Abstract
Sequence-specific RNA–protein interactions, though commonly used in biological systems to regulate translation, are challenging to selectively modulate. Here, we demonstrate the use of a chemically-inducible RNA–protein interaction to regulate eukaryotic translation. By genetically encoding Tet Repressor protein (TetR)-binding RNA elements into the 5′-untranslated region (5′-UTR) of an mRNA, translation of a downstream coding sequence is directly controlled by TetR and tetracycline analogs. In endogenous and synthetic 5′-UTR contexts, this system efficiently regulates the expression of multiple target genes, and is sufficiently stringent to distinguish functional from non-functional RNA–TetR interactions. Using a reverse TetR variant, we illustrate the potential for expanding the regulatory properties of the system through protein engineering strategies.
INTRODUCTION
Translational regulation mediated by specific RNA–protein interactions is pervasive in shaping diverse biological processes including metabolism (1), early development (1,2), neuronal plasticity (2) and immunity (3). However, it remains challenging to recapitulate this mode of regulation in a way that is transcript-specific and readily modulated experimentally. Such selective control represents a valuable tool for studying and designing translational regulation mechanisms based on RNA–protein interactions. Using naturally occurring RNA–protein interactions for this purpose is problematic because many are directly toggled by protein modifications such as phosphorylation (2–4), which are in turn shaped by complex cellular signaling events that cannot be precisely manipulated.
Several systems for regulating eukaryotic gene expression post-transcriptionally have been previously described, and include RNA interference (RNAi) (5), small molecule-regulated cis-acting ribozymes and riboswitches (6). Although RNAi has proven a highly useful tool in many contexts, its use is limited to organisms expressing the necessary machinery. Even in contexts where RNAi is well established as a routine tool, the introduction of interfering RNAs can produce unintended off-target effects (7). Furthermore, gene expression knockdown with RNAi cannot be rapidly switched off, and the targeted protein may not return to its initial level for days as the catalytic siRNA is slowly depleted (8). Complementary to RNAi, small molecule-regulated cis-acting ribozymes and riboswitches can act in the absence of organism-specific RNA degradation pathways, allowing their application in a broader variety of contexts. Riboswitches, in particular, circumvent target transcript degradation and are likely to provide highly dynamic regulation. However, these ligand-regulated systems do not immediately support a direct path toward integration with endogenous translation regulation mechanisms. Such natural systems frequently couple the use of specific primary RNA–protein interactions with secondary protein–protein interactions to modulate translation in different contexts. For example, the cellular programs controlling inflammation and its resolution (3), as well as the establishment of asymmetric protein distributions critical for neuronal development (4) and cell migration (9), make use of this paradigm to achieve translational control over effector genes. For this reason, it is highly desirable to construct translational regulation schemes that can interface directly with biologically relevant, protein-based signaling networks. Therefore, we sought to construct a system for eukaryotic translational regulation using well defined, ligand-regulated and specific RNA–protein interactions that can be directly controlled in a straightforward and cell type-independent way.
Attempts at achieving such a ligand-regulated RNA–protein interaction have been described previously (10–15). However, in these examples, the interaction either cannot be modulated in vivo (12), requires a ubiquitous and stringently regulated metabolite ligand such as iron (11), or relies on inducible transcription to control the RNA-binding protein (14,15). Thus, despite the widespread nature of RNA–protein interactions in regulating eukaryotic translation, no methods exist for reversibly and precisely reproducing such regulation in vivo. We expect such methods to be broadly applicable and invaluable for reconstructing and interrogating native translation control mechanisms, as well as for designing novel cellular functions.
MATERIALS AND METHODS
RNA aptamers
Secondary structure predictions, such as the one shown in Figure 1b, were made using mfold (16). Equilibrium binding constants were determined using a cytometric bead binding assay as described (17).
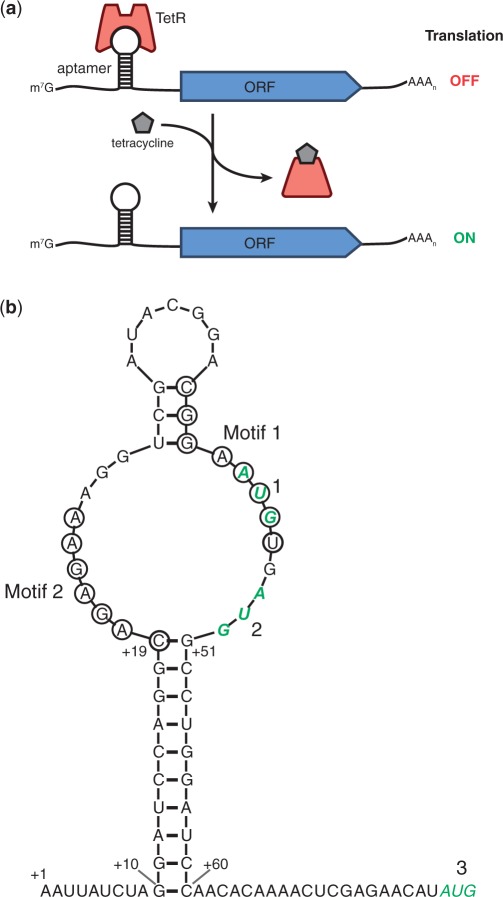
Figure 1.
Placement of TetR-binding RNA aptamers within the 5′-UTR. (a) Illustration of the regulation scheme used to demonstrate functionality of the TetR–aptamer module. TetR binding to the aptamer element within the 5′-UTR of the regulated ORF inhibits its translation. Tetracycline analogs (e.g. aTc and Dox) induce translation by disrupting the TetR–aptamer interaction. (b) The primary and predicted secondary structures of aptamer 5–1.2 (bases 10–60) within the 5′-UTR context used in this study are shown. Residues comprising two conserved regions (Motifs 1 and 2) indispensable to TetR binding are circled. Three potential start codons, 1–3, are shown in green bold italics. Start codon 1 (within Motif 1) is out of frame, whereas 2 is in frame with 3, the downstream ORF’s native start codon.
Plasmid construction
Unless otherwise noted, cloning procedures were performed by standard techniques. Reporter plasmids were based on YCp22FL1, which contains a Saccharomyces cerevisiae TEF1 promoter driving firefly luciferase (FLuc) (18). Alternative reporter plasmids containing Venus yellow fluorescent protein (vYFP) (19) were created by replacing the XhoI/XbaI FLuc fragment of YCp22FL1 with vYFP PCR amplified using primers BJBOL268/269 (Supplementary Table S2). PCR (25 cycles) was performed with Phusion DNA polymerase (New England Biolabs) according to the manufacturer's instructions, annealing at 55°C for 30 s. Aptamer-encoding sequences were inserted by annealing oligonucleotide pairs (Supplementary Table S1), extending with Klenow fragment (3′→5′ exo-, New England Biolabs), digesting with BglII/XhoI and ligating to BamHI/XhoI-digested plasmid.
The URA3-marked TetR(B) plasmid, YCpSUP-TetR, was created by PCR amplifying tetR(B) fused to sequences encoding 5′T7 and 3′His6 tags from pET24-TetR (17), using primers BJBOL111/112, digesting with SalI/AvrII and ligating to XhoI/XbaI-digested YCpSUP-IRF1 (18). The HIS3-marked TetR plasmid pRS413-TetR was created by subcloning the ClaI fragment from YCpSUP-TetR containing the PGK1/GAL promoter through the PGK1 terminator into ClaI-digested pRS413 (American Type Culture Collection, ATCC). The URA3-marked revTetR plasmid pSG116 was created by PCR amplifying revTetR-S2 (20) (without affinity tags or transcriptional activator domains) using primers BJBOL295/296 and cloning as described for YCpSUP-TetR. The LEU2-marked plasmid pSG95, encoding tetR(B) under the control of the TDH3 (GPD) promoter, was created by inserting tetR(B) into pRS415-GPD (ATCC) by yeast gap repair cloning (21). Yeast strains expressing chromosomally-integrated reporters were constructed by cloning aptamer-regulated reporter constructs into pRS404 (ATCC), linearizing by digestion with EcoRV, transforming into S. cerevisiae W303-1B and selecting on media lacking tryptophan. Reporter integration at the TRP1 locus was confirmed by PCR with primers BJBOL411/412 and BJBOL410/413, which flank the 5′ and 3′ integration sites, respectively.
In vitro translation experiments
Templates for in vitro transcription containing 5–1.2 and 5–1.2m2 were amplified from yeast reporter plasmids using SG151/SG137 and SG291/SG137, respectively (Supplementary Table S3). All other DNA templates were constructed by PCR amplifying the FLuc gene from YCp22FL1 with the reverse primer SG137 (encoding an A60 tail) and a combination of overlapping forward primers (Supplementary Table S3) to generate the desired 5′-UTR with an upstream T7 promoter. PCR mixtures contained the outermost primers at 0.5 µM and all additional overlapping primers at 0.05 µM. Assembly PCR (30 cycles) was performed with Phusion DNA polymerase (New England Biolabs) according to the manufacturer's instructions, annealing at 65°C for 20 s and extending at 72°C for 25 s. Unpurified PCR products were transcribed with the MEGAScript T7 kit (Ambion), precipitated with an equal volume of 7.5 M LiCl/50 mM EDTA, washed with 70% ethanol and redissolved in water. The mRNA products were capped with the ScriptCap m7G capping system (Epicentre Biotechnologies) or the Vaccinia Capping System (New England Biolabs), precipitated with an equal volume of 7.5 M LiCl/50 mM EDTA, washed with 70% ethanol and redissolved in RBB [50 mM Tris–HCl, pH 8.0, 50 mM KCl, 5 mM MgCl2, 5% (v/v) glycerol, 0.05% (v/v) Tween-20]. mRNA concentrations were determined by measuring absorbance at 260 nm on a Nanodrop ND-1000 spectrophotometer (ThermoFisher Scientific).
For cell-free translation with rabbit reticulocyte lysate (RRL), the mRNA concentration was adjusted to 133 nM in RBBD (RBB plus 1 mM dithiothreitol and 10 μg/ml bovine serum albumin, New England Biolabs) and refolded by heating at 65°C for 2 min and incubating at room temperature for 10 min. TetR(B) with N-terminal T7 and C-terminal His6 tags, and revTetR-S2 with a C-terminal His6 tag were purified from Escherichia coli as previously described (17). TetR and revTetR-S2 dilutions were prepared in RBBD with or without 22 µM doxycycline (Dox). For each translation reaction, 1.5 μl mRNA was mixed with 2 μl TetR or revTetR-S2, allowed to equilibrate for 30 min at room temperature, and then mixed with 0.5 µl of a complete amino acid solution (0.5 mM each amino acid, Promega) and 6 μl of nuclease-treated RRL (Promega). The final reactions contained 20 nM mRNA, 0–300 nM repressor protein and 25 µM amino acids in 0.6 × RRL. Reactions were incubated at 30°C for 20 min. For the screening experiment in Figure 2b, reactions were stopped by adding 200 μl Stop Buffer [20 mM GlyGly-NaOH, pH 7.8, 8 mM magnesium acetate, 0.13 mM EDTA, 500 µM cycloheximide]. FLuc activity was determined by mixing 90 μl of the reaction mixture with 40 μl FLuc Assay Buffer [20 mM GlyGly–NaOH, pH 7.8, 90 mM dithiothreitol (DTT), 8 mM Mg(OAc)2, 0.13 mM EDTA, 1.5 mM ATP, 0.8 mM coenzyme A, 1.4 mM D-luciferin] in a 96-well microplate and measuring luminescence with a Spectramax L plate reader (Molecular Devices). Translation activity was calculated as the intensity of FLuc signal. For the TetR and RevTetR(S2) titration experiments in Figures 2c and 5a, a Renilla luciferase mRNA lacking an aptamer was included in each translation reaction as a reference. Reactions were performed as described above, and stopped with the addition of 200 μl of 500 μM cycloheximide in water. Four microliters (4µ L) of the reaction mixture was mixed with 20 μl of passive lysis buffer (Promega) and dual luciferase activity was measured by the sequential addition of 100 μl DLB1 (75 mM HEPES-K, 20 mM DTT, 4 mM MgSO4, 0.1 mM EDTA, 0.53 mM ATP, 0.27 mM coenzyme A, 0.47 mM D-luciferin, pH 8.0) and 100 μl DLB2 [15 mM Na4P2O7, 7.5 mM sodium acetate, 10 mM EDTA, 400 mM Na2SO4, 1% (v/v) methanol, 10 μM 2-(4′-(dimethylamino)phenyl)-6-methyl-benzothiazole, 5 mM KI, 12 μM benzyl coelenterazine, pH 5.0]. Translation was calculated as the ratio of FLuc (aptamer-regulated) to RLuc (aptamer-independent) signal.
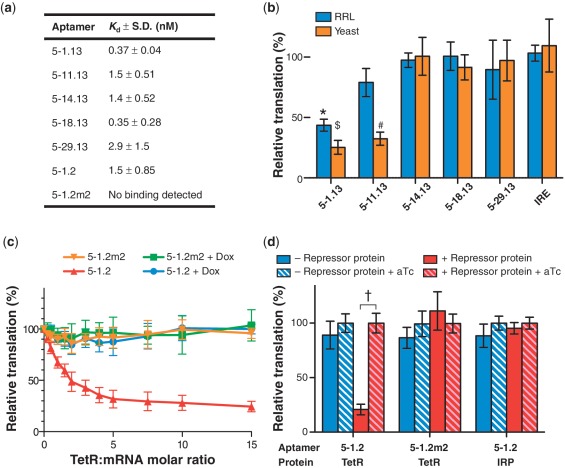
Figure 2.
Genetically encoded TetR aptamers enable translational regulation in RRL and in yeast. (a) Aptamers bind TetR with high affinity. The aptamer 5–1.2m2 contains two point mutations that eliminate specific binding to TetR. (b) Only aptamers 5–1.13 and 5–11.13 function to repress translation in RRL in vitro and in yeast in vivo. Relative translation was calculated by dividing FLuc signal in the absence of aTc by FLuc signal in the presence of aTc. (c) TetR dose-dependently represses RRL translation of synthetic FLuc mRNA containing the minimized aptamer 5–1.2, but not the mutant 5–1.2m2. Addition of 1μM Dox completely relieves repression. (d) Inducible expression in yeast requires a functional TetR-binding aptamer and TetR expression. In all figures, data represent the mean ± s.d. of at least four experiments. A two-tailed, unpaired t-test was used to calculate the significance (α = 0.005) of the difference between induced and uninduced conditions. *P = 5.1 × 10−7; $P = 2.0 × 10−7; #P = 4.9 × 10−12 †P = 2.2 × 10−11.
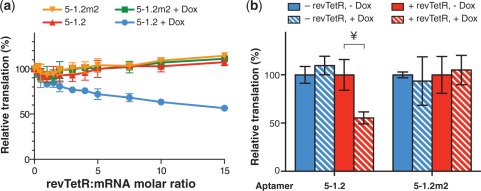
Figure 5.
Inversion of Dox-inducible phenotype with a mutant TetR. (a) A revTetR variant dose-dependently represses RRL translation of synthetic FLuc mRNA as in Figure 2c, but only in the presence of Dox. (b) A revTetR variant specifically represses translation as in Figure 2d, but only in the presence of Dox. In all figures, data represent the mean ± SD of at least four experiments. A two-tailed, unpaired t-test was used to calculate the significance (α = 0.005) of the difference between induced and uninduced conditions. ¥P = 8.1 × 10−5.
Yeast inducible expression assays
S. cerevisiae W303-1B cells harboring both a repressor (YCpSUP-TetR or pSG116) and reporter plasmid were grown to saturation at 30°C in Synthetic Defined Media #1 (SD1) [6.7 g/l yeast nitrogen base without amino acids (RPI), 20 mg/l adenine, 30 mg/l lysine, 100 mg/l leucine, 20 mg/l histidine] + 20 g/l glucose. Cells were diluted 1:80 into SD1 + 20 g/l raffinose and grown for 4 h. Glucose (to repress TetR or revTetR-S2 expression) or galactose (to induce TetR or revTetR-S2 expression) was added to 20 g/l, and cells were grown 16 h at 30°C with shaking before measurement. For FLuc activity measurements, luminescence values were normalized to the OD600 of each culture as determined using a Spectramax M2 plate reader (Molecular Devices). Eighty microliters (80µ L) of yeast culture was added to 20 µl of 5 × Passive Lysis Buffer (Promega) and incubated for 10 s. Ten microliters (10µ L) of this suspension was added to 100 μl of FLuc Assay Buffer in a 96-well microplate and measured as described for cell-free translation experiments. For vYFP measurement by flow cytometry, cells were grown as above and analyzed on a C6 Flow Cytometer (Accuri). For each sample, 5 × 104 events were captured and vYFP fluorescence was measured in the FL1 channel.
Plate-based yeast selection to identify functional aptamers
S. cerevisiae W303-1B cells harboring both pRS413-TetR and a URA3 plasmid were grown to saturation at 30°C in Synthetic Defined Media #2 (SD2) (6.7 g/l YNB, 20 mg/l adenine, 30 mg/l leucine, 20 mg/l lysine, 50 mg/l uracil) + 20 g/l glucose. Cells were diluted 1:40 into SD2 + 20 g/l raffinose and grown for 4 h. Galactose (to induce TetR expression) was added to 20 g/l and cells were grown for 4 h. The cultures were washed once in SD2 without uracil and serially diluted 10-fold into SD2 without uracil. Cell dilutions were spotted onto agar plates as indicated and grown for 3 days before visualization. Uracil dropout plates for positive selection contained SD2 without uracil, 20 g/l agar and 20 g/l galactose, with or without 1 μM anhydrotetracycline (aTc). Negative selection plates contained SD2, 20 g/l agar, 20 g/l galactose and 0.25 g/l 5-fluoroorotic acid (RPI).
cDNA preparation and quantitative PCR
Cells were grown to mid-exponential phase as described above. Total RNA was extracted using an RNeasy Mini Kit (Qiagen). Contaminating genomic and plasmid DNA were removed by treating with 10 U of TURBO DNase (Ambion) for 2 h at 37°C, followed by phenol/chloroform extraction and LiCl precipitation. In total, 2 µg of RNA was reverse transcribed to cDNA using random hexamer primers (Fermentas) and RevertAid M-MuLV reverse transcriptase (Fermentas). The reaction mixture was incubated at 25°C for 10 min, 42°C for 60 min, then 70°C for 10 min. The cDNA was diluted 1:100 in water to produce a template solution. Thermocycling was performed in a PTC-200 Peltier Thermal Cycler (MJ Research) equipped with a Chromo4 Detector (Bio-Rad). FLuc cDNA levels were quantitated and compared with levels of the endogenous ACT1 gene. The primer pairs used for FLuc and ACT1 amplification were BJBOL233/234 and ACT1F/ACT1R, respectively (Supplementary Table S2). The qPCR reactions were performed in 20 µl and contained: 1 × Standard Taq Buffer (New England Biolabs), 2.5 mM MgCl2, 200 µM each dNTP, 100 nM each primer, 0.4 × SYBR Green I (Invitrogen), 0.4 U Taq DNA polymerase (New England Biolabs) and 10 µl template solution. Reactions were performed by denaturing at 95°C for 2 min, followed by 40 cycles of: 95°C for 30 s, 60°C for 30 s and 72°C for 30 s. SYBR Green I fluorescence was measured at the end of each cycle, and relative quantitation was performed as described (22).
Preparation of polysome fractions
S. cerevisiae W303-1B cells harboring both a repressor (YCpSUP-TetR) and an integrated 5–1.2-vYFP reporter were grown to saturation at 30°C in SD1 + 20 g/l glucose. Cells were diluted 1:300 into 250 ml SD1 + 20 g/l galactose, in the presence or absence of 22 µM Dox, and grown until the cultures reached OD600 = 0.6. Prior to cell lysis, flow cytometry analysis was performed to confirm that vYFP expression under induced and uninduced conditions was identical to previous experiments. Once the proper cell density was reached, cycloheximide was added to 0.1 mg/ml and incubated at 30°C with shaking for 2 min. Cells were pelleted by centrifugation (12 000g, 4°C, 5 min) and resuspended in 40 ml of cold Polysome Lysis Buffer (PLB, 20 mM HEPES-K, pH 7.4, 2 mM magnesium acetate, 100 mM potassium acetate, 0.1 mg/ml cycloheximide, 3 mM DTT, 10 ml/l Triton X-100). Cells were pelleted again (2200g, 4°C, 5 min), resuspended in 30 ml PLB and pelleted again. The supernatant was removed and cells were weighed. For each gram of cell mass, 1.5 ml of PLB and 5 g of 0.5-mm glass beads were added. The cells were lysed by vortexing for 2 min. The crude lysate was centrifuged (2200g, 4°C, 5 min) and the supernatant subsequently centrifuged again (15 800g, 4°C, 20 min). The final supernatant was frozen in liquid nitrogen, and stored at −80°C until fractionation.
For polysome fractionation, 15 A260 units of yeast lysate were loaded onto a 10–50% sucrose gradient in Polysome Gradient Buffer (20 mM HEPES-K, pH 7.4, 2 mM magnesium acetate, 100 mM potassium acetate, 0.1 mg/ml cycloheximide, 3 mM DTT). The samples were centrifuged in a Beckman SW-41 rotor for 3 h at 35 000 rpm at 4°C. Individual fractions were collected on a Gradient Station (BioComp Instruments) and frozen at −20°C until further analysis.
cDNA preparation and qPCR analysis of polysome fractions
RNA from each polysome fraction was isolated with an RNeasy kit (Qiagen) according to the manufacturer’s protocol. Prior to RNeasy purification, 1 fmol of in vitro transcribed FLuc RNA was added to each sample as an internal control. RNA was DNase-treated (TURBO DNA-free kit, Ambion) and reverse transcribed as described above for other qPCR experiments. The qPCR measurements were performed with the PrimeTime primer and 5′ hydrolysis probe sets listed in Supplementary Table S6 (Integrated DNA Technologies). qPCR measurement of FLuc was duplexed with measurement of either vYFP or ACT1. Each reaction included 1×Thermopol Buffer (New England Biolabs), 0.2 mM dNTPs, 500 nM each primer, 250 nM each probe, 1 µl cDNA, and 0.1 µl Taq DNA polymerase (New England Biolabs). Thermocycling was performed on a Roche LightCycler 480 II for 40 cycles according to the following protocol: initial denaturation: 95°C for 20 s; denature: 95°C for 3 s; anneal/extend: 60°C for 30 s; fluorescence measurement. Raw data were color compensated and threshold cycle values were determined. Standard curves were generated from purified vYFP, ACT1 or FLuc DNA to calculate amplification efficiencies and to ensure that FLuc internal standard and cDNA template concentrations were within the linear detection range. The vYFP or ACT1 qPCR measurement from each sample was then normalized to the duplexed FLuc qPCR measurement from the same sample.
Western blot analysis
Yeast lysates were prepared by heating cell pellets in 2× Laemmli sample buffer for 10 min at 95°C. Proteins were then separated by 12% SDS–PAGE and transferred to a PVDF membrane. TetR was detected using an anti-His6 tag antibody (Abcam #ab18184). An anti-GAPDH antibody (Genscript #A00191) was used to quantify glyceraldehyde 3-phosphate dehydrogenase levels as a loading control.
RESULTS AND DISCUSSION
Introduction of TetR aptamers into a eukaryotic 5′-UTR context
Previously, we described a series of RNA aptamers that bind TetR tightly, but only in the absence of tetracycline (17). Here, we report using TetR aptamers to regulate eukaryotic translation in a mammalian cell-free extract [Rabbit reticulocyte lysate, (RRL)] and S. cerevisiae (yeast). Translation repression based on the TetR–aptamer system is directly relieved by aTc or Dox, which disrupts the TetR–aptamer interaction (Figure 1a), and no manipulation at the transcriptional level is required. To determine the regulatory effect of TetR–aptamer interaction in the 5′-UTR, we chose five RNA aptamers derived from a library selected for tight, Tc-regulated TetR binding (17). We eliminated an AUG codon present in a dispensable region of these aptamers to obtain 5–1.13, 5–11.13, 5–14.13, 5–18.13 and 5–29.13 (Supplementary Table S1). We were unable to modify the AUG located within the conserved Motif 1, known to be crucial for TetR binding (17), without sacrificing high binding affinity (Figure 1b). The modified aptamers bound TetR with low nanomolar affinity, similarly to their parents (Figure 2a).
In order to test translation repression with the system, we inserted TetR aptamers into a short 5′-UTR upstream of the firefly luciferase gene (FLuc), a context used previously to study protein–UTR interactions (23). We co-transformed yeast with a plasmid encoding TetR under the control of a galactose-inducible promoter, and a plasmid containing FLuc regulated by either a TetR aptamer or the iron-responsive element (IRE) as a structured, non-TetR-binding control. We then measured FLuc activity after growth in the presence or absence of aTc. We also prepared each mRNA construct by in vitro transcription and tested for translational regulation in RRL supplemented with recombinant TetR in the presence and absence of aTc (Figure 2b). Although all five aptamers bound TetR with similarly high affinity (Figure 2a), they did not similarly regulate translation. In the presence of TetR, only 5–1.13 inhibited FLuc synthesis in both RRL (∼50% repression) and yeast (∼80%), while 5–11.13 inhibited FLuc synthesis only in yeast (∼67%). Regulation, when observed, was always fully aTc-modulated as expected based on Figure 1a.
Aptamer minimization and functional validation
Since 5–1.13 regulated translation most effectively in both test systems, we selected it for further optimization. We constructed the more minimal aptamer 5–1.2 by retaining the conserved loop region and flanking it with a predicted RNA stem that lowered the folding stability relative to the starting aptamer (ΔGfold = −22.6 versus −25.2 kcal/mol). This potentially reduces the negative impact that structured RNA within the 5′-UTR can have on translational efficiency(24). We also introduced two point mutations in the conserved Motif 2 to produce 5–1.2m2, which has an identical predicted secondary structure, but no longer binds TetR.
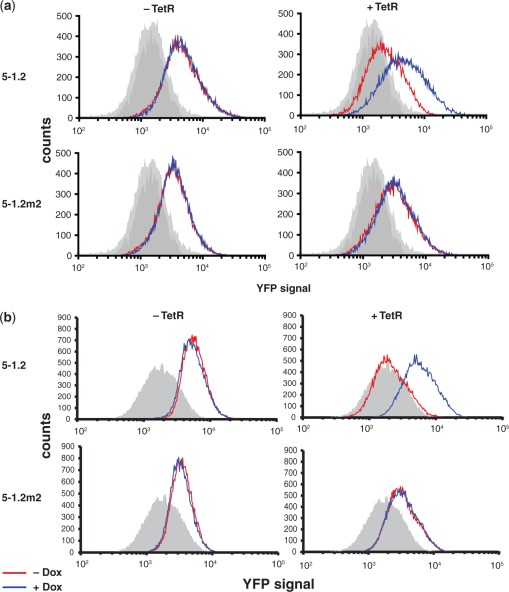
We performed RRL translation regulation experiments using fixed mRNA and titrated TetR concentrations. Translation of mRNA containing aptamer 5–1.2, but not 5–1.2m2, was dose-dependently repressed by TetR, reaching ∼70% repression at a 5:1 TetR:mRNA ratio (Figure 2c). This indicates that TetR-dependent translational repression is specific to mRNA containing an aptamer competent for binding TetR and definitively occurs post-transcriptionally. Repression was fully relieved by Dox (22 μM), consistent with inducibility of the TetR–aptamer interaction. Next, we tested the specificity of inducible translation in yeast using the experimental design described above, but including either a non-functional RNA (5–1.2m2) or an unrelated protein (the iron-responsive element binding protein, IRP) as substitutes for 5–1.2 and TetR, respectively (Figure 2d). We observed aTc-inducible regulation of FLuc synthesis (∼80%) only in strains simultaneously carrying 5–1.2 and expressing TetR. Importantly, TetR was only expressed in galactose-containing media and its abundance was not decreased by aTc, as confirmed by western blot (Supplementary Figure S1). For the above experiments, we used galactose-inducible transcription to control TetR expression, allowing us to test isogenic strains. However, even when TetR was expressed constitutively from the TDH3 promoter, we found identical Tc-dependent translational regulation (Supplementary Figure S2). These data underscore the potential for using this system in biological contexts where transcriptional control is not accessible. qPCR analysis showed that increased FLuc reporter expression in the presence of aTc was not accompanied by an increase in FLuc mRNA, as would be expected if a transcriptional response or a significant change in mRNA stability were responsible for inducible expression (Supplementary Figure S3). Altogether, these data demonstrate that the observed in vivo regulation occurs at the translational level and is due to a specific interaction between TetR and an RNA aptamer competent for binding TetR. Lastly, we replaced FLuc with Venus yellow fluorescent protein (vYFP), and this reporter was either expressed episomally or integrated at the TRP1 locus. Flow cytometry showed quantitatively that inducible expression is homogeneous across a yeast cell population, and similar in dynamic range irrespective of the gene being regulated (Figure 3).
Figure 3.
Population measurements of yeast gene expression regulated by TetR aptamers.Flow cytometry histograms show population-wide expression levels of aptamer-regulated vYFP. The aptamer located within the 5′-UTR of vYFP and the expression status of TetR are indicated. In (a), the vYFP reporter is episomal, and in (b) integrated at the TRP1 locus. Shaded gray histograms represent the auto-fluorescence of the background yeast strain. Measurements for yeast grown in the absence (red) and presence (blue) of Dox are indicated. Data for each histogram are representative of four independent experiments.
In vivo selection of functional aptamer–protein interactions
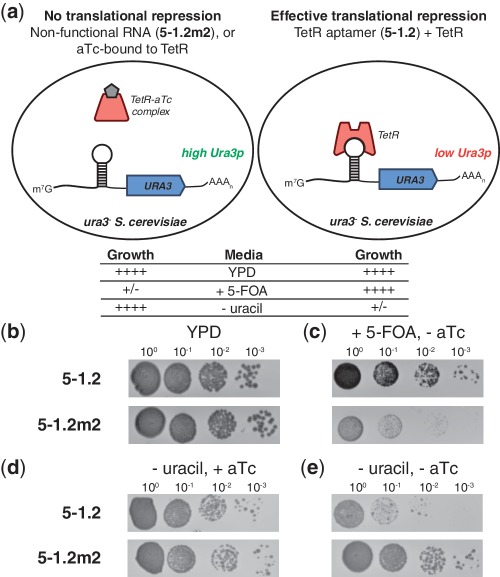
Different implementations of the translation regulatory system may necessitate discovering TetR aptamers with unique binding characteristics. Therefore, it is important to define a strategy for rapidly identifying new functional aptamer variants. To support this effort, we devised a positive/negative selection scheme capable of distinguishing between TetR-binding and non-binding RNA sequences (Figure 4a). To establish proof-of-concept, we constructed plasmids containing the yeast URA3 gene (encoding orotidine-5′-phosphate decarboxylase, Ura3p) with either 5–1.2 or 5–1.2m2 within the 5′-UTR context used earlier. We co-transformed each separately with a TetR-encoding plasmid into a yeast ura3 mutant auxotrophic for uracil. These strains exhibited similar growth on nutrient-rich YPD media (Figure 4b). For the negative selection, we plated cells on media containing 5-fluoroorotic acid (5-FOA), which is converted to the cytotoxic 5-fluorouracil by Ura3p. These conditions only permit growth of cells containing a functional TetR/5–1.2 interaction that can repress Ura3p synthesis (Figure 4c). For the positive selection, we plated cells in the presence of aTc on uracil-deficient media. Cells surviving this selection step have been induced to synthesize Ura3p to complement the uracil auxotrophy. This ensures that interactions identified by negative selection are aTc inducible (Figure 4d). As expected, the cells containing a functional TetR–aptamer system exhibit a significant growth defect when plated in the absence of aTc on uracil-deficient media (Figure 4e). To further establish the utility of this selection scheme, we mixed TetR-expressing cells containing URA3 controlled by either 5–1.2 or 5–1.2m2 in a ratio of 1:104, respectively. From this mixture, we plated ∼1.5 × 105 cells on media containing 5-FOA, and 17 large colonies were grown. Sequencing the 5′-UTR of plasmid DNA isolated from 10 of these colonies revealed that all carried the 5–1.2 sequence. These data demonstrate that this selection strategy can specifically recover functional TetR aptamers from a large non-functional background, which should prove useful for identifying aptamer or TetR variants with novel regulatory characteristics.
Figure 4.
Positive/negative selection of yeast mediated by TetR aptamers. (a) Illustration of the URA3 positive/negative selection scheme showing the predicted growth phenotypes of the strains under indicated conditions. (b) TetR-expressing strains in which Ura3p synthesis is controlled by either 5–1.2 or 5–1.2m2 grow similarly on YPD. (c) Negative selection on 5-FOA only permits growth of strains capable of repressing Ura3p translation. (d, e) When 5–1.2 controls Ura3p translation, aTc is required for growth in the absence of uracil. In each condition, 10-fold dilutions are shown from left to right. Plates were imaged after growth at 30°C for 2 days (YPD) or 3 days (synthetic defined media).
Inversion of Tc dependence with a TetR variant
Several engineered RNA regulatory schemes are based on direct interactions between small molecule ligands and RNA(25–27). However, a unique and compelling basis for using protein–RNA interactions is the potential to take advantage of protein engineering strategies to expand the scope of regulatory behavior achievable while maintaining the aptamer as a validated and defined component. To illustrate this principle, we chose revTetR-S2, a TetR variant previously derived through protein mutagenesis (20) that binds the cognate tetO DNA operator, but only in the presence of Dox. Our earlier work established that TetR aptamers compete with tetO for binding to TetR, indicating that both likely interact with the TetR nucleic acid binding domain (17). Therefore, provided that the amino acid residues involved in the interaction between TetR and its aptamers are retained in revTetR-S2, the latter is reasonably expected to bind these aptamers, but with an inverse dependence on Dox. We first determined that purified revTetR-S2 bound 5–1.2 tightly in vitro (Kd = 3.1 ± 1.0 nM). Similarly to TetR, revTetR-S2 repressed translation of aptamer-containing mRNA in RRL, but as expected, only in the presence of Dox (Figure 5a). Upon expressing revTetR-S2 in yeast with 5–1.2-regulated vYFP, Dox enabled 50% repression of vYFP (Figure 5b). Expectedly, 5–1.2m2 did not regulate vYFP expression, emphasizing the retained requirement for a specific revTetR-S2–aptamer interaction. Interestingly, another previously reported revTetR based on the TetR(BD) hybrid repressor, revTetR r1.7 (28), did not demonstrate aptamer- and aTc- dependent translation repression activity (data not shown). The mutations present in the nucleic acid binding domains of revTetR-S2 and revTetR r1.7 are distinct. Whereas both proteins support tetO binding, our data suggest that the aptamer 5–1.2 discriminates between these two TetR variants. Thus, in cases where modifying TetR can potentially disrupt the binding interface with the aptamer, it is important to confirm that the protein–aptamer interaction is preserved.
Aptamer engineering to reduce inhibition of translation in the induced state
A noteworthy challenge associated with placing an aptamer in the 5′-UTR is that this may decrease the maximal protein expression levels of the regulated ORF. Indeed, we observe that aptamer 5–1.2 causes a significant decrease in maximal reporter gene expression levels compared with when no aptamer is present. Therefore, in optimizing our system to ensure its broadest utility, it is desirable to define functional aptamer elements that minimally impact the maximum protein output attainable for any given 5′-UTR while preserving regulatory dynamic range. In addressing this need, we surmised that the observed translation inhibition caused by aptamer 5–1.2 could be due to: (i) the stability of its stem structure and/or (ii) the putative start codon and its sequence context within the aptamer (29). To test the effect of secondary structure stability on translation levels, we scrambled the first 25 bases of 5–1.2 to obtain 5–1.2half (Supplementary Table S4). This removed substantial predicted secondary structure within the 5′-UTR, but left the start codon sequence context intact. Placing 5–1.2half upstream of vYFP minimally increased basal vYFP expression compared with 5–1.2, suggesting that the stability of the aptamer stem was not the major determinant of reduced expression of the downstream reporter (Supplementary Figure S4).
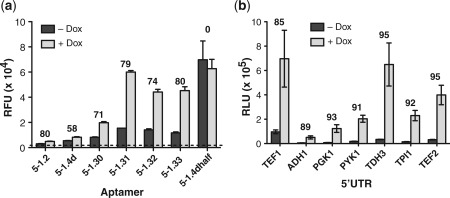
The 5–1.2 aptamer contains an AUG start codon followed immediately by a stop codon and a second start codon that is out of frame with the first, but in frame with the downstream reporter gene. Because previous studies have demonstrated that short upstream ORFs can inhibit translation of a downstream ORF (29), we investigated whether the sequence 43AUGUGAUG50 within a predicted loop region of 5–1.2 (Figure 1a) could be primarily responsible for the translation efficiency of a downstream ORF. While keeping The rest of 5–1.2 constant, we introduced an A→G mutation at position 48 in the loop region above to generate 5–1.4d containing the sequence 43AUGUGGUG50. This change simultaneously eliminated the stop codon and the second start codon within the aptamer loop. Replacing 5–1.2 with 5–1.4d (and placing the regulated ORF in frame with the single start codon in 5–1.4d) resulted in modestly higher expression levels, but maintained TetR-dependent regulation (Figure 6a). To further increase expression of the aptamer-regulated ORF, we systematically reduced aptamer stem strength by successively eliminating base–pair interactions at the stem base while retaining sequence downstream of the initiator AUG within the aptamer (Supplementary Table S4). When we used these aptamers (5–1.30, 5–1.31, 5–1.32, 5–1.33) to control translation as described previously, we measured a large increase in maximal expression level. Furthermore, TetR-dependent regulation was preserved at the ≥80% repression level previously observed (Figure 6a). To determine the expected upper limit of expression when using 5–1.4d, we scrambled the first 25 bases of the aptamer to remove substantial predicted secondary structure (5–1.4dhalf). When used to control gene expression, this modification produced a maximal expression level comparable with that of 5–1.31, but with no TetR-dependent regulation (Figure 6a), indicating that further destabilization of the aptamer was unlikely to yield further increases in maximal expression levels. Overall, replacing 5–1.2 with 5–1.31 increases maximal expression by ∼25-fold, and with no adverse impact on the magnitude of Dox inducible, TetR-dependent regulation.
Figure 6.
Systematic optimization of an aptamer variant that improves maximal expression levels and demonstration of robust regulation when an aptamer is placed in different 5′-UTR contexts. (a) Several aptamer variants generated by introducing a point mutation within Motif 1 and reducing the stem length of 5-1.2 were tested for their ability to regulate vYFP. These modifications allowed for expression level maximization while preserving regulatory range. (b) Aptamer 5-1.31 was placed within the context of several endogenous 5′-UTRs and used to control FLuc expression. In (a) and (b), numbers above the bars indicate the percent repression observed in the − Dox condition relative to the + Dox condition. In all cases, cells expressed TetR. These data show that regulation is achieved in all contexts tested.
Assessing the modularity of the system
Lastly, to validate the utility of the TetR-aptamer system in different native 5′-UTR contexts, we transcriptionally fused 5–1.31 with several yeast promoters of varied strength. We retained the sequence downstream of the aptamer used in previous experiments. Despite the expected variation in the maximal expression levels due to differences in promoter strength, FLuc expression was regulated with over 85% repression in all contexts (Figure 6b and Supplementary Table S5). We found similar, albeit slightly reduced, regulation when using vYFP as the reporter. Similarly, 5–1.2 in the same 5′-UTR contexts yielded strong, inducible repression. In these experiments, the TDH3 5′-UTR context produced 97% repression when paired with the 5–1.2 aptamer. This led us to hypothesize that the TDH3 5′-UTR/5–1.2 combination might be a superior sequence context module that increases the regulatory efficiency of our TetR-aptamer system. To test this, we fused the combined TDH3 5′-UTR–5–1.2 sequence downstream of the other 5′-UTR contexts tested above. Although we did not achieve equally high repression levels as in the original TDH3 5′-UTR context, robust regulation was nonetheless seen in every case (Supplementary Table S5). Altogether, these data strongly support the feasibility of using our optimized TetR–aptamer regulatory system in endogenous 5′-UTR contexts of diverse RNA sequence, even when little information about the targeted 5′-UTR's size, sequence and structural characteristics is available.
Polysome analysis
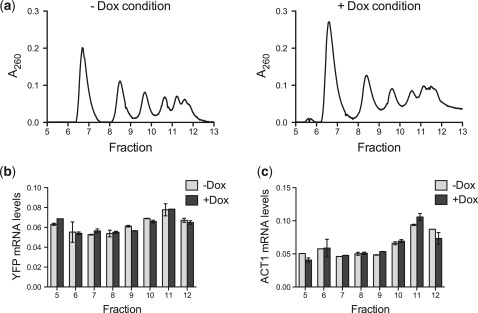
Our data from cell-free translation and qPCR experiments firmly support a post-transcriptional regulatory mechanism that does not act via a decrease in mRNA levels. Therefore, using polysome analysis, we sought to define whether the aptamer–TetR interaction modulates initiation or some downstream step in the translation process. For these experiments, vYFP regulated by 5–1.2 was used as a representative target transcript. If TetR interaction with 5–1.2-vYFP mRNA predominantly inhibits translation initiation, in the absence of Dox, this should reduce 5–1.2-vYFP mRNA ribosome occupancy and lead to the transcript's accumulation in non-polysomal fractions. Conversely, disrupting the TetR–5–1.2 interaction by adding Dox would result in more efficient translation initiation and increased accumulation of 5–1.2-vYFP mRNA in polysomal fractions. However, we consistently found no significant difference in 5–1.2-vYFP mRNA ribosome occupancy between the condition where the TetR–5–1.2 interaction is intact (− Dox) or disrupted (+ Dox) (Figure 7 and Supplementary Figure S5). This suggests that either: (i) standard polysome profiling is insufficiently sensitive to detect a small but functionally important shift in ribosome occupancy that may be occurring and/or (ii) the aptamer–TetR interaction inhibits translation mainly downstream of initiation. Our polysome profiling results indicate that both the translationally repressed and actively translated 5–1.2-vYFP mRNA are similarly associated with the polysomal fractions. While polysome-associated mRNAs are generally considered to be actively translated, some of these mRNAs are known to be translationally repressed (30–34). The specific molecular details underlying repression of polysome-associated mRNA are still generally unclear, but they could involve mRNA decapping, mRNA deadenylation, altered elongation kinetics, nascent polypeptide degradation and impaired ribosome release (35). Understanding exactly how the aptamer–TetR interaction and its disruption by Dox facilitate differential partitioning of aptamer-containing target transcripts between translationally repressed and actively translated pools is an intriguing problem that will require more detailed study beyond the scope of the present work. However, such efforts could provide additional insight into regulation mechanisms downstream of translation initiation, which are increasingly being recognized to be of broad biological importance (33,35–38). Furthermore, this knowledge can further enable engineering improved versions of our presently described system for inducibly regulating protein expression.
Figure 7.
Polysome profiles of aptamer-containing mRNA indicate regulation is independent of translation initiation. (a) Polysomes were fractionated from yeast expressing both TetR and a 5-1.2-containing vYFP reporter mRNA, which were grown in the absence or presence of Dox. Polysome profiles for both the − Dox and + Dox growth conditions are shown. (b) qPCR measurements of the relative amounts of reporter mRNA within each polysome fraction, both for the − Dox and + Dox conditions. (c) qPCR measurements of relative amount of ACT1 mRNA in each polysome fraction, under − Dox and + Dox conditions. For both (b) and (c) error bars indicate the range of values for technical replicates. The data are representative of two independent, biological replicates.
SUMMARY
We have developed a new system for directly and transcript-specifically controlling protein expression that requires a small number of defined, genetically-encoded components and no knowledge of transcriptional regulation. Induction is achieved using inexpensive, cell permeable and well-tolerated tetracycline analogs. Both the magnitude and stringency of the regulation attainable permit modulation of a survival phenotype in yeast under highly selective growth conditions, indicating that this approach is sufficiently robust so as to be biologically useful. Furthermore, this system functions equally well within the context of several natural 5′-UTRs, and we have optimized it such that translational repression due to the presence of structured RNA within the 5′-UTR is minimal. Because key aspects of translation are well conserved among eukaryotes, and as we have demonstrated translation control in both yeast and mammalian contexts, this system may be broadly applicable in controlling gene expression. We anticipate this system could be specifically useful in organisms with poorly understood transcriptional regulatory mechanisms and few inducible gene expression options. Overall, we have described a modular, inducible framework for biological control that provides a direct interface with protein synthesis. We foresee that the minimal nature of this system will enable investigation of previously intractable problems in cell biology, such as the role of gene-specific translational regulation in early development and neurobiology. Additionally, the modularity and host cell independence of the system make it particularly suited for use in the construction of synthetic biological circuits that operate independently of transcription.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–6 and Supplementary Figures 1–5.
FUNDING
National Institutes of Health, NIH Director's New Innovator Award Program (grant number 1DP2OD007124 to J.C.N.); National Institute of Environmental Health Sciences, Predoctoral Training Grant (grant number 5-T32-ES007020 to S.J.G.); National Science Foundation, Research Experiences for Undergraduates (grant number 1005055 to A.M.P.); MIT startup funds. Funding for open access charge: National Institutes of Health, NIH Director's New Innovator Award Program (grant number 1DP2OD007124).
Conflict of interest statement: S.J.G., B.J.B. and J.C.N. are co-inventors of the genetically encoded protein-binding RNA aptamer technology described and have filed patent applications with another co-inventor.
Supplementary Material
ACKNOWLEDGEMENTS
We thank John E. McCarthy (University of Manchester) for providing the YCpSUP and YCp22FL1 plasmids, and we acknowledge the use of analytical instrumentation in the MIT Biotechnology Process Engineering Center. We also want to extend our gratitude to Josh Arribere and Wendy Gilbert for insightful discussions and their assistance with the polysome profiling experiments and analysis.
REFERENCES
- 1.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat. Rev. Mol. Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Besse F, Ephrussi A. Translational control of localized mRNAs: restricting protein synthesis in space and time. Nat. Rev. Mol. Cell Biol. 2008;9:971–980. doi: 10.1038/nrm2548. [DOI] [PubMed] [Google Scholar]
- 3.Anderson P. Post-transcriptional regulons coordinate the initiation and resolution of inflammation. Nat. Rev. Immunol. 2010;10:24–35. doi: 10.1038/nri2685. [DOI] [PubMed] [Google Scholar]
- 4.Hüttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438:512–515. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- 5.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Topp S, Gallivan JP. Emerging applications of riboswitches in chemical biology. ACS Chem. Biol. 2010;5:139–148. doi: 10.1021/cb900278x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson AL, Linsley PS. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat. Rev. Drug Discov. 2010;9:57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- 8.Matsukura S, Jones PA, Takai D. Establishment of conditional vectors for hairpin siRNA knockdowns. Nucleic Acids Res. 2003;31:e77. doi: 10.1093/nar/gng077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Condeelis J, Singer RH. How and why does beta-actin mRNA target? Biol. Cell. 2005;97:97–110. doi: 10.1042/BC20040063. [DOI] [PubMed] [Google Scholar]
- 10.Harvey I, Garneau P, Pelletier J. Forced engagement of a RNA/protein complex by a chemical inducer of dimerization to modulate gene expression. Proc. Natl Acad. Sci. USA. 2002;99:1882–1887. doi: 10.1073/pnas.042693399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macchi P, Hemraj I, Goetze B, Grunewald B, Mallardo M, Kiebler MA. A GFP-based system to uncouple mRNA transport from translation in a single living neuron. Mol. Biol. Cell. 2003;14:1570–1582. doi: 10.1091/mbc.E02-08-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paraskeva E, Atzberger A, Hentze MW. A translational repression assay procedure (TRAP) for RNA-protein interactions in vivo. Proc. Natl Acad. Sci. USA. 1998;95:951–956. doi: 10.1073/pnas.95.3.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plummer KA, Carothers JM, Yoshimura M, Szostak JW, Verdine GL. In vitro selection of RNA aptamers against a composite small molecule-protein surface. Nucleic Acids Res. 2005;33:5602–5610. doi: 10.1093/nar/gki867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saito H, Kobayashi T, Hara T, Fujita Y, Hayashi K, Furushima R, Inoue T. Synthetic translational regulation by an L7Ae-kink-turn RNP switch. Nat. Chem. Biol. 2010;6:71–78. doi: 10.1038/nchembio.273. [DOI] [PubMed] [Google Scholar]
- 15.Ausländer D, Wieland M, Ausländer S, Tigges M, Fussenegger M. Rational design of a small molecule-responsive intramer controlling transgene expression in mammalian cells. Nucleic Acids Res. 2011;39:e155. doi: 10.1093/nar/gkr829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belmont BJ, Niles JC. Engineering a direct and inducible protein-RNA Interaction to regulate RNA biology. ACS Chem. Biol. 2010;5:851–861. doi: 10.1021/cb100070j. [DOI] [PubMed] [Google Scholar]
- 18.Koloteva N, Müller PP, McCarthy JEG. The position dependence of translational regulation via RNA-RNA and RNA-protein interactions in the 5′-untranslated region of eukaryotic mRNA is a function of the thermodynamic competence of 40 S ribosomes in translational initiation. J. Biol. Chem. 1997;272:16531–16539. doi: 10.1074/jbc.272.26.16531. [DOI] [PubMed] [Google Scholar]
- 19.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 20.Urlinger S, Baron U, Thellmann M, Hasan MT, Bujard H, Hillen W. Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc. Natl Acad. Sci. USA. 2000;97:7963–7968. doi: 10.1073/pnas.130192197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma H, Kunes S, Schatz PJ, Botstein D. Plasmid construction by homologous recombination in yeast. Gene. 1987;58:201–216. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- 22.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oliveira CC, Goossen B, Zanchin NIT, McCarthy JEG, Hentze MW, Stripecke R. Translational repression by the human iron-regulatory factor (IRF) in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:5316–5322. doi: 10.1093/nar/21.23.5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oliveira CC, Heuvel JJ, McCarthy JEG. Inhibition of translational initiation in Saccharomyces cerevisiae by secondary structure: the roles of the stability and position of stem-loops in the mRNA leader. Mol. Microbiol. 1993;9:521–532. doi: 10.1111/j.1365-2958.1993.tb01713.x. [DOI] [PubMed] [Google Scholar]
- 25.Kötter P, Weigand JE, Meyer B, Entian K-D, Suess B. A fast and efficient translational control system for conditional expression of yeast genes. Nucleic Acids Res. 2009;37:e120. doi: 10.1093/nar/gkp578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suess B, Fink B, Berens C, Stentz R, Hillen W. A theophylline responsive riboswitch based on helix slipping controls gene expression in vivo. Nucleic Acids Res. 2004;32:1610–1614. doi: 10.1093/nar/gkh321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Win MN, Smolke CD. Higher-order cellular information processing with synthetic RNA devices. Science. 2008;322:456–460. doi: 10.1126/science.1160311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scholz O, Henssler E-M, Bail J, Schubert P, Bogdanska-Urbaniak J, Sopp S, Reich M, Wisshak S, Köstner M, Bertram R, et al. Activity reversal of Tet repressor caused by single amino acid exchanges. Mol. Microbiol. 2004;53:777–789. doi: 10.1111/j.1365-2958.2004.04159.x. [DOI] [PubMed] [Google Scholar]
- 29.McCarthy JE. Posttranscriptional control of gene expression in yeast. Microbiol. Mol. Biol. Rev. 1998;62:1492–1553. doi: 10.1128/mmbr.62.4.1492-1553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berry JO, Carr JP, Klessig DF. mRNAs encoding ribulose-1,5-bisphosphate carboxylase remain bound to polysomes but are not translated in amaranth seedlings transferred to darkness. Proc. Natl Acad. Sci. USA. 1988;85:4190–4194. doi: 10.1073/pnas.85.12.4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ch'ng JL, Shoemaker DL, Schimmel P, Holmes EW. Reversal of creatine kinase translational repression by 3′ untranslated sequences. Science. 1990;248:1003–1006. doi: 10.1126/science.2343304. [DOI] [PubMed] [Google Scholar]
- 32.Kaspar RL, Gehrke L. Peripheral blood mononuclear cells stimulated with C5a or lipopolysaccharide to synthesize equivalent levels of IL-1 beta mRNA show unequal IL-1 beta protein accumulation but similar polyribosome profiles. J. Immunol. 1994;153:277–286. [PubMed] [Google Scholar]
- 33.Mootz D, Ho DM, Hunter CP. The STAR/Maxi-KH domain protein GLD-1 mediates a developmental switch in the translational control of C. elegans PAL-1. Development. 2004;131:3263–3272. doi: 10.1242/dev.01196. [DOI] [PubMed] [Google Scholar]
- 34.Theodorakis NG, Banerji SS, Morimoto RI. HSP70 mRNA translation in chicken reticulocytes is regulated at the level of elongation. J. Biol. Chem. 1988;263:14579–14585. [PubMed] [Google Scholar]
- 35.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 36.Maroney PA, Yu Y, Fisher J, Nilsen TW. Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nat. Struct. Mol. Biol. 2006;13:1102–1107. doi: 10.1038/nsmb1174. [DOI] [PubMed] [Google Scholar]
- 37.Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 38.Seggerson K, Tang L, Moss EG. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev. Biol. 2002;243:215–225. doi: 10.1006/dbio.2001.0563. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.