Abstract
The DNA-dependent protein kinase (DNA-PK) and Poly(ADP-ribose) polymerase-1 (PARP1) are critical enzymes that reduce genomic damage caused by DNA lesions. They are both activated by DNA strand breaks generated by physiological and environmental factors, and they have been shown to interact. Here, we report in vivo evidence that DNA-PK and PARP1 are equally necessary for rapid repair. We purified a DNA-PK/PARP1 complex loaded on DNA and performed electron microscopy and single particle analysis on its tetrameric and dimer-of-tetramers forms. By comparison with the DNA-PK holoenzyme and fitting crystallographic structures, we see that the PARP1 density is in close contact with the Ku subunit. Crucially, PARP1 binding elicits substantial conformational changes in the DNA-PK synaptic dimer assembly. Taken together, our data support a functional, in-pathway role for DNA-PK and PARP1 in double-strand break (DSB) repair. We also propose a NHEJ model where protein–protein interactions alter substantially the architecture of DNA-PK dimers at DSBs, to trigger subsequent interactions or enzymatic reactions.
INTRODUCTION
DNA double-strand breaks (DSBs) are the most cytotoxic form of DNA damage. If unrepaired or incorrectly repaired, they can lead to apoptosis or genome instability. Two major DSB repair pathways exist: homologous recombination (HR) and non-homologous end joining (NHEJ) (1). In the presence of sister chromatids acting as templates for repair, DSBs can be repaired by HR. In NHEJ, the DNA broken ends are resected and/or processed and the DNA backbones ligated to restore strand continuity without the need for a template.
The DNA-dependent protein kinase (DNA-PK) heterotrimeric enzyme is the early player in mammalian NHEJ (2). DNA-PK is formed by a DSB recognition module, called Ku (preassembled as a heterodimer of the Ku70 and Ku80 proteins) (3) and a large catalytic subunit (DNA-PKcs, ∼0.5 MDa) (4,5). After recognizing and binding to a DSB, Ku recruits the catalytic subunit via the C-terminal domain of the Ku80 subunit. DNA-PKcs is a serine/threonine kinase belonging to the phosphatidylinositol-3-OH kinase (PI3K)-related (PIKK) family. DNA-PK assembles on DNA as a bridging complex, where two heterotrimers maintain the two DNA broken ends in close proximity, providing a scaffolding platform to recruit further NHEJ enzymes (6). These factors include Artemis and PNK, which are required to process the broken ends; X family polymerases, which promote microhomology and cohesion between the broken ends; DNA ligase IV-XRCC4, which closes the phospho-diester backbone on both strands, and the XLF/Cernunnos factor (7). Autophosphorylation sites, crucial to NHEJ regulation, have been identified in DNA-PKcs at two main clusters, as well as within its catalytic domain (7). A recent electron microscopy study visualized a substantial remodeling of DNA-PK on autophosphorylation (8).
PARP enzymes use nicotinamide as a substrate to polymerize ADP-ribose moieties onto target proteins, a process called poly-ADP-ribosylation (or PARylation) (9). The best-studied PARP enzyme is PARP1, which has a key role in DNA repair and in particular single-strand break repair/base excision repair (10). PARP1 detects and binds single strand DNA breaks and then poly-ADP-ribosylates itself (auto-PARylation) and other proteins (e.g. histones). It recognizes DNA strand breaks and binds to them, both in vivo and in vitro (11). PARP1 is a 113 kDa enzyme with three functional domains: an N-terminal DNA-binding domain, a central automodification domain, and a C-terminal catalytic domain.
Genetic interaction between DNA-PK and PARP was initially related to recombinational events (12). The cross-talk between DNA-PK or its component Ku70:80 and PARP1 within NHEJ and V(D)J recombination has been described in various independent studies (13–21). A recent study, focusing on the role of PARP1 in V(D)J recombination, reports that the immunoprecipitation of the BRCT domain of PARP1 pulls down Ku70 and the DNA-PK complex in a DNA-independent manner (22). This finding suggests that PARP1 modulates DNA-PK in gene conversion, and that this is mediated through BRCT domain-mediated interactions. Alternatively, an important and DNA-PK-independent role of PARP1 in NHEJ has recently emerged. PARP1 is proposed to operate in an alternative NHEJ pathway, which backs up the classical pathway and is particularly active in microhomology-facilitated NHEJ (23,24). Recent data indicate that PARP1 can modulate competition between HR and NHEJ -PARP inhibition alone causes cell death in HR-defective, e.g. BRCA1 mutant cells. However it now appears that co-inactivation of NHEJ (by inactivation of DNA-PKcs or 53BP1) rescues BRCA1 mutant cells from PARP inhibitor cytotoxicity (25,26). These roles of PARP1 are not mutually exclusive, however they need a much finer characterization to fully understand the interplay of PARP1 and other repair factors.
DNA-PKcs and DNA-PK have been at the centre of numerous structural studies in the last 15 years. X-ray crystallography, NMR, electron microscopy (EM) and SAXS have all contributed to our understanding of these proteins, either in isolation or in complex (6,27–37). We were the first in visualizing DNA-PK synaptic dimers by electron microscopy and single particle analysis (6). Our findings were later supported by a SAXS study, where DNA-PK synaptic dimers loaded on a Y-shaped DNA were shown to arrange in the same way in solution as in our single particle analysis (37). DNA-PK dimers loaded on hairpin DNA produced a different architecture in solution, with DNA-PKcs heads (and therefore kinase domains) in close proximity. This shows the plasticity of DNA-PK, which responds to different macromolecular interactions with substantial architectural rearrangements. Recently, we showed how autophosphorylation has a dramatic structural effect on DNA-PK dimers, causing disassembly and structural heterogeneity (8).
The structural data available for PARP1 are so far limited to its isolated domains, probably due to its size and flexibility. The human PARP1 enzyme is a modular protein of 113 kDa. Three zinc finger domains, Zn1–Zn3, are located at its extreme N-terminus. An internal automodification region contains a BRCT domain involved in mediating protein–protein interactions and three lysines that are targeted for automodification. The catalytic domain is located at the extreme C-terminus of the enzyme. The crystal structure of the catalytic domain (38) led to the development of a major breakthrough in structure-based drug design: PARP inhibitors are currently in clinical trials for the treatment of breast cancer (39). A crystal structure has recently been solved for the Zn3 domain (40). Early DNA binding studies using a Zn1–Zn2 domain fragment suggested that PARP1 binds to DNA as a dimer (41), and kinetic analysis of PARP1 activity indicated that PARP1 is a catalytic dimer (42). However, more recent data suggest that the Zn1–Zn2 fragment recognizes DNA single-strand breaks as a monomer (43). A direct interaction between Ku and PARP1 has been described (22), however the stoichiometry of the Ku/PARP1 assembly has not been investigated to date.
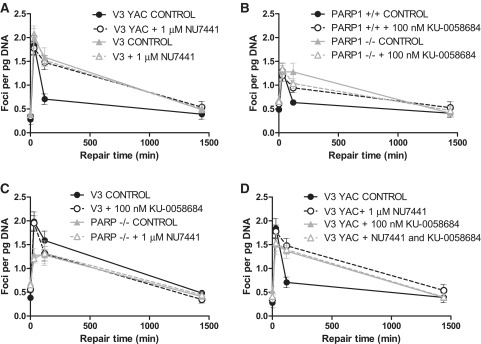
Here, we present in vivo data on the effect of DNA-PK and PARP1 inhibitors in repair competent cell lines, as well as in cells deficient in either DNA-PK or PARP1. Our data highlight that, upon exposure to a clinically relevant dose of radiation (2Gy) (44), γH2AX foci, which mark DNA DSBs (45), were formed. We show lack of additivity of the two inhibitors in DNA-PK proficient V3-YAC cells, and a PARP inhibitor KU-0058684 having no further impact on the repair of DSBs in DNA-PK deficient V3 cells. The same trends were observed with PARP1 deficient and proficient cells treated with a DNA-PK inhibitor or combination of inhibitors.
Although a DNA-PK/PARP complex has been described in biochemical studies, no structural data is currently available for it. To understand the nature of the DNA-PK/PARP1 interplay, we performed structural studies of the purified DNA-PK/PARP1 complex loaded on DNA, analysed by electron microscopy and single particle analysis. We identified and analysed the structure of DNA-PK/PARP1 heterotetramers and dimers-of-tetramers. A tight interaction surface between the Ku dimer and PARP1 within these complexes is apparent. Fitting of the PARP1 catalytic domain into the EM map is consistent with it not being in direct contact with Ku, supporting the fact that the BRCT domain may be its interactor. Our fitting also supports a model where one PARP1 subunit is involved in the complex with DNA-PK, opposite to the homodimerization occurring in free PARP1. The architecture of DNA-PK/PARP1 dimers is strikingly different from the DNA-PK assemblies we analysed in the past (6,8,46) and from those reported in SAXS studies on Y-shaped DNA (37). In the presence of PARP1, the catalytic domain of one of the DNA-PKcs molecules involved in the complex is in direct contact with the arm domain of its DNA-PKcs counterpart, supporting the possibility of an autophosphorylation in trans (47). The DNA-PKcs catalytic domains are not in the same orientation as in the SAXS analysis of DNA-PK loaded on hairpin DNA. This would suggest a role for PARP1 in modulating DNA repair by eliciting a major architectural rearrangement of the DNA-PK-mediated synapsis, as well as supporting a high degree of conformational articulation of DNA-PK in response to diverse stimuli.
MATERIALS AND METHODS
Inhibitors
The DNA-PK inhibitor NU7441 (48) and the PARP1 inhibitor KU-0058684 (49), synthesized by Newcastle University, UK, and KuDOS Pharmaceuticals (Cambridge, UK), respectively, were dissolved in 100% (v/v) dimethyl sulfoxide (DMSO) at stock concentrations of 2 mM and 200 mM, respectively, and stored at −20°C. Drugs were added in 1% (v/v) final DMSO concentration, and control cells were exposed to 1% DMSO. NU7441 (IC50 for DNA-PK inhibition = 14 nM) was used at 1 μM as previously described (50). KU-0058684 (IC50 for PARP1 inhibition = 3.2 nM) was used at 100 nM.
Cell lines and culture
Primary PARP1+/+ and PARP1−/− mouse embryonic fibroblasts (MEFs) were a gift from Professor Gilbert de Murcia, France (51). Chinese hamster ovary cell lines V3 (mutated in DNA-PKcs) and V3 YAC (V3 transfected with yeast artificial chromosome containing human DNA-PKcs cDNA) were provided by Dr Penny Jeggo, UK (52). Cell lines were cultured in RPMI 1640 + 10% (v/v) fetal calf serum and 1% (v/v) penicillin–streptomycin. V3 YAC cells were maintained under antibiotic selection with 500 mg/ml geneticin to ensure YAC retention. PARP and DNA-PK activities of these cell lines have been described previously (20,53).
DNA double strand break determination by gH2AX immunofluorescence
DSBs were detected by γH2AX immunofluorescence assays utilizing an anti-phospho H2AX antibody as previously published (anti ser139, clone JBW301, Upstate, now Millipore Corp., USA) (54). NU7441 and KU-0058684 were added to cells 1 h before 2 Gy X-irradiation (Gulmay D3300 X-Ray System, Gulmay Medical Ltd., UK) as previously described (50,54). Image capture was performed as previously described (54), recording 2 viewfields each containing 20 nuclei. Mean focus number per nucleus was converted to foci/pg DNA present to account for different cellular DNA content (determined by picogreen DNA quantitation assay, Qiagen, UK). DNA repair was calculated as percent focus loss.
Isolation of the DNA-bound DNA-PKcs/Ku70/Ku80/PARP1 complexes
The complex containing DNA-PKcs, Ku70, Ku80 and PARP1 was purified from HeLa nuclear extract (CilBiotech, Belgium) with a buffer system based on 20 mM HEPES (pH 7.5), 1 mM DTT, 0.5 mM EDTA, 0.001% β-octylglucoside and 10% glycerol, with NaCl concentrations ranging from 50 to 1000 mM in different chromatographic steps. After fractionation on heparin and dsDNA cellulose resins (GE Healthcare, Sweden), fractions containing DNA-PKcs, Ku70 and Ku80 were loaded onto a 1 ml HiTrapQ column (GE Healthcare, Sweden) and eluted with a linear NaCl gradient. This differs from our previous protocol for the purification of DNA-PK heterotrimer (6), since we used a HiTrapQ column instead of a Resource Q column, therefore obtaining a less stringent fractionation of proteins interacting with Q resin. Fractions containing all three protein components were incubated with λ-phosphatase for 1 h at 4°C. The sample was incubated for an additional hour at 4°C with dsDNA iminobiotinylated at the 5′-end of the longer oligo (IBA GmbH, Germany). The sequences of the oligos (with the linear duplex segments in uppercase) are shown in Table 1.
Table 1.
Oligos used to assemble the structured DNA on which the DNA-PK/PARP1 complex is loaded
| Oligo A | Biotin-5′-CGCGCCCAgctttcccagctaATAAACTAAAAACTATTATTATGGCCGCACGCGTCCACCATGGGGTACAACT-3′ |
| Oligo B | 5′-AGTTGTACCCCATGGTGGACGCGTGCGGCCATAATAATAGTTTTTAGTTTATTGGGCGCG-3′ |
Upper case: bases annealing to for dsDNA. Lower case: bases forming a loop.
The samples were then loaded on a 18–60% glycerol gradient, with the buffer system 20 mM HEPES (pH 7.5), NaCl 200 mM, 1 mM DTT, 0.5 mM EDTA, 0.001% β-octylglucoside. Further separation was achieved by using a Beckman SW28 rotor, spinning for 72 h at 25 000 rpm. Fractions were collected from the bottom and analysed by means of SDS–PAGE on 4–12% Bis-Tris NOVEX gradient gels (Invitrogen, UK). This analysis highlighted the co-migration of stoichiometric quantities of an additional protein with the three protein components of DNA-PK (Figure 2B). The fourth protein species was identified as PARP1 by mass spectrometry and immunoblotting (data not shown).
Figure 2.
Purification of the DNA-PK/PARP1 complex. (A) Schematics of purification protocol. (B) Silver stained SDS–PAGE analysis of the purified complex, showing four apparently stoichiometric bands for the DNA-PKcs, PARP1, Ku70 and Ku80 polypeptides. Electron microscopy and single particle analysis of the DNA-PK/PARP1 complex. (C) Raw micrograph, showing heterogeneity in the sample (monomeric and dimeric complexes coexist, in analogy with the DNA-PK complex in isolation). White circles: monomers; white squares: dimers. (D and E) Reference-free class averages of monomeric complex show heterogeneity, since related views can either have or not have a strong density (indicated by the white arrow in (E). (F) Class averages used in the final 3D reconstruction. (G) Reprojections of the 3D model. (H) 3D model calculated by single particle analysis, accounting for the volume of the sum of DNA-PKcs, Ku heterodimers and PARP1. I. Fourier Shell Correlation plot.
Electron microscopy and single-particle analysis of negatively stained DNA-PKcs/Ku70/Ku80/PARP1 complexes
The DNA-PKcs/Ku70/Ku80/PARP1 sample was applied to carbon-coated grids, negatively stained with 1% uranyl acetate and observed in a JEOL 1200EX electron microscope operating at 100 kV. Micrographs were recorded at a calibrated magnification of 34 000× under low-dose conditions and digitized with a Nikon Coolscan scanner. A total of 21 000 molecular images were extracted with the Boxer program from the EMAN package, processed and refined with a combination of EMAN (55), IMAGIC-5 (56) and Spider (57) routines. 3D fitting experiments were performed using Chimera (58).
RESULTS
DNA-PK and PARP1 lack additivity in DSB repair
We studied the role of PARP1 in the cellular response to clinically relevant IR doses by measuring DNA DSB repair in DNA-PK+/+ (V3-YAC), DNA-PK−/− (V3), PARP1+/+ and PARP1−/− cells. After irradiation of PARP1 proficient cells, DNA DSBs were rapidly induced as determined by γH2AX focus formation after 2 Gy IR (Figure 1). In repair-competent cell lines and consistent with our previous data with these cells (19) and independent data (59), DSB resolution followed a biphasic pattern. There was a rapid resolution of DSBs during the first 2 h with 80 ± 24% and 71 ± 10% being repaired in PARP1+/+ and V3-YAC cells, respectively. However, PARP1−/− MEFs and DNA-PKcs deficient V3 cells followed a different repair kinetics, with a much slower rate such that only 27 ± 21% and 25 ± 9%, respectively, had been repaired.
Figure 1.
The effect of DNA-PK and PARP inhibitors on DSB repair. The recovery from IR-induced DSBs in the presence, or absence, of the DNA-PK inhibitor NU7441 or the PARP1 inhibitor KU-0058684, or both, was measured with g2HAX assays. (A) The effect on NU7441 on DNA-PK proficient and deficient cells. (B) The effect of KU-0058648 on PARP1 proficient and deficient cells. (C) The effect of NU7441 on PARP1 deficient cells and KU-0058684 on DNA-PK deficient cells. (D) The combinatorial effect of DNA-PK and PARP1 inhibitors on DNA-PK proficient cells. Cells were exposed to 2 Gy ionizing radiation and allowed to recover. Data are the mean ± standard error of three independent experiments over a 24-h time course.
We have previously shown that the DNA-PK inhibitor, NU7441, at 1 µM radiosensitized V3-YAC cells but not V3 cells and inhibited DSB repair in SW620 cells (50). Investigation of the specificity of the inhibitors for their target at the DNA repair level revealed that NU7441 inhibited repair in the V3-YAC cells (20.6 ± 20.3% repair) but not V3 cells, such that there was no difference in repair between V3-YAC + NU7441 and V3 cells (± NU7441) at 2 h (Figure 1 A). Similarly, we previously showed that PARP inhibition radiosensitized PARP+/+ cells but not PARP1−/− cells (53). In the current study, the PARP inhibitor, KU-0058684, at 100 nM, a concentration that completely inhibited PARP activity in permeabilized cells (data not shown) retarded repair in the PARP1+/+ cells but not the PARP1−/− cells (Figure 1B). We found that inhibiting PARP in cells lacking DNA-PK, or inhibiting DNA-PK in cells lacking PARP1 had no further impact on repair (Figure 1C). This finding, that inhibition of one enzyme does not further retard repair in a cell line already lacking the second enzyme, suggests that DNA-PK and PARP1 may function in the same epistatic pathway or in a co-operating manner in DSB repair. This was supported by a non-additive effect of KU-0058684 and NU7441 when utilized in combination in V3 YAC cells (Figure 1D). Similar effects were observed in PARP1+/+ cells (data not shown). Previous research utilizing PARP inhibitors in DNA-PK-defective V3 cells has been ambiguous—early research showed V3 cells were hypersensitive to the PARP inhibitor, 4-amino-1,8-naphthalimide (60) but these have not been replicated using the more potent and specific inhibitor, AG14361 (61), suggesting a compound-specific rather than class-effect.
Electron microscopy analysis of the DNA-PK/PARP1 complex reveals additional density compared to the DNA-PK complex
DNA-PK and PARP1 have been shown to interact in several studies in the past. We purified to homogeneity a human DNA-PKcs/Ku70/Ku80/PARP1 complex assembled on DNA from non-irradiated HeLa cell nuclear extracts by batch chromatography, column chromatography and glycerol gradient centrifugation (Figure 2A and B). Consistently with earlier independent data (17), our sample had PARylation and kinase activities (data not shown).
Electron micrographs of negatively stained DNA-PK/PARP1 complex showed clear and detailed molecular images characterized by different apparent sizes (Figure 2C), consistent with our previous work on DNA-PK (6). The observation of individual particles and their classification revealed that the sample was heterogeneous due to the presence of both monomeric and dimeric assemblies, similar to those previously observed by analysis of DNA-PK preparations (6). We performed an initial reference-free classification using the IMAGIC-5 software. The dimeric assemblies were mainly characterized by ‘T-shape’ architecture. Classification of a sub-set of images representing the monomeric assemblies revealed that some of these had an extra density adjacent to the region assigned to Ku in our previous analysis of the DNA-PK complex (6) (Figure 2D and E).
MSA classification, performed using the IMAGIC-5 software and based on classification using two eigenimages, was used to separate molecular images according to their dimensions, so that the original image dataset was divided in two stacks corresponding to 14 049 monomeric and 6099 dimeric complexes (62). The sub-set of monomeric complexes was further subjected to successive rounds of alignment and classification in order to improve the resulting image class-averages. Selected class-averages were used to calculate a starting 3D volume by common lines using the Euler program in the IMAGIC-5 package. During the subsequent refinement of the 3D map a number of class averages were identified which lacked the extra density adjacent to Ku and therefore correlated more closely with the DNA-PK structure previously determined (6). The images forming these class averages were excluded from the data set used for the final refinement by projection matching performed in IMAGIC-5. The final data set was composed of the 2099 particles, which had the best fit for the bigger complex. The class averages used for the final reconstruction (Figure 2F) are in good agreement with their corresponding reprojections, indicating consistency in the analysis (Figure 2G). The resolution of the final map (Figure 2H) was estimated as ∼35 Å at 0.5 FSC (Figure 2I). The method used to determine resolution involves splitting a data set into two halves and calculating two independent reconstructions.
The resulting 3D map of the DNA-PK/PARP1 complex is ∼230 Å long, 160 Å wide and 120 Å deep. Within the map, regions can be readily recognized that are highly compatible with our previous analyses of DNA-PKcs, DNA-PK and Ku (6,27–29,63) as well as with the crystallographic structures of isolated components of the complex (35,64) (Figure 3). An extra density compatible with the size of full-length PARP1 can be visually located in contact with Ku (Figures 2H and 3). This observation is consistent with previous data showing genetic interaction between DNA-PK and PARP1 and the formation of a Ku/PARP1 complex (12–18), as well as for a physical role of PARP1 in modulating NHEJ.
Figure 3.
Docking known X-ray structures into the DNA-PK/PARP1 density map. We used the UCSF Chimera software (58) to dock known structures for human DNA-PKcs (red), truncated human Ku heterodimer (green) and chicken PARP1 catalytic subunit (2PAW, blue) into the DNA-PK/PARP1 3D map.
We performed docking of known X-ray structures for DNA-PKcs (3KGV), truncated Ku (1JEY) and PARP1 catalytic domain (2PAW) using UCSF Chimera (58). To facilitate the manual fitting of crystallographic structures into portions of the map, we segmented the DNA-PK/PARP1 3D map in 13 sections using the Segger routine, and we then hand-grouped the segments into three regions accounting for DNA-PKcs, the Ku heterodimer and a density whose size is compatible with a protein of the molecular weight of one PARP1 molecule. We fitted the crystallographic structures for DNA-PKcs and Ku using the Fit-to-Segment command in Chimera (Figure 3, DNA-PKcs is coloured red, Ku is coloured green). We then re-segmented the PARP1 density and tested the Fit-to-segment routine on the 2PAW pdb entry (chicken PARP1 catalytic domain, 361 residues out of 1011, 87% identity between the human and chicken catalytic domains). This fits well in the density represented with a blue mesh in Figure 3. The density represented solid and coloured blue is compatible with the size of the rest of the human PARP1 protein (1014 residues full-length). The density accounting for PARP1 is 110 Å long and 86 Å wide. The catalytic domain fits in the region of PARP1, which is not in direct contact with Ku. This is consistent with the biochemical observation that the pull-down of Ku is mediated by the PARP1 BRCT domain (22), since the BRCT domain is located N-terminally to the catalytic site, with the latter being at the C-terminus of the enzyme.
As a control experiment, we also aligned the monomeric particles to our cryo-EM reconstruction of DNA-PKcs (29). We then classified them using a local MSA mask centered on the PARP1 density. The particles belonging to classes, which contained the density, were extracted to one subset. The particles belonging to classes, which did not contain the density, were extracted to a second subset. Particles belonging to classes representing top views, which are very similar for both complexes, were combined to both sub-data sets. We then processed these two subsets (DNA-PK/PARP = 5163 particles; DNA-PK = 4903 particles) separately, using the cryoEM reconstruction for DNA-PKcs as a starting model and an Eman-based projection matching refinement. As illustrated in Supplementary Figure S1, these reconstructions are in agreement with the overall features of the two complexes. In particular, the additional density for the PARP1-containing complex is shown in Supplementary Figure S1B, and the comparison between this DNA-PK/PARP1 reconstruction and the more detailed one obtained with a more stringent choice of particles is shown in Supplementary Figure S1C.
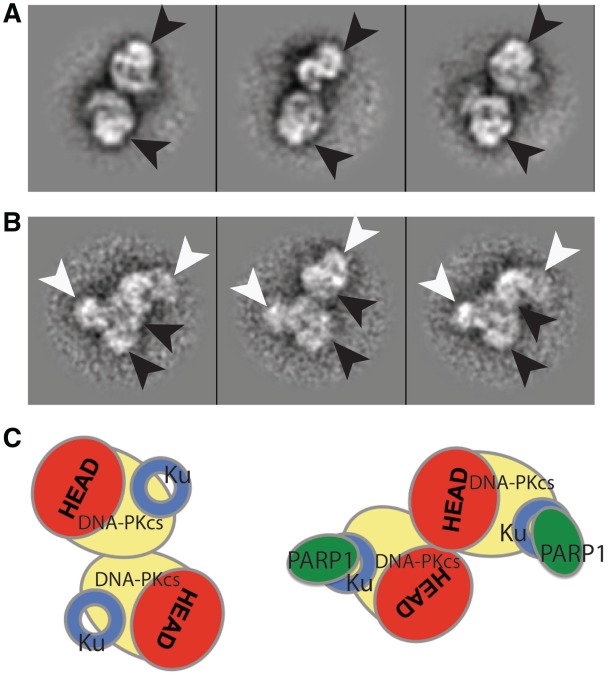
Synaptic DNA-PK/PARP1 dimers show orientation changes compared to DNA-PK dimers
A total of 3125 dimeric appearances of the DNA-PK/PARP1 complex were manually extracted to a sub-data set. They were then analysed in 2D by alignment and classification, revealing a ‘T-shaped’ architecture. Each DNA-PK/PARP1 assembly within class averages of this dimer-of-heterotetramers retains the general features of the smaller DNA-PK/PARP1 particles, with a clear added density attached to the Ku end of the DNA-PK complex. Importantly, in contrast to DNA-PK dimers loaded on Y-shaped DNA (Figure 4A) and similarly to DNA-PK dimers loaded on hairpin DNA, one DNA-PKcs head domain is located in close contact with the DNA-PKcs arm domain within the DNA-PKcs opposite (Figure 4B). The orientation of the two halves of the complex is however different from the one of DNA-PK dimers on hairpin DNA (Figure 4C). This orientation is compatible with the ability to auto-phosphorylate in trans (47), since the two main DNA-PKcs autophosphorylation clusters have been mapped in the HEAT repeats region, forming the arm and palm domain of DNA-PKcs (64). The dimers-of-tetramers were analysed as a single stack of particles, which all seemed to contain the PARP1 density, since all the class averages calculated present a strong signal at the same position of the PARP1 density in the tetramer (white arrowhead in Figure 4B, while the black arrowhead points to the DNA-PKcs head domain).
Figure 4.
The dynamic nature of DNA-PKcs in the formation of dimers, as assessed by electron microscopy and single particle analysis. (A) DNA-PK dimers on Y-shaped DNA, with head domains facing opposite. (B) DNA-PK/PARP1 dimers on Y-shaped DNA, with head domains in close contact. (C) Schematics of the relative arrangement of DNA-PK head domains in DNA-PK dimers and DNA-PK/PARP1 dimers, both loaded on Y-shaped DNA. DNA-PKcs is coloured yellow, apart from the head domain, which is coloured red. Ku is represented as a blue ring and PARP1 as a green oval.
DISCUSSION
The interplay between the PARP1 and DNA-PK enzymes seems to be quite intricate. On one hand, an interaction between the two enzymes has been reported by several studies (12–22). On the other hand, other work implies that they have separate roles within the NHEJ DNA repair pathway (23,24). However, it may well be that different roles for PARP1 and DNA-PK coexist in NHEJ, since these molecules respond to a number of macromolecular interactions and post-translational modifications (36,65), which can seriously affect their conformation and functional properties.
Here, we describe in vivo data suggesting that DNA-PK and PARP1 lack additivity in DSB repair. We purified a DNA-PK/PARP1 complex, which was tested for both kinase and PARP activities. This complex is organized in both ‘monomer-of-heterotetramers’ and ‘dimer-of-heterotetramers’ assemblies. In both cases, tight direct interactions of PARP1 with the Ku DNA recognition module are apparent. This is consistent with independent biochemical characterizations of a PARP1-Ku interaction, and with the observation that Ku is needed for PARP activation by 5′-overhang DNA DSBs (19). By analogy with the DNA-PK heterotrimer and dimer-of-heterotrimers (6), these complexes do not form in the absence of DNA. This supports a mechanism of assembly of the complex on its substrate, in the same way as a number of chromatin remodeling factors. Opposite to the architecture of PARP1 in isolation, our 3D fitting experiments show that PARP1 interacts with DNA-PK in a monomeric form, with the catalytic domain distal to Ku. In fact, the density for the extra protein is really well defined, accounting for the volume of only one PARP1 molecule (not smaller or bigger lumps) with no blurring which might suggest flexibility in the PARP1 region.
Importantly to our understanding of the functional mechanisms of this complex, and in contrast to our previous analysis of DNA-PK dimers-of-trimers loaded on Y-shaped DNA (6), the DNA-PKcs heads are close in space, suggesting that in this complex autophosphorylation may take place in trans, as reported in earlier biochemical studies (47). Our study does not however answer the question of how DNA assembles in this complex. This might be addressed by future cryo-EM or small angle neutron scattering studies. This orientation of the DNA-PK/PARP1 dimer-of-tetramers has some analogies with the conformation of DNA-PK dimers loaded on hairpin DNA recently analysed by SAXS (37), since DNA-PKcs head domains are close in space. However, the overall architecture of the dimer-of-heterotetramers is different, as it is arranged in ‘T-shaped’ assemblies. This observation is crucial towards understanding the function of DNA-PK, since it shows that this dynamic enzyme can adopt substantially different conformations not only upon recognition of different DNA structures (37) or upon autophosphorylation (8), but also upon interaction with other DNA repair factors. This further structural snapshot within the DNA-PK interacting network emphasizes the need to characterize the structures of the full range of DNA-PK assemblies such as the DNA-PK/XRCC4/LigaseIV complex in order to fully understand the role of a pivotal enzyme in the DNA damage response.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figure S1.
FUNDING
Cancer Research UK (Programme Grant to L.H.P.; Project Grant C17335/A10470 to L.S.); Scottish Universities Life Sciences Alliance (to The Electron Microscopy Facility at Edinburgh); Wellcome Trust (087658/Z/08/Z) (to The Electron Microscopy Facility at Edinburgh). Funding for open access charge: Wellcome Trust grant 087658/Z/08/Z.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Paula C.A. da Fonseca for her advice.
REFERENCES
- 1.Pardo B, Gomez-Gonzalez B, Aguilera A. DNA repair in mammalian cells: DNA double-strand break repair: how to fix a broken relationship. Cell Mol. Life Sci. 2009;66:1039. doi: 10.1007/s00018-009-8740-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson SP. DNA-dependent protein kinase. Int. J. Biochem. Cell Biol. 1997;29:935. doi: 10.1016/s1357-2725(97)00006-x. [DOI] [PubMed] [Google Scholar]
- 3.Downs JA, Jackson SP. A means to a DNA end: the many roles of Ku. Nat. Rev. Mol. Cell Biol. 2004;5:367. doi: 10.1038/nrm1367. [DOI] [PubMed] [Google Scholar]
- 4.Hartley KO, Gell D, Smith GC, Zhang H, Divecha N, Connelly MA, Admon A, Lees-Miller SP, Anderson CW, Jackson SP. DNA-dependent protein kinase catalytic subunit: a relative of phosphatidylinositol 3-kinase and the ataxia telangiectasia gene product. Cell. 1995;82:849. doi: 10.1016/0092-8674(95)90482-4. [DOI] [PubMed] [Google Scholar]
- 5.Ochi T, Sibanda BL, Wu Q, Chirgadze DY, Bolanos-Garcia VM, Blundell TL. Structural biology of DNA repair: spatial organisation of the multicomponent complexes of nonhomologous end joining. J. Nucleic Acids. 2010 doi: 10.4061/2010/621695. Article ID 621695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spagnolo L, Rivera-Calzada A, Pearl LH, Llorca O. Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair, Mol. Cell. 2006;22:511. doi: 10.1016/j.molcel.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 7.Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem. J. 2009;417:639. doi: 10.1042/BJ20080413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morris EP, Rivera-Calzada A, da Fonseca PC, Llorca O, Pearl LH, Spagnolo L. Evidence for a remodelling of DNA-PK upon autophosphorylation from electron microscopy studies. Nucleic Acids Res. 2011;39:5757. doi: 10.1093/nar/gkr146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 2006;7:517. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 10.Dantzer F, Schreiber V, Niedergang C, Trucco C, Flatter E, De La Rubia G, Oliver J, Rolli V, Menissier-de Murcia J, de Murcia G. Involvement of poly(ADP-ribose) polymerase in base excision repair. Biochimie. 1999;81:69. doi: 10.1016/s0300-9084(99)80040-6. [DOI] [PubMed] [Google Scholar]
- 11.Malanga M, Althaus FR. The role of poly(ADP-ribose) in the DNA damage signaling network. Biochem. Cell Biol. 2005;83:354. doi: 10.1139/o05-038. [DOI] [PubMed] [Google Scholar]
- 12.Morrison C, Smith GC, Stingl L, Jackson SP, Wagner EF, Wang ZQ. Genetic interaction between PARP and DNA-PK in V(D)J recombination and tumorigenesis. Nat. Genet. 1997;17:479. doi: 10.1038/ng1297-479. [DOI] [PubMed] [Google Scholar]
- 13.Henrie MS, Kurimasa A, Burma S, Menissier-de Murcia J, de Murcia G, Li GC, Chen DJ. Lethality in PARP-1/Ku80 double mutant mice reveals physiological synergy during early embryogenesis. DNA Repair. 2003;2:151. doi: 10.1016/s1568-7864(02)00199-4. [DOI] [PubMed] [Google Scholar]
- 14.Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 2000;275:40974. doi: 10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
- 15.Galande S, Kohwi-Shigematsu T. Caught in the act: binding of Ku and PARP to MARs reveals novel aspects of their functional interaction. Crit. Rev. Eukaryot. Gene Expr. 2000;10:63. [PubMed] [Google Scholar]
- 16.Galande S, Kohwi-Shigematsu T. Poly(ADP-ribose) polymerase and Ku autoantigen form a complex and synergistically bind to matrix attachment sequences. J. Biol. Chem. 1999;274:20521. doi: 10.1074/jbc.274.29.20521. [DOI] [PubMed] [Google Scholar]
- 17.Ruscetti T, Lehnert BE, Halbrook J, Le Trong H, Hoekstra MF, Chen DJ, Peterson SR. Stimulation of the DNA-dependent protein kinase by poly(ADP-ribose) polymerase. J. Biol. Chem. 1998;273:14461. doi: 10.1074/jbc.273.23.14461. [DOI] [PubMed] [Google Scholar]
- 18.Hossain MB, Ji P, Anish R, Jacobson RH, Takada S. Poly(ADP-ribose) polymerase 1 interacts with nuclear respiratory factor 1 (NRF-1) and plays a role in NRF-1 transcriptional regulation. J. Biol. Chem. 2009;284:8621. doi: 10.1074/jbc.M807198200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitchell J, Smith GC, Curtin NJ. Poly(ADP-Ribose) polymerase-1 and DNA-dependent protein kinase have equivalent roles in double strand break repair following ionizing radiation. Int. J. Radiat. Oncol. Biol. Phys. 2009;75:1520. doi: 10.1016/j.ijrobp.2009.07.1722. [DOI] [PubMed] [Google Scholar]
- 20.Veuger SJ, Curtin NJ, Smith GC, Durkacz BW. Effects of novel inhibitors of poly(ADP-ribose) polymerase-1 and the DNA-dependent protein kinase on enzyme activities and DNA repair. Oncogene. 2004;23:7322. doi: 10.1038/sj.onc.1207984. [DOI] [PubMed] [Google Scholar]
- 21.Dong F, Soubeyrand S, Hache RJ. Activation of PARP-1 in response to bleomycin depends on the Ku antigen and protein phosphatase 5. Oncogene. 2010;29:2093. doi: 10.1038/onc.2009.492. [DOI] [PubMed] [Google Scholar]
- 22.Paddock MN, Bauman AT, Higdon R, Kolker E, Takeda S, Scharenberg AM. Competition between PARP-1 and Ku70 control the decision between high-fidelity and mutagenic DNA repair. DNA Repair. 2011;10:338. doi: 10.1016/j.dnarep.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perrault R, Wang H, Wang M, Rosidi B, Iliakis G. Backup pathways of NHEJ are suppressed by DNA-PK. J. Cell Biochem. 2004;92:781. doi: 10.1002/jcb.20104. [DOI] [PubMed] [Google Scholar]
- 24.Wang H, Perrault AR, Takeda Y, Qin W, Iliakis G. Biochemical evidence for Ku-independent backup pathways of NHEJ. Nucleic Acids Res. 2003;31:5377. doi: 10.1093/nar/gkg728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patel AG, Sarkaria JN, Kaufmann SH. Nonhomologous end joining drives poly(ADP-ribose) polymerase (PARP) inhibitor lethality in homologous recombination-deficient cells. Proc. Natl Acad. Sci. USA. 2011;108:3406. doi: 10.1073/pnas.1013715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boskovic J, Rivera-Calzada A, Maman JD, Chacon P, Willison KR, Pearl LH, Llorca O. Visualization of DNA-induced conformational changes in the DNA repair kinase DNA-PKcs. EMBO J. 2003;22:5875. doi: 10.1093/emboj/cdg555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rivera-Calzada A, Spagnolo L, Pearl LH, Llorca O. Structural model of full-length human Ku70-Ku80 heterodimer and its recognition of DNA and DNA-PKcs. EMBO Rep. 2007;8:56. doi: 10.1038/sj.embor.7400847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rivera-Calzada A, Maman JD, Spagnolo L, Pearl LH, Llorca O. Three-dimensional structure and regulation of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) Structure. 2005;13:243. doi: 10.1016/j.str.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 30.Chiu CY, Cary RB, Chen DJ, Peterson SR, Stewart PL. Cryo-EM imaging of the catalytic subunit of the DNA-dependent protein kinase. J. Mol. Biol. 1998;284:1075. doi: 10.1006/jmbi.1998.2212. [DOI] [PubMed] [Google Scholar]
- 31.Williams DR, Lee KJ, Shi J, Chen DJ, Stewart PL. Cryo-EM structure of the DNA-dependent protein kinase catalytic subunit at subnanometer resolution reveals alpha helices and insight into DNA binding. Structure. 2008;16:468. doi: 10.1016/j.str.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leuther KK, Hammarsten O, Kornberg RD, Chu G. Structure of DNA-dependent protein kinase: implications for its regulation by DNA. EMBO J. 1999;18:1114. doi: 10.1093/emboj/18.5.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brewerton SC, Dore AS, Drake AC, Leuther KK, Blundell TL. Structural analysis of DNA-PKcs: modelling of the repeat units and insights into the detailed molecular architecture. J. Struct. Biol. 2004;145:295. doi: 10.1016/j.jsb.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 34.Harris R, Esposito D, Sankar A, Maman JD, Hinks JA, Pearl LH, Driscoll PC. The 3D solution structure of the C-terminal region of Ku86 (Ku86CTR) J. Mol. Biol. 2004;335:573. doi: 10.1016/j.jmb.2003.10.047. [DOI] [PubMed] [Google Scholar]
- 35.Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 36.Dobbs TA, Tainer JA, Lees-Miller SP. A structural model for regulation of NHEJ by DNA-PKcs autophosphorylation. DNA Repair. 2010;9:1307. doi: 10.1016/j.dnarep.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hammel M, Yu Y, Mahaney BL, Cai B, Ye R, Phipps BM, Rambo RP, Hura GL, Pelikan M, So S, et al. Ku and DNA-dependent protein kinase dynamic conformations and assembly regulate DNA binding and the initial non-homologous end joining complex. J. Biol. Chem. 2010;285:1414. doi: 10.1074/jbc.M109.065615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruf A, de Murcia G, Schulz GE. Inhibitor and NAD+ binding to poly(ADP-ribose) polymerase as derived from crystal structures and homology modeling. Biochemistry. 1998;37:3893. doi: 10.1021/bi972383s. [DOI] [PubMed] [Google Scholar]
- 39.Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O'Connor MJ, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 2009;361:123. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 40.Langelier MF, Planck JL, Roy S, Pascal JM. Crystal structures of poly(ADP-ribose) polymerase-1 (PARP-1) zinc fingers bound to DNA: Structural And Functional Insights Into Dna-Dependent PARP-1 activity. J. Biol. Chem. 2011;286:10690. doi: 10.1074/jbc.M110.202507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pion E, Ullmann GM, Ame JC, Gerard D, de Murcia G, Bombarda E. DNA-induced dimerization of poly(ADP-ribose) polymerase-1 triggers its activation. Biochemistry. 2005;44:14670. doi: 10.1021/bi050755o. [DOI] [PubMed] [Google Scholar]
- 42.Mendoza-Alvarez H, Alvarez-Gonzalez R. Poly(ADP-ribose) polymerase is a catalytic dimer and the automodification reaction is intermolecular. J. Biol. Chem. 1993;268:22575. [PubMed] [Google Scholar]
- 43.Eustermann S, Videler H, Yang JC, Cole PT, Gruszka D, Veprintsev D, Neuhaus D. The DNA-binding domain of human PARP-1 interacts with DNA single-strand breaks as a monomer through its second zinc finger. J. Mol. Biol. 2011;407:149. doi: 10.1016/j.jmb.2011.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jones B, Dale RG. Radiobiologically based assessments of the net costs of fractionated focal radiotherapy. Int. J. Radiat. Oncol. Biol. Phys. 1998;41:1139. doi: 10.1016/s0360-3016(98)00155-2. [DOI] [PubMed] [Google Scholar]
- 45.Rothkamm K, Lobrich M. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low x-ray doses. Proc. Natl Acad. Sci. USA. 2003;100:5057. doi: 10.1073/pnas.0830918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morris E, Rivera-Calzada A, da Fonseca P, Llorca O, Pearl L, Spagnolo L. Evidence for a remodelling of DNA-PK upon autophosphorylation from electron microscopy studies. Nucleic Acids Res. 2011;39:5757. doi: 10.1093/nar/gkr146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meek K, Douglas P, Cui X, Ding Q, Lees-Miller SP. trans Autophosphorylation at DNA-dependent protein kinase's two major autophosphorylation site clusters facilitates end processing but not end joining. Mol. Cell Biol. 2007;27:3881. doi: 10.1128/MCB.02366-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leahy JJ, Golding BT, Griffin RJ, Hardcastle IR, Richardson C, Rigoreau L, Smith GC. Identification of a highly potent and selective DNA-dependent protein kinase (DNA-PK) inhibitor (NU7441) by screening of chromenone libraries. Bioorg. Med. Chem. Lett. 2004;14:6083. doi: 10.1016/j.bmcl.2004.09.060. [DOI] [PubMed] [Google Scholar]
- 49.Loh VM, Jr, Cockcroft XL, Dillon KJ, Dixon L, Drzewiecki J, Eversley PJ, Gomez S, Hoare J, Kerrigan F, Matthews IT, et al. Phthalazinones. Part 1: The design and synthesis of a novel series of potent inhibitors of poly(ADP-ribose)polymerase. Bioorg. Med. Chem. Lett. 2005;15:2235. doi: 10.1016/j.bmcl.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 50.Zhao Y, Thomas HD, Batey MA, Cowell IG, Richardson CJ, Griffin RJ, Calvert AH, Newell DR, Smith GC, Curtin NJ. Preclinical evaluation of a potent novel DNA-dependent protein kinase inhibitor NU7441. Cancer Res. 2006;66:5354. doi: 10.1158/0008-5472.CAN-05-4275. [DOI] [PubMed] [Google Scholar]
- 51.de Murcia JM, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver FJ, Masson M, Dierich A, LeMeur M, et al. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc. Natl Acad. Sci. USA. 1997;94:7303. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blunt T, Finnie NJ, Taccioli GE, Smith GC, Demengeot J, Gottlieb TM, Mizuta R, Varghese AJ, Alt FW, Jeggo PA, et al. Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell. 1995;80:813. doi: 10.1016/0092-8674(95)90360-7. [DOI] [PubMed] [Google Scholar]
- 53.Veuger SJ, Curtin NJ, Richardson CJ, Smith GC, Durkacz BW. Radiosensitization and DNA repair inhibition by the combined use of novel inhibitors of DNA-dependent protein kinase and poly(ADP-ribose) polymerase-1. Cancer Res. 2003;63:6008. [PubMed] [Google Scholar]
- 54.Cowell IG, Durkacz BW, Tilby MJ. Sensitization of breast carcinoma cells to ionizing radiation by small molecule inhibitors of DNA-dependent protein kinase and ataxia telangiectsia mutated. Biochem. Pharmacol. 2005;71:13. doi: 10.1016/j.bcp.2005.09.029. [DOI] [PubMed] [Google Scholar]
- 55.Ludtke SJ. 3-D structures of macromolecules using single-particle analysis in EMAN. Methods Mol. Biol. 2010;673:157. doi: 10.1007/978-1-60761-842-3_9. [DOI] [PubMed] [Google Scholar]
- 56.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J. Struct. Biol. 1996;116:17. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 57.Shaikh TR, Gao H, Baxter WT, Asturias FJ, Boisset N, Leith A, Frank J. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat. Protocols. 2008;3:1941. doi: 10.1038/nprot.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J. Struct. Biol. 2007;157:281. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 59.Staszewski O, Nikolova T, Kaina B. Kinetics of gamma-H2AX focus formation upon treatment of cells with UV light and alkylating agents. Environ. Mol. Mutagen. 2008;49:734. doi: 10.1002/em.20430. [DOI] [PubMed] [Google Scholar]
- 60.Bryant HE, Helleday T. Inhibition of poly (ADP-ribose) polymerase activates ATM which is required for subsequent homologous recombination repair. Nucleic Acids Res. 2006;34:1685. doi: 10.1093/nar/gkl108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith LM, Willmore E, Austin CA, Curtin NJ. The novel poly(ADP-Ribose) polymerase inhibitor, AG14361, sensitizes cells to topoisomerase I poisons by increasing the persistence of DNA strand breaks. Clin. Cancer Res. 2005;11:8449. doi: 10.1158/1078-0432.CCR-05-1224. [DOI] [PubMed] [Google Scholar]
- 62.White HE, Saibil HR, Ignatiou A, Orlova EV. Recognition and separation of single particles with size variation by statistical analysis of their images. J. Mol. Biol. 2004;336:453. doi: 10.1016/j.jmb.2003.12.015. [DOI] [PubMed] [Google Scholar]
- 63.Llorca O, Pearl LH. Electron microscopy studies on DNA recognition by DNA-PK. Micron. 2004;35:625. doi: 10.1016/j.micron.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 64.Sibanda BL, Chirgadze DY, Blundell TL. Crystal structure of DNA-PKcs reveals a large open-ring cradle comprised of HEAT repeats. Nature. 2010;463:118. doi: 10.1038/nature08648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kawaichi M, Ueda K, Hayaishi O. Multiple autopoly(ADP-ribosyl)ation of rat liver poly(ADP-ribose) synthetase. Mode of modification and properties of automodified synthetase. J. Biol. Chem. 1981;256:9483. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.