Abstract
The exploration of the non-protein-coding RNA (ncRNA) transcriptome is currently focused on profiling of microRNA expression and detection of novel ncRNA transcription units. However, recent studies suggest that RNA processing can be a multi-layer process leading to the generation of ncRNAs of diverse functions from a single primary transcript. Up to date no methodology has been presented to distinguish stable functional RNA species from rapidly degraded side products of nucleases. Thus the correct assessment of widespread RNA processing events is one of the major obstacles in transcriptome research. Here, we present a novel automated computational pipeline, named APART, providing a complete workflow for the reliable detection of RNA processing products from next-generation-sequencing data. The major features include efficient handling of non-unique reads, detection of novel stable ncRNA transcripts and processing products and annotation of known transcripts based on multiple sources of information. To disclose the potential of APART, we have analyzed a cDNA library derived from small ribosome-associated RNAs in Saccharomyces cerevisiae. By employing the APART pipeline, we were able to detect and confirm by independent experimental methods multiple novel stable RNA molecules differentially processed from well known ncRNAs, like rRNAs, tRNAs or snoRNAs, in a stress-dependent manner.
INTRODUCTION
The exploding repertoire of recently identified functional non-protein-coding RNAs (ncRNAs) in all three domains of life suggests their fundamental role in the regulation of gene expression (1). The major difficulties for estimating the complete ncRNA catalog of an organism relate to the complex biogenesis of ncRNA, which include multiple processing steps. Recent finding suggest that a single RNA molecule can function in distinct ways depending on different post-transcriptional ncRNA processing events. It has been shown that some functional snoRNAs which are initially processed from mRNA introns can give rise to microRNAs after further processing events took place (2,3). It is assumed that such alternative ncRNA processing can provide genome complexity comparable to the alternative splicing phenomena of pre-mRNA transcripts. Since several years the rising interest in revealing such hidden layers of the transcriptome can be observed (4,5).
One of the most straightforward experimental methods for studying ncRNA processing is the use of deep-sequencing techniques. In these approaches the RNA content of a cell, a tissue or an organism is converted into cDNA and subsequently subjected to deep-sequencing analysis. Although preparations of cDNA libraries enriched in functional small ncRNA species have been already well documented (6–9), the recent development of bioinformatic tools for deep-sequencing data analysis was focused on the estimation of expression profiles of known genes (10–14), the detection of novel splicing variants (15–16) or the identification of novel microRNA genes (17,18). Some of the methods are indeed able to detect novel ncRNA transcripts; however the potential of differential RNA processing has not been adequately addressed.
Here we address this challenge of ncRNA genome research by providing a complete workflow allowing for detection of stable ncRNA species, including novel ncRNA transcripts and RNA processing products. It is based on a novel computational pipeline, named APART (for Automated Pipeline for Analysis of RNA Transcripts). This bioinformatic tool provides an automated assembly and annotation of deep-sequencing data including the identification of novel stable ncRNA species. As proof of principle we have applied APART on a specialized cDNA library derived from the yeast Saccharomyces cerevisiae. In our experimental system, we have used one of the key cellular macromolecular complexes, namely the ribosome, as bait for the selection of a potentially functional small RNA interactome. Predicted processing of several ncRNA candidates was experimentally verified, thus highlighting the potential of APART for correctly revealing the so far enigmatic processing of ncRNA transcriptomes.
MATERIALS AND METHODS
Strain and growth conditions
Saccharomyces cerevisiae strain BY4741 (MATa; his3Δ 1; leu2Δ 0; met15Δ 0; ura3Δ 0) was grown in synthetic complete (SC) yeast medium supplemented with 2% carbon source at 30°C, as described (19). The strain was transformed with pGAL1-RPL25-FH (BIT757) carrying the gene for a full-length C-terminal FH-tagged form of RPL25 (20).
Cells were grown in 12 different growth conditions as described (21–23). Stress treatments were performed as follows: cells were grown to mid-log phase (optical density at 600 nm 0.7), the stress was applied for 15 min, the cells were harvested by centrifugation, frozen in liquid nitrogen and stored at −80°C. The temperature shifts to 37°C (heat shock) or to 15°C (cold shock) were carried out by the addition of an equal volume of SC pre-warmed to 49°C or chilled to 4°C, respectively. The cultures were either supplemented with 1 M NaCl (high salt conditions), with 0.1 M Tris–HCl pH 8.3 resulting in a final pH of 7.9 (high pH conditions) or with 1 M citric acid (low pH conditions of pH 4.0). In the UV stress, the cell suspension was irradiated with a UV dose of 120 J/m2. To induce hyper-osmotic shock the medium was supplemented with 1 M sorbitol. For hypo-osmotic conditions the cells were grown to mid-log phase in SC supplemented with 1 M sorbitol, then collected by centrifugation and resuspended in SC without sorbitol. For amino acid and sugar starvation stresses, cells were collected by centrifugation at mid-log phase and further grown in medium lacking amino acids or sugar, respectively. In parallel, anaerobic and normal growth of S. cerevisiae was performed.
Generation of a S. cerevisiae cDNA library
Saccharomyces cerevisiae ribosomes of unstressed and stressed cells were isolated as described (20). In short, cells were lysed in the presence of glass beads and the lysates were affinity-purified with anti-FLAG M2-agarose resin. Ribosome-associated RNA was extracted with phenol and precipitated with ethanol. Subsequently, equal amounts of ribosome-associated RNAs were size-fractionated by denaturing 8% PAGE. RNAs in the size range between 15 and 500 nt were excised from the gel, passively eluted into 0.3 M NaOAc and ethanol precipitated. RNAs were subsequently C-tailed at their 3′-ends using poly(A) polymerase and ligated to a 5′-adaptor (GTCAGCAATCCCTAACGAG) by T4 RNA ligase as described (6). RNAs from the library were subsequently converted into cDNAs by RT–PCR, employing primers complementary to the linkers (6) and subjected to 454 pyrosequencing (GATC Biotech AG). Original sequencing data have been submitted to NCBI SRA archive with the accession number SRP008250.1.
Northern blot analysis
Total RNA from S. cerevisiae grown under selected conditions (optimal, UV radiation, anaerobic, high pH, low pH, amino acid starvation or sugar starvation) was isolated using the Master PureTM Yeast Purification kit (Epicentre), separated on 8% denaturing polyacrylamide gel, transferred onto nylon membranes and probed with 5′-[32P]-end-labeled antisense DNA probes as described (24).
Semi-quantitative stem–loop RT–PCR assays
Stem–loop reverse transcription (RT) was followed by PCR as described (25) with minor modifications. The stem–loop RT primer used to initiate RT (5′-GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACTACTCCTACC-3′) was designed to be complementary to the last 6 nt of the target RNA. Subsequently, the RT product was amplified during 15 PCR cycles using an RNA-specific forward primer (5′-GCGGCGGTTGACCTCAAATCA-3′) and the stem–loop reverse primer. The template for this RT–PCR was size-selected (10–50 nt) total RNA.
Pre-processing and cleaning by APART
The APART pipeline supports data obtained with the vast majority of next-generation sequencing platforms available on the market, including 454, solexa, illumina, ion torrent and SOLID. Prior to cleaning, large datasets are divided into subsections according to available memory limits and read names are changed to follow the pipeline scheme. First step is the search for the used adaptor sequences within the reads using the patmatch (26) program. By default two changes (mismatches, insertions or deletions) are allowed for the 5′-adaptor and three changes for the 3′-adaptor (due to the usually lower quality of 3′-end of the reads), however values can be adjusted to meet specific needs. Estimated adaptor positions are used next for trimming of the reads. The trimming procedure includes the possibility of additional removal of the homopolymeric tract (poly-C tails) from the 3′-end of the reads. Due to sequencing length limitations, it is possible that the 3′-adaptor is not fully represented in the reads, thus reads, lacking a full length 3′-adaptor according to patmatch, are subjected for trimming of the partial 3′-adaptors using the perl regular expressions. This is followed by quality filtering, including the rejection of reads with mean quality values below the limit provided by the user (default is 25 in phred scale) and trimming of low-quality 3′-ends. If the length of the reads after the above steps is higher or equal to the minimal read lengths defined by the user (default is 18 bases), reads are retained and classified to one of the classes: trimmed at both ends, trimmed at 5′-end only, trimmed at 3′-end only or untrimmed. All rejected reads are grouped into a separate file allowing for further investigation of the library quality issues.
Genome mapping and contig assembly
For the genome mapping the bowtie (27) aligner is used. By default APART uses the ‘-n’ alignment strategy and allows for one mismatch. The reported alignments are limited to those within the best ‘stratum’. Results are obtained in SAM format. Contig assembly is fulfilled separately for plus and minus genomic strands. First, SAM alignments are sorted and converted to pileup format using the samtools (28) software. Next, during the pileup output parsing APART identify the contigs with their genomic positions writing them in a BED format, create the contig coverage plots and writing them in a WIG format and calculates the read counts and maximal coverage for every contig. Additionally the uniqueness of the contig is estimated as the mean number of genomic hits observed for the reads within the contig. APART calls also a consensus for every base of the contig and calculates a consensus quality value which corresponds to the frequency of the most abundant base in a defined position which is also selected as the consensus letter.
Identification of the RNA processing products
The identification of the RNA processing products is performed as part of the contig assembly process. During the parsing of the pileup output, the changes in coverage between the neighboring bases are stored. Once the contig is completed, the coverage shifts are inspected and those which are higher than one-third of the maximal contig coverage are assigned as putative processing sites. The assignment of the putative RNA processing product requires two of such processing sites (characterized by rise and drop of the coverage) to be in a distance equal or higher than a minimal length of the reads used for genome mapping. Single, ‘orphan’ processing sites are discarded. Additionally the expression levels for the RNA processing products are assigned. They are calculated as the maximal coverage detected within a particular product subtracted by the background coverage (the coverage observed next to 3′- or 5′-end of the predicted processing product—the higher value is used).
Clustering of the contigs
The name-based clustering is fulfilled by comparison of the lists of reads mapped within the contigs characterized by genomic uniqueness value >1. First, contigs are sorted by decreasing read number, and then by length. Clustering is realized in two runs. First the fast scanning for contigs of identical read composition is performed by regular expression search. The contig of highest read number is designated as a representative and the contigs of the same or lower read content are compared to it. In the second run, representative contigs are compared in a detailed way to estimate the ratio of shared reads. In order to reduce the search space, the contigs association matrix is created during the assembly. In this way only the contigs which share at least one read are compared.
The sequence-based clustering is fulfilled using the cd-hit (29) software. In this approach, the consensus sequences of the contigs are clustered with a threshold of 95% of identity. The representative contig is the longest one within the cluster.
Annotation and output generation
The annotation process is divided into three steps. First, the genomic repeats overlapping the contigs are identified. This is fulfilled using the repeat bed files obtained from UCSC genome browser (30) and bedtools (31) package. The minimal requirement is that at least 10% of the contig overlap the repeat. Based on the results extensive statistics of identified repeat types are created. Next, known genes mapped to the reference genome overlapping the contigs are identified. For this purpose, annotation tables of internal format are used. The genome annotations are based on Ensemble gene predictions (32), including ‘known’ and ‘novel’ subdivisions. The annotation process includes the recognition of the intron/exon features and relative orientation (sense/antisense). In case when contigs span multiple structural features (e.g. only part of the contig maps to a gene or exon) the structural feature ‘junction’ is introduced. If the contig spans over multiple genes, all of them are recognized and listed in an annotation file and final table. In the last annotation step the intergenic, intronic and antisense to known genes contigs are used. It is based on the NCBI blast+ (33) comparison of the contigs consensus sequences to sequences within the Functional RNA Database (34). The requirements for annotation in this case are 80% of identity and gap content <10% of the alignment length. At the final step all the information gathered during the analysis are combined into a series of the static html files.
Availability
The APART pipeline can be obtained from http://apart.sf.net.
RESULTS
Construction of the cDNA library containing stable ncRNA species
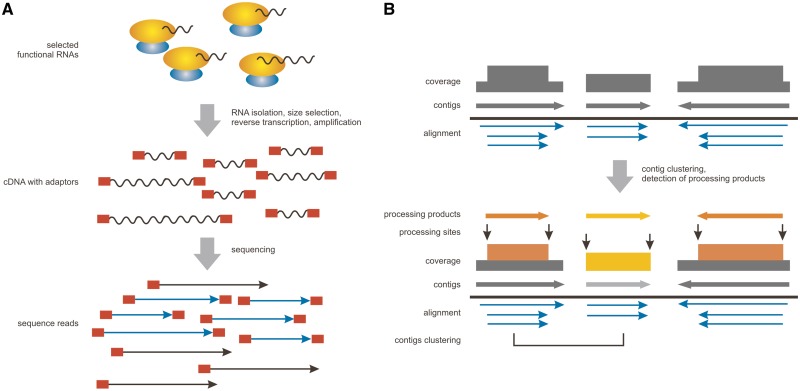
The major assumption behind the construction of a cDNA library aiming at identifying stable ncRNA species is that merely functional RNAs are expected to be protected from degradation. In order to enrich for functional ncRNAs, it has previously been shown that construction of libraries from ribonucleoprotein (RNP) particles rather than from purified total RNA is beneficial (7). Following the same logic, we have generated a cDNA library enriched for small RNAs (sized 20–500 nt) that co-purified with S. cerevisiae ribosomes under 12 different growth conditions. The rationale for choosing yeast was the lack of the miRNA pathway, since miRNAs are very abundant in other organisms and often mask other small RNAs in transcriptomic data (4). The employed procedure did not include a random RNA fragmentation step, resulting in cDNAs with ends correspond to the natural ends of the RNA species. Moreover, we have used amplification adaptors attached to both the 5′- and 3′-ends of the cDNA (see ‘Materials and Methods’ section for details) in order to validate if sequencing spans the full length of the cDNAs (Figure 1). Before addition of the 5′-adaptor, we have treated the RNAs with tobacco acid pyrophosphatase in order to enable the adaptor ligation to both, processed and primary transcripts. However, by omitting this step, it would be possible to select exclusively for processed RNAs, as it is commonly used for micro RNA identification (35).
Figure 1.
Schematic representation of the key steps of the experimental workflow leading to identification of RNA processing products. (A) Experimental preparation of the cDNA library. In order to select for functional RNAs, yeast ribosomes have been used here as bait. The next important step is the size selection of ribosome-associated RNAs and the subsequent attachment of 5′- and 3′-adaptors which are marking the natural ends of the RNAs. After deep-sequencing of the library, adaptor sequences are used to select for the reads covering the full length of the original RNA molecule (both adaptors are observed). (B) Computational analysis of the data with the APART pipeline. First, reads are aligned to the reference genome and contigs together with respective coverage plots are created. Next, contigs derived from the same read sets are clustered and only non-representative contigs (marked by lighter colors) are removed from the main results list. Processing products are predicted by scanning of the coverage plots and their abundance is estimated by subtraction of the background coverage from the maximal coverage within the predicted product (abundance correspond to the area of coverage plot marked with color).
APART workflow
Our computational pipeline is based on reference genomes as a guide for assembly and annotation of the high-throughput cDNA sequences. In contrast to existing methods, the analysis is based on contigs composed from overlapping reads, instead of genes. Thus, all the characteristics which usually are assigned to genes, like read number or expression level, are not summarized among the gene, but are calculated for individual contigs within the gene. Such an approach enables the detection of abundant fragments of known primary transcripts as well as novel intergenic RNAs.
The first step in analysis of the raw data set is a pre-processing and cleaning procedure (for a summary of the workflow see Supplementary Figure S1). The major filters used include assessment of read quality and length. During the cleaning, it is also possible to remove any adaptors and polynucleotide tails that have been added during the library preparation. Next, reads are mapped to the reference genome using the bowtie aligner (27). The assembly of the contigs is based on overlapping positions of the reads. During this process, the pipeline is calling a read-based consensus sequence for every contig. Additionally, for every base a score describing the ambiguity of the consensus letter is calculated. Next, contigs are annotated by overlap with known genes and repeat units. Additionally all intergenic, intronic and antisense contigs are subjected to a sequence search for similarity with known ncRNAs deposited in the Functional RNA database [fRNAdb (36)]. As the output of the analysis, APART is generating a number of html files containing all the gathered information about the contigs and number of files allowing for an interactive investigation of the results in the genome browsers (for an overview see Supplementary Figure S2).
Handling of non-unique reads
In order to support the identification of highly repeated ncRNAs, like snoRNAs or tRNAs, one of the priorities in the design of APART was to implement an efficient handling of non-unique reads. In most of the so far available approaches for expression profiling, only minor redundancy is allowed in order not to interfere with statistical testing for differential expression of the genes. Such a procedure is amenable for cDNA libraries derived from mRNAs or microRNAs, genes which are typically present only in low copy number in eukaryotic genomes. However, in case of libraries containing highly repeated ncRNAs, exclusion of repeat-derived reads would result in removal of those transcripts from the data set. By default, the parameters of the bowtie aligner are set to identify up to 100 locations for every read restricted to hits within best ‘stratum’. To identify contigs composed of the reads which map to more than one location on the reference genome the pipeline estimates the ‘genomic uniqueness’, calculated as the average number of hits obtained for every read. Thus, a value of 1 means that a contig is unique. Any higher value suggests that at least some portion of the reads forming a contig can align to multiple genomic loci.
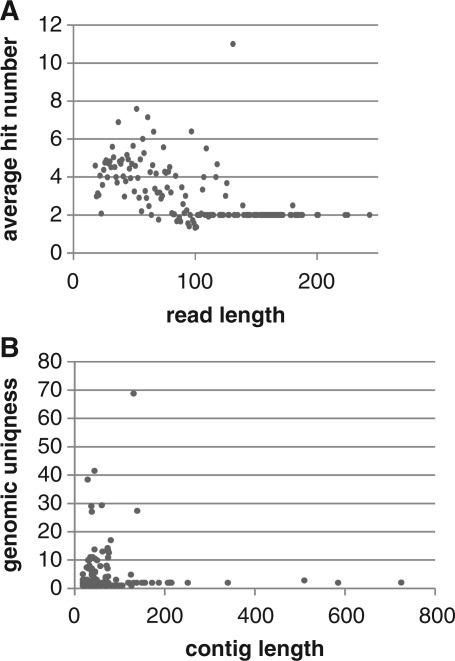
The second reason for multiple mapping of the reads to the reference genome is the random similarity of short sequence blocks across the genome. The shorter a particular read is, the higher is the probability of a random match outside of the loci of origin of the transcript. Such spurious matching could influence the calculation of the genomic uniqueness. To prevent such random alignments, the minimum length of the reads used for analysis is set by default to 18. The analysis of our yeast ribosome-derived library shows that at such a read length cut-off there is no strict dependence between the read length and the number of genomic matches (Figure 2A). Similarly, there is no correlation between genomic uniqueness values and contigs length (Figure 2B). Higher variability observed in the lower length range seems to be rather caused by higher number of reads/contigs of such length originating from various types of genes than increased spurious matching (shortest are not the most variable).
Figure 2.
The length dependence of multiple mapping events on the level of reads and contigs observed in the ribosome-associated cDNA library. (A) Distribution of the average genomic hit numbers for reads of different lengths. No significant increase of hit numbers is observed for shorter reads. (B) Distribution of genomic uniqueness values for contigs identified in the study. Although for shorter contigs (<150 nt) higher variability of uniqness is observed, there is no strict dependence on the contig length.
As a consequence of including non-unique reads in the analysis, multiple identical contigs are generated. This is caused by the manifold mappings of the same read sets across the genome. In order to remove such a redundancy, APART performs clustering using one of two distinct methods. The first is based on the comparison of the lists of read names between the contigs. In this approach all contigs that share at least 95% reads are joined into groups and next the representative contig is selected based on the highest read number. The benefit of this method is that all the distinguishable loci will be displayed separately based on even minor divergence between the duplicated genes. For the opposite behavior, we have implemented the clustering of the contigs based on sequence comparison using the cd-hit software (29). In this case, all the contigs with consensus sequence identity of 95% or higher will be clustered. This includes the contigs which are derived from different sets of reads but with high similarity.
The clustering of the contigs not only clarifies the result list but also solves the problem of normalization of the read count. In existing pipelines for annotation of ncRNA deep-sequencing data, like DARIO (37), the multiple matching reads are normalized by number of mappings. By employing the read name-based contig clustering the spurious read mappings are removed, thus such artificial normalization is no longer necessary. This results in more accurate read counts which is not affected by the uniqueness of the reads.
Detection of stable RNA species
The major emphasis in the APART pipeline is the identification of stable RNA species. Detection is achieved by scanning of the contig coverage plot in search for significant changes (Figure 1B). APART is considering a position as a putative processing site when the coverage shift between 2 nt is larger than one-third of the maximum coverage of the contig. For assigning a region as a potentially processed RNA fragment, APART requires the presence of a clear 5′-processing site (indicated by a sharp increase of sequence read coverage at a particular nucleotide position), a 3′- processing site (sharp decrease of the sequence read stack at a particular genomic location) and a minimum distance between them which is larger or equal to the minimum length of the mapped reads. In case of single ‘orphan’ processing sites followed by progressive coverage changes, it is not possible to estimate the real length of the RNA product, since it can be located even outside of the read span. Thus, contigs harboring such ‘orphan’ processing sites are not taken into account. Additionally, for every putative processing product APART is assigning the expression value corresponding to the maximal coverage observed within the processing product after subtraction of surrounding precursor-derived background coverage.
Read number versus maximal coverage
The ample processing of RNA transcripts let us also reconsidering the use of read counts for the estimation of RNA abundance. RNA transcripts, especially ncRNAs, are frequently post-transcriptionally processed, thus the use of read counts in next-generation-sequencing projects for the estimation of the RNA abundance is problematic. Read count works well as a starting point for analyses focused on expression profiling of known genes. In this case, after proper normalization [for review see (38)], such number correspond to the number of the observed transcripts of a particular gene. However characteristics of cDNA libraries aimed for the identification of RNA processing products differ from those used for either mRNA or miRNA profiling. The main divergence is that the library preparation procedure does not involve a random fragmentation step (unlike mRNA profiling projects do) and no strict length separation is performed (unlike in the miRNA profiling approaches) resulting in a collection of transcript of various lengths. Thus, the same read count can be obtained for long contigs with random read distribution and for a short one with clear processing pattern. In this case, the analysis of the read distribution among the contig is crucial. Assuming that two non-overlapping reads mapped within a single contig can be derived from a single primary transcript by a processing event, the ultimate measure for RNA level will be the maximum coverage observed within the transcript. This measure corresponds to the number of overlapping reads observed for the assembled contig and reflects the minimal number of separate transcript copies in the cell. The use of the maximal coverage value is even more important when differential processing patterns of similar RNAs are taken into consideration, like in the case of tRNA-His and tRNA-Ser (Figure 5A and B). In this particular case, the use of read count for tRNA-His doubles the expression value comparing to tRNA-Ser. Thus, additionally to the raw count of reads, APART also calculates the maximal coverage values for contigs and identified stable RNA species.
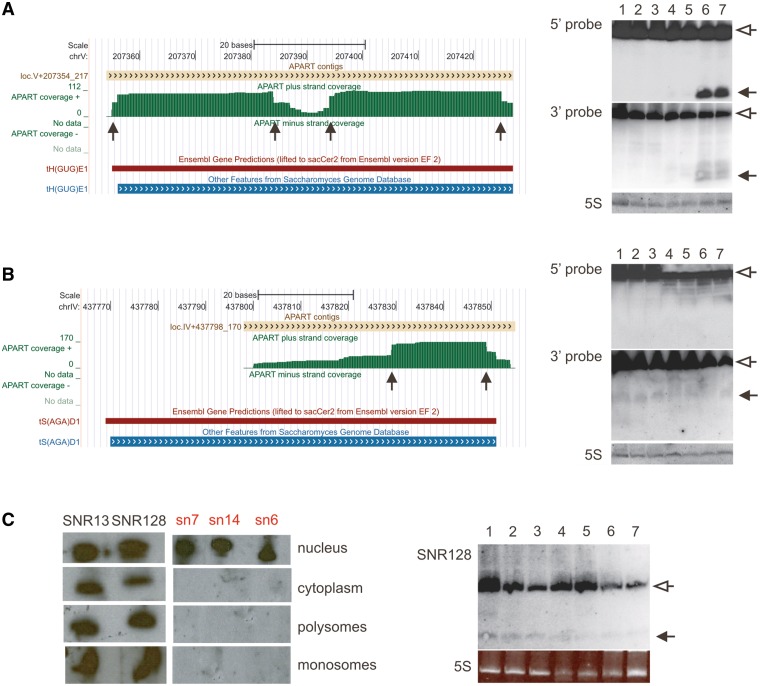
Figure 5.
Experimental validation of the APART-predicted putative processing products. (A) Processing of the tRNA-His(GUG). On the left, UCSC Genome Browser visualization of the APART tracks (green) showing two possible processing products (processing sites marked with arrows). On the right, results of the northern blot experiment using total RNA isolated from S. cerevisiae grown in different environmental conditions (lanes: 1-UV radiation, 2-anaerobic, 3-optimal, 4-high pH, 5-low pH, 6-amino acid starvation, 7-sugar starvation) with probes against 5′- and 3′-halves of the tRNA-His. Full length tRNA is marked with open arrows, processing products are indicated by filled arrows. Differential stability of both parts can be observed. (B) Processing of the tRNA-Ser(AGA) (labeling as above). The inexact ends of the contig displayed on UCSC Genome Browser visualization suggest decreased stability of the 3′-derived processing product, comparing to tRNA-His, which is reflected by the northern blot results (right). (C) Cytoplasmic localization and processing of snoRNAs. On the left, northern blot presenting subcellular localization of snoRNA 128 (identified in this study) and snoRNA13 (not found in our cDNA library). The localization of the small nuclear RNAs sn7, sn14 and sn6 in the particular cellular fractions is also shown. For the northern analysis, total RNAs prepared either from the nuclear fraction, the cytoplasmic fraction, or from the mono- or polysomal fraction were blotted. The observed northern blot signals in the polysomal samples suggest that snoRNAs are associated with translating ribosomes in yeast. On the right, identification of the processing products derived from snoRNA 128 by northern blot using total RNA isolated from yeast grown under different environmental conditions. In all panels 5S rRNA served as internal loading control.
Presentation of the results
At the final stage, APART is generating a number of html files containing all the gathered information about the contigs and number of files allowing for an interactive investigation of the results in the genome browsers. It is accompanied with extensive statistics of the analyzed library including contig and read length distributions, annotated gene numbers and others (Supplementary Figure S2). The main table contains all the representative contigs derived from clustering together with description including read count, maximal coverage values, positions of detected processing products and overlapping annotation features. Moreover, for every contig a detail page is generated, complementing the information with the consensus sequence with the quality values and the alignment to the genomic reference sequence. Furthermore, the sequences of the putative RNA processing products are shown as well as a list of contigs is given which has been clustered together and was not presented in the main table. Additionally, it delivers links to the fasta file of reads corresponding to the contig and a detailed sequence alignment in the SAM format (28).
Performance
The APART pipeline has been optimized for low memory usage. APART can be successfully run on a machine with memory of only 2 GB; however, this causes an extension of the running time. The computationally most demanding and memory-intensive procedure is the name-based clustering of the contigs. The memory consumption and running time at this step depends on read numbers, contig numbers and repetitiveness of the reads, thus cannot be estimated before the contig assembly. However, on low-end machines there is still the possibility of performing the sequence-based clustering utilizing the cd-hit software, which is memory and time-efficient. The running time for the described S. cerevisiae cDNA library analysis on a four-core 2.40 GHz 64-bit Linux workstation with 16 GB of RAM was 20 s (1.47 s CPU time). By using cDNA library reads from Haloferax volcanii (our unpublished data), we have measured the dependence of the APART running time versus read numbers. For this purpose, we have used the whole set of 72 million reads as well as subsets of 36 million, 18 million, 9 million and 4.5 million reads. We have used default APART settings including removal of the 3′-adaptors and the C-tails. The APART running time of the smallest data set was <10 min (100.86 s CPU time), and of the largest 4.5 h (5323.61 s CPU time), showing a linear correlation between analyzed read numbers and required time (Supplementary Figure S3).
Ribosome-associated ncRNAs
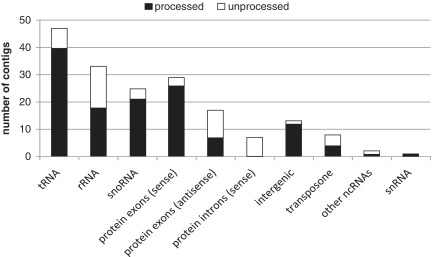
To highlight all the above mentioned features, we have applied APART to deep-sequencing data of a specialized S. cerevisiae cDNA library. We have generated a cDNA library from small RNAs (sized 20–500 nt) that co-purify with ribosomes under different environmental conditions. After sequencing, we have obtained a pool of 125 868 raw reads containing amplification adapters on both the 5′- and 3′-ends and an additional poly-C-tail at the 3′-end which has been used for initial RT [‘Materials and Methods’ section and (6)]. During the cleaning procedure, 81 790 reads were discarded due to length exclusions (<18 nt) or due to the read quality filters. For downstream analysis, we have used 18 679 reads for which adapters on both ends could be detected to ensure that all the reads are derived from full-length cellular RNAs. In total, 12 494 reads (66.89%) were mapped to the reference yeast genome with a maximum of 100 genomic hits. These reads were assembled into 716 contigs each containing at least two reads. After read name-based clustering, we have obtained 174 representative contigs. For 131 of those, the APART pipeline has detected at least one possible stable RNA species. Most of them were processing products derived predominantly from rRNAs, tRNAs and snoRNAs (Figure 3).
Figure 3.
A summary of the genomic features identified in the ribosome-derived cDNA library. As indicated, for most of the contigs putative processing products were observed.
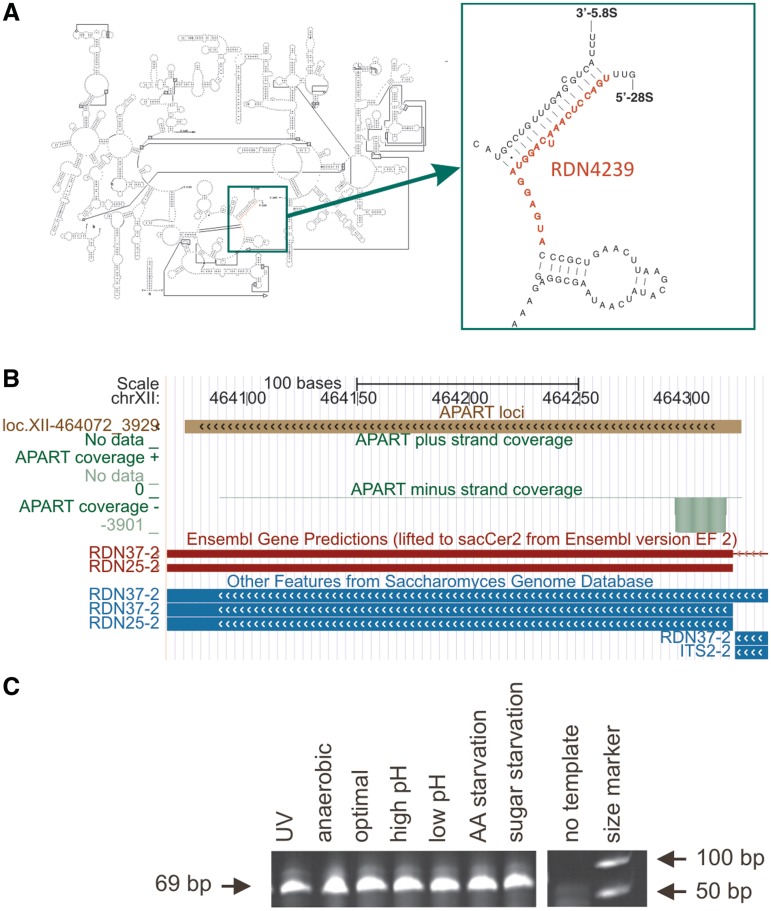
To substantiate these bioinformatically predicted RNA processing events, we have experimentally verified the presence of selected RNA fragments. For the most abundant class of ribosomal RNAs, we have selected a 23-nt long piece derived from the 5′-part of 25S rRNA found in 3901 copies that showed almost exactly identical ends (Figure 4). Experimental investigations confirmed the presence of this particular rRNA fragment under all investigated growth conditions (Figure 4C). Similar rRNA cleavage (however, primarily from the 3′-end of the 25S rRNA) has been already observed in S. cerevisiae during oxidative stress as well as during entry into stationary phase (39).
Figure 4.
Processing of a 23-mer from 5′-end of 25S ribosomal RNA. (A) The location of the detected processing product on the secondary structure diagram of large ribosomal subunit rRNA is depicted. (B) UCSC Genome Browser visualization of the APART tracks (green) within the region of contig loc.XII-464072_3929 containing the 23-mer. (C) Semi-quantitative RT–PCR with primers specific for the 23-mer using size-selected (10–50 nt) total RNA as template results in a 69-nt long PCR product. By using 10- to 50-nt long RNAs as template amplification of this 23-mer from the unprocessed full-length 25S rRNA is avoided.
The second most abundant class of processed RNAs identified in our screen were tRNAs. In addition to previously reported cleavage in the anticodon loop in yeast tRNAs (5), we detected also other breakage points (e.g. in the D- and T-loop regions), reminiscent to those observed previously in higher eukaryotes (40). Moreover, we have noticed an obvious differential stability of tRNA halves. Northern blot analysis confirmed the presence of two stable processing products derived from tRNA-His and revealed that cleavage is stress-dependent (Figure 5A). Similar to previous findings tRNA processing occurs mainly during amino acid and sugar starvation conditions. On the contrary, experimental results obtained for tRNA-Ser suggest that only the 3′-part of this tRNA is stable (Figure 5B). Probes directed against the 5′-end of the tRNA-Ser revealed a series of products likely deriving from degradation rather than processing. Moreover, the band observed for 3′-probe on the northern blot is less defined than in the case of tRNA-His. This is in agreement with the APART results, which show a rather dispersed coverage at the tRNA-Ser 3′-end suggesting the processed fragment to be less well preserved and stable.
The APART analysis of our cDNA library revealed also a number of snoRNA fragments to be associated with ribosomes. Since snoRNAs are supposed to be specifically localized within the nucleolus, we have confirmed their cytoplasmic localization with northern blot analysis. To exclude a possible nuclear contamination, we have used probes against nuclear-specific snRNAs. While we could detect the snRNAs solely in the nuclear fraction, the tested snoRNAs were indeed also present in the cytoplasm and moreover also in the mono- and polysomal fractions (Figure 5C). The presence of snoRNA processing products predicted by APART could be experimentally verified (Figure 5C). The results confirmed the presence of the shortened version of the snoRNA under most growth conditions. Similar processing of snoRNAs into smaller functional RNAs has been described before in mammalian cells (3) as well as in the primitive eukaryote Giardia lamblia (41), but this is to our knowledge the first report of a putative snoRNA processing event in a microorganism.
DISCUSSION
APART represents a user-friendly bioinformatic tool for obtaining a full overview on the global transcriptome of a cell or an entire organism. Due to the contig-based analysis instead of profiling of the known genes, APART can be used for the identification of novel stable RNA species, including both intergenic transcripts and RNA processing products. Other benefits over existing pipelines include efficient handling of non-unique reads, a novel measure for transcript abundance assigned to the contigs and putative processing products and a convenient presentation of the results. These features are especially important for genome-wide ncRNA studies in higher eukaryotes since the recent past clearly revealed the ncRNA transcriptomes to be far more complex than initially anticipated (1,42).
The proposed workflow for detection of stable RNA species consist of two steps. First is the appropriate experimental preparation of the cDNA library enriched in functional stable RNAs. The enrichment methodology has been already well documented (6–7,43), thus the major challenge was the development of a novel computational tool for analysis of the deep-sequencing data. The key feature of the APART pipeline is detection of novel stable RNA molecules. Although the employed method is very simple, it follows the idea that stable RNA transcripts and processing products should be protected against endo- and exo-nucleases. Thus, the strict limitation of the exact 5′- and 3′-ends has been introduced. This is opposite to the method used in the blockbuster algorithm (44), where reads with non-identical ends are joined into ‘blocks’. The blockbuster approach was however initially developed for separation of microRNA and microRNA* blocks of reads in order to enable the assignment of separate expression values. The major difference between our approach and microRNA profiling experiments is that read data obtained from microRNA profiling experiments contain almost no background reads derived from precursor hairpins (due to specific amplification of exclusively RNA processing products). In our dataset the amount of background reads was in many cases substantial, thus read distribution analysis proposed in blockbuster failed to separate the putative processing products.
The predominant limitations of the APART pipeline are related to the mapping procedure. Some classes of ncRNAs, like tRNAs, contain a number of post-transcriptional modifications, including modified nucleotides and non-encoded nucleotides, e.g. CCA at the 3′-ends. Especially nucleobase modifications can potentially lead to incorrect cDNA synthesis during RT. These putative cDNA errors would subsequently lead to additional mismatches during the alignment process to the reference genome. To address this issue, we have compared the number of the reads aligning to the most intensely modified RNA species—rRNA and tRNA allowing one, two, or three mismatches. It turned out that only a minor portion of the reads can be additionally aligned when the number of allowed mismatches is increased (Supplementary Figure S4). Moreover, the observed difference is similar when hyper-modified RNA species (rRNA and tRNA) or total reads from the library are considered. Thus, we decided to allow only one mismatch as the default parameter for APART. However in higher eukaryotes, where the ratio of RNA modification is higher, the value should be adjusted accordingly. In cases of libraries composed predominantly from tRNAs or other hyper-modified RNA species, we suggest to use other alignment tools, like segemehl (45) which allow also for insertions and deletions. This feature will allow not only more efficient handling of modified nucleotides, but also non-encoded CCA tails of tRNAs. In such cases, the work with APART would start from read alignment file in SAM format.
Also detection of the stable RNA can be potentially hampered by the experimental procedure employed for generation of the cDNA library. The main step causing potential bias is the amplification of the cDNA. During this procedure, a preference for short molecules is observed (46). The unequal amplification can lead to multiplication of single cDNA molecules, resulting in a false sharp increase of the coverage of some genomic regions. Such cases can lead to false predictions of processing events. However, our experimental data suggest that such events are very rare, since the presence of all of the tested processing products has been experimentally confirmed.
One also has to keep in mind that not every sharp shift in read coverage is related to an RNA processing event. It could also be caused by an RNA structure-dependent drop-off of the reverse transcriptase (47) or by preferential amplification of some of the short cDNA sequences during library generation. However, such cases cannot be distinguished based on the sole analysis of cDNA sequences.
In order to estimate the performance of the APART pipeline, we have used it for the analysis of a previously published dataset. For this purpose we have used the small RNA library generated by Kawaji et al. (4) in which numerous types of RNA processing products were observed. APART was able to detect and annotate the processing products using a fully automated mode in a similar way (Table 1). The only remarkable exceptions were miRNAs. In Kawaji et al., 821 contigs corresponding to miRNAs have been identified, whereas the default APART analysis resulted in annotation of only 230 miRNAs. Such a high difference could arise from the different approach for analysis. The authors of the original work used a hierarchical mapping of the reads to different ncRNA classes, using the threshold of 80% sequence identity. During such an approach, reads are aligned to different classes of transcripts not simultaneously, but in a specified order. Reads mapped to the first category are not considered for downstream categories. As a result, reads which could map to downstream transcript types with higher identity can be assigned to a false category and lead to overestimation of the categories placed in the beginning of the list. In contrast, APART by using the reference genome for read mapping always picks the best aligning genomic loci, resulting in a more unbiased analysis. Additionally, implemented in APART clustering of the redundant contigs derived from multiple alignments of the same stets of reads lead also to a reduction of the final number of the reported contigs.
Table 1.
Assessment of the APART performance in comparison to data published by Kawaji et al. (4)
| number of contigs (genes) identified in Kawaji et al. (4) | number of contigs (genes) identified by APART | number of processing products identified by APART | |
|---|---|---|---|
| miRNA | 821 (n.d.) | 230 (186) | 210 |
| tRNA | 1138 (20) | 141 (18) | 88 |
| snoRNA | 20 (n.d.) | 15 (14) | 15 |
| snRNA | 110 (3) | 15 (7) | 12 |
| rRNA | 233 (4) | 45 (4) | 8 |
Number of genes for some of the gene types were not determined in Kawaji et al. study (indicated as n.d. in the table). The lower numbers of contigs reported by APART is a consequence of clustering of redundant contigs derived from the same set of reads and due to an overestimation of miRNA and tRNA contigs in the Kawaji et al. study by using a hierarchical annotation procedure.
A high number of novel processing products and novel intergenic ncRNAs suggested by the analysis of cDNA library constructed from ribosome-associated small RNAs reveal the potential of the presented methodology. Due to the APART features we were able to predict and experimentally verify the differential and stress-dependent processing of tRNAs, rRNAs and snoRNAs. Furthermore, our data suggest that these ncRNA processing products are associated with yeast ribosomes under different environmental growth conditions. Beside the presented yeast cDNA library, APART has also been successfully applied on archaeal, mouse and human ncRNA libraries (M.Z., N.P., unpublished data), as well as on libraries generated by genomic SELEX or CLIP (M.Z., Renee Schroeder, Andrea Barta, unpublished data) approaches employing complex eukaryal model organisms (M.Z., Alexander Hüttenhofer, unpublished data). This emphasizes the general potential of APART for efficient de novo assembly and annotation of short read libraries.
ACCESSION NUMBER
SRP008250.1.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online: Supplementary Figures 1–4.
FUNDING
Austrian Science Foundation FWF (grant number Y315 to N.P.); the Austrian Ministry of Science and Research (GenAU project consortium ‘non-coding RNAs’, grant number D-110420-012-012 to N.P.); Lise-Meitner program from the FWF (grant number M1074-B11 to K.B.-Z.). Funding for open access charge: Austrian Science Foundation FWF.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Isabella Moll is acknowledged for stimulating discussions and Toshifumi Inada for providing plasmids. Alexander Hüttenhofer, Renee Schroeder and Andrea Barta are thanked for sharing unpublished deep-sequencing data used for optimization of the APART pipeline.
REFERENCES
- 1.Mattick JS. Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep. 2001;2:986–991. doi: 10.1093/embo-reports/kve230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brameier M, Herwig A, Reinhardt R, Walter L, Gruber J. Human box C/D snoRNAs with miRNA like functions: expanding the range of regulatory RNAs. Nucleic Acids Res. 2011;39:675–686. doi: 10.1093/nar/gkq776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol. Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 4.Kawaji H, Nakamura M, Takahashi Y, Sandelin A, Katayama S, Fukuda S, Daub CO, Kai C, Kawai J, Yasuda J, et al. Hidden layers of human small RNAs. BMC Genomics. 2008;9:157. doi: 10.1186/1471-2164-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thompson DM, Parker R. Stressing out over tRNA cleavage. Cell. 2009;138:215–219. doi: 10.1016/j.cell.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Mrazek J, Kreutmayer SB, Grasser FA, Polacek N, Huttenhofer A. Subtractive hybridization identifies novel differentially expressed ncRNA species in EBV-infected human B cells. Nucleic Acids Res. 2007;35:e73. doi: 10.1093/nar/gkm244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rederstorff M, Bernhart SH, Tanzer A, Zywicki M, Perfler K, Lukasser M, Hofacker IL, Huttenhofer A. RNPomics: defining the ncRNA transcriptome by cDNA library generation from ribonucleo-protein particles. Nucleic Acids Res. 2010;38:e113. doi: 10.1093/nar/gkq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tougan T, Okuzaki D, Nojima H. Chum-RNA allows preparation of a high-quality cDNA library from a single-cell quantity of mRNA without PCR amplification. Nucleic Acids Res. 2008;36:e92. doi: 10.1093/nar/gkn420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang W, Ying D, Lau YL. In-depth cDNA library sequencing provides quantitative gene expression profiling in cancer biomarker discovery. Genomics Proteomics Bioinformatics. 2009;7:1–12. doi: 10.1016/S1672-0229(08)60028-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buermans HP, Ariyurek Y, van Ommen G, den Dunnen JT, t Hoen PA. New methods for next generation sequencing based microRNA expression profiling. BMC Genomics. 2010;11:716. doi: 10.1186/1471-2164-11-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang PJ, Liu YC, Lee CC, Lin WC, Gan RR, Lyu PC, Tang P. DSAP: deep-sequencing small RNA analysis pipeline. Nucleic Acids Res. 2010;38:W385–W391. doi: 10.1093/nar/gkq392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiang DY, Getz G, Jaffe DB, O'Kelly MJ, Zhao X, Carter SL, Russ C, Nusbaum C, Meyerson M, Lander ES. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat. Methods. 2009;6:99–103. doi: 10.1038/nmeth.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 14.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedlander MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nat. Biotechnol. 2008;26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 18.Hendrix D, Levine M, Shi W. miRTRAP, a computational method for the systematic identification of miRNAs from high throughput sequencing data. Genome Biol. 2010;11:R39. doi: 10.1186/gb-2010-11-4-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guthrie C, Fink G. Guide to Yeast Genetics and Molecular Biology. San Diego, California: Academic Press; 1991. [Google Scholar]
- 20.Inada T, Winstall E, Tarun SZ, Jr, Yates JR, 3rd, Schieltz D, Sachs AB. One-step affinity purification of the yeast ribosome and its associated proteins and mRNAs. RNA. 2002;8:948–958. doi: 10.1017/s1355838202026018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, Lee TI, True HL, Lander ES, Young RA. Remodeling of yeast genome expression in response to environmental changes. Mol. Biol. Cell. 2001;12:323–337. doi: 10.1091/mbc.12.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conconi A, Paquette M, Fahy D, Bespalov VA, Smerdon MJ. Repair-independent chromatin assembly onto active ribosomal genes in yeast after UV irradiation. Mol. Cell. Biol. 2005;25:9773–9783. doi: 10.1128/MCB.25.22.9773-9783.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang TH, Polacek N, Zywicki M, Huber H, Brugger K, Garrett R, Bachellerie JP, Huttenhofer A. Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol. Microbiol. 2005;55:469–481. doi: 10.1111/j.1365-2958.2004.04428.x. [DOI] [PubMed] [Google Scholar]
- 25.Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP. Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods. 2007;3:12. doi: 10.1186/1746-4811-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan T, Yoo D, Berardini TZ, Mueller LA, Weems DC, Weng S, Cherry JM, Rhee SY. PatMatch: a program for finding patterns in peptide and nucleotide sequences. Nucleic Acids Res. 2005;33:W262–W266. doi: 10.1093/nar/gki368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 30.Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 34.Kin T, Yamada K, Terai G, Okida H, Yoshinari Y, Ono Y, Kojima A, Kimura Y, Komori T, Asai K. fRNAdb: a platform for mining/annotating functional RNA candidates from non-coding RNA sequences. Nucleic Acids Res. 2007;35:D145–D148. doi: 10.1093/nar/gkl837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hafner M, Landgraf P, Ludwig J, Rice A, Ojo T, Lin C, Holoch D, Lim C, Tuschl T. Identification of microRNAs and other small regulatory RNAs using cDNA library sequencing. Methods. 2008;44:3–12. doi: 10.1016/j.ymeth.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mituyama T, Yamada K, Hattori E, Okida H, Ono Y, Terai G, Yoshizawa A, Komori T, Asai K. The Functional RNA Database 3.0: databases to support mining and annotation of functional RNAs. Nucleic Acids Res. 2009;37:D89–D92. doi: 10.1093/nar/gkn805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fasold M, Langenberger D, Binder H, Stadler PF, Hoffmann S. DARIO: a ncRNA detection and analysis tool for next-generation sequencing experiments. Nucleic Acids Res. 2011;39:W112–W117. doi: 10.1093/nar/gkr357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyer SU, Pfaffl MW, Ulbrich SE. Normalization strategies for microRNA profiling experiments: a 'normal' way to a hidden layer of complexity? Biotechnol. Lett. 2010;32:1777–1788. doi: 10.1007/s10529-010-0380-z. [DOI] [PubMed] [Google Scholar]
- 39.Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA. 2008;14:2095–2103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saraiya AA, Wang CC. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 2008;4:e1000224. doi: 10.1371/journal.ppat.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huttenhofer A, Schattner P, Polacek N. Non-coding RNAs: hope or hype? Trends Genet. 2005;21:289–297. doi: 10.1016/j.tig.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 43.Huttenhofer A, Vogel J. Experimental approaches to identify non-coding RNAs. Nucleic Acids Res. 2006;34:635–646. doi: 10.1093/nar/gkj469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langenberger D, Bermudez-Santana C, Hertel J, Hoffmann S, Khaitovich P, Stadler PF. Evidence for human microRNA-offset RNAs in small RNA sequencing data. Bioinformatics. 2009;25:2298–2301. doi: 10.1093/bioinformatics/btp419. [DOI] [PubMed] [Google Scholar]
- 45.Hoffmann S, Otto C, Kurtz S, Sharma CM, Khaitovich P, Vogel J, Stadler PF, Hackermuller J. Fast mapping of short sequences with mismatches, insertions and deletions using index structures. PLoS Comput. Biol. 2009;5:e1000502. doi: 10.1371/journal.pcbi.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oshlack A, Wakefield MJ. Transcript length bias in RNA-seq data confounds systems biology. Biol. Direct. 2009;4:14. doi: 10.1186/1745-6150-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Renalier MH, Nicoloso M, Qu LH, Bachellerie JP. SnoRNA U21 is also intron-encoded in Drosophila melanogaster but in a different host-gene as compared to warm-blooded vertebrates. FEBS Lett. 1996;379:212–216. doi: 10.1016/0014-5793(95)01511-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.