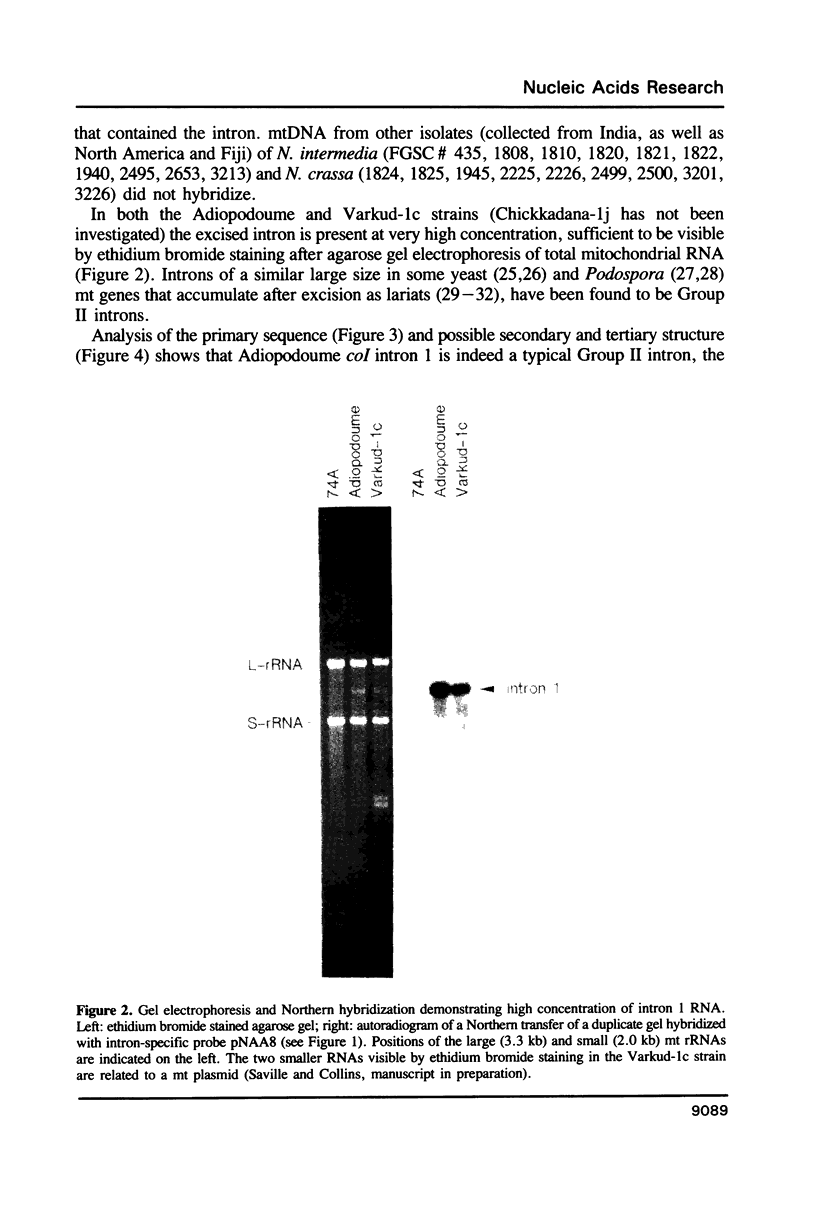
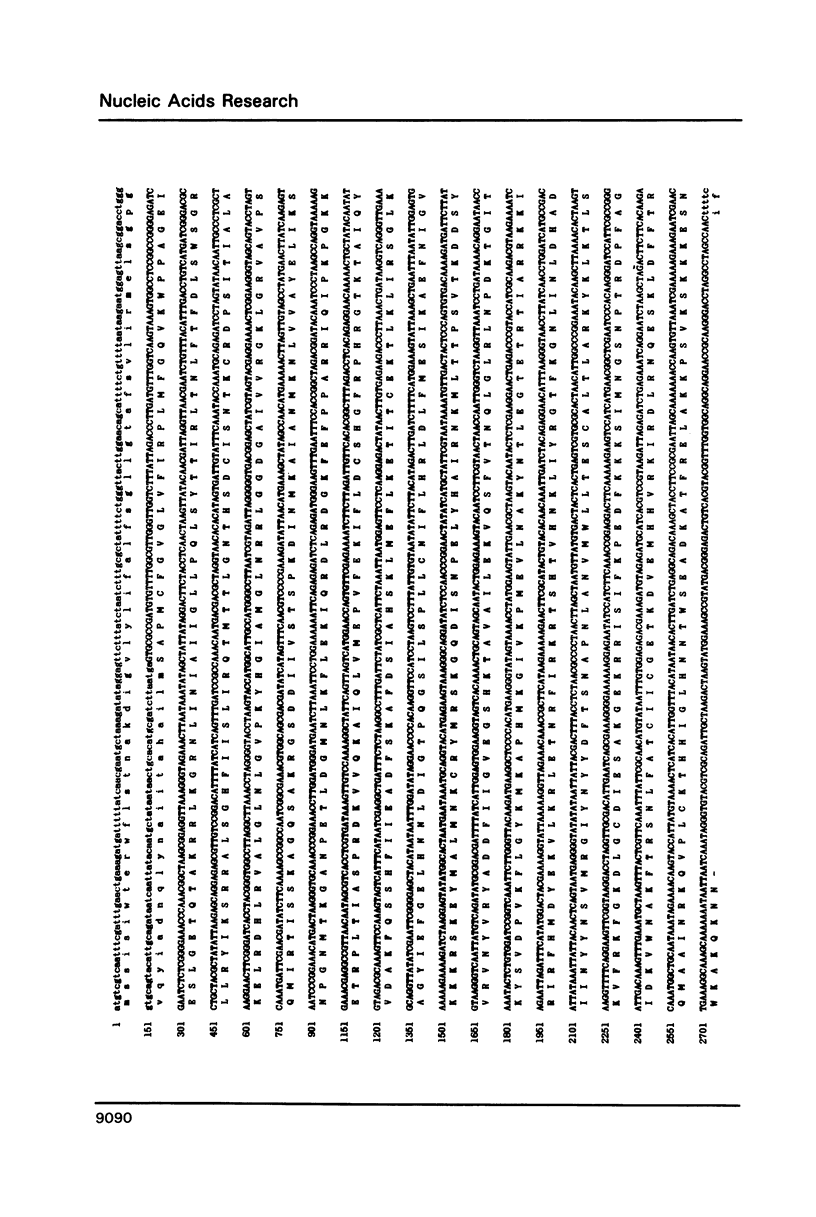
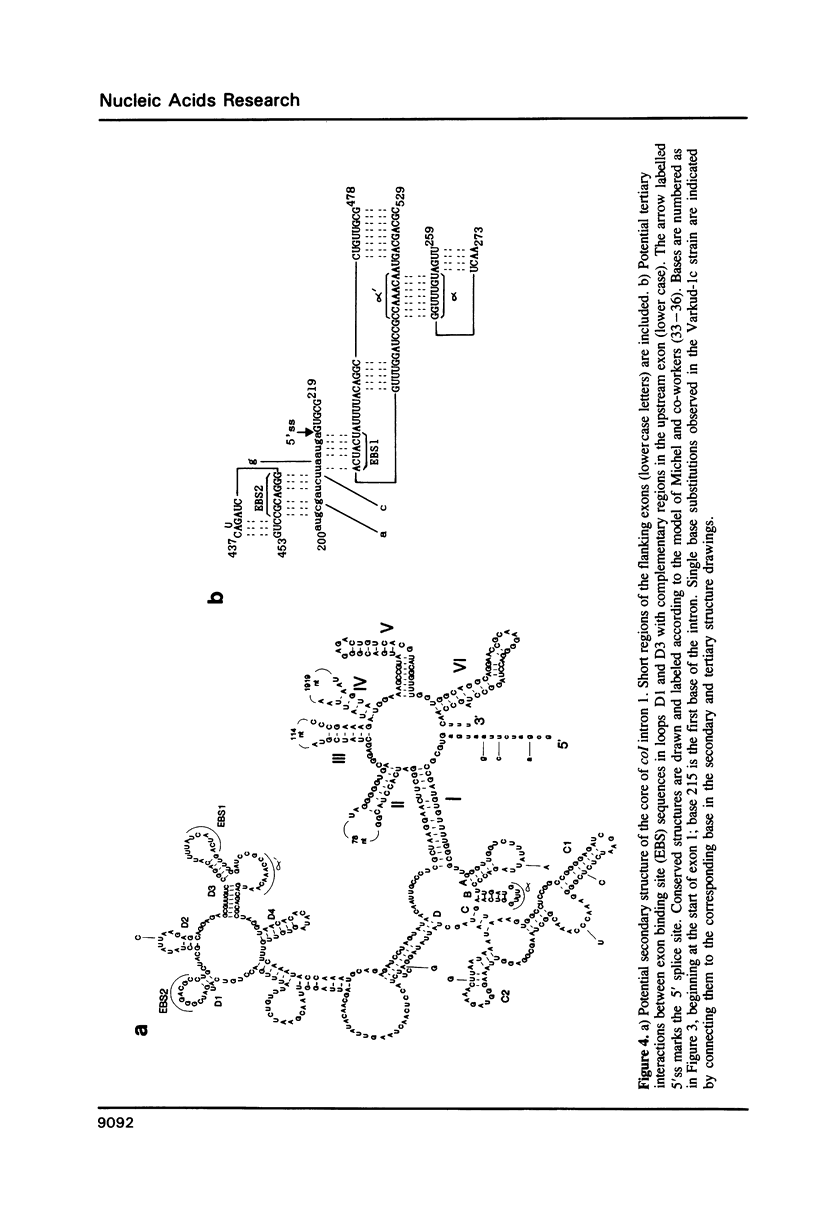
Abstract
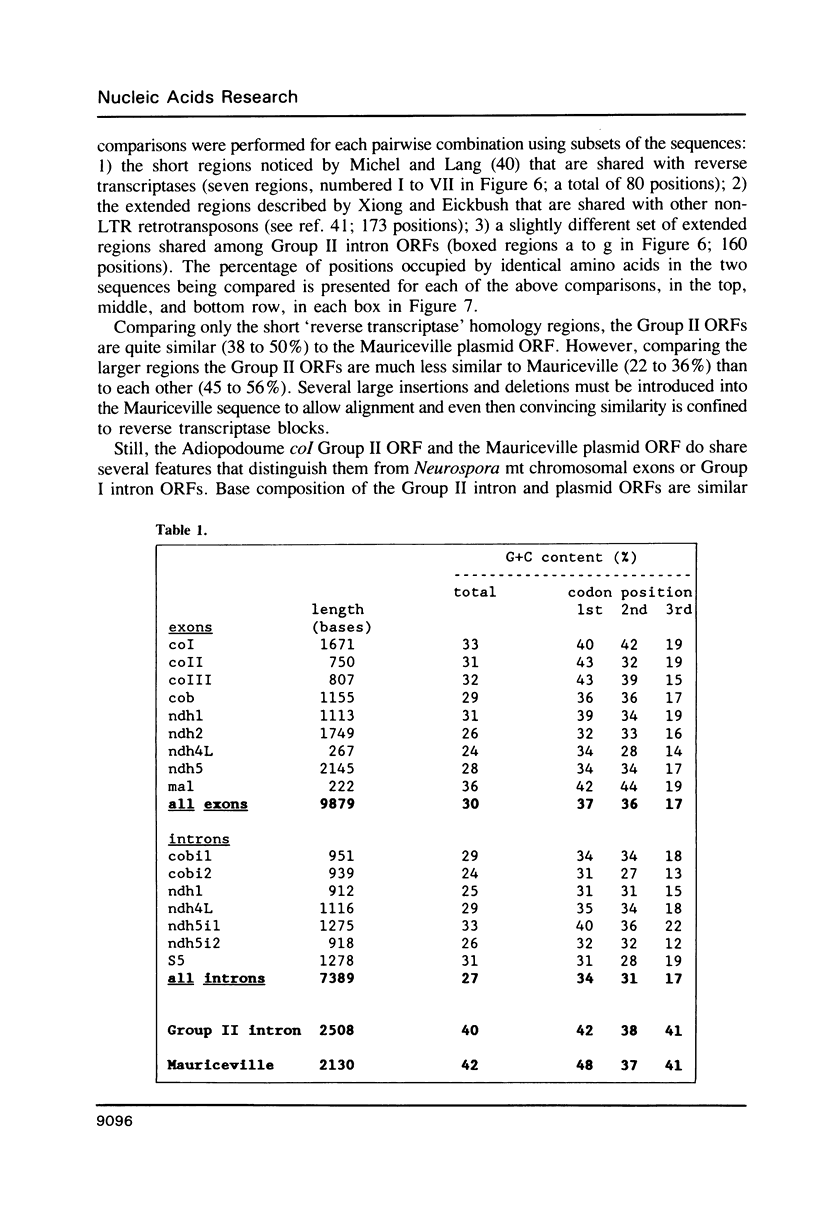
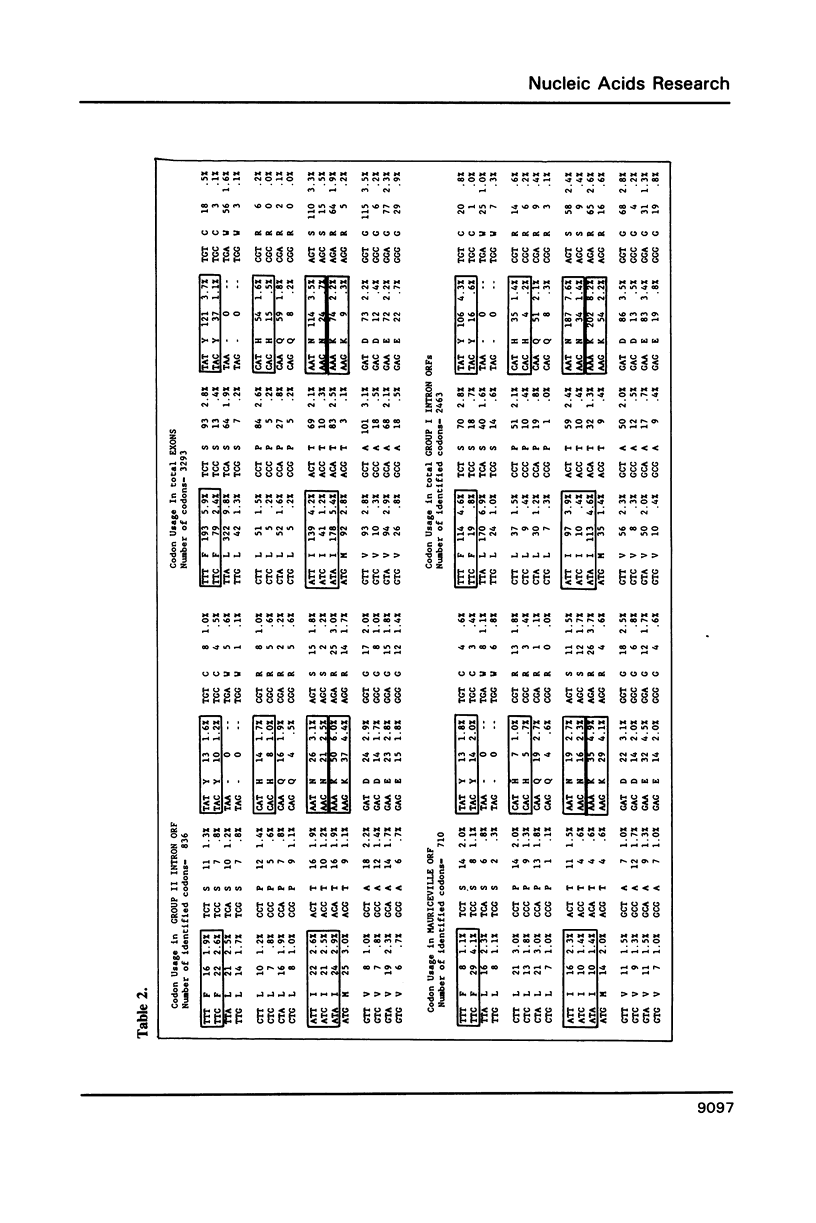
The temperature-sensitive Neurospora nuclear mutant cyt18-1 is deficient in splicing many Group I mitochondrial introns when grown at its non-permissive temperature; however, splicing of intron 1 in the coI gene of the Adiopodoume (formerly called North Africa) strain is unaffected (R.A. Collins and A.M. Lambowitz, J. Mol. Biol. 184: 413-428, 1985). Here we show that coI intron 1 is a typical Group II intron, the only one identified to date in Neurospora. The differential effect of the cyt18-1 mutation suggests that splicing of certain introns could be regulated independently of others by nuclear-encoded proteins. The intron contains a long open reading frame (ORF) resembling that of the Neurospora Mauriceville mitochondrial plasmid. The intron and plasmid ORFs share unusual features of codon usage that suggest both evolved outside of the Neurospora mitochondrial genetic system.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnberg A. C., Van Ommen G. J., Grivell L. A., Van Bruggen E. F., Borst P. Some yeast mitochondrial RNAs are circular. Cell. 1980 Feb;19(2):313–319. doi: 10.1016/0092-8674(80)90505-x. [DOI] [PubMed] [Google Scholar]
- Bonitz S. G., Coruzzi G., Thalenfeld B. E., Tzagoloff A., Macino G. Assembly of the mitochondrial membrane system. Structure and nucleotide sequence of the gene coding for subunit 1 of yeast cytochrme oxidase. J Biol Chem. 1980 Dec 25;255(24):11927–11941. [PubMed] [Google Scholar]
- Burger G., Scriven C., Machleidt W., Werner S. Subunit 1 of cytochrome oxidase from Neurospora crassa: nucleotide sequence of the coding gene and partial amino acid sequence of the protein. EMBO J. 1982;1(11):1385–1391. doi: 10.1002/j.1460-2075.1982.tb01327.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke J. M., Breitenberger C., Heckman J. E., Dujon B., RajBhandary U. L. Cytochrome b gene of Neurospora crassa mitochondria. Partial sequence and location of introns at sites different from those in Saccharomyces cerevisiae and Aspergillus nidulans. J Biol Chem. 1984 Jan 10;259(1):504–511. [PubMed] [Google Scholar]
- Burke J. M. Molecular genetics of group I introns: RNA structures and protein factors required for splicing--a review. Gene. 1988 Dec 20;73(2):273–294. doi: 10.1016/0378-1119(88)90493-3. [DOI] [PubMed] [Google Scholar]
- Burke J. M., RajBhandary U. L. Intron within the large rRNA gene of N. crassa mitochondria: a long open reading frame and a consensus sequence possibly important in splicing. Cell. 1982 Dec;31(3 Pt 2):509–520. doi: 10.1016/0092-8674(82)90307-5. [DOI] [PubMed] [Google Scholar]
- Cech T. R. Conserved sequences and structures of group I introns: building an active site for RNA catalysis--a review. Gene. 1988 Dec 20;73(2):259–271. doi: 10.1016/0378-1119(88)90492-1. [DOI] [PubMed] [Google Scholar]
- Collins R. A., Lambowitz A. M. RNA splicing in Neurospora mitochondria. Defective splicing of mitochondrial mRNA precursors in the nuclear mutant cyt18-1. J Mol Biol. 1985 Aug 5;184(3):413–428. doi: 10.1016/0022-2836(85)90291-8. [DOI] [PubMed] [Google Scholar]
- Collins R. A., Lambowitz A. M. Structural variations and optional introns in the mitochondrial DNAs of Neurospora strains isolated from nature. Plasmid. 1983 Jan;9(1):53–70. doi: 10.1016/0147-619x(83)90031-8. [DOI] [PubMed] [Google Scholar]
- Cummings D. J., MacNeil I. A., Domenico J., Matsuura E. T. Excision-amplification of mitochondrial DNA during senescence in Podospora anserina. DNA sequence analysis of three unique "plasmids". J Mol Biol. 1985 Oct 20;185(4):659–680. doi: 10.1016/0022-2836(85)90052-x. [DOI] [PubMed] [Google Scholar]
- Dale R. M., McClure B. A., Houchins J. P. A rapid single-stranded cloning strategy for producing a sequential series of overlapping clones for use in DNA sequencing: application to sequencing the corn mitochondrial 18 S rDNA. Plasmid. 1985 Jan;13(1):31–40. doi: 10.1016/0147-619x(85)90053-8. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Garriga G., Bertrand H., Lambowitz A. M. RNA splicing in Neurospora mitochondria: nuclear mutants defective in both splicing and 3' end synthesis of the large rRNA. Cell. 1984 Mar;36(3):623–634. doi: 10.1016/0092-8674(84)90342-8. [DOI] [PubMed] [Google Scholar]
- Garriga G., Lambowitz A. M. Protein-dependent splicing of a group I intron in ribonucleoprotein particles and soluble fractions. Cell. 1986 Aug 29;46(5):669–680. doi: 10.1016/0092-8674(86)90342-9. [DOI] [PubMed] [Google Scholar]
- Garriga G., Lambowitz A. M. RNA splicing in neurospora mitochondria: self-splicing of a mitochondrial intron in vitro. Cell. 1984 Dec;39(3 Pt 2):631–641. doi: 10.1016/0092-8674(84)90470-7. [DOI] [PubMed] [Google Scholar]
- Helmer-Citterich M., Morelli G., Macino G. Nucleotide sequence and intron structure of the apocytochrome b gene of Neurospora crassa mitochondria. EMBO J. 1983;2(8):1235–1242. doi: 10.1002/j.1460-2075.1983.tb01575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hensgens L. A., Bonen L., de Haan M., van der Horst G., Grivell L. A. Two intron sequences in yeast mitochondrial COX1 gene: homology among URF-containing introns and strain-dependent variation in flanking exons. Cell. 1983 Feb;32(2):379–389. doi: 10.1016/0092-8674(83)90457-9. [DOI] [PubMed] [Google Scholar]
- Jacquier A., Michel F. Multiple exon-binding sites in class II self-splicing introns. Cell. 1987 Jul 3;50(1):17–29. doi: 10.1016/0092-8674(87)90658-1. [DOI] [PubMed] [Google Scholar]
- Korneluk R. G., Quan F., Gravel R. A. Rapid and reliable dideoxy sequencing of double-stranded DNA. Gene. 1985;40(2-3):317–323. doi: 10.1016/0378-1119(85)90055-1. [DOI] [PubMed] [Google Scholar]
- Lambowitz A. M. Preparation and analysis of mitochondrial ribosomes. Methods Enzymol. 1979;59:421–433. doi: 10.1016/0076-6879(79)59103-4. [DOI] [PubMed] [Google Scholar]
- Lang B. F., Ahne F., Bonen L. The mitochondrial genome of the fission yeast Schizosaccharomyces pombe. The cytochrome b gene has an intron closely related to the first two introns in the Saccharomyces cerevisiae cox1 gene. J Mol Biol. 1985 Aug 5;184(3):353–366. doi: 10.1016/0022-2836(85)90286-4. [DOI] [PubMed] [Google Scholar]
- Mannella C. A., Collins R. A., Green M. R., Lambowitz A. M. Defective splicing of mitochondrial rRNA in cytochrome-deficient nuclear mutants of Neurospora crassa. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2635–2639. doi: 10.1073/pnas.76.6.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMaster G. K., Carmichael G. G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Dujon B. Conservation of RNA secondary structures in two intron families including mitochondrial-, chloroplast- and nuclear-encoded members. EMBO J. 1983;2(1):33–38. doi: 10.1002/j.1460-2075.1983.tb01376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel F., Jacquier A., Dujon B. Comparison of fungal mitochondrial introns reveals extensive homologies in RNA secondary structure. Biochimie. 1982 Oct;64(10):867–881. doi: 10.1016/s0300-9084(82)80349-0. [DOI] [PubMed] [Google Scholar]
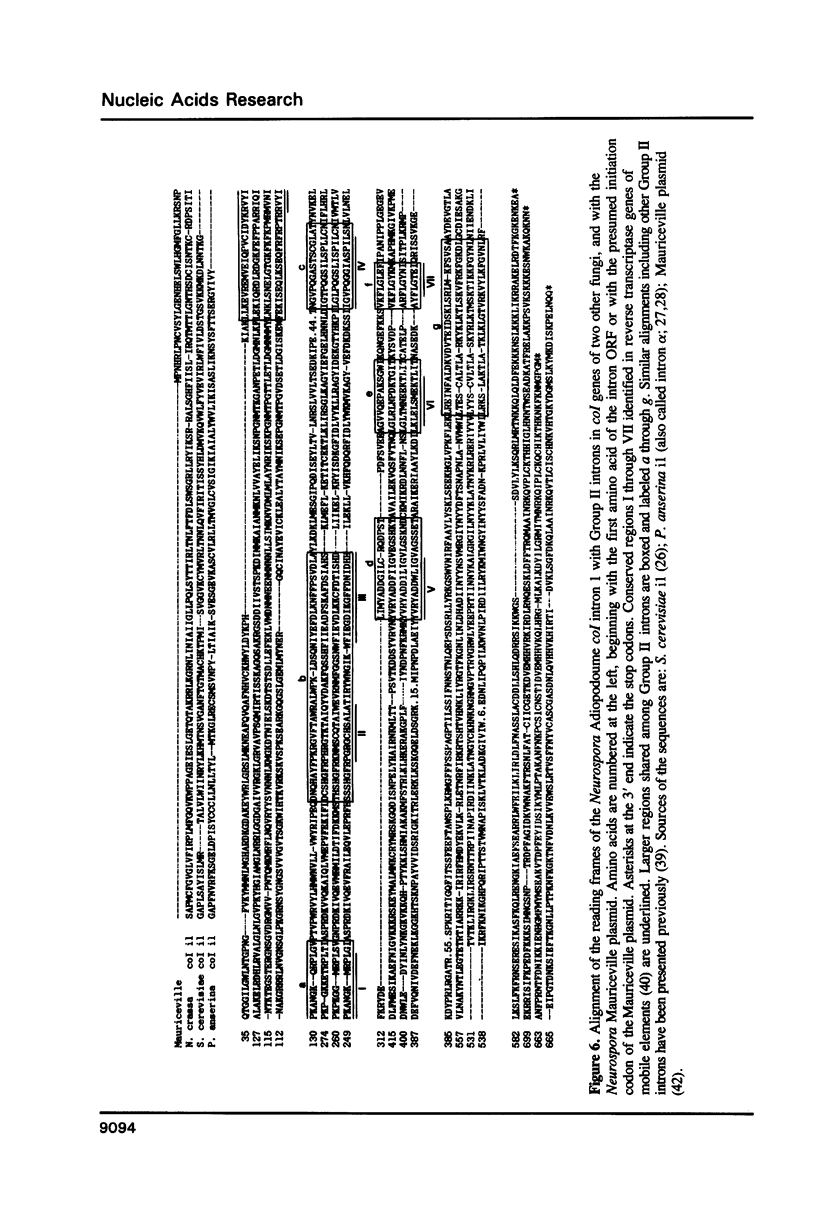
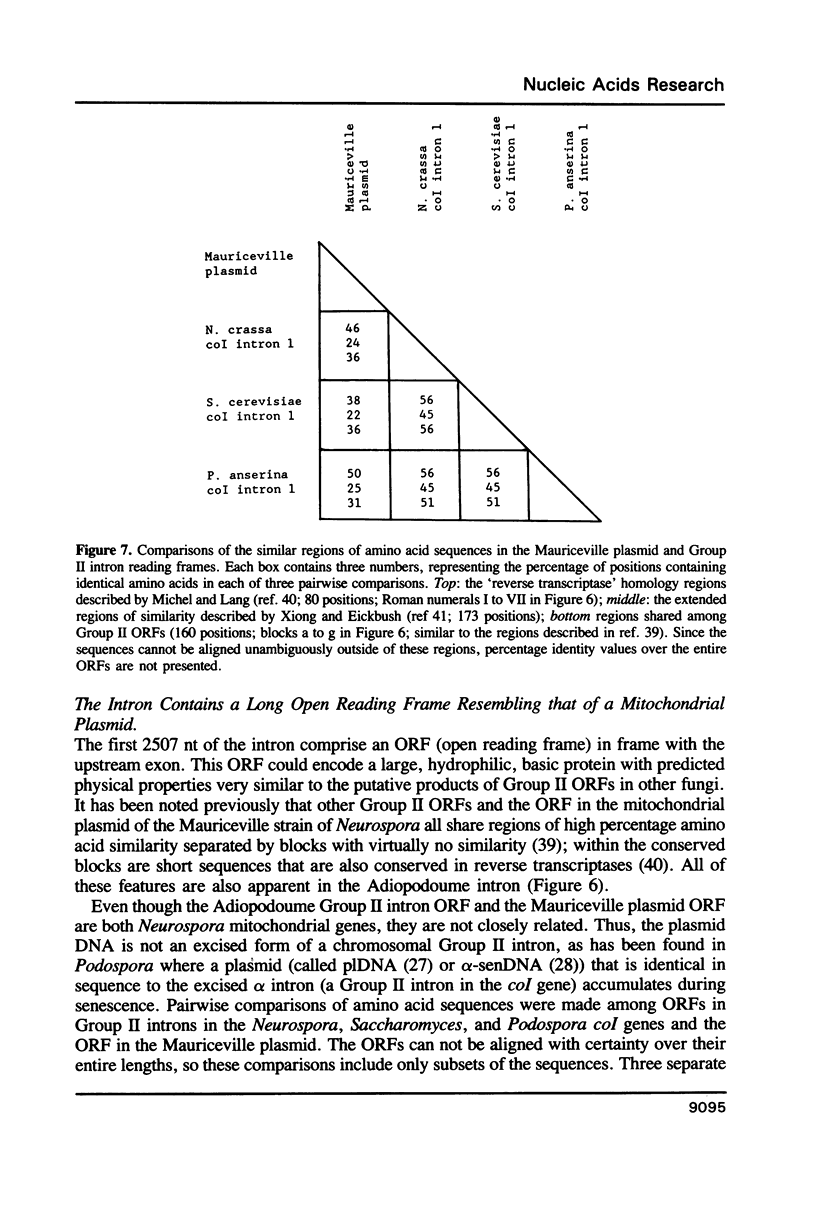
- Michel F., Lang B. F. Mitochondrial class II introns encode proteins related to the reverse transcriptases of retroviruses. Nature. 1985 Aug 15;316(6029):641–643. doi: 10.1038/316641a0. [DOI] [PubMed] [Google Scholar]
- Mizusawa S., Nishimura S., Seela F. Improvement of the dideoxy chain termination method of DNA sequencing by use of deoxy-7-deazaguanosine triphosphate in place of dGTP. Nucleic Acids Res. 1986 Feb 11;14(3):1319–1324. doi: 10.1093/nar/14.3.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peebles C. L., Perlman P. S., Mecklenburg K. L., Petrillo M. L., Tabor J. H., Jarrell K. A., Cheng H. L. A self-splicing RNA excises an intron lariat. Cell. 1986 Jan 31;44(2):213–223. doi: 10.1016/0092-8674(86)90755-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzer C., Schweyen R. J. Self-splicing of group II introns in vitro: mapping of the branch point and mutational inhibition of lariat formation. Cell. 1986 Aug 15;46(4):557–565. doi: 10.1016/0092-8674(86)90881-0. [DOI] [PubMed] [Google Scholar]
- Welser F., Wolf K. Strain-dependent variation of intron-exon pairings in a group II intron of Schizosaccharomyces pombe. Nucleic Acids Res. 1988 Feb 25;16(4):1629–1629. doi: 10.1093/nar/16.4.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollenzien P. L., Cantor C. R., Grant D. M., Lambowitz A. M. RNA splicing in neurospora mitochondria: structure of the unspliced 35S precursor ribosomal RNA detected by psoralen cross-linking. Cell. 1983 Feb;32(2):397–407. doi: 10.1016/0092-8674(83)90459-2. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Eickbush T. H. Similarity of reverse transcriptase-like sequences of viruses, transposable elements, and mitochondrial introns. Mol Biol Evol. 1988 Nov;5(6):675–690. doi: 10.1093/oxfordjournals.molbev.a040521. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- van der Veen R., Arnberg A. C., van der Horst G., Bonen L., Tabak H. F., Grivell L. A. Excised group II introns in yeast mitochondria are lariats and can be formed by self-splicing in vitro. Cell. 1986 Jan 31;44(2):225–234. doi: 10.1016/0092-8674(86)90756-7. [DOI] [PubMed] [Google Scholar]