Abstract
Ataxia telangiectasia mutated (ATM), a PI-3 kinase essential for maintaining genomic stability, has been shown to regulate TRF1, a negative mediator of telomerase-dependent telomere extension. However, little is known about ATM-mediated TRF1 phosphorylation site(s) in vivo. Here, we report that ATM phosphorylates S367 of TRF1 and that this phosphorylation renders TRF1 free of chromatin. We show that phosphorylated (pS367)TRF1 forms distinct non-telomeric subnuclear foci and that these foci occur predominantly in S and G2 phases, implying that their formation is cell cycle regulated. We show that phosphorylated (pS367)TRF1-containing foci are sensitive to proteasome inhibition. We find that a phosphomimic mutation of S367D abrogates TRF1 binding to telomeric DNA and renders TRF1 susceptible to protein degradation. In addition, we demonstrate that overexpressed TRF1-S367D accumulates in the subnuclear domains containing phosphorylated (pS367)TRF1 and that these subnuclear domains overlap with nuclear proteasome centers. Taken together, these results suggest that phosphorylated (pS367)TRF1-containing foci may represent nuclear sites for TRF1 proteolysis. Furthermore, we show that TRF1 carrying the S367D mutation is unable to inhibit telomerase-dependent telomere lengthening or to suppress the formation of telomere doublets and telomere loss in TRF1-depleted cells, suggesting that S367 phosphorylation by ATM is important for the regulation of telomere length and stability.
INTRODUCTION
Telomeres are specialized heterochromatic structures found at the ends of linear eukaryotic chromosomes. Mammalian telomeric DNA consists of TTAGGG tandem repeats that are coated with shelterin, a telomere-specific protein complex composed of TRF1, TRF2, TIN2, Rap1, TPP1 and POT1 (1–3). The shelterin complex functions not only to protect telomeres from being recognized as double strand breaks (1,4) but also to maintain telomere length homeostasis, which is intimately associated with tumorigenesis and aging. Disruption of shelterin proteins has been shown to induce the de-protection of telomeres, resulting in telomere abnormalities including telomere end-to-end fusions, telomere loss and telomere doublets/fragile telomeres (more than one telomeric signal at a single chromatid end) (4–12). These dysfunctional telomeres are recognized as damaged DNA (8–10,12,13) and can contribute to genomic instability.
TRF1, a component of the shelterin complex, binds specifically to duplex telomeric DNA (14), and is implicated in telomere replication, telomere protection and telomere length maintenance. Deletion of TRF1 promotes the formation of fragile telomeres in S phase, a phenomena thought to be associated with replication-dependent defects (6,9). It has been suggested that TRF1 is required to prevent fork stalling, allowing efficient replication of telomeric DNA (9). Loss of TRF1 from telomeres has been shown to induce telomerase-dependent telomere lengthening whereas overexpression of TRF1 results in telomere shortening, suggesting that TRF1 negatively regulates telomerase-dependent telomere extension, perhaps by restricting the access of telomerase to the ends of telomeres (15–17).
TRF1 interacts with ataxia telangiectasia mutated (ATM) (18), a PI-3 kinase essential for maintaining genomic stability. Mutations in ATM give rise to ataxia–telangiectasia (AT), an autosomal recessive disorder characterized by immunodeficiency, spontaneous chromosomal instability, hypersensitivity to ionizing irradiation and a predisposition to cancer (19). Primary fibroblasts derived from AT patients accumulate telomere abnormalities and show an elevated rate of telomere shortening as compared to cells from normal individuals (20–25). In agreement with these findings, inhibition of ATM has been shown to induce telomere shortening in telomerase-expressing cancer cells, indicative of the role of ATM as a positive mediator of telomere length maintenance (26). It has been suggested that ATM promotes telomerase-dependent telomere elongation by negatively regulating TRF1 association with telomeric DNA (26). However, little is known about the ATM phosphorylation site(s) of TRF1 important for telomere length maintenance.
In this report, we demonstrate that ATM phosphorylates S367 of TRF1 both in vivo and in vitro. We show that a phosphomimic mutation of S367D abrogates TRF1 interaction with telomeric DNA. Using a phosphospecific antibody, we demonstrate that phosphorylated (pS367)TRF1 forms distinct subnuclear foci that are not associated with telomeric DNA. These results reveal that the phosphorylation of S367 by ATM prevents TRF1 binding to telomeric DNA. We show that phosphorylated (pS367)TRF1-containing foci predominantly occur in S and G2 phases, implying that the formation of these foci is cell cycle regulated. In addition, we find that phosphorylated (pS367)TRF1-containing foci are sensitive to proteasome inhibition. We show that the phosphomimic mutation of S367D renders mutant TRF1 susceptible to protein degradation and promotes its localization in phosphorylated (pS367)TRF1-containing subnuclear domains, which overlap with nuclear proteasome centers. These results suggest that phosphorylated (pS367)TRF1-containing foci may represent nuclear proteolytic sites for TRF1 degradation. Furthermore, we demonstrate that the phosphomimic mutation of S367D impairs the ability of TRF1 not only to inhibit telomerase-dependent telomere elongation but also to suppress the formation of telomere doublets and telomere loss in TRF1-depleted cells, suggesting that S367 phosphorylation by ATM plays an important role in regulating telomere length and stability.
MATERIALS AND METHODS
DNA constructs, cell culture and retroviral infection
The oligonucleotides encoding siRNAs directed against TRF1 have been previously described (12). The annealed oligonucleotides were ligated into pRetroSuper (pRS) vector (kindly provided by Titia de Lange, Rockefeller University), giving rise to pRetroSuper-shTRF1. The QuickChange site-directed mutagenesis kit (Strategene) was used to create TRF1 mutations of S367A and S367D as well as silent mutations resistant to shTRF1 as previously described (12). Wild-type TRF1 and TRF1 mutants (S367A and S367D) carrying silent mutations resistant to shTRF1 were then subcloned into either the retroviral vector pWZL-N-Myc (27) or the bacterial expression vector pHis-Parallel-2 (28).
Cells were grown in DMEM medium with 10% fetal bovine serum (FBS) for GM09607 (Coriell), GM05849 (Coriell), GM16666 (Coriell), GM16667 (Coriell), HeLaI.2.11, HeLaII, GM847, Phoenix cells and 15% FBS for IMR90 cells, supplemented with non-essential amino acids, glutamine, 100 U/ml penicillin and 0.1 mg/ml streptomycin. The ATM-deficient AT22IJE cell line was transfected with either an ATM expression construct or an empty vector, giving rise to ATM-corrected (GM16667) and ATM-deficient (GM16666) stable cell lines, respectively (29). HeLaI.2.11 and HeLaII are two sublines of HeLa cells of different telomere length (4,30). Retroviral gene delivery was carried out as described (31–33) to generate stable cell lines. HeLaII cells expressing pRS/pWZL, shTRF1/pWZL, shTRF1/TRF1, shTRF1/S367A or shTRF1/S367D were maintained in the selection medium containing either puromycin (2 µg/ml) or hygromycin (90 µg/ml) alternating every 2 weeks for the entirety of the experiments.
Cell cycle analysis
Synchronization of HeLaI.2.11 with a double thymidine block was carried out essentially as described (34) with the exception that 2 mM thymidine instead of aphidicolin was used. For FACS analysis, two million cells were fixed in 80% ethanol, digested with RNase A (2 mg/ml), stained with 50 µg/ml propidium iodide, and analyzed using a Becton–Dickinson LSRII located at the SickKids-UHN flow cytometry facility, Toronto, ON, Canada.
Protein extracts, differential salt extraction of chromatin and immunoblotting
Differential salt extraction of chromatin was performed as described (35). Protein extracts and immunoblotting were carried out essentially as described (27,34). Rabbit polyclonal anti-pS367 antibody was developed by Biosynthesis Inc. against a TRF1 peptide containing phosphorylated serine 367 (VSK-pS-QPVTPEKHRARKR). Antibodies used were anti-TRF1 (a kind gift from Titia de Lange), anti-proteasome (PA1-1962, Pierce), anti-Myc (9E10, Calbiochem), anti-H3K9m3 (Upstate) and anti-γ-tubulin (GTU88, Sigma).
Production of recombinant TRF1 proteins
Production of 6xHis-tagged TRF1 proteins was carried out as described (10,12). Briefly, induction of wild-type and mutant TRF1 proteins was carried out overnight with 0.1 mM Isopropyl β-D-1-thiogalactopyranoside at room temperature. The cell pellet was resuspended in binding buffer [20 mM Tris–HCl (pH 7.9) and 500 mM NaCl] and then lysed by sonication. Following centrifugation, the supernatant was incubated with nickel-resin (Qiagen) for 2 h at 4°C. The beads were washed three times with 60 mM imidazole and bound proteins were eluted with a buffer containing 1 M imidazole, 20 mM Tris–HCl (pH 7.9) and 0.5 M NaCl.
Immunoprecipitation of ATM and in vitro kinase assays
Immunoprecipitation of ATM was performed as previously described (26). Briefly, cell lysates were made in lysis buffer [50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 10% Glycerol, 1% Tween-20, 50 mM NaF, 1 mM NaVO4, 0.1 mM DTT, 0.5 mM PMSF, 0.5 µg/ml leupeptin], followed by sonication (50% duty cycle, nine pulses and output of three). For each ATM IP, 4 µg anti-ATM (Ab-3) antibody and 300 µl cell lysate (equivalent to 6 × 106 cells) were used. Following 1 h incubation on ice, 25 µl protein G sepharose slurry (GE Healthcare) was added to each IP and continued incubation for 1 h at 4°C. The IP pellet was washed twice in lysis buffer, once in LiCl buffer (0.5 M LiCl and 0.1 M Tris–HCl (pH 7.5)], and twice in kinase buffer [(10 mM Hepes pH 7.9, 50 mM NaCl, 10 mM MgCl2, 10 mM MnCl2, 5 µM ATP and 1 mM DTT). For ATM kinase assays, the final IP pellet was resuspended in kinase buffer, mixed with bacterial-derived recombinant wild-type TRF1 (2 µg) or TRF1-S367A (2 µg) in the presence of 10 µCi γ-32P-ATP in a final volume of 15 µl. For ATM kinase assays followed by in vitro gel-shift assays, cold ATP (1.8 mM) was used. For DNA-PKcs kinase assays, recombinant wild-type TRF1 (2 µg) or TRF1-S367A (2 µg) was incubated with 20 units of purified DNA-PKcs (Promega, V5811) in the presence of γ-32P-ATP according to the manufacturer's instruction.
Immunofluorescence and fluorescence in situ hybridization
Immunofluoresence (IF) was performed essentially as described (10,32,34). IF–FISH (fluorescence in situ hybridization) analysis was conducted as described (9). Briefly, cells grown on coverslips were fixed at RT for 10 min in PBS-buffered 2% paraformaldehyde, washed in PBS twice for 5 min each, followed by incubation at RT for 30 min in blocking buffer containing 1 mg/ml BSA, 3% goat serum, 0.1% Triton X-100 and 1 mM EDTA in PBS. Blocked coverslips were incubated with anti-pS367 antibody in blocking buffer at RT for 1 h. After three washes in PBS, coverslips were incubated with fluorescein isothiocyanate (FITC)-conjugated donkey anti-rabbit (1:100, Jackson Laboratories) at RT for 30 min. Subsequently, cells on coverslips were fixed again in PBS-buffered 2% paraformaldehyde for 5 min and followed by dehydration in a series of 70, 85 and 100% ethanol. The air-dried coverslips were denatured at 80°C for 10 min and hybridized with 0.5 µg/ml tetramethyl rhodamine isothiocyanate (TRITC)-conjugated-(TTAGGG)3 peptide nucleic acid (PNA) probe (Biosynthesis Inc.) for 2 h in the dark at RT. Following incubation, cover slips were washed with 70% formamide and 10 mM Tris–HCl (pH 7.2) twice for 15 min. After three washes in PBS, DNA was counter-stained with 4,6-diamidino-2-phenylindole (DAPI; 0.2 µg/ml) and embedded in 90% glycerol/10% PBS containing 1 mg/ml p-phenylene diamine (Sigma). All cell images were recorded on a Zeiss Axioplan 2 microscope with a Hammamatsu C4742-95 camera and processed in Open Lab.
Metaphase chromosome spreads
Metaphase chromosome spreads were essentially prepared as described (4,32). TRF1-depleted HeLaII cells expressing various TRF1 alleles or the vector alone were arrested in nocodazole (0.1 µg/ml) for 90 min. Following arrest, cells were harvested by trypsinization, incubated for 7 min at 37°C in 75 mM KCl, and fixed in freshly made methanol/glacial acidic acid (3:1). Cells were stored overnight at 4°C, dropped onto slides and air-dried overnight in a chemical hood.
FISH analysis on metaphase chromosome spreads was carried out essentially as described (32,36). Slides with chromosome spreads were incubated with 0.5 µg/ml FITC-conjugated-(CCCTAA)3 PNA probe (Biosynthesis Inc.) for 2 h at room temperature. Following incubation, slides were washed, counter-stained with 0.2 µg/ml DAPI and embedded in 90% glycerol/10% PBS containing 1 mg/ml p-phenylene diamine (Sigma). All cell images were recorded on a Zeiss Axioplan 2 microscope with a Hammamatsu C4742-95 camera and processed in Open Lab.
In vitro gel-shift assays
In vitro gel-shift assays were done essentially as described (26,37) with an end-labeled 188-bp BglII–XhoI fragment from plasmid pTH12 (37). Bacteria-derived recombinant wild-type or mutant TRF1 protein was incubated with the end-labeled DNA (1 ng) at room temperature for 20 min in a 20 -µl reaction containing 20 mM HEPES–KOH (pH7.9), 150 mM KCl, 5% (v/v) glycerol, 4% (w/v) Ficoll, 1 mM EDTA, 0.1 mM MgCl2, 0.5 mM DTT, 70 µg BSA, 2 µg sheared Escherichia coli DNA and 50 ng β-casein. The DNA–protein complexes were fractionated on a 0.7% agarose gel run in 0.1 × TBE (8.9 mM Tris-base, 8.9 mM boric acid, 0.2 mM EDTA) at 130 V for 1 h at room temperature. Gels were dried and exposed to PhosphorImager screens.
Telomere length analysis
Genomic DNA isolated from cells was digested with RsaI and HinfI and loaded onto a 0.7% agarose gel in 0.5× TBE. Blotting for telomeric fragments was carried out according to standard protocols (15). The average telomeric restriction fragment length was determined by PhosphorImager analysis using ImageQuant and MS Excel as described (38).
RESULTS
ATM phosphorylates S367 of TRF1 both in vivo and in vitro
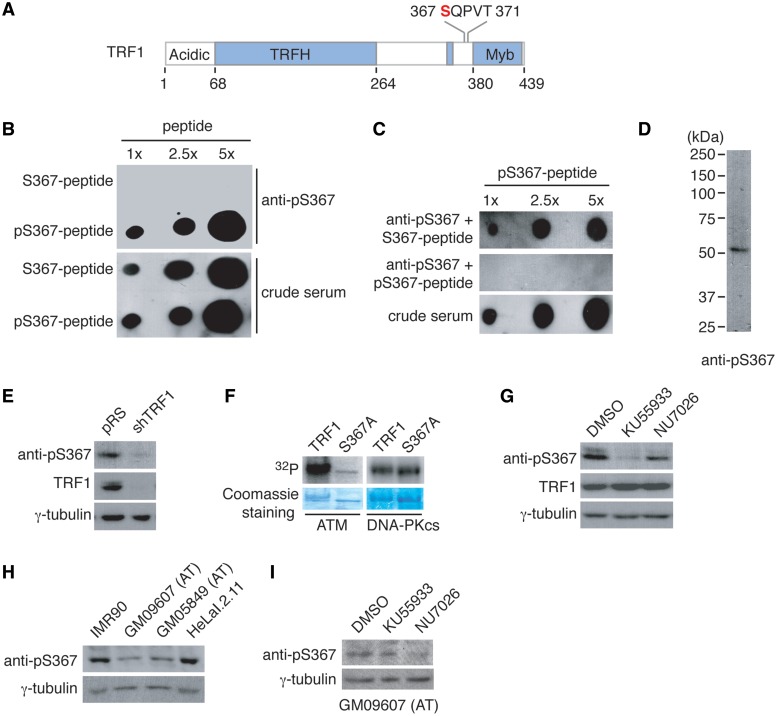
Phosphorylation has been shown to play an important role in modulating the function of TRF1 in telomere metabolism (12,26,39–41). In an effort to identify TRF1 phosphorylation sites in vivo, we generated an HT1080 cell line stably expressing Flag-tagged TRF1. Mass spectrometry analysis of immunoprecipitated Flag-TRF1 indicated serine at position 367 of TRF1 to be a candidate phosphorylation site in vivo (Figure 1A and data not shown).
Figure 1.
ATM phosphorylates S367 of TRF1 both in vivo and in vitro. (A) Schematic diagram of TRF1 domain structures. S367 of TRF1 in red was identified by mass spectrometry analysis to be a candidate phosphorylation site. (B) Affinity-purified anti-pS367 antibody specifically recognizes TRF1 peptide containing phosphorylated S367 (pS367-peptide). An increasing amount of peptide either carrying unmodified S367 (S367-peptide) or phosphorylated S367 (pS367-peptide) was spotted on a nitrocellulose membrane, followed by immunoblotting with affinity-purified anti-pS367 antibody or crude serum. The amount of peptide spotted from left to right is 0.7, 1.75 and 3.5 µg. (C) Peptide competition assays. Affinity-purified anti-pS367 antibody was incubated with 5.5 µg of either unmodified (S367-peptide) or phosphorylated peptide (pS367-peptide) prior to immunoblotting. Crude serum was used to show the presence of pS367-peptide on the nitrocellulose membrane. The amount of pS367-peptide spotted from left to right is 0.7, 1.75 and 3.5 µg. (D) Western analysis of phosphorylated TRF1. The whole cell extract (20 µg) from HeLaII cells was immunoblotted with affinity-purified anti-pS367 antibody. (E) Depletion of endogenous TRF1 leads to loss of phosphorylated TRF1 recognized by anti-pS367 antibody. HeLaII cells were infected with retrovirus expressing shTRF1 or the vector pRS alone. Western analysis was performed with anti-pS367 or anti-TRF1 antibody. The γ-tubulin blot was used as a loading control. (F) In vitro kinase assays. Bacterial-expressed his-tagged wild-type TRF1 (2 µg) or TRF1 mutant S367A (2 µg) was incubated with either ATM immunoprecipitated from HeLa cells or purified DNA-PKcs in the presence of γ-32P-ATP. (G) ATM inhibition leads to a diminished anti-pS367 staining. HeLaI.2.11 cells were treated with DMSO, KU55933 (an ATM inhibitor) or NU7026 (a DNA-PKcs inhibitor) for 90 min, followed by western analysis. Immunoblotting was performed with anti-pS367, anti-TRF1 or anti-γ-tubulin antibody. (H) Loss of anti-pS367 staining in ATM-deficient cells. Western analysis of cell extracts from IMR90 cells, HeLaI.2.11 cells and ATM-deficient cells (GM09607 and GM05849). Immunoblotting was performed with anti-pS367 or anti-γ-tubulin antibody. (I) NU7026 leads to a further reduction in S367 phosphorylation in ATM-deficient cells. GM09607 cells were treated with DMSO, KU55933 or NU7026 for 90 min, following by immunoblotting with anti-pS367 or anti-γ-tubulin antibody.
To further investigate the phosphorylation of S367 of TRF1 in vivo, we raised an antibody against a TRF1 peptide containing phosphorylated S367, referred to as anti-pS367. Anti-pS367 antibody specifically recognized the phosphorylated TRF1 peptide but not the unphosphorylated TRF1 peptide (Figure 1B). The ability of anti-pS367 to specifically recognize the phosphorylated (pS367)TRF1 peptide was further demonstrated by peptide competition assays. While pre-incubation of unmodified peptide had no effect on binding of anti-pS367 antibody to the phosphorylated peptide, pre-incubation of the phosphorylated peptide completely abrogated its ability to bind the phosphorylated peptide (Figure 1C). We showed that anti-pS367 antibody predominantly recognized a protein band with an apparent molecular weight indistinguishable from that of endogenous TRF1 in HeLa cell lysate (Figure 1D). Depletion of TRF1 resulted in a loss of TRF1 recognized by anti-pS367 antibody in vivo (Figure 1E). Taken together, these results suggest that anti-pS367 antibody specifically recognizes phosphorylated S367 of TRF1 in vivo.
S367 (S367Q) of TRF1 matches the consensus sequence for ATM and we decided to investigate whether ATM may be involved in phosphorylating S367 of TRF1. To address this question, we performed in vitro ATM kinase assays with bacterial-derived recombinant wild-type TRF1 or mutant TRF1 carrying a single amino acid substitution at S367 (TRF1-S367A). We found that while ATM immunoprecipitated from HeLa cells was able to phosphorylate wild-type TRF1, consistent with previous findings (18,26), the alanine substitution of S367 impaired TRF1 phosphorylation by ATM in vitro (Figure 1F). However, the S367A mutation had no effect on TRF1 phosphorylation by DNA-PKcs in vitro (Figure 1F), indicative of the specificity of S367 phosphorylation by ATM.
Using the phospho-specific anti-pS367 antibody, we examined the effect of ATM inhibition on S367 phosphorylation in vivo. HeLa cells were treated with either dimethyl sulfoxide (DMSO), KU55933, a highly specific inhibitor of ATM (42) or NU7026, a specific inhibitor of DNA-PKcs (43). We found that treatment with KU55933 but not NU7026 led to a reduction in S367 phosphorylation (Figure 1G). Furthermore, we detected a substantial reduction in S367 phosphorylation in ATM-deficient GM09607 and GM05849 cells compared to ATM-proficient IMR90 and HeLaI.2.11 cells (Figure 1H). Treatment of GM09607 cells with the DNA-PKcs inhibitor NU7026 led to a further decrease in S367 phosphorylation and such decrease was not observed in KU55933-treated GM09607 cells (Figure 1I), implying that DNA-PKcs may phosphorylate S367 in vivo in the absence of functional ATM. Taken together, these results demonstrate that ATM phosphorylates S367 of TRF1 both in vitro and in vivo.
Phosphorylated (pS367)TRF1 forms distinct non-telomeric nuclear foci that are cell cycle regulated
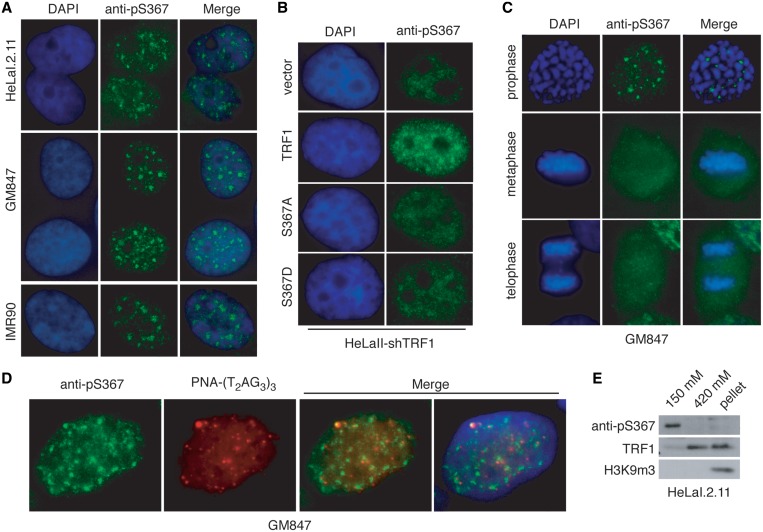
We have shown that TRF1 is phosphorylated at S367 in vivo and we decided to examine the nuclear localization of phosphorylated (pS367)TRF1. Using the phospho-specific anti-pS367 antibody, we performed indirect IF on both primary and transformed cell lines. We found that in interphase cells, anti-pS367 staining formed distinct nuclear foci in both primary and transformed cell lines (Figure 2A) and that these foci were very heterogenous in size, a feature distinct from telomere staining. We found that depletion of TRF1 led to a severe reduction in anti-pS367 staining and the formation of anti-pS367-containing foci (Figure 2B). On the other hand, anti-pS367-containing foci were fully restored in TRF1-depleted cells expressing wild-type TRF1 but not in TRF1-depleted cells expressing TRF1 carrying either a non-phosphorylatable mutation (S367A) or a phosphomimic mutation (S367D) (Figure 2B), suggesting that these foci represent phosphorylated (pS367)TRF1. The lack of rescue of anti-pS367-containing foci by TRF1 mutants demonstrates that phospho-specific anti-pS367 antibody does not recognize TRF1 carrying a single amino acid substitution of S367. Phosphorylated (pS367)TRF1-containing foci were also visible in early prophase cells (Figure 2C); however, anti-pS367 staining became very diffuse in the rest of mitosis from metaphase to telophase (Figure 2C). These results suggest that the formation of phosphorylated (pS367)TRF1-containing foci may be cell cycle regulated.
Figure 2.
Phosphorylated (pS367)TRF1 forms distinct nuclear foci that are not associated with telomere chromatin. (A) Indirect IF using anti-pS367 antibody was performed on human primary IMR90 cells as well as transformed HeLaI.2.11 and GM847 cells. Cell nuclei were stained with DAPI shown in blue. (B) Indirect IF using anti-pS367 antibody was performed on TRF1-depleted HeLaII cells expressing the vector alone, wild-type TRF1, TRF1-S367A or TRF1-S367D. Wild-type TRF1, TRF1-S367A or TRF1-S367D were engineered to be resistant to shTRF1. Cell nuclei were stained with DAPI shown in blue. (C) Phosphorylated (pS367)TRF1-containing foci are visible in early prophase but become diffuse in the rest of mitosis. Indirect IF using anti-pS367 antibody was performed on GM847 cells. Cell nuclei were stained with DAPI shown in blue. (D) Phosphorylated (pS367)TRF1 is not associated with telomeric DNA. IF–FISH analysis was performed on fixed GM847 cells with anti-pS367 antibody (green) in conjunction with TRITC-conjugated telomeric DNA-containing PNA probe (red). Cell nuclei were stained with DAPI shown in blue. (E) Phosphorylated (pS367)TRF1 is predominantly free of chromatin. Differential salt (KCl) extraction of chromatin was performed on HeLaI.2.11 cells. Immunoblotting was carried out with anti-pS367, anti-TRF1 or anti-H3K9m3 antibody. The anti-H3K9m3 blot was used as control to assess the extraction of chromatin.
To further investigate the nature of phosphorylated (pS367)TRF1-containing foci, we conducted IF–FISH analysis with anti-pS367 antibody in conjunction with a TRITC-conjugated telomeric DNA-containing PNA probe. We observed very little overlap between the majority of bright foci stained by anti-pS367 and telomeric DNA (Figure 2D). Occasionally, we observed an overlap between one or two bright anti-pS367 foci and telomeric DNA (Figure 2D). While the nature of this overlap is unknown, it may be insignificant due to the high number of anti-pS367 foci (Figure 2D). Analysis of a differential salt extraction of chromatin revealed that while TRF1 was predominantly found in the chromatin-bound fraction (420 mM KCl), consistent with previous findings (35), phosphorylated (pS367)TRF1 was overwhelmingly associated with the chromatin-free fraction (150 mM KCl) (Figure 2E). Taken together, these results demonstrate that phosphorylated (pS367)TRF1 is not associated with telomere chromatin. We estimated that 1–5% of endogenous TRF1 may be phosphorylated at S367 (Figure 2E).
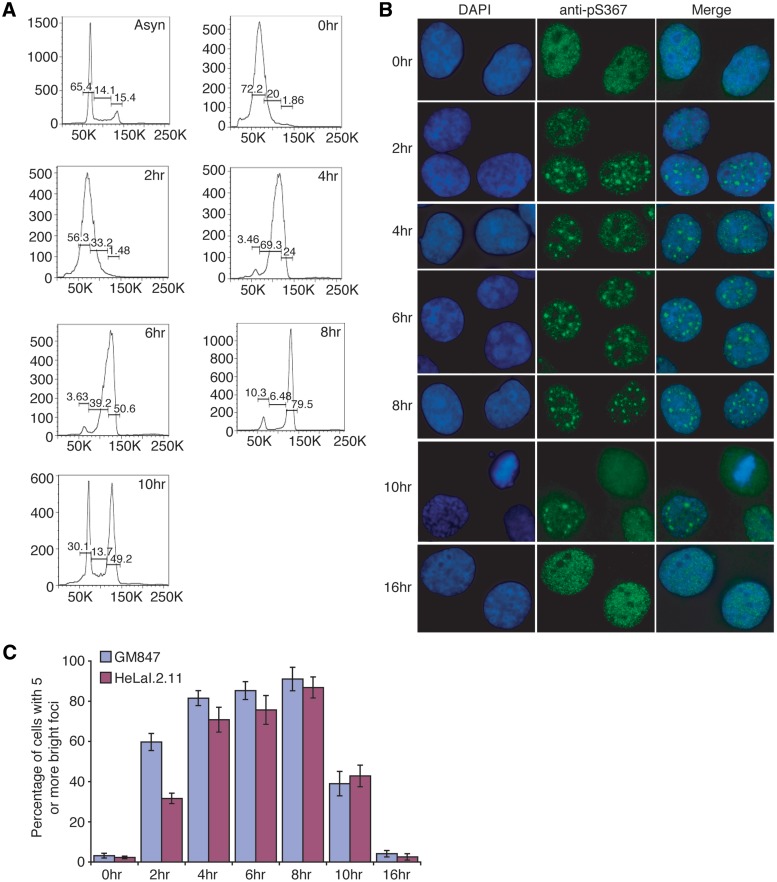
To address the cell cycle-dependent nature of anti-pS367 staining, we arrested HeLaI.2.11 and GM847 cells at the G1/S boundary with a double thymidine block and then released them into fresh media for 0, 2, 4, 6, 8, 10 or 16 h. Consistent with our previous findings (34), we found that HeLa cells progressed through S phase 2–6 h post release and enter mitosis 10 h post release (Figure 3A). Analysis of immunofluoresence showed that the bright foci stained by anti-pS367 antibody were predominantly seen in cells fixed 2–8 h post release from a double thymidine block whereas very few cells arrested at G1/S or in G1 (16 h post release from a double thymidine block) displayed such foci (Figure 3B and C). These results suggest that phosphorylated (pS367)TRF1-containing foci are cell cycle regulated, occuring in S and G2 phase.
Figure 3.
Phosphorylated (pS367)TRF1 forms distinct non-telomeric foci in a cell cycle regulated manner, enriched in S and G2 phases. (A) FACS analysis of synchronized HeLaI.2.11 cells. y-axis, cell numbers; x-axis, relative DNA content on the basis of staining with propidium iodine. Asyn, asynchronous population; 0–10 h, cells were released for 0–10 h from a double thymidine block. (B) Indirect IF using anti-pS367 antibody was performed on GM847 cells released for 0–16 h from a double thymidine block. Cell nuclei were stained with DAPI shown in blue. (C) Quantification of percentage of GM847 and HeLaI.2.11 cells exhibiting five or more phosphorylated (pS367)TRF1-containing foci. For each of the indicated time-points post-release from a double thymidine block, a total of at least 1500 cells from three independent experiments were scored in blind for both GM847 and HeLaI.2.11 cells. Standard deviations from three independent experiments are indicated.
ATM is required for the subnuclear localization of phosphorylated (pS367)TRF1
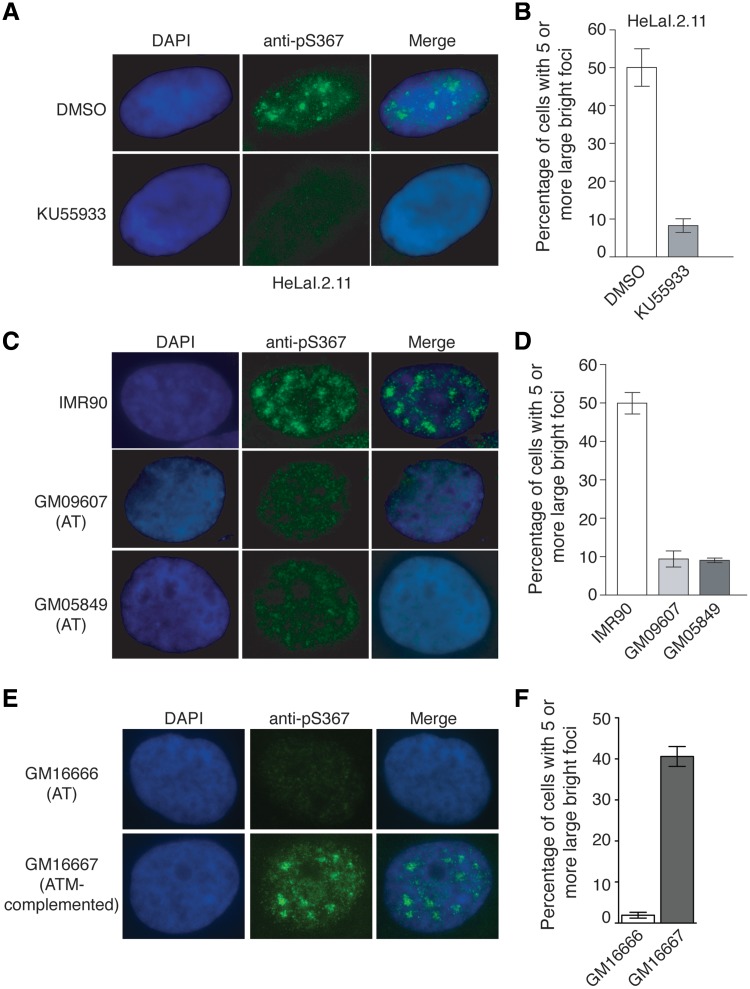
To investigate whether phosphorylated (pS367)TRF1-containing foci might be ATM-dependent, indirect IF with anti-pS367 antibody was performed on HeLaI.2.11 cells treated with either DMSO or KU55933. We observed a severe loss of anti-pS367 staining along with the disappearance of distinct nuclear foci of phosphorylated (pS367)TRF1 in KU55933-treated cells as compared to DMSO-treated cells (Figure 4A). Treatment with KU55933 led to a 6-fold reduction in the percentage of cells exhibiting phosphorylated (pS367)TRF1-containing foci (Figure 4B). The absence of distinct anti-pS367 nuclear foci was also evident in ATM-deficient GM09607 and GM05849 cells when compared to ATM-proficient IMR90 cells (Figure 4C and 4D). In addition, we found that introduction of ATM into ATM-deficient cells restored phosphorylated (pS367)TRF1-containing foci (Figure 4E and F). Taken together, these results suggest that the subnuclear localization of phosphorylated (pS367)TRF1 is dependent upon functional ATM.
Figure 4.
ATM is required for the subnuclear localization of phosphorylated (pS367)TRF1. (A) ATM inhibition leads to loss of phosphorylated (pS367)TRF1-containing foci. Indirect IF was performed with anti-pS367 antibody on HeLaI.2.11 cells treated with either DMSO or KU55933. Cell nuclei were stained with DAPI in blue. (B) Quantification of DMSO-treated or KU55933-treated HeLaI.2.11 cells showing five or more phosphorylated (pS367)TRF1-containing foci. A total of at least 1500 cells from three independent experiments were scored in blind for both DMSO-treated and KU55933-treated cells. Standard deviations from three independent experiments are indicated. (C) ATM-deficient cells lack phosphorylated (pS367)TRF1-containing foci. IF was performed with anti-pS367 antibody on IMR90 cells as well as ATM-deficient GM09607 and GM05849 cells. Cell nuclei were stained with DAPI in blue. (D) Quantification of ATM-proficient or ATM-deficient cells showing five or more phosphorylated (pS367)TRF1-containing foci. A total of at least 1500 cells from three independent experiments were scored in blind for IMR90, GM09607 and GM05849 cells. Standard deviations from three independent experiments are indicated. (E) Introduction of ATM into ATM-deficient cells restores phosphorylated (pS367)TRF1-containing foci. IF was conducted with anti-pS367 antibody on ATM-deficient cells expressing either the vector alone (GM16666) or complemented with ATM (GM16667). Cell nuclei were stained with DAPI in blue. (F) Quantification of GM16666 or GM16667 cells showing five or more phosphorylated (pS367)TRF1-containing foci. A total of 1500 cells from three independent experiments were scored. Standard deviations from three independent experiments are indicated.
The phosphomimic mutation S367D abrogates TRF1 association with telomeres and renders it susceptible to degradation
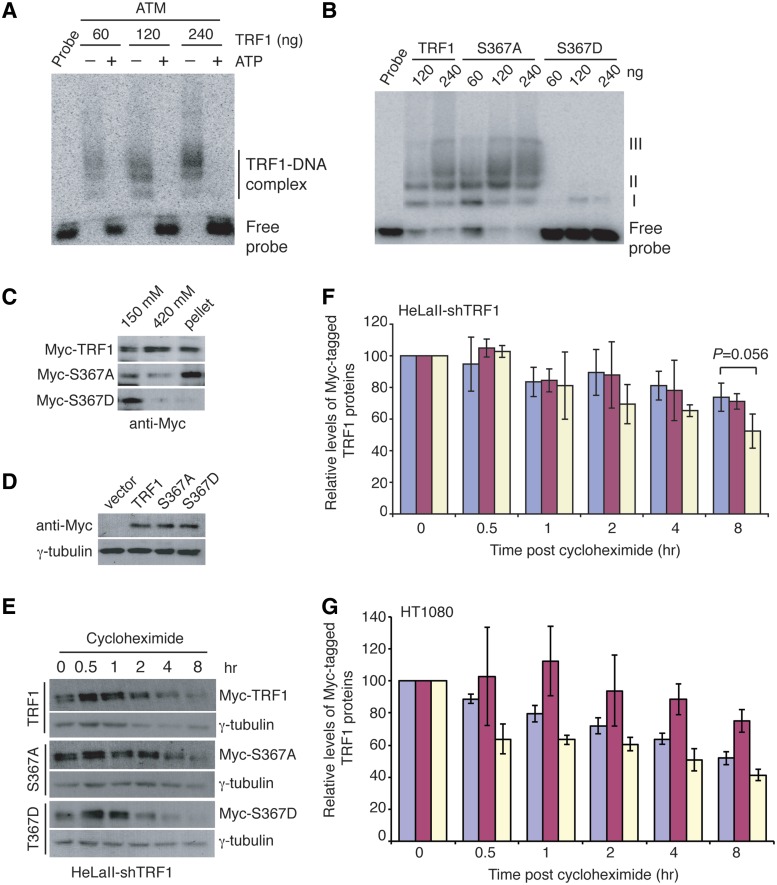
We have shown that phosphorylation of S367 by ATM renders TRF1 free of telomere chromatin in vivo. Therefore we decided to investigate whether prephosphorylation of TRF1 by ATM might affect TRF1 binding to telomeric DNA in vitro. Bacteria-derived recombinant wild-type TRF1 was pre-incubated with ATM either in the presence or absence of cold ATP prior to gel-shift assays. We found that pre-incubation of TRF1 with ATM in the presence of cold ATP completely abrogated TRF1 binding to telomeric DNA (Figure 5A), consistent with our previous findings that ATM negatively regulates TRF1 binding to telomeric DNA (26).
Figure 5.
A phosphomimic mutation of S367D not only impairs the ability of TRF1 to interact with telomeric DNA but also renders it susceptible to degradation. (A) Prephosphorylation of TRF1 by ATM abrogates its ability to bind telomeric DNA. Bacterial-derived recombinant TRF1 of varying amouts as indicated above the lanes was pre-incubated with ATM immunoprecipiated from HeLa cells in the presence or absence of cold ATP, followed by in vitro gel-shift assays. (B) TRF1-S367D is defective in binding to telomeric DNA. Bacterial-derived recombinant TRF1 proteins were used in the gel-shift assays as indicated above the lanes. The positions of three TRF1-containing complexes (I, II and III) are indicated on the right. The concentrations of recombinant TRF1 used were indicated above the lanes. (C) Differential salt (KCl) extraction of chromatin was performed on TRF1-depleted HeLaII cells expressing Myc-tagged wild-type TRF1, Myc-tagged TRF1-S367A or Myc-tagged TRF1-T367D. Immunoblotting was carried out with anti-Myc antibody. (D) Western analysis of expression of various Myc-tagged TRF1 proteins in TRF1-depleted HeLaII cells. Immunoblotting was carried out with anti-Myc or anti-γ-tubulin antibody. (E) Cycloheximide chase experiments. TRF1-depleted HeLaII cells stably expressing Myc-TRF1, Myc-TRF1-S367A or Myc-TRF1-S367D were treated with 100 µg/ml cycloheximide for the indicated times, followed by immunoblotting of the lysates with anti-Myc or anti-γ-tubulin antibody. (F) Quantification of Myc-tagged wild-type TRF1 (blue bars), Myc-tagged TRF1-S367A (burgundy bars) and Myc-tagged TRF1-S367D (light yellow bars) from (E). The signals from the western blots were quantified with densitometry. The level of Myc-tagged TRF1 proteins is represented in arbitrary units after their signals were normalized relative to those of γ-tubulin. Standard deviations from three independent experiments are indicated. (G) Quantification of cycloheximide chase experiments from HT1080 cells stably expressing Myc-tagged wild-type (blue bars), Myc-tagged TRF1-S367A (burgundy bars) and Myc-tagged TRF1-S367D (light yellow bars). Quantification was conducted as described in 5 F. Standard deviations from three independent experiments are indicated.
To further investigate the role of S367 phosphorylation in TRF1 binding and stability, we changed S367 of TRF1 to either alanine (S367A, non-phosphorylatable) or aspartic acid (S367D, phosphomimic). Using bacterial-derived recombinant wild-type TRF1, TRF1-S367A and TRF1-S367D, we performed in vitro gel-shift assays and found that while TRF1-S367A bound telomeric DNA at a level indistinguishable from wild-type TRF1, TRF1-S367D displayed a severe defect in its interaction with telomeric DNA (Figure 5B). To gain evidence that the phosphomimic mutation of S367D might also affect TRF1 association with telomeres in vivo, we performed differential salt extraction of chromatin on TRF1-depleted HeLaII cells stably expressing Myc-TRF1, Myc-TRF1-S367A or Myc-TRF1-S367D. We found that the majority of overexpressed Myc-TRF1 or Myc-TRF1-S367A was associated with chromatin whereas very little of overexpressed Myc-TRF1-S367D was associated with chromatin (Figure 5C), consistent with our earlier finding that phosphorylated (pS367)TRF1 is not associated with chromatin (Figure 2E). Expression of Myc-TRF1-S367D was comparable to that of Myc-TRF1 or Myc-TRF1-S367A (Figure 5D), suggesting that it is unlikely that the lack of Myc-TRF1-T367D association with chromatin might have arisen from a difference in protein expression. These results support the notion that S367 phosphorylation by ATM prevents TRF1 association with telomeric DNA.
To investigate whether S367 phosphorylation might be involved in regulating TRF1 stability, we used cycloheximide to inhibit protein translation in TRF1-depleted HeLaII cells stably expressing various TRF1 alleles. We found that Myc-TRF1-S367D was less stable than Myc-TRF1 or Myc-TRF1-T367A (Figure 5E). At 8 h post-cycloheximide chase, ∼50% of Myc-TRF1-S367D was degraded whereas <30% of degradation was observed for Myc-TRF1-T367A or Myc-TRF1 (Figure 5F). Myc-TRF1-S367D was also observed to be less stable than Myc-TRF1 or Myc-TRF1-S367A when overexpressed in HT1080 cells (Figure 5G). These results suggest that S367 phosphorylation by ATM renders TRF1 susceptible to protein degradation.
The subnuclear localization of phosphorylated (pS367)TRF1 is sensitive to proteasome inhibition
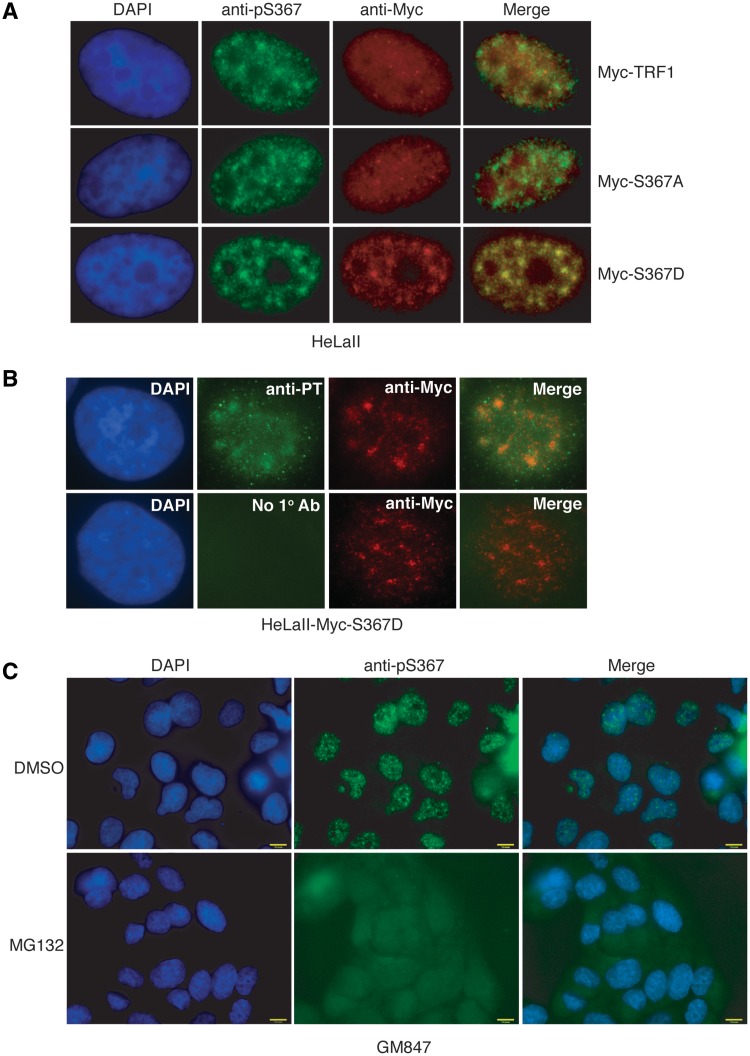
Analysis of dual indirect IF with anti-pS367 antibody in conjunction with anti-Myc antibody revealed that when transiently expressed in HeLa cells, both Myc-TRF1 and Myc-TRF1-S367A exhibited punctate nuclear staining (Figure 6A), indicative of their association with telomeric DNA (Figure 6A). While overexpressed Myc-TRF1 or Myc-TRF1-S367A did not exhibit any overlap with endogenous phosphorylated (pS367)TRF1 (Figure 6A), overexpressed Myc-TRF1-S367D was found to co-localize with phosphorylated (pS367)TRF1 (Figure 6A). Earlier we have shown that a very small amount (1–5%) of TRF1 is phosphorylated at S367 (Figure 2E) and therefore it is likely that the vast majority of overexpressed Myc-TRF1 is not phosphorylated at S367, which may account for the apparent lack of an overlap between anti-Myc and anti-pS367 staining in Myc-TRF1-overexpressing HeLaII cells. In addition, we observed an overlap between Myc-TRF1-S367D-containing foci and proteasome-containing nuclear centers (Figure 6B), suggesting that Myc-TRF1-S367D-containing foci are part of nuclear proteasome centers. Consistent with this notion, we found that treatment of cells with MG132, a potent proteasome inhibitor, completely abrogated the formation of phosphorylated (pS367)TRF1-containing foci (Figure 6C). Taken together, these results suggest that phosphorylated (pS367)TRF1-containing foci may represent the nuclear proteolytic sites for TRF1 degradation.
Figure 6.
(A) Nuclear localization of overexpressed various Myc-tagged TRF1 proteins. Dual indirect IF with anti-pS367 in conjunction with anti-Myc antibody was performed on HeLa cells transiently overexpressing Myc-TRF1, Myc-TRF1-S367A or Myc-TRF1-S367D. Cell nuclei were stained with DAPI shown in blue. (B) Overexpressed TRF1-S367D colocalizes with nuclear proteasomes. Dual IF was conducted on HeLaII cells transiently expressing Myc-S367D with anti-Myc antibody (red) in conjunction with either rabbit anti-proteasome (anti-PT) antibody (green) or no primary antibody (no 1° Ab). Cell nuclei were stained with DAPI shown in blue. (C) Phosphorylated (pS367)TRF1-containing foci are sensitive to proteasome inhibition. GM847 cells were treated with either DMSO or MG132 (12.5 µM) for 4 hr prior to IF analysis with anti-pS367 antibody. Cell nuclei were stained with DAPI shown in blue.
TRF1 carrying a phosphomimic mutation of S367D is unable to inhibit telomerase-dependent telomere lengthening
To investigate the role of S367 phosphorylation in telomere maintenance, we infected TRF1-depleted HeLaII cells with retrovirus expressing the vector pWZL alone, Myc-tagged wild-type TRF1, Myc-tagged TRF1-S367A or Myc-tagged TRF1-S367D, giving rise to four stable cell lines (shTRF1/pWZL, shTRF1/TRF1, shTRF1/S367A, shTRF1/S367D). These stable cell lines contained pools of cells and they were not single cell clones. As a control, we also generated a HeLaII cell line stably expressing two vectors pRetroSuper (pRS) and pWZL. Myc-tagged wild-type TRF1, Myc-tagged TRF1-S367A and Myc-tagged TRF1-S367D were resistant to shTRF1 due to engineered silent mutations. We chose to use TRF1-depleted cells for analysis of TRF1 mutants to minimize the interference from endogenous TRF1.
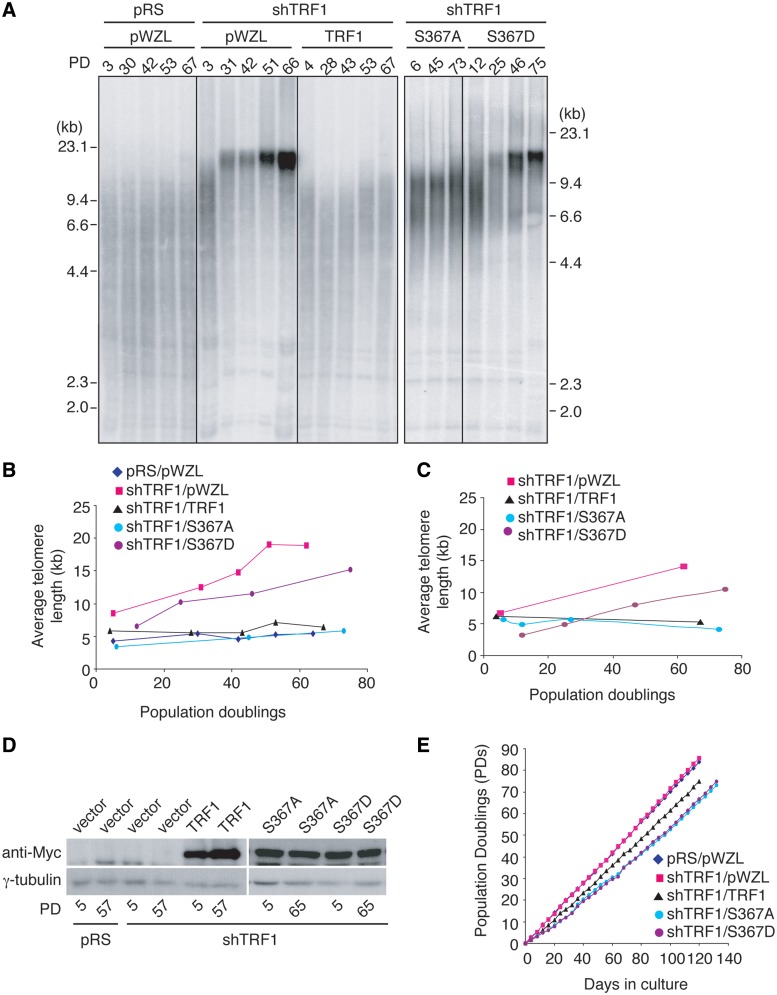
HeLaII cells stably expressing pRS/pWZL, shTRF1/pWZL, shTRF1/TRF1, shTRF1/S367A, shTRF1/S367D were cultured for over 66 population doublings (PDs). Depletion of TRF1 resulted in telomere elongation (Figure 7A and B). Introduction of shTRF1-resistant wild-type TRF1 into TRF1-depleted HeLaII cells fully reversed shTRF1-mediated telomere lengthening (Figure 7A–C). We found that TRF1-S367A was able to reverse shTRF1-mediated telomere lengthening in a manner similar to wild-type TRF1 (Figure 7A–C). In contrast, a severe defect in suppressing shTRF1-mediated telomere lengthening was reproducibly observed in TRF1-depleted cells overexpressing TRF1-S367D (Figure 7A–C). This defect was unlikely due to the lack of protein expression (Figure 7D). TRF1-S367D-expressing cells grew at a rate indistinguishable from TRF1-S367A-expressing cells (Figure 7E), arguing against the possibility that the difference in cell proliferation may account for the inability of TRF1-S367D to inhibit telomere length maintenance. These results altogether suggest that S367 phosphorylation negatively regulates the ability of TRF1 to modulate telomerase-dependent telomere lengthening.
Figure 7.
TRF1 carrying a phosphomimic mutation is defective in the inhibition of telomerase-dependent telomere lengthening. (A) Genomic blots of telomeric restriction fragments from TRF1-depleted HeLaII cells (shTRF1) expressing either the vector alone (pWZL) or various TRF1 alleles as indicated above the lanes. HeLaII cells expressing both pRS and pWZL were used as a control. PD are indicated above the lanes whereas DNA molecular weight markers are shown on both left and right of the blots. About 3 µg of RsaI/HinfI-digested genomic DNA from each sample was used for gel electrophoresis. (B) Average telomere length of indicated cell lines was plotted against PD. (C) Average telomere length of indicated cell lines was plotted against PD from a repeated experiment. (D) Western analysis of expression of various Myc-tagged TRF1 proteins in early and late PDs. Immunoblotting was performed with anti-Myc antibody. The anti-γ-tubulin blots were used as a loading control. (E) Growth curve of HeLaII cells expressing various constructs as indicated. The number of PDs was plotted against days in culture.
TRF1 carrying a phosphomimic mutation of S367D fails to suppress shTRF1-induced telomere doublets and telomere loss
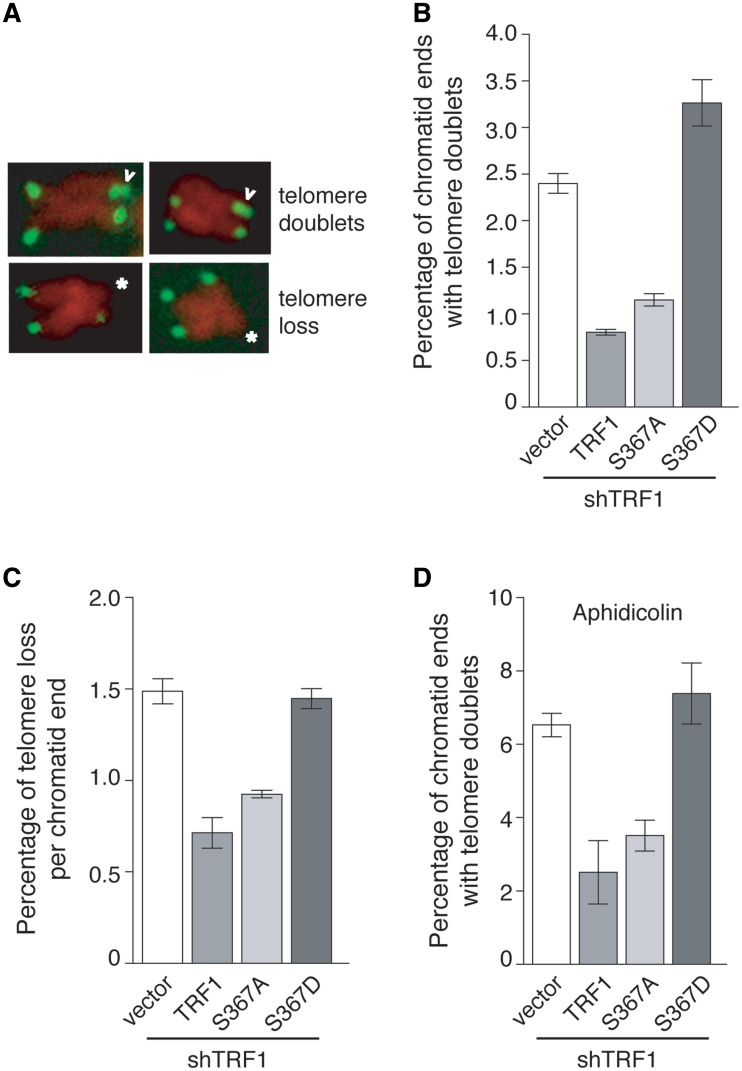
TRF1 has been shown to be important for telomere replication (6,9), the defect of which can give rise to fragile telomeres (6,9), also known as telomere doublets. Previously we have shown that depletion of TRF1 induces the formation of telomere doublets and telomere loss, both of which can be suppressed by wild-type TRF1 (12). We asked whether S367 phosphorylation might affect the ability of TRF1 to suppress these abnormalities. FISH analysis was performed on metaphase cells derived from HeLaII cells stably expressing shTRF1/pWZL, shTRF1/TRF1, shTRF1/S367A or shTRF1/S367D. Consistent with previous findings (12), we found that overexpression of wild-type TRF1 was able to suppress the formation of both telomere doublets and telomere loss in TRF1-depleted cells (Figure 8A–C). Introduction of TRF1-S367A into TRF1-depleted cells was also able to suppress these telomere abnormalities (Figure 8A–C). On the other hand, we found that overexpression of TRF1-S367D failed to result in any reduction in telomere doublets or telomere loss in TRF1-depleted HeLaII cells (Figure 8B and C). In fact, we detected a further increase in the formation of telomere doublets as a result of TRF1-S367D expression in TRF1-depleted HeLaII cells (Figure 8B). Aphidicolin, an inhibitor of DNA replication, has been shown to induce telomere doublets (6,9). We observed a substantial increase in telomere doublets in TRF1-depleted cells upon treatment with aphidicolin (Figure 8D), consistent with a previous report that the effect of aphidicolin was additive with the loss of TRF1 (9). Such a increase was also observed in TRF1-S367D-expressing cells (Figure 8D). Taken together, these results suggest that the phosphomimic mutation of S367D impairs TRF1 function in telomere replication.
Figure 8.
Overexpression of TRF1-S367D promotes the formation of telomere doublets in TRF1-depleted HeLaII cells. (A) Images of metaphase chromosomes depicting telomere abnormalities. Metaphase chromosomes were stained with DAPI and false-colored in red. Telomeric DNA was detected by FISH using a FITC-conjugated (CCCTAA)3-containing PNA probe in green. Open arrows represent telomere doublets whereas asterisks indicate telomere loss. (B) Quantification of telomere doublets from TRF1-depleted cells expressing indicated constructs. For each cell line, a total of 3150–3750 chromosomes from 60 to 69 metaphase cells were scored in a blind manner for the presence of telomere doublets in B and D and telomere loss in C. Standard deviations derived from three independent experiments are indicated. (C) Quantification of telomere loss from TRF1-depleted cells expressing indicated constructs. Telomere loss refers to chromatids without a detectable telomere signal. The total number of chromatid ends without a detectable telomere signal was divided by the total number of chromatid ends scored, giving rise to the percentage of telomere loss per chromatid end. (D) Quantification of telomere doublets from aphidicolin-treated cells. TRF1-depleted cells expressing indicated constructs were treated with aphidicolin (0.2 µM) for 16 h prior to FISH analysis.
DISCUSSION
In this report, we have demonstrated that ATM phosphorylates S367 of TRF1 both in vivo and in vitro. Using multiple approaches including IF, differential salt extraction and mutational analysis, we have shown that the phosphorylation of S367 by ATM prevents TRF1 association with telomere chromatin, consistent with previous findings that ATM acts as a negative mediator of TRF1 binding to telomeric DNA (26). These results suggest that S367 may be a major phosphorylation site for ATM to control TRF1 interaction with telomeric DNA in vivo.
Previous work suggests that TRF1 undergoes phosphorylation at S219 by ATM in response to ionizing irradiation (18). We did not observe any increase in the level of S367 phosphorylation in response to ionizing irradiation, nor any association of S367 phosphorylation with sites of DNA damage (M. McKerlie and X.-D. Zhu, unpublished data). These findings altogether suggest that ATM may regulate TRF1 function through phosphorylation at distinctive sites.
Using a phospho-specific antibody, we have shown that phosphorylated (pS367)TRF1 forms proteasome-dependent subnuclear foci. Our finding that TRF1 carrying the phosphomimic mutation of S367D accumulates in these foci and overlaps with nuclear proteasome centers suggests that these foci may represent the nuclear proteolytic sites for TRF1 degradation. Our findings are consistent with previous studies that several nuclear proteins are found to be detectable at proteasome-associated nuclear foci while being degraded including histones, splicing factor SC35, splicesomal components (U1-70 k, SmB/B′) and promyelocytic leukemia protein (PML) (44). Further studies are required to investigate the kinetics of TRF1 turnover in these centers.
Proteasomes are known to localize in both the cytoplasm and the nucleus (45). It has been reported that proteasome-mediated nuclear proteolysis takes place in distinct nucleoplasmic foci that can partially overlap with subnuclear structures such as PML and nuclear speckles (46). However, we observed little overlap between phosphorylated (pS367)TRF1-containing foci and PML bodies, nuclear speckles or Cajal bodies (M. McKerlie and X.-D. Zhu, unpublished data), suggesting that these foci may represent a novel type of subnuclear domain.
Phosphorylation plays an important role in regulating TRF1 interaction with telomeric DNA (12,26,39–41). Previously we have shown that TRF1 is phosphorylated on T371 by Cdk1 during mitosis and this phosphorylation plays an important role in the resolution of sister chromatids (12). S367 is located four amino acids upstream from T371 in the linker region immediately preceding the DNA binding domain (Myb domain) of TRF1 (Figure 1A). The fact that S367 phosphorylation prevents TRF1 from binding to telomeric DNA, along with our previous finding that phosphorylation of T371 also keeps TRF1 away from telomere chromatin (12) suggest that phosphorylation in this portion of the linker region may interfere with TRF1 binding to telomeric DNA, perhaps due to the creation of unfavorable electrostatic interactions. We have shown that S367 phosphorylation by ATM targets TRF1 to proteasome-dependent nuclear foci, which is associated with its increased susceptibility to protein degradation. This contrasts with Cdk1-mediated T371 phosphorylation, which has been shown to increase the half-life of TRF1 (12). Perhaps the temporal separation of these two phosphorylation events (at different stages of the cell cycle) may account for the differential role that these two closely positioned sites play at telomeres. These results highlight the tight regulation of TRF1 function and the important role of post-translational modifications in this control.
We have shown that phosphorylated (pS367)TRF1-containing foci are cell cycle regulated, appearing predominantly in S and G2 phases. ATM has been shown to be associated with human telomeres in a cell cycle regulated manner, peaking in S and G2 phases (47,48). Perhaps when ATM is recruited to telomeres in S and G2 phase, it phosphorylates TRF1 at S367, promoting its dissociation from telomeric DNA and its accumulation in proteasome-dependent subnuclear domains. However, we did not observe any significant change in the level of S367 phosphorylation throughout the cell cycle (data not shown), raising the possibility that phosphorylation of S367 by ATM per se may not be cell cycle regulated. How phosphorylated (pS367)TRF1-containing foci are predominantly formed in S and G2 phases requires further investigation.
We have shown that TRF1 carrying the phosphomimic mutation of S367D (TRF1-S367D) fails to suppress the formation of telomere doublets and telomere loss in TRF1-depleted cells. While telomere doublets are thought to arise from a defect in telomere replication (6,9), telomere loss can arise from homologous recombination-mediated events (8). Our findings suggest that S367 phosphorylation might play a role in modulating the pathway(s) responsible for the formation of telomere doublets or telomere loss.
TRF1 is required for efficient telomere replication (6,9); however, it needs to be removed from telomeres to support telomerase-dependent telomere extension (15–17,26). We have shown that TRF1-S367D is not only defective in supporting telomere replication but also unable to inhibit telomerase-dependent telomere extension. We have estimated that 1–5% of endogenous TRF1 is phosphorylated at S367 in a cell cycle-dependent manner. These results raise the possibility that the action of ATM on S367 of TRF1 may occur only at a few telomeres, which might have a severe replication defect and require the repair from telomerase. Taken together, these results suggest that S367 phosphorylation by ATM is important for the regulation of telomere length and stability.
FUNDING
Ontario Early Researcher Award (ER07-04-157), Canadian Institutes of Health Research New Investigator Award (MSH-76599) and Canadian Institutes of Health Research (MOP-86620) (to X.-D.Z.) Funding for open access charge: Canadian Institutes of Health Research (MOP-86620).
Conflict of interest statement. None declared.
ACKNOWLDEGEMENTS
We are grateful for Titia de Lange (Rockefeller University) for providing various reagents, including TRF1 cDNA, retroviral vectors, antibodies to TRF1 as wells as cell lines HeLaI.2.11, HeLaII and phoenix. Yili Wu and Kajaparan Jeyanthan are thanked for immunoprecipitation of Flag-tagged TRF1 and bacterial expression of recombinant wild-type TRF1. We thank John R. Walker for providing critical comments.
REFERENCES
- 1.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 2.Liu D, O'Connor MS, Qin J, Songyang Z. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J. Biol. Chem. 2004;279:51338–51342. doi: 10.1074/jbc.M409293200. [DOI] [PubMed] [Google Scholar]
- 3.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 4.van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 5.Iwano T, Tachibana M, Reth M, Shinkai Y. Importance of TRF1 for functional telomere structure. J. Biol. Chem. 2004;279:1442–1448. doi: 10.1074/jbc.M309138200. [DOI] [PubMed] [Google Scholar]
- 6.Martinez P, Thanasoula M, Munoz P, Liao C, Tejera A, McNees C, Flores JM, Fernandez-Capetillo O, Tarsounas M, Blasco MA. Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev. 2009;23:2060–2075. doi: 10.1101/gad.543509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell. 2009;35:403–413. doi: 10.1016/j.molcel.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang RC, Smogorzewska A, de Lange T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004;119:355–368. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 9.Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitchell TR, Glenfield K, Jeyanthan K, Zhu XD. Arginine methylation regulates telomere length and stability. Mol. Cell. Biol. 2009;29:4918–4934. doi: 10.1128/MCB.00009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat. Cell. Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- 12.McKerlie M, Zhu XD. Cyclin B-dependent kinase 1 regulates human TRF1 to modulate the resolution of sister telomeres. Nat. Commun. 2011;2:371. doi: 10.1038/ncomms1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr. Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- 14.Chong L, van Steensel B, Broccoli D, Erdjument-Bromage H, Hanish J, Tempst P, de Lange T. A human telomeric protein. Science. 1995;270:1663–1667. doi: 10.1126/science.270.5242.1663. [DOI] [PubMed] [Google Scholar]
- 15.van Steensel B, de Lange T. Control of telomere length by the human telomeric protein TRF1 [see comments] Nature. 1997;385:740–743. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- 16.Smogorzewska A, van Steensel B, Bianchi A, Oelmann S, Schaefer MR, Schnapp G, de Lange T. Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol. 2000;20:1659–1668. doi: 10.1128/mcb.20.5.1659-1668.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ancelin K, Brunori M, Bauwens S, Koering CE, Brun C, Ricoul M, Pommier JP, Sabatier L, Gilson E. Targeting assay to study the cis functions of human telomeric proteins: evidence for inhibition of telomerase by TRF1 and for activation of telomere degradation by TRF2. Mol. Cell. Biol. 2002;22:3474–3487. doi: 10.1128/MCB.22.10.3474-3487.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kishi S, Zhou XZ, Ziv Y, Khoo C, Hill DE, Shiloh Y, Lu KP. Telomeric protein Pin2/TRF1 as an important ATM target in response to double strand DNA breaks. J. Biol. Chem. 2001;276:29282–29291. doi: 10.1074/jbc.M011534200. [DOI] [PubMed] [Google Scholar]
- 19.Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle DA, Smith S, Uziel T, Sfez S, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 20.Metcalfe JA, Parkhill J, Campbell L, Stacey M, Biggs P, Byrd PJ, Taylor AM. Accelerated telomere shortening in ataxia telangiectasia. Nat. Genet. 1996;13:350–353. doi: 10.1038/ng0796-350. [DOI] [PubMed] [Google Scholar]
- 21.Ranganathan V, Heine WF, Ciccone DN, Rudolph KL, Wu X, Chang S, Hai H, Ahearn IM, Livingston DM, Resnick I, et al. Rescue of a telomere length defect of Nijmegen breakage syndrome cells requires NBS and telomerase catalytic subunit. Curr. Biol. 2001;11:962–966. doi: 10.1016/s0960-9822(01)00267-6. [DOI] [PubMed] [Google Scholar]
- 22.Pandita TK. ATM function and telomere stability. Oncogene. 2002;21:611–618. doi: 10.1038/sj.onc.1205060. [DOI] [PubMed] [Google Scholar]
- 23.Hande MP, Balajee AS, Tchirkov A, Wynshaw-Boris A, Lansdorp PM. Extra-chromosomal telomeric DNA in cells from Atm(-/-) mice and patients with ataxia-telangiectasia. Hum. Mol. Genet. 2001;10:519–528. doi: 10.1093/hmg/10.5.519. [DOI] [PubMed] [Google Scholar]
- 24.Pandita TK, Pathak S, Geard CR. Chromosome end associations, telomeres and telomerase activity in ataxia telangiectasia cells. Cytogenet. Cell. Genet. 1995;71:86–93. doi: 10.1159/000134069. [DOI] [PubMed] [Google Scholar]
- 25.Smilenov LB, Morgan SE, Mellado W, Sawant SG, Kastan MB, Pandita TK. Influence of ATM function on telomere metabolism. Oncogene. 1997;15:2659–2665. doi: 10.1038/sj.onc.1201449. [DOI] [PubMed] [Google Scholar]
- 26.Wu Y, Xiao S, Zhu XD. MRE11-RAD50-NBS1 and ATM function as co-mediators of TRF1 in telomere length control. Nat. Struct. Mol. Biol. 2007;14:832–840. doi: 10.1038/nsmb1286. [DOI] [PubMed] [Google Scholar]
- 27.Wu Y, Zacal NJ, Rainbow AJ, Zhu XD. XPF with mutations in its conserved nuclease domain is defective in DNA repair but functions in TRF2-mediated telomere shortening. DNA Repair. 2007;6:157–166. doi: 10.1016/j.dnarep.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 28.Sheffield P, Garrard S, Derewenda Z. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr. Purif. 1999;15:34–39. doi: 10.1006/prep.1998.1003. [DOI] [PubMed] [Google Scholar]
- 29.Ziv Y, Bar-Shira A, Pecker I, Russell P, Jorgensen TJ, Tsarfati I, Shiloh Y. Recombinant ATM protein complements the cellular A-T phenotype. Oncogene. 1997;15:159–167. doi: 10.1038/sj.onc.1201319. [DOI] [PubMed] [Google Scholar]
- 30.Saltman D, Morgan R, Cleary ML, de Lange T. Telomeric structure in cells with chromosome end associations. Chromosoma. 1993;102:121–128. doi: 10.1007/BF00356029. [DOI] [PubMed] [Google Scholar]
- 31.Karlseder J, Smogorzewska A, de Lange T. Senescence induced by altered telomere state, not telomere loss. Science. 2002;295:2446–2449. doi: 10.1126/science.1069523. [DOI] [PubMed] [Google Scholar]
- 32.Zhu XD, Niedernhofer L, Kuster B, Mann M, Hoeijmakers JH, de Lange T. ERCC1/XPF removes the 3' overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol. Cell. 2003;12:1489–1498. doi: 10.1016/s1097-2765(03)00478-7. [DOI] [PubMed] [Google Scholar]
- 33.Wu Y, Mitchell TR, Zhu XD. Human XPF controls TRF2 and telomere length maintenance through distinctive mechanisms. Mech. Ageing Dev. 2008;129:602–610. doi: 10.1016/j.mad.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 34.Zhu XD, Kuster B, Mann M, Petrini JH, Lange T. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat. Genet. 2000;25:347–352. doi: 10.1038/77139. [DOI] [PubMed] [Google Scholar]
- 35.Ye JZ, de Lange T. TIN2 is a tankyrase 1 PARP modulator in the TRF1 telomere length control complex. Nat. Genet. 2004;36:618–623. doi: 10.1038/ng1360. [DOI] [PubMed] [Google Scholar]
- 36.Lansdorp PM, Verwoerd NP, van de Rijke FM, Dragowska V, Little MT, Dirks RW, Raap AK, Tanke HJ. Heterogeneity in telomere length of human chromosomes. Hum. Mol. Genet. 1996;5:685–691. doi: 10.1093/hmg/5.5.685. [DOI] [PubMed] [Google Scholar]
- 37.Bianchi A, Smith S, Chong L, Elias P, de Lange T. TRF1 is a dimer and bends telomeric DNA. EMBO J. 1997;16:1785–1794. doi: 10.1093/emboj/16.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li B, de Lange T. Rap1 affects the length and heterogeneity of human telomeres. Mol. Biol. Cell. 2003;14:5060–5068. doi: 10.1091/mbc.E03-06-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu ZQ, Yang X, Weber G, Liu X. Plk1 phosphorylation of TRF1 is essential for its binding to telomeres. J. Biol. Chem. 2008;283:25503–25513. doi: 10.1074/jbc.M803304200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee TH, Tun-Kyi A, Shi R, Lim J, Soohoo C, Finn G, Balastik M, Pastorino L, Wulf G, Zhou XZ, et al. Essential role of Pin1 in the regulation of TRF1 stability and telomere maintenance. Nat. Cell. Biol. 2009;11:97–105. doi: 10.1038/ncb1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim MK, Kang MR, Nam HW, Bae YS, Kim YS, Chung IK. Regulation of telomeric repeat binding factor 1 binding to telomeres by casein kinase 2-mediated phosphorylation. J. Biol. Chem. 2008;283:14144–14152. doi: 10.1074/jbc.M710065200. [DOI] [PubMed] [Google Scholar]
- 42.Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GC. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 43.Veuger SJ, Curtin NJ, Richardson CJ, Smith GC, Durkacz BW. Radiosensitization and DNA repair inhibition by the combined use of novel inhibitors of DNA-dependent protein kinase and poly(ADP-ribose) polymerase-1. Cancer Res. 2003;63:6008–6015. [PubMed] [Google Scholar]
- 44.Dino Rockel T, von Mikecz A. Proteasome-dependent processing of nuclear proteins is correlated with their subnuclear localization. J. Struct. Biol. 2002;140:189–199. doi: 10.1016/s1047-8477(02)00527-0. [DOI] [PubMed] [Google Scholar]
- 45.Wojcik C, DeMartino GN. Intracellular localization of proteasomes. Int. J. Biochem. Cell. Biol. 2003;35:579–589. doi: 10.1016/s1357-2725(02)00380-1. [DOI] [PubMed] [Google Scholar]
- 46.Rockel TD, Stuhlmann D, von Mikecz A. Proteasomes degrade proteins in focal subdomains of the human cell nucleus. J. Cell. Sci. 2005;118:5231–5242. doi: 10.1242/jcs.02642. [DOI] [PubMed] [Google Scholar]
- 47.Verdun RE, Crabbe L, Haggblom C, Karlseder J. Functional human telomeres are recognized as DNA damage in G2 of the cell cycle. Mol. Cell. 2005;20:551–561. doi: 10.1016/j.molcel.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 48.Verdun RE, Karlseder J. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell. 2006;127:709–720. doi: 10.1016/j.cell.2006.09.034. [DOI] [PubMed] [Google Scholar]