Abstract
A simple approach for creating libraries of circularly permuted proteins is described that is called PERMutation Using Transposase Engineering (PERMUTE). In PERMUTE, the transposase MuA is used to randomly insert a minitransposon that can function as a protein expression vector into a plasmid that contains the open reading frame (ORF) being permuted. A library of vectors that express different permuted variants of the ORF-encoded protein is created by: (i) using bacteria to select for target vectors that acquire an integrated minitransposon; (ii) excising the ensemble of ORFs that contain an integrated minitransposon from the selected vectors; and (iii) circularizing the ensemble of ORFs containing integrated minitransposons using intramolecular ligation. Construction of a Thermotoga neapolitana adenylate kinase (AK) library using PERMUTE revealed that this approach produces vectors that express circularly permuted proteins with distinct sequence diversity from existing methods. In addition, selection of this library for variants that complement the growth of Escherichia coli with a temperature-sensitive AK identified functional proteins with novel architectures, suggesting that PERMUTE will be useful for the directed evolution of proteins with new functions.
INTRODUCTION
In nature, chromosomal rearrangements can break genes into pieces and rearrange their coding sequence so that they have architectures that are circularly permuted (1,2). At the protein level, this permutation leads to the covalent attachment of a protein's original termini, the creation of new termini elsewhere in the primary sequence, and altered contact order in the tertiary structure. In the laboratory, circularly permuted proteins have been created to study how changes in protein contact order affect topology (3), thermostability (4), oligomerization (5), ligand binding (6), catalytic activity (7), folding rates (8) and folding pathways (9). More recently, libraries of circularly permuted proteins have been constructed and used for laboratory evolution to engineer proteins with novel functions (10). Selections and screens of these libraries have yielded proteins with increased catalytic activity (11), altered fluorescence (12), decreased proteolytic susceptibility (13) and enhanced crystallization (14). Libraries of circularly permuted proteins also have the potential to accelerate the construction of biosensors and molecular switches for synthetic biology (15). Domain insertion studies have revealed that the functions of two domains can be allosterically coupled when circularly permuted variants of one domain are inserted at different locations within the primary sequence of a second domain (16).
Libraries of vectors that express circularly permuted variants of a protein are typically constructed by digesting a closed circular gene with the non-specific nuclease DNAse I, whose activity is hard to control (17,18). This reaction yields an ensemble of linear permuted genes with an assortment of termini (single stranded and blunt) and internal nicks (17,18), because DNAse I catalyzes both double-stranded breaks and single-stranded nicks (19). To facilitate cloning into expression vectors, linear genes generated by DNAse I digestion are treated with DNA ligase and polymerase which repair nicks and blunt termini. After repair, a majority of the DNAse-digested genes encode proteins with deletions of primary sequence proximal to their new termini, and many of the genes lacking deletions contain sequence duplications (20,21). These deletions and duplications vary in size, so the sequence diversity in these libraries is the product of the number of possible permuted variants and the number of deletions and duplications that are layered onto each permuted variant.
One way to minimize deletions and duplications when fragmenting a circular gene is to randomly insert a unique restriction site into the gene using a transposase and digest the products of the transposase reaction at the inserted restriction site (22). Transposases have been leveraged to introduce a diverse array of mutations into proteins, including tripeptide insertions (23), single amino acid deletions (24), truncations (25), hexahistidine insertions (26) and single amino acid substitutions (27,28). In addition, transposases have been used to construct domain insertion libraries (29) and libraries that express fragmented protein variants (30). Herein, we present a new method termed PERMutation Using Transposase Engineering (PERMUTE) that leverages transposase-mediated gene fragmentation to create a combinatorial library of vectors that express circularly permuted variants of a protein. We demonstrate that PERMUTE produces protein variants with distinct sequence diversity from the existing approach used to build libraries (17,18), and we show that PERMUTE can be coupled to a bacterial selection to discover circularly permuted variants of an enzyme that retain catalytic activity.
MATERIALS AND METHODS
Materials
Escherichia coli XL1-Blue was from Stratagene, E. coli MegaX DH10B was from Invitrogen and E. coli CV2 (31) was from the Yale Coli Genetic Stock Center. Synthetic oligonucleotides were from Integrated DNA Technologies. Kits for DNA purification were from Qiagen and Zymo Research. All other enzymes were from Epicentre Biotechnologies and New England Biolabs.
Construction of the target vector
A temperature-sensitive origin of replication (repAts) and chloramphenicol acetyltransferase gene (cat) were PCR amplified from pKO3 (32) using Vent Polymerase and primers that add NotI restrictions sites at the termini of the amplicon. This amplicon was digested with NotI and self-ligated to generate pKO3-NotI. The adk gene encoding Thermotoga neapolitana adenylate kinase (TnAK) was PCR amplified from pTNAK2::Km (33) using Vent Polymerase and primers that add a single adenine before the start codon, remove the stop codon and incorporate flanking NotI restriction sites on both sides of the gene. This amplicon was digested with NotI and subcloned into pKO3-NotI to create pMM1, whose sequences are provided in the Supplementary Data.
Minitransposon synthesis
DNA containing the pBR322 origin of replication and kanamycin nucleotidyltransferase resistance (kanR) cassette was PCR amplified from pET-24d using primers that add a ribosomal binding site and start codon adjacent to the kanR, a transcription terminator adjacent to the origin of replication and portions of the MuA-binding sites (R1R2 and R2R1) at both termini (22). The resulting PCR product was used as template for a second amplification reaction, which added the full MuA binding sites flanked by BglII restriction sites to both ends of the DNA. This synthetic minitransposon was digested with BglII and ligated to adk flanked by BglII sites to create pMT2, a vector that expresses full-length TnAK. To obtain minitransposon to use in transposase reactions, pMT2 was digested with BglII, the minitransposon was separated from adk and uncut vector using agarose gel electrophoresis, and the minitransposon was isolated and purified using a Zymo Gel DNA Recover kit. pMT2 and minitransposon sequences are provided in the Supplementary Data.
Library construction
Minitransposon insertion reactions (20 µl) containing HyperMu buffer, 300 ng of pMM1, 100 ng minitransposon and 1 U of HyperMu (Epicentre) were incubated at 37°C for 16 h. Reactions were terminated by adding 2 µl of HyperMu 10× Stop Solution, gently mixing and incubating each reaction at 70°C for 10 min. Total DNA was purified using a Zymo DNA Clean & Concentrator kit and electroporated into DH10B E. coli. Cells were allowed to recover for 1 h at 37°C, plated onto LB agar medium containing 15 µg/ml chloramphenicol and 25 µg/ml kanamycin, and incubated for 24 h at 30 and 43°C, respectively. Libraries selected at 30°C (MM1-MiniT-30 library) and 43°C (MM1-MiniT-43 library) were harvested by scraping plates, pooling cells and purifying DNA using a Qiagen Miniprep kit. To obtain adk genes containing a minitransposon integrated at different locations, the MM1-MiniT1-43 library (100 ng) was digested with NotI, the digestion products were separated using agarose gel electrophoresis and the band having a molecular weight corresponding to adk with a single integrated minitransposon was purified using a Zymoclean Gel DNA Recovery Kit. The size-selected DNA was circularized through ligation using T4 DNA ligase, desalted using a Zymo DNA Clean & Concentrator kit and electroporated into E. coli DH10B. Cells were allowed to recover for 60 min at 37°C, spread onto multiple LB agar plates containing 25 µg/ml kanamycin and incubated for 24 h. DNA was purified from the cells harvested from plates, and transformed into E. coli CV2 using electroporation. After allowing cells to recover for 10–60 min, cells were spread onto LB agar plates containing 15 or 50 µg/ml kanamycin and incubated at 40°C for 48–72 h. Colonies selected at 40°C were used to inoculate LB liquid cultures (4 ml) containing 25 µg/ml kanamycin, plasmid DNA was purified using a Qiagen Miniprep kit and functional permuted AK were identified through DNA sequencing.
Library sequence diversity was estimated by simulating random sampling of a population consisting of target vectors harboring a single inserted minitransposon at different locations. This calculation was repeated 1000 times, and the mean value was used to estimate library sampling at each step of the procedure. The number of possible variants used for our simulations (5500) was lower than the maximal number of variants created by MuA-mediated insertion reaction (6828 = number bp in target vector × two possible minitransposon orientations) because only a subset of all possible target vector variants were expected to produce colonies on agar plates. Twenty percent of the possible insertion sites within the target vector occur within the cat gene and promoter, whose functions was predicted to be disrupted by minitransposon insertion, thereby preventing these variants from yielding colonies on agar plates during library construction. In each simulation, the number of vector variants sampled in our protocol was the number of colony forming units observed on agar plates corrected for the number of doublings that occurred during the outgrowth portion of the transformation protocol.
Complementation strength
Each sequenced plasmid was transformed into E. coli CV2 and cells were grown on LB agar plates containing kanamycin (50 µg/ml) and incubated at 30°C for 24 h. Single colonies were used to inoculate LB liquid cultures containing 25 µg/ml kanamycin, and cultures were grown for 18–24 h to a stationary phase at 30°C. Cells were diluted to an A600 = 2, serial dilutions (1×, 10×, 100× and 1000×) of cells (10 µl) were spotted LB plates, and plates were incubated at 30 and 40°C, respectively. After 24 h, growth at each spot was imaged (Supplementary Figures S1–S17). Linear minitransposon was circularized through ligation and used as a negative control, and a minitransposon expressing native TnAK was used as a positive control (pMT2). Experiments were performed in triplicate using three distinct colonies of E. coli CV2 transformed with each sequenced vector.
RESULTS
Strategy for creating libraries
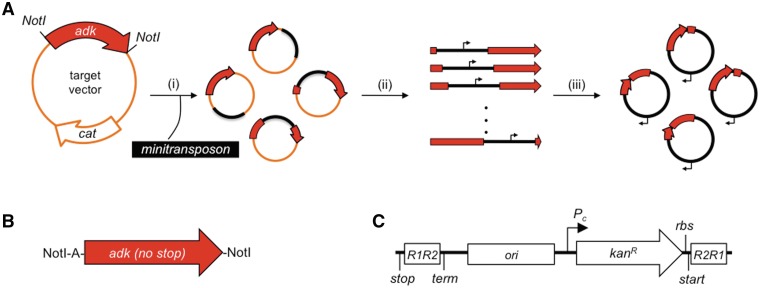
Figure 1A illustrates how PERMUTE creates libraries of vectors that express circularly permutated variants of a protein. First, the gene encoding the protein of interest, adk herein, is cloned into a vector containing a chloramphenicol-resistance cassette (catR) and a temperature-sensitive origin of replication (repAts) (32). In this vector, the target adk gene is flanked at the 5′-end by an adenine and NotI site (Figure 1B), whereas the 3′-end lacks a stop codon and is abutted by a NotI site. These flanking sequences are destined to encode the tripeptide linker (Ala-Ala-Ala) that connects the N- and C-termini of the target protein in each of the variants produced by PERMUTE. Second, the transposase MuA is used to randomly integrate an engineered minitransposon into the target plasmid containing the gene being permuted. The minitransposon developed for PERMUTE (Figure 1C) contains all of the attributes of a bacterial protein expression vector, including (i) an origin of replication, (ii) an antibiotic selection marker, (iii) a promoter for driving the transcription of permuted genes, (iv) a ribosomal-binding site to initiate translation of the permuted proteins, (v) a stop codon to terminate translation of permuted proteins and (vi) a terminator for ending transcription of permuted genes. Third, the ensemble of target vectors containing an integrated minitransposon are selectively amplified by transforming the DNA products from the MuA reaction into E. coli and growing cells under conditions where the target vector does not replicate efficiently unless it contains an integrated minitransposon. Finally, adk genes harboring a minitransposon are excised from the vector ensemble using NotI, size selected using agarose gel electrophoresis and circularized through intramolecular ligation.
Figure 1.
Scheme for creating PERMUTE libraries. (A) In this method: (i) MuA integrates a synthetic minitransposon (black) into a target vector (orange) containing the adk gene (red), (ii) adk genes harboring an integrated minitransposon are excised from modified target vectors using NotI and (iii) these genes are self-ligated to create a library of vectors that express the different circularly permuted AK variants. (B) The target gene lacks a stop codon and is flanked by identical restriction sites (NotI), which become joined within the circularly permuted genes, where they encode the linker that connects the original N- and C-termini of TnAK. An additional adenine was inserted between the initial NotI site and the adk gene to keep the linker in frame upon permutation. Larger linkers could be incorporated by adding additional codons between the NotI and adenine. (C) The minitransposon contains a stop codon (stop), MuA recognition sites (R1R2 and R2R1), a terminator (term), a pBR322-derived origin of replication (ori), a constitutive promoter (Pc) a kanR selectable marker, a ribosomal binding site (rbs) and a start codon (start).
Library sequence diversity
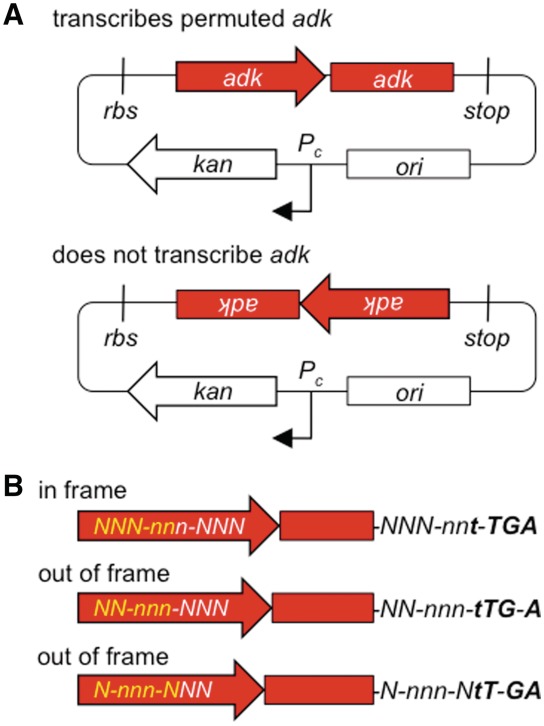
For a target gene encoding a protein of length N, PERMUTE generates up to 6N unique vectors. This occurs because the synthetic minitransposon can be integrated into the target gene in two orientations after each base pair (Figure 2A). Only one of these orientations has the minitransposon oriented so that the target gene is transcribed. The minitransposon can also be integrated at different locations within each codon of the target gene (Figure 2B). Among the vectors with the minitransposon integrated in an orientation that leads to transcription of the target gene, only one-third of the possible vectors have a minitransposon integrated in the codon frame and orientation that leads to translation of a circularly permuted protein. This subset of vectors expresses permuted proteins with a peptide (MGFRIYRETLSRFSCAAQ) fused to their N-terminus, because translation is initiated within the minitransposon before the MuA-binding site (R2R1) that precedes the permuted gene. Two residues are also added to the C-terminus of these circularly permuted proteins, whose identity depends on the location of minitransposon integration within the original gene. Among the other vectors that transcribe the permuted gene, the target gene is out of frame with respect to the start codon. A majority of these vectors are not expected to express a circularly permuted protein. However, some of these vectors could express portions of the target protein using alternative start codons or translational frameshifting (34).
Figure 2.
Vector types present in a PERMUTE library. (A) The minitransposon can be integrated in two orientations within the target vector, only one of which is expected to transcribe the permuted adk genes (top) using the promoter Pc. (B) Among the vectors that transcribe permuted adk, one-third are predicted to have their codons (NNN and nnn) in frame such that they translate a circularly permuted TnAK. The initial five base pairs of each permuted gene (yellow) are duplicated and fused to the 3′-end of each gene by the minitransposon integration reaction. The first four base pairs of the integrated minitransposon (Figure 1C) that become fused to circularly permuted genes are shown in bold, illustrating how the stop codon (TGA) is only in frame within the vectors that transcribe and translate circularly permuted proteins.
Construction of an AK library
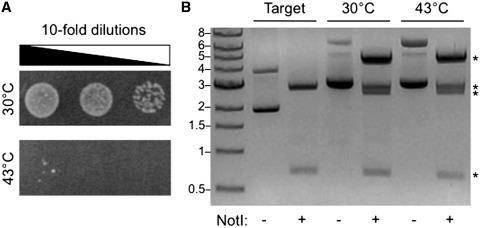
We tested PERMUTE by applying it to AK from Thermotoga neapolitana (33), a thermostable AK whose phosphotransferase activity (ADP + ADP ⇔ AMP + ATP) can be assessed using E. coli complementation (35). To obtain a target vector for performing PERMUTE, we cloned the gene encoding TnAK into a vector with a temperature-sensitive origin of replication (32). Figure 3A shows that the target vector containing the TnAK gene complements E. coli growth on LB agar medium containing chloramphenicol at 30°C but not at 43°C. To obtain an ensemble of target vectors containing an integrated minitransposon, this target vector was incubated with MuA and the synthetic minitransposon (Figure 1C), the DNA products of this reaction were transformed in E. coli and cells were grown on LB agar plates containing chloramphenicol and kanamycin at a temperature (43°C) where the target vector does not confer resistance to chloramphenicol. Plating two-thirds of a single transformation reaction yielded approximately 9000 colonies. Assuming the cells doubled once during the outgrowth after transformation, simulation of our procedure estimated that our single transformation sampled 6750 variants. This number is greater than the total number of possible vectors that can be created by random minitransposon insertion into the target vector (n = 5500), which we calculated as the number of possible insertion sites in the target vector that do not disrupt chloramphenicol resistance times the number of possible minitransposon insertion orientations at each site. Using these values for sample size and number of possible unique variants, we estimated that our reaction sampled 71% of the permuted AK variants. Furthermore, we calculated that this sampling could be increased to 91% by simply running two insertion reactions in parallel and >99% by running four reactions in parallel.
Figure 3.
Temperature selection of the target vector. (A) Cells transformed with the target vector (pMM1) complement bacterial growth at 30°C but not 43°C on LB agar medium containing chloramphenicol (34 µg/ml). (B) NotI treatment of the target vector before and after performing the MuA reaction. For MuA reaction products, NotI digestion was performed on DNA that was purified from E. coli that had been selected for growth at 30 or 43°C on LB agar plates containing kanamycin (25 µg/ml) and chloramphenicol (15 µg/ml). NotI cleaved the target vector into two products: adk (669 bp) and target vector backbone (2745 bp). In contrast, NotI digestion of MuA reaction products amplified in E. coli at 30 and 43°C yielded four products (asterisk), whose weights correspond to adk (669 bp), adk with a single integrated minitransposon (2483 bp), target vector backbone (2745 bp) and target vector backbone containing a single integrated minitransposon (4559). A band corresponding to the minitransposon alone (1809 bp) was not observed.
To determine if the colonies selected at 43°C were enriched in target vectors containing a single integrated minitransposon, we harvested colonies from plates, purified plasmids from the mixed colonies and treated the purified DNA with NotI, a restriction endonuclease that cuts adjacent to both termini of the parental adk gene (Figure 1A). Figure 3B shows that NotI digestion produced four distinct bands. These bands occur at molecular weights consistent with that expected from an ensemble of target vectors containing a single minitransposon integrated at different locations, including bands corresponding to adk (669 bp), adk containing a single integrated minitransposon (2483 bp), target vector backbone (2745 bp) and target vector backbones containing an integrated minitransposon (4559 bp). This can be contrasted with NotI digestion of purified target vector, which yielded two bands at the molecular weights expected for adk and the target vector backbone. To create the final PERMUTE library, adk with an integrated minitransposon was excised from the agarose gel, purified, and circularized through ligation. Transformation of the ligation products into E. coli yielded more than 100 000 colonies on LB agar plates containing kanamycin, from which the final vector library was purified. Simulation of the full PERMUTE protocol using colony counts after each step involving a transformation estimated that the final library contained 70% of the possible permuted AK variants.
To determine if the temperature selection of the transposase reaction at 43°C affects the yield of target vector containing an integrated minitransposon, we also transformed the DNA from our transposase reaction into E. coli, plated cells on LB agar medium containing chloramphenicol and kanamycin and selected for bacterial growth at 30°C. This low temperature selection yielded a similar number of colonies after 48 h as selections performed at 43°C. In addition, Figure 3B shows that NotI restriction digestion of the library purified from colonies grown at 30°C yields four bands with molecular weights and relative intensities similar to that observed when digesting DNA purified from cells selected at 43°C.
Characterization of the unselected library
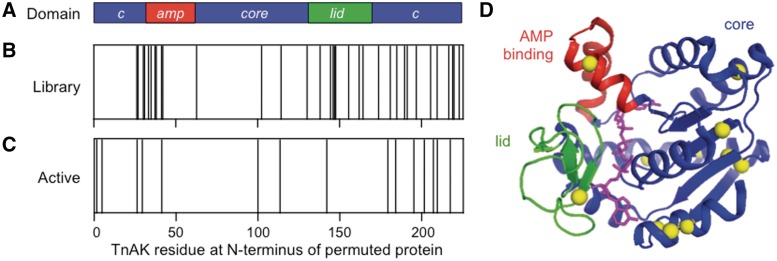
To evaluate how the diversity of vectors created by PERMUTE relates to the TnAK domain structure (Figure 4A), we sequenced individual clones from our final unselected library. Figure 4B shows that among the vectors successfully sequenced (n = 55), unique permuted adk were discovered whose first codon corresponds to residues within all three domains (AMP binding, core and lid) of the parental TnAK. Insertion sites were also observed in all three possible codon frames (with 21% in frame and 79% out of frame) in these unselected variants. In addition, minitransposons were observed in both possible orientations relative to the permuted adk genes (Figure 2B), with 36% inserted in the orientation that allows for productive transcription of permuted variants and 64% in the opposite orientation.
Figure 4.
Sequences of permuted TnAK from the unselected and selected libraries. (A) Color coding of the core (blue) AMP binding (red) and lid (green) domains in TnAK sequence helps the reader map the location of new N- and C-termini in the permuted variants. (B) The TnAK residues encoded by the first codon in unselected adk are indicated with a line, as well as (C) selected variants that complement E. coli CV2. (D) The TnAK residues encoded by the first codon in each functional variant (yellow spheres) are mapped onto the structure of Bacillus subtilis AK (36), which displays 48% sequence identity with TnAK. The substrate analog P1,P5-di(adenosine-5) pentaphosphate is shown in magenta. The image was created using PyMOL.
Selection of functional AK
Our final library was mined for functional TnAK by selecting for variants that complement E. coli CV2 at 40°C on LB agar plates containing kanamycin. Colonies obtained from this selection were used to inoculate LB–kanamycin cultures, and plasmids encoding the complementing TnAK were purified and sequenced. Selected vectors were also rescreened for activity by transforming sequenced vectors into E. coli CV2 and assessing complementation strength on LB agar plates at 40°C as previously described (30). Sequence analysis of complementing vectors revealed that our library contains diverse functional permuted TnAK (Table 1). All the vectors that complemented E. coli CV2 at 40°C contain an adk with a single minitransposon integrated in the orientation that drives transcription (upper vector, Figure 2A), compared with 40% in the unselected library. In addition, the vectors encoding active permuted TnAK all have minitransposons integrated within a codon frame that leads to translation of the permuted TnAK encoded by the vector (upper gene, Figure 2B), compared with 21% in the unselected library. Statistical analysis of this frame distribution in selected variants indicates a significant difference from the distribution observed in our unselected variants (binomial test, P < 0.05). Figure 4C and D compare the architecture of the functional permuted TnAK with the domain structure of a native AK. While a majority of the circularly permuted TnAK have primary sequences that begin with residues in the core domain, variants were identified whose sequences start with residues in the mobile lid and AMP binding domains.
Table 1.
Circularly permuted TnAK that retain in vivo phosphotransferase activity
| Gene sequence (base pairs from adk) | Primary sequence (TnAK residues) | Number of occurrences |
|---|---|---|
| 4-660 fused to 1-4 | 2-220 fused to 1-2 | 1 |
| 13-660 fused to 1-18 | 5-220 fused to 1-5 | 1 |
| 76-660 fused to 1-80 | 26-220 fused to 1-26 | 5 |
| 85-660 fused to 1-89 | 29-220 fused to 1-29 | 3 |
| 121-660 fused to 1-125 | 41-220 fused to 1-41 | 13 |
| 307-660 fused to 1-311 | 103-220 fused to 1-103 | 1 |
| 337-660 fused to 1-341 | 113-220 fused to 1-113 | 4 |
| 424-660 fused to 1-424 | 142-220 fused to 1-142 | 2 |
| 535-660 fused to 1-539 | 179-220 fused to 1-179 | 1 |
| 550-660 fused to 1-554 | 184-220 fused to 1-184 | 18 |
| 583-660 fused to 1-587 | 195-220 fused to 1-195 | 3 |
| 601-660 fused to 1-605 | 201-220 fused to 1-201 | 1 |
| 619-660 fused to 1-623 | 207-220 fused to 1-207 | 2 |
| 625-660 fused to 1-629 | 209-220 fused to 1-209 | 4 |
| 649-660 fused to 1-653 | 217-220 fused to 1-217 | 1 |
DISCUSSION
Our results show for the first time that transposon mutagenesis can be used to construct a combinatorial library of vectors that express circularly permuted variants of a protein, extending the types of mutagenesis that can be achieved using transposase engineering (23–30). Sequence analysis of unselected TnAK vectors revealed that PERMUTE creates an ensemble of permuted genes (and proteins) with the sequence diversity predicted in Figure 2. Selection of our TnAK library further showed that many of the variants in this library retain enzymatic function upon expression in E. coli, demonstrating that our transposase method can be used to engineer enzymes with new architectures that retain parent-like function.
The adk target vector used to validate PERMUTE contains a selectable marker (catR) that differs from the marker built into the artificial minitransposon (kanR). In addition, this vector contains a temperature-sensitive origin of replication (32), unlike the artificial minitransposon, whose origin is functional at 43°C. This experimental design allowed the use of simultaneous antibiotic (chloramphenicol and kanamycin) and temperature (43°C) selections of our transposase reactions in the first step of PERMUTE to amplify target vectors containing integrated minitransposons. We chose this approach because we had previously found that a two antibiotic selection is not effective at selecting against bacteria cotransformed with separate circular vector (catR) and linear minitransposon (kanR) when these two DNA sequences both contain replication origins that function at 43°C (data not shown). Surprisingly, selection of our transposase reaction products in E. coli at 30°C yielded the same level of adk containing an integrated minitransposon as selection at 43°C. The strong selection at 30°C is interpreted as arising because minitransposon integration increases the copy number of the target vector at this temperature, enhances the concentrations of chloramphenicol acetyltransferase in cells containing the modified target vector and boosts the bacterial fitness of these cells compared to cells cotransformed with separate circular target vector and linear minitransposon. This finding suggests that PERMUTE could be performed using our artificial minitransposon and target vectors whose replication origins are not temperature sensitive, provided that minitransposon integration increases target vector copy number to a level that enhances the antibiotic resistance conferred by the target vector.
Although PERMUTE completely avoids deletions of the amino acid sequence in the protein being circularly permuted, which occur with existing methods used for this type of mutagenesis (17,18), the vectors produced by PERMUTE express circularly permuted proteins with peptides fused to their N- and C-termini. Each variant begins with an 18 amino acid peptide, whose sequence is encoded by the R2R1 transposase-binding site that separates the start codon within the minitransposon from the first codon in the permuted gene (Figure 1C). In addition, each variant has two amino acids fused to its C-terminus. The sequence of these two residues varies among the different permuted variants, because they are generated by the five base pair duplication that occurs during minitransposon integration within the target gene (22). As illustrated in Figure 2B, transposon integration produces circularly permuted genes that begin and end with the same five base pairs.
Our discovery of 15 unique permuted AK that retain in vivo function demonstrates that many backbone locations tolerate addition of peptide tags to the new termini created by primary sequence permutation. This functional robustness to backbone cleavage and tag addition has been previously observed. A recent study found that T. neapolitana AK tolerates backbone cleavage without permutation at nine of the sites discovered herein (30). In this previous study, peptide tags were added to both protein termini created by backbone fragmentation, and these tags were tolerated when incorporated at sites that exhibit different levels of accessible surface area within the AK structure (36). Future experiments will be required to determine if libraries encoding permuted AK without tags contain a higher fraction of permuted AK that retain function. This could be achieved by PCR amplifying the permuted genes from the final PERMUTE library using primers that incorporate Type IIS restriction sites adjacent to the permuted gene (37), digesting these amplicons with Type IIS restriction endonucleases to remove the undesired sequences and cloning the ensemble into an expression vector.
PERMUTE will be useful for future studies that explore protein fitness landscapes through directed evolution (38). Libraries of circularly permuted proteins generated by PERMUTE should be useful for evaluating the effect of protein thermostability on protein tolerance to permutation type mutations (39), since this method can be applied to homologous proteins and used to create pairs of libraries that express structurally-related ensembles of protein variants. PERMUTE libraries should also be useful for examining how conservation of structure and function in permuted proteins is affected by the peptide used to link the N- and C-termini in the parental protein. At the DNA level, libraries can be built with linkers having variable sequences and sizes by simply adding codons to the termini of the target gene. Sequence diversity built into the linker sequences within PERMUTE libraries should be more readily accessible through selections and screens than existing library methods (17,18), because PERMUTE creates fewer permuted variants than existing methods by avoiding deletions and duplications of varying length (20,21). Finally, PERMUTE should help simplify the construction of molecular switches created using domain insertion (40). PERMUTE libraries can be subcloned into different sites within other proteins, and variants with allosterically-coupled functions can be screened and selected to obtain new molecular switches for synthetic biology (15).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–17, Supplementary Dataset 1.
FUNDING
National Aeronautics and Space Administration (grant number: NNX08AO20G); Robert A. Welch Foundation (grant number: C-1614). Funding for open access charge: Robert A. Welch Foundation.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Lindqvist Y, Schneider G. Circular permutations of natural protein sequences: structural evidence. Curr. Opin. Struct. Biol. 1997;7:422–427. doi: 10.1016/s0959-440x(97)80061-9. [DOI] [PubMed] [Google Scholar]
- 2.Schmidt-Goenner T, Guerler A, Kolbeck B, Knapp EW. Circular permuted proteins in the universe of protein folds. Proteins. 2010;78:1618–1630. doi: 10.1002/prot.22678. [DOI] [PubMed] [Google Scholar]
- 3.Luger K, Hommel U, Herold M, Hofsteenge J, Kirschner K. Correct folding of circularly permuted variants of a beta alpha barrel enzyme in vivo. Science. 1989;243:206–210. doi: 10.1126/science.2643160. [DOI] [PubMed] [Google Scholar]
- 4.Zhang T, Bertelsen E, Benvegnu D, Alber T. Circular permutation of T4 lysozyme. Biochemistry. 1993;32:12311–12318. doi: 10.1021/bi00097a006. [DOI] [PubMed] [Google Scholar]
- 5.Yang YR, Schachman HK. Aspartate transcarbamoylase containing circularly permuted catalytic polypeptide chains. Proc. Natl Acad. Sci. USA. 1993;90:11980–11984. doi: 10.1073/pnas.90.24.11980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uversky VN, Kutyshenko VP, Protasova N, Rogov VV, Vassilenko KS, Gudkov AT. Circularly permuted dihydrofolate reductase possesses all the properties of the molten globule state, but can resume functional tertiary structure by interaction with its ligands. Protein Sci. 1996;5:1844–1851. doi: 10.1002/pro.5560050910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pieper U, Hayakawa K, Li Z, Herzberg O. Circularly permuted beta-lactamase from Staphylococcus aureus PC1. Biochemistry. 1997;36:8767–8774. doi: 10.1021/bi9705117. [DOI] [PubMed] [Google Scholar]
- 8.Viguera AR, Blanco FJ, Serrano L. The order of secondary structure elements does not determine the structure of a protein but does affect its folding kinetics. J. Mol. Biol. 1995;247:670–681. doi: 10.1006/jmbi.1994.0171. [DOI] [PubMed] [Google Scholar]
- 9.Bulaj G, Koehn RE, Goldenberg DP. Alteration of the disulfide-coupled folding pathway of BPTI by circular permutation. Protein Sci. 2004;13:1182–1196. doi: 10.1110/ps.03563704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu Y, Lutz S. Circular permutation: a different way to engineer enzyme structure and function. Trends Biotechnol. 2011;29:18–25. doi: 10.1016/j.tibtech.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 11.Qian Z, Lutz S. Improving the catalytic activity of Candida antarctica lipase B by circular permutation. J. Am. Chem. Soc. 2005;127:13466–13467. doi: 10.1021/ja053932h. [DOI] [PubMed] [Google Scholar]
- 12.Carlson HJ, Cotton DW, Campbell RE. Circularly permuted monomeric red fluorescent proteins with new termini in the beta-sheet. Protein Sci. 2010;19:1490–1499. doi: 10.1002/pro.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whitehead TA, Bergeron LM, Clark DS. Tying up the loose ends: circular permutation decreases the proteolytic susceptibility of recombinant proteins. Protein Eng. Des. Sel. 2009;22:607–613. doi: 10.1093/protein/gzp034. [DOI] [PubMed] [Google Scholar]
- 14.Schwartz TU, Walczak R, Blobel G. Circular permutation as a tool to reduce surface entropy triggers crystallization of the signal recognition particle receptor beta subunit. Protein Sci. 2004;13:2814–2818. doi: 10.1110/ps.04917504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grünberg R, Serrano L. Strategies for protein synthetic biology. Nucleic Acids Res. 2010;38:2663–2675. doi: 10.1093/nar/gkq139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guntas G, Mansell TJ, Kim JR, Ostermeier M. Directed evolution of protein switches and their application to the creation of ligand-binding proteins. Proc. Natl Acad. Sci. USA. 2005;102:11224–11229. doi: 10.1073/pnas.0502673102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graf R, Schachman HK. Random circular permutation of genes and expressed polypeptide chains: application of the method to the catalytic chains of aspartate transcarbamoylase. Proc. Natl Acad. Sci. USA. 1996;93:11591–11596. doi: 10.1073/pnas.93.21.11591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hennecke J, Sebbel P, Glockshuber R. Random circular permutation of DsbA reveals segments that are essential for protein folding and stability. J. Mol. Biol. 1999;286:1197–1215. doi: 10.1006/jmbi.1998.2531. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 20.Guntas G, Ostermeier M. Creation of an allosteric enzyme by domain insertion. J. Mol. Biol. 2004;336:263–273. doi: 10.1016/j.jmb.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 21.Hida K, Won SY, Di Pasquale G, Hanes J, Chiorini JA, Ostermeier M. Sites in the AAV5 capsid tolerant to deletions and tandem duplications. Arch. Biochem. Biophys. 2010;496:1–8. doi: 10.1016/j.abb.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haapa S, Taira S, Heikkinen E, Savilahti H. An efficient and accurate integration of mini-Mu transposons in vitro: a general methodology for functional genetic analysis and molecular biology applications. Nucleic Acids Res. 1999;27:2777–2784. doi: 10.1093/nar/27.13.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poussu E, Vihinen M, Paulin L, Savilahti H. Probing the alpha-complementing domain of E. coli beta-galactosidase with use of an insertional pentapeptide mutagenesis strategy based on Mu in vitro DNA transposition. Proteins. 2004;54:681–692. doi: 10.1002/prot.10467. [DOI] [PubMed] [Google Scholar]
- 24.Jones DD. Triplet nucleotide removal at random positions in a target gene: the tolerance of TEM-1 beta-lactamase to an amino acid deletion. Nucleic Acids Res. 2005;33:e80. doi: 10.1093/nar/gni077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poussu E, Jantti J, Savilahti H. A gene truncation strategy generating N- and C-terminal deletion variants of proteins for functional studies: mapping of the Sec1p binding domain in yeast Mso1p by a Mu in vitro transposition-based approach. Nucleic Acids Res. 2005;33:e104. doi: 10.1093/nar/gni102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoeller BM, Reiter B, Abad S, Graze I, Glieder A. Random tag insertions by transposon integration mediated mutagenesis (TIM) J. Microbiol. Methods. 2008;75:251–257. doi: 10.1016/j.mimet.2008.06.017. [DOI] [PubMed] [Google Scholar]
- 27.Baldwin AJ, Busse K, Simm AM, Jones DD. Expanded molecular diversity generation during directed evolution by trinucleotide exchange (TriNEx) Nucleic Acids Res. 2008;36:e77. doi: 10.1093/nar/gkn358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daggett KA, Layer M, Cropp TA. A general method for scanning unnatural amino acid mutagenesis. ACS Chem. Biol. 2009;4:109–113. doi: 10.1021/cb800271f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edwards WR, Busse K, Allemann RK, Jones DD. Linking the functions of unrelated proteins using a novel directed evolution domain insertion method. Nucleic Acids Res. 2008;36:e78. doi: 10.1093/nar/gkn363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Segall-Shapiro TH, Nguyen PQ, Dos Santos ED, Subedi S, Judd J, Suh J, Silberg JJ. Mesophilic and hyperthermophilic adenylate kinases differ in their tolerance to random fragmentation. J. Mol. Biol. 2011;406:135–148. doi: 10.1016/j.jmb.2010.11.057. [DOI] [PubMed] [Google Scholar]
- 31.Haase GH, Brune M, Reinstein J, Pai EF, Pingoud A, Wittinghofer A. Adenylate kinases from thermosensitive Escherichia coli strains. J. Mol. Biol. 1989;207:151–162. doi: 10.1016/0022-2836(89)90446-4. [DOI] [PubMed] [Google Scholar]
- 32.Link AJ, Phillips D, Church GM. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 1997;179:6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vieille C, Krishnamurthy H, Hyun HH, Savchenko A, Yan H, Zeikus JG. Thermotoga neapolitana adenylate kinase is highly active at 30 degrees C. Biochem. J. 2003;372:577–585. doi: 10.1042/BJ20021377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seligmann H. Cost minimization of ribosomal frameshifts. J. Theor. Biol. 2007;249:162–167. doi: 10.1016/j.jtbi.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen PQ, Liu S, Thompson JC, Silberg JJ. Thermostability promotes the cooperative function of split adenylate kinases. Protein Eng. Des. Sel. 2008;21:303–310. doi: 10.1093/protein/gzn005. [DOI] [PubMed] [Google Scholar]
- 36.Bae E, Phillips GN., Jr Structures and analysis of highly homologous psychrophilic, mesophilic, and thermophilic adenylate kinases. J. Biol. Chem. 2004;279:28202–28208. doi: 10.1074/jbc.M401865200. [DOI] [PubMed] [Google Scholar]
- 37.Pingoud A, Fuxreiter M, Pingoud V, Wende W. Type II restriction endonucleases: structure and mechanism. Cell Mol. Life Sci. 2005;62:685–707. doi: 10.1007/s00018-004-4513-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Romero PA, Arnold FH. Exploring protein fitness landscapes by directed evolution. Nat. Rev. Mol. Cell Biol. 2009;10:866–876. doi: 10.1038/nrm2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bloom JD, Silberg JJ, Wilke CO, Drummond DA, Adami C, Arnold FH. Thermodynamic prediction of protein neutrality. Proc. Natl Acad. Sci USA. 2005;102:606–611. doi: 10.1073/pnas.0406744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ostermeier M. Engineering allosteric protein switches by domain insertion. Protein Eng. Des. Sel. 2005;18:359–364. doi: 10.1093/protein/gzi048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.