Abstract
Human telomerase RNA (hTR) contains several guanine tracts at its 5′-end that can form a G4-quadruplex structure. Previous evidence suggests that a G4-quadruplex within this region disrupts the formation of an important structure within hTR known as the P1 helix, a critical element in defining the template boundary for reverse transcription. RNA associated with AU-rich element (RHAU) is an RNA helicase that has specificity for DNA and RNA G4-quadruplexes. Two recent studies identify a specific interaction between hTR and RHAU. Herein, we confirm this interaction and identify the minimally interacting RNA fragments. We demonstrate the existence of multiple quadruplex structures within the 5′ region of hTR and find that these regions parallel the minimal sequences capable of RHAU interaction. We confirm the importance of the RHAU-specific motif in the interaction with hTR and demonstrate that the helicase activity of RHAU is sufficient to unwind the quadruplex and promote an interaction with 25 internal nucleotides to form a stable P1 helix. Furthermore, we have found that a 5′-terminal quadruplex persists following P1 helix formation that retains affinity for RHAU. Finally, we have investigated the functional implications of this interaction and demonstrated a reduction in average telomere length following RHAU knockdown by small interfering RNA (siRNA).
INTRODUCTION
RNA associated with AU-rich element (RHAU), also known as DHX36 and G4R1, is an adenosine triphosphate (ATP)-dependent RNA helicase that belongs to the DExH/D family of RNA modifying enzymes. Proteins in this family participate in a wide range of functions including RNA splicing, messenger RNA (mRNA) stability, ribosome assembly, microRNA processing, ribonucleoprotein remodeling and RNA trafficking (1–4). RHAU is primarily expressed in the nucleus and this localization is dependent upon a region within the N-terminus of the protein (5). In 2004, Tran et al. (6) identified DHX36 as a protein from HeLa cell lysates that bound with high affinity to the AU-rich element (ARE) of urokinase plasminogen activator mRNA and therefore renamed the protein RHAU (RNA helicase associated with ARE). Their work established a role for RHAU in destabilizing mRNAs containing an ARE in the 3′-untranslated region via exosome recruitment.
Although RHAU was initially identified as a DEAH box RNA helicase that binds an ARE, a pivotal study in 2005 shifted the focus of RHAU to its unique ability to preferentially bind and unwind G4-quadruplexes (7). G4-quadruplexes are four-stranded tetrad structures mediated by Hoogsteen hydrogen bonds formed between four guanines in a single plane stabilized by a monovalent cation (8). Quadruplexes are found in both DNA and RNA and can be formed both intermolecularly and intramolecularly. G4-quadruplexes are hypothesized to play a regulatory role in a number of contexts, particularly in the regulation of RNA stability and gene transcription (9). Guanine rich nucleic acid sequences have been demonstrated to form highly stable G4-quadruplexes in vitro for nearly 30 years, but were only recently demonstrated to exist in vivo with the identification of stable quadruplexes within human telomeres (10–12). Along with this initial report, several subsequent studies suggest a widespread prevalence of both DNA and RNA quadruplexes, particularly within regulatory regions of the human genome and untranslated regions of mRNAs (10,13). The prevalence and biological implications of these unique structural motifs is now only beginning to be unravelled.
RHAU was initially identified as a protein with high affinity for G4-quadruplex DNA and was further characterized as capable of resolving intermolecular DNA quadruplexes into free single strands (7). RHAU was later demonstrated to unwind intramolecular quadruplexes derived from the c-Myc and Zic1 promoters (14). Immunodepletion of RHAU from HeLa cell lysates resulted in >50% reduction of total G4-DNA resolvase activity (15). In the same study, RHAU was also found to bind and resolve RNA quadruplexes with similar efficiency as DNA quadruplexes. This finding greatly expanded the potential functions of the DHX36 gene (15). RHAU specific knockdowns resulted in a significant impact, both positively and negatively, on the mRNA levels of multiple genes as measured by mRNA expression microarrays, indicating an important role in both transcriptional regulation and mRNA stability (5). Supporting these results, RHAU was recently demonstrated to control gene expression through direct binding to quadruplexes within the promoters of the TNAP and YY1 genes (16,17).
Investigation of the primary amino acid sequence of RHAU revealed a region, consisting of the first 105 amino acids, that contains a conserved RNA binding motif. This region is referred to as the RHAU-specific motif (RSM) and is necessary for direct RNA interactions as well as localization of RHAU to stress granules in HeLa cells (18). A highly conserved 13 amino acid core sequence contained within this region was essential for interaction with both RNA and DNA quadruplexes. While a truncation containing the first 105 amino acids of RHAU was sufficient for specific binding to G4-quadruplexes, the full length protein demonstrated higher affinity and was necessary for helicase activity (19).
Telomerase reverse transcriptase is an enzyme that has gained considerable interest in the context of human disease as it plays a critical role in telomere extension and immortalization of tumor cells (20). Telomerase is a reverse transcriptase composed of two main subunits, the catalytic protein TERT as well as the telomerase RNA (human telomerase RNA [hTR]/human telomerase RNA component [hTERC]). In addition to TERT and hTR, several accessory proteins regulate the activity of the enzyme (21). Telomerase catalyzes the extension of telomeres through interactions with telomere ends followed by generation of telomeric DNA from the hTR RNA template. As telomerase activity is essential for most tumor types, the telomerase enzyme has become an attractive therapeutic target (21–23).
The 5′ region of the telomerase RNA has been shown to adopt a quadruplex structure that may play a regulatory role in enzyme function (24,25). This quadruplex forming domain coincides with a region that participates in a long-range base-paired helix (P1 helix) (26). To confine reverse transcription to only the 6-nt RNA template sequence, a physical boundary in the RNA must be established to prevent the misincorporation of incorrect nucleotides. In hTR, the P1 helix serves as this boundary (27). Specific alteration of the distance between P1 and the 6-nt RNA template results in aberrant template copying both in vitro and in vivo, highlighting the importance of a proper boundary condition (27,28). Given the central role that the P1 helix plays in ensuring accurate reverse transcription, it is expected that modifications of this RNA structure would significantly impact the ability of hTR to present a functional RNA template for the synthesis of telomeric DNA. As quadruplex formation within this region was shown to disrupt the P1 helix (24,29) it is reasonable to hypothesize that quadruplexes within this region would require resolution to form the proper template boundary and RHAU is an ideal candidate helicase for this process.
A recent article by Sexton et al. (30) demonstrated an interaction between RHAU and TERT that was dependent upon overexpression of the hTR RNA in HEK293 cells. G to C substitutions within the putative quadruplex forming region resulted in reduced binding of RHAU. While the mutations did not demonstrate a substantial impact on telomerase activity in 293 cells, overexpression of mutant versions of hTR in a cell line deficient in endogenous hTR expression showed reduced incorporation of the RNA into the telomerase enzyme. Mutations in the quadruplex forming region resulted in reduced telomerase activity as well as a reduction in average telomere length (30). The authors propose that the quadruplex forming region of hTR acts as a protective 5′ cap and that RHAU resolves the quadruplex to favour incorporation into the telomerase holoenzyme. This study is supported in part by a recent paper by Lattmann et al. (31) in which an interaction dependent upon the RSM of RHAU and the 5′ region of TERC is confirmed.
In this study, we confirm an interaction between the hTR RNA and the N-terminal domain of RHAU. We characterize this interaction in vitro and have validated the results with the endogenously expressed RHAU and hTR. We have established the existence of multiple quadruplex structures within the first 43 nt of hTR and identified the minimal sequences of hTR sufficient for quadruplex formation and RHAU interaction. We demonstrate that an internal quadruplex comprising nucleotides 11–28 inhibits P1 helix formation and that following P1 helix formation, a quadruplex forms within the first 17 non-base pairing nucleotides. Finally, we have investigated the biological implications of the interaction and have defined an important role for RHAU in the promotion of P1 helix assembly. We present data for the first time supporting a helicase function of RHAU on an intramolecular RNA quadruplex and show that the enzyme is capable of completely converting quadruplex RNA to a stable duplex. Preliminary evidence supporting an impact of RHAU on telomerase function is provided in that RHAU knockdown by small interfering RNA (siRNA) results in a significant reduction in average telomere lengths.
MATERIALS AND METHODS
Cell culture and reagents
The HEK293T cell line was a gift from Dr Thomas Klonisch and the HeLa cell line was a gift from Dr Spencer Gibson. Cells were maintained in a humidified incubator at 37°C with 5% CO2 and passaged every 3 days. All cells were grown in Dulbecco’s Modified Eagle’s Medium (Invitrogen, Burlington, ON, Canada) supplemented with 10% fetal calf serum (Invitrogen), 100 U/ml penicillin and 100 µg/ml streptomycin (Invitrogen). Monoclonal anti-RHAU (clone 12F33) was purified from a hybridoma, which was a kind gift from Dr Yoshikuni Nagamine. Briefly, the hybridoma was adapted to hybridoma serum-free media (Invitrogen) and cells were expanded in a high surface area corning HYPERFlask (Fisher Scientific, Pittsburgh, PA, USA). Cells were grown to confluence and separated from media by centrifugation. The monoclonal antibody was purified on a HiTrap protein G column (GE Life Sciences, Piscataway, NJ, USA) and stored at −20°C in 10 mM sodium 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES), pH 7.5 with 150 mM NaCl, 50% glycerol and 100 µg/ml bovine serum albumin (BSA). The polyclonal rabbit anti-RHAU antibody was purified from rabbit serum, a kind gift from Dr Yoshikuni Nagamine, by peptide affinity chromatography and stored in the same manner as above. The following additional antibodies were used: Mouse anti-synthetic hapten (isotype control; Abcam), rabbit anti-protein kinase R (PKR) (3072 Cell Signaling Technology, Boston, MA, USA), mouse anti-α-tubulin (T6074 Sigma-Aldrich, Oakville, ON, Canada). Synthetic RNAs and DNA primers were purchased from Integrated DNA Technologies (Coralville, IA, USA). Adenylyl-imidodiphosphate (AMP-PNP) was purchased from MP Biomedicals (Solon, OH, USA).
Quadruplex formation and disruption
Quadruplex formation was carried out by dissolving synthetic RNAs in 20 mM Tris pH 7.6, 100 mM KCl, 1 mM Ethylenediaminetetraacetic acid (EDTA) at a concentration of 5 µM. RNAs were heated to 95°C for 5 min and allowed to passively cool to room temperature. To disrupt quadruplex formation, 100 mM LiCl was substituted for 100 mM KCl.
In-gel quadruplex staining
The quadruplex specific dye n-methyl mesoporphyrin IX was dissolved in 0.2 N HCl at a concentration of 5 mg/ml and stored at 4°C (Frontier Scientific, Logan, UT, USA) (32). For in-gel staining, 200 pmols of RNA was separated by native Tris–borate EDTA (TBE) polyacrylamide gel electrophoresis. Following electrophoresis, the gel was immersed in 10 ml staining solution (1 µg/ml n-methyl mesoporphyrin IX in 20 mM Tris pH 7.6, 100 mM KCl, 1 mM EDTA) for 15 min at room temperature on a rocking platform. Gels were visualized on a Fluorchem Q imaging system with the Cy3 excitation and emission filters.
Western blotting
All whole-cell lysates were prepared with radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris pH 8.0, 150 mM NaCl, 2 mM EDTA, 1% non-ident P-40, 0.5% sodium deoxycholate, 0.1% SDS) supplemented with protease inhibitors (Complete mini, Roche, Laval, QC, Canada) and phosphatase inhibitors (phosphatase inhibitor cocktails 1 and 2, Sigma-Aldrich). Equal amounts of protein were resolved by sodium dodecyl sulfate/polyacrylamide gel electrophoresis (SDS/PAGE) and transferred to polyvinyl difluoride membranes. Membranes were blocked in Tris-buffered saline containing 0.1% Tween-20 (TBS-T) and 5% skimmed milk powder. Primary antibodies were incubated overnight at 4°C in 5% milk TBS-T. Following incubation, membranes were washed 3-fold in TBS-T and then incubated with the appropriate secondary antibody conjugated to horse radish peroxidise (HRP) for 1 h at room temperature in 5% milk TBS-T. Proteins were visualized on a FluorChem Q imaging system (Cell Biosciences, Santa Clara, CA, USA) by enhanced chemiluminescence (GE Healthcare, Piscataway, NJ, USA).
siRNA transfections
siRNA transfections were carried out using lipofectamine RNAiMAX (Invitrogen) using the reverse transfection protocol supplied by the manufacturer. For long-term siRNA knockdowns, cells were retransfected and split every 3 days throughout the experiment. Prevalidated siRNA duplexes were obtained from Invitrogen. The sequence of the DHX36 siRNA is as follows: GGUGUUCGGAAAAUAGUAA. As a control, all experiments were performed using an equal amount of the Medium GC StealthRNAi Universal Negative Control duplex (12 935–300) from Invitrogen Corporation (control siRNA).
RNA gel shift assays
RNA–protein binding reactions were prepared in a 25 -µl volume in 50 mM Tris–acetate pH 7.8 with 100 mM KCl, 10 mM NaCl, 3 mM MgCl2, 70 mM glycine and 10% glycerol (19). RNA concentrations were kept constant at 200 nM with protein concentrations ranging from 0 to 1 µM. The binding reactions were incubated at room temperature for 15 min and then resolved by native electrophoresis on a 12% TBE gel. RNA was stained with the SYBR Gold fluorescent nucleic acid stain (Invitrogen) and visualized on a FluorChem Q imaging system (Cell Biosciences).
RNA immunoprecipitation
To perform RNA immunoprecipitations, cells were harvested from confluent 150 mm dishes. The cell pellet was resuspended in cytoplasmic lysis buffer [25 mM HEPES pH 7.9, 5 mM KCl, 0.5 mM MgCl2, 0.5% NP-40 supplemented with protease inhibitors (Roche Complete Mini)]. The cells were lysed on ice for 10 min and centrifuged at 5000 rpm in a bench-top microfuge for 5 min. The supernatant was set aside on ice and the cell pellet was resuspended in nuclear lysis buffer [25 mM HEPES pH 7.9, 10% (w/v) sucrose, 350 mM NaCl, 0.01% NP-40 supplemented with protease inhibitors (Roche Complete Mini)]. To lyse nuclei, the tubes were vortexed for 30 s followed by a 30-min incubation at 4°C with end-over-end mixing. The nuclear and cytoplasmic fractions were combined and insoluble material was removed by centrifugation at 14 000 rpm in a bench-top microfuge at 4°C followed by passage through a 0.45 -µm syringe filter. The cell lysate was then precleared for 30 min with 50 µL Pierce protein-G agarose (Fisher Scientific). For immunoprecipitations, 10 µg of each antibody was added to 1 mg of precleared cell lysate and incubated for 2 h at 4°C with end-over-end mixing. After 2 h, 50 µL preequilibriated protein G-agarose was added to each sample and mixing was continued for 1 h at 4°C. Protein G-agarose beads were pelleted and washed 4-fold with RIPA buffer. Beads were resuspended in buffer RLT from the Qiagen RNeasy Minelute Cleanup kit and heated to 95°C for 3 min to denature the protein–RNA complexes. Immunoprecipitated RNA was then purified according to the manufacturer’s protocol. For real-time PCR analysis, 2 µl of the purified RNA was used directly with the iScript one-step RT-PCR kit (Biorad Laboratories, Hercules, CA, USA) according to the manufacturer’s protocol with an annealing temperature of 55°C. For gel based analysis, 200 ng of purified RNA was reverse transcribed to cDNA using avian myeloblastosis virus (AMV) reverse transcriptase (New England Biolabs, Ipswich, MA, USA) with template specific primers in a 20 µl volume according to the manufacturer’s protocol. cDNA of 4 µl was amplified by PCR with primers specific to the hTR or glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as a negative control. Cycling conditions were as follows: 95°C for 5 min followed by 25 cycles of 95°C for 10 s, 55°C for 30 s, 72°C for 30 s, followed by a final elongation step at 72°C for 10 min. PCR products were analyzed by agarose gel electrophoresis. The primer sequences used are as follows: hTR-forward: TCTAACCCTAACTGAGAAGGGCGT, hTR-reverse: TGCTCTAGAATGAACGGTGGAAGG, GAPDH-forward: ACCCACTCCTCCACCTTTG, GAPDH-reverse: CTCTTGTGCTCTTGCTGGG.
Streptavidin pull-down assays
To assess RHAU interactions with various truncations of the hTR RNA, a streptavidin pull-down assay was performed with biotin labeled hTR RNAs. Synthetic RNAs were biotinylated with the Pierce 3′ RNA biotinylation kit (Fisher Scientific) according to the manufacturer’s protocol. Biotinylation efficiency was assessed by dot blotting on a positively charged nylon membrane and detection was performed with streptavidin-HRP and enhanced chemiluminescence on a FluorChem Q imaging system. Whole-cell extraction was performed on 20 × 106 cells per time point. Whole-cell extracts were precleared with 50 µl streptavidin agarose beads (Invitrogen) for 30 min at 4°C with rotation. Following preclearance, a binding reaction was prepared that contained 500 µg whole-cell extract, 50 ng/µl poly I-C, 1/2 volume binding buffer (50 mM Tris–acetate pH 7.8, 100 mM KCl, 10 mM NaCl, 3 mM MgCl2, 70 mM glycine and 10% glycerol) and 100 nM biotin-labelled probe. Binding reactions were incubated for 30 min at room temperature after which 50 µl streptavidin–agarose beads were added. Following a 30-min incubation, beads were spun down at 3000 rpm for 1 min and washed three times for 5 min in lysis buffer followed by three additional washes in phosphate-buffered saline. After washing, the beads were resuspended in 50 µl 2 × SDS loading dye, boiled for 5 min and the isolated proteins were separated by SDS/PAGE and western blotting was performed to assess protein binding.
Cloning, expression and purification of RHAU53–105
The RHAU53–105 fragment was generated by PCR from the RHAU-FLAG vector (RHAU1–1008 in pIRES-EGFP-FLAG-N1), a generous gift from Dr Y. Nagamine (FMI, Basel, Switzerland). The fragment was cloned into the pET15b vector. The final RHAU53–105 construct contained an N-terminal hexahistidine tag and a thrombin cleavage site for tag removal. The protein was expressed in the Escherichia coli strain BL21(DE3) and purified using a cobalt affinity resin. The cells were lysed in 20 mM sodium phosphate, 2 mM imidazole, 6 M guanidine hydrochloride, pH 7.0 and the clarified lysate was applied to an equilibrated affinity column. Refolding was achieved on column, with 20 mM sodium phosphate, 2 mM imidazole, pH 7.0, 286 mM sodium chloride. Following washing and elution, the hexahistidine tag was cleaved with thrombin and the eluate was dialysed into 10 mM HEPES, pH 7.5, 150 mM sodium chloride. Thrombin was removed by applying the retentate to a HiTrap benzamidine FF column (GE Healthcare).
Full-length RHAU purification and activity assays
The full-length RHAU protein was purified as previously described (19). RHAU helicase assays were performed by combining 200 nM hTR RNA with 400 nM 25P1 RNA in 1× buffer (50 mM Tris–acetate, pH 7.8 with 100 mM KCl, 10 mM NaCl, 3 mM MgCl2, 70 mM glycine, 10% glycerol and 1 mM ATP) in a 25 -µl reaction volume. RHAU (50 nM) was added and the helicase assay was carried out for 30 min at 30°C. Reaction products were resolved on a 10% native TBE polyacrylamide gel.
Induced formation of P1 helix
To induce P1 helix formation, 40 µM of hTR RNA was combined with 80 µM 25P1 RNA in a 25 µl volume and heated at 95°C for 5 min. Following heating, the samples were allowed to cool to room temperature for 15 min.
Measurements of telomere length
For relative telomere length measurements, genomic DNA was isolated from 1 × 106 cells per sample using the DNeasy Blood and Tissue Kit (Qiagen) and quantified by measurement of the absorbance at 260 nm. Relative telomere lengths were determined using a previously published protocol (33) and referenced to the single copy gene albumin. Assay validation over a range of genomic DNA concentrations is presented in Supplementary Figure 8. RT-PCR was performed on a MyIQ thermal cycler and data analysis was performed with IQ5 Optical System Software (Biorad).
RESULTS
RHAU interacts with the hTR
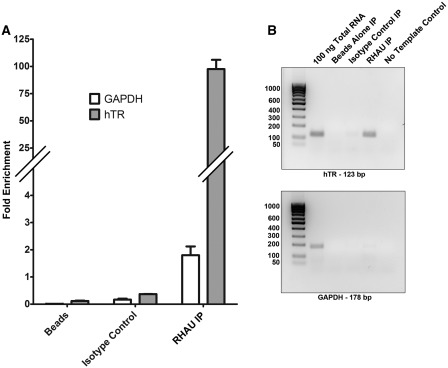
RHAU has previously been demonstrated to exhibit specificity for both DNA and RNA G4-quadruplex structures in terms of affinity and helicase activity (7,15,19). hTR contains several guanine runs in the 5′ region that are known to form quadruplex in vitro. These guanine tracts are diagrammed in the context of the hTR RNA secondary structure in Figure 2F. Quadruplex formation in this region is reported to interfere with formation of the P1 helix (24). We hypothesized that RHAU interacts with the quadruplex forming region of hTR and recently published data (30,31) as well as our own data support this hypothesis. We analyzed the interaction between the endogenously expressed protein and RNA by RNA coimmunoprecipitation experiments. To confirm the interaction between RHAU and hTR, RHAU was immunoprecipitated from HEK293T whole-cell lysates without crosslinking. Copurified RNA was isolated, followed by detection and quantification by reverse transcription and RT-PCR. Results were expressed relative to the amplification of 10 ng of a total RNA extraction. Figure 1A confirms an ∼100-fold enrichment of hTR by RHAU immunoprecipitation when compared with the GAPDH mRNA, which is only marginally enriched (<2-fold). Immunoprecipitation controls with no antibody (beads alone) or an isotype matched antibody fail to enrich either hTR or the GAPDH mRNA. A similar experiment was performed by reverse transcription followed by 25 cycles of standard PCR to allow for visualization of the products on an agarose gel. Figure 1B demonstrates a single product produced for each primer set with specific enrichment of hTR by RHAU immunoprecipitation with no enrichment observed for any of the negative controls. These results support and confirm an interaction between endogenously expressed RHAU and hTR.
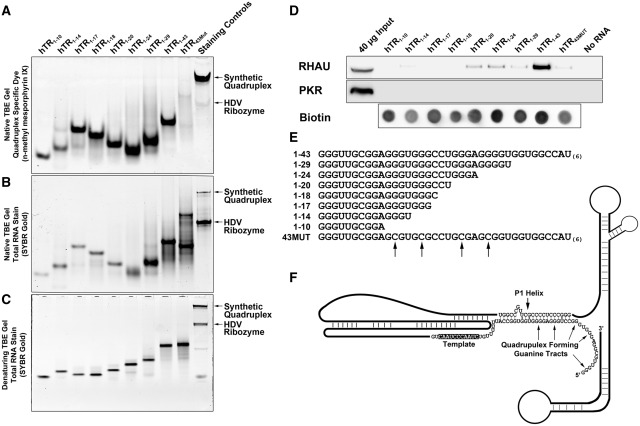
Figure 2.
hTR1–17 is the minimal sequence capable of forming a G4-quadruplex and hTR1–20 is the smallest truncation that demonstrates affinity for endogenous RHAU. (A) Approximately 200 pmols of each hTR RNA truncation was separated by native TBE polyacrylamide gel electrophoresis and stained with 1 µg/ml n-methyl mesoporphyrin IX diluted in 20 mM Tris pH 7.5, 100 mM KCl and 1 mM EDTA for 15 min at room temperature. The gel was imaged on a Fluorchem Q system using the Cy3 excitation and emission filters. (B) Approximately 5 pmols of each hTR RNA truncation was separated as in (A) and the gel was stained with the total RNA stain SYBR Gold according to the manufacturer’s protocol. (C) Approximately 5 pmols of each hTR RNA truncation was heated at 95°C for 5 min in 1× denaturing load dye and separated by denaturing TBE polyacrylamide gel electrophoresis and stained with the total RNA stain SYBR Gold according to the manufacturer’s protocol. (D) Western blot of proteins enriched by streptavidin pull-down assays performed with biotinylated hTR truncations. 3′ biotinylated hTR truncations were incubated with HEK293T whole cell extracts for 30 min and protein/RNA complexes were pulled-down with 50 µl streptavidin agarose beads. The beads were boiled for 5 min in 1× SDS loading dye and the binding of RHAU to each RNA was assessed by performing SDS/PAGE and western blotting. As a control for binding specificity, blots were reprobed with anti-PKR antibodies and to control for non-specific interactions with the streptavidin agarose, a beads alone (no RNA) control was performed. Biotinylation efficiency for each RNA is demonstrated by a dot blot of 5 pmols of each RNA detected with streptavidin-HRP. (E) Sequences of each of the RNAs used for the streptavidin pull-down assay. G to C substitutions made in 43MUT are indicated by arrows. (F) Schematic of the hTR RNA based upon the published proposed secondary structure highlighting the P1 helix and overlapping quadruplex forming guanine tracts (26).
Figure 1.
RHAU interacts with hTR. (A) RT-PCR quantification of RNA immunoprecipitations. RHAU was immunoprecipitated from 500 µg of HEK293T whole cell lysate and the coprecipitated RNA was isolated and purified using the Qiagen RNEasy MinElute kit. As a negative control, an immunoprecipitation was performed with no antibody (beads alone) as well as an isotype matched antibody. Reverse transcription and RT-PCR were performed with primers specific for hTR or, as a control for specificity, GAPDH. Coprecipitated RNA of 2 µL (10%) was analyzed and measured in triplicate. RHAU immunoprecipitation resulted in a ∼100-fold enrichment of the hTR RNA, whereas the GAPDH RNA was enriched ∼2-fold. Fold enrichment was calculated by the comparative CT method relative to 10 ng of a total RNA extraction. Data represents the mean + standard error. (B) A similar experiment as in (A); with the exception that standard PCR was carried out following reverse transcription for 25 cycles and products were analyzed by agarose gel electrophoresis. Total RNA of 100 ng was used as a positive control.
Nucleotides 1–17 comprise the minimal length of the 5′ region of hTR capable of forming quadruplex
While several reports have identified quadruplex formation within the 5′ region of hTR (24), seven conserved guanine runs exist within the first 40 nt that could participate in the structure (Figure 2E). As four guanine tracts represent the hypothetical number capable of forming an intramolecular quadruplex, multiple quadruplex structures are possible within this region of hTR. While one publication suggests only the first 17 nt are involved in quadruplex formation (25), other groups conclude that internal guanine runs corresponding to the region necessary to base-pair in the P1 helix play a critical role (24,30,31). To clearly define the minimal sequence of the hTR RNA capable of forming a quadruplex, we generated synthetic RNAs with successive deletions from the 3′-end (Figure 2E) and analyzed equimolar quantities on native and denaturing TBE polyacrylamide gels followed by staining for both quadruplex and total RNA. The longest RNA studied (hTR1–43) consists of the first 43 nt of hTR and contains seven guanine runs that could potentially participate in quadruplex formation. Truncations hTR1–29, hTR1–24 and hTR1–20 each successively remove one guanine tract, leaving a minimal number of runs to form a quadruplex in hTR1–20. As is shown in Figure 2A, each of these RNAs demonstrate similar staining intensity for the quadruplex dye n-methyl mesoporphyrin IX. Deletion of the terminal nucleotides of hTR1–20 to generate the hTR1–18 and hTR1–17 RNAs resulted in significant alteration of migration on the gel, presumably due to a loss of charge, but no loss in staining intensity with the quadruplex specific dye. Truncation 1–14, which contains only three guanine runs, shows a marked decrease in staining intensity, with residual staining possibly due to the formation of a small population of intermolecular quadruplexes. G to C substitutions made in the hTR43MUT RNA abolish staining with n-methyl mesporphyrin IX. To control for staining specificity, a sample containing both an intermolecular quadruplex (A15G5A15) control as well as a negative control, the HDV Ribozyme was examined. Each RNA of 5 pmols were also run out on a native gel (Figure 2B) and denaturing TBE–urea gel (Figure 2C) and stained with SYBR Gold to demonstrate the presence of RNA in each lane. Unlike n-methyl mesoporphyrin IX, which is expected to stain each RNA containing a similar quadruplex with equal intensity regardless of total RNA length, the SYBR Gold intensity is based on total nucleic acid quantity with some variation due to secondary structure elements.
To assess whether these RNAs could interact with RHAU in cell lysates, a streptavidin pull-down assay was performed with each of the RNA truncations. As is shown in Figure 2D, the minimal sequence capable of enriching the endogenous RHAU protein consists of the first 20 nt. Similar binding is observed for hTR1–24 and hTR1–29; however, enrichment is significantly increased when the pull down is performed with hTR1–43. Whether this represents an increased affinity or enhanced biotin accessibility remains to be determined. To control for specificity, the blot was reprobed with antibodies to PKR, an established double-stranded RNA binding protein, which demonstrated no enrichment with any of the RNAs tested. Biotinylation efficiency for each RNA was similar, as is shown by a dot blot of 5 pmols of each RNA with detection by streptavidin-HRP (Figure 2D).
An N-terminal truncation of RHAU containing the RSM interacts with hTR1–43
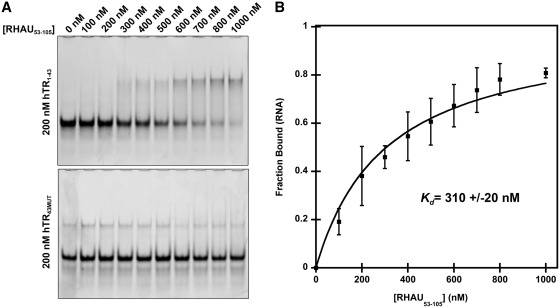
It has been well established that a region within the N-terminus of RHAU, referred to as the RSM, plays an essential role in the recognition and binding to G4-quadruplexes (19). To determine if the region corresponding to the RSM was sufficient for interacting with hTR, we performed electrophoretic mobility shift assays with an N-terminal truncation of RHAU consisting of amino acids 53–105 (RHAU53–105). Interactions were probed with the first 43 nt of hTR (hTR1–43) as well as a version of hTR1–43 that contained four G to C substitutions to disrupt quadruplex formation (hTR43MUT). RHAU53–105 was selected based upon predictions indicating minimal impact on secondary structure elements as well as elimination of an N-terminal glycine-rich region which may impede future structural analysis. hTR was maintained at a constant concentration of 200 nM in a 25 -µl binding reaction with increasing concentrations of RHAU53–105. Figure 3 demonstrates an interaction between hTR1–43 with ∼80% of the RNA in complex when RHAU53–105 is present at a 4-fold molar excess. Similar interactions with RHAU53–105 were obtained with the truncations hTR1–29, hTR1–24, hTR1–20, hTR1–18 and hTR1–17 (data not shown). This is in contrast to hTR43MUT in which no significant binding takes place over the range of RHAU53–105 concentrations. Quantification data (Figure 3B) is based upon densitometry analysis of three independent experiments (Supplementary Figure 1). Using a previously published method, the data reveal a dissociation constant of 310 nM for the hTR1–43-RHAU53–105 interaction (34). This suggests the interaction between RHAU and hTR is both quadruplex dependent and mediated through the RSM.
Figure 3.
hTR1–43 interacts with an N-terminal RHAU truncation (RHAU53–105) containing the RSM. (A) Electrophoretic mobility shift assay demonstrating a specific interaction of RHAU53–105 with hTR1–43. 200 nM hTR RNA was incubated in a binding reaction with increasing concentrations of RHAU53–105 for 15 min at room temperature and the free RNA and RNA/protein complexes were resolved by native TBE polyacrylamide gel electrophoresis and stained with the nucleic acid dye SYBR Gold. The lower gel demonstrates a loss of interaction when G to C substitutions are introduced into the RNA sequence (hTR43MUT). (B) Quantification of the free RNA band for hTR1–43. Data represent the mean of three independent experiments ± standard deviation. Curve fitting and calculation of Kd was performed by a previously published method (34). Additional gel images are provided in Supplementary Figure S1.
RHAU promotes P1 helix formation in hTR1–43
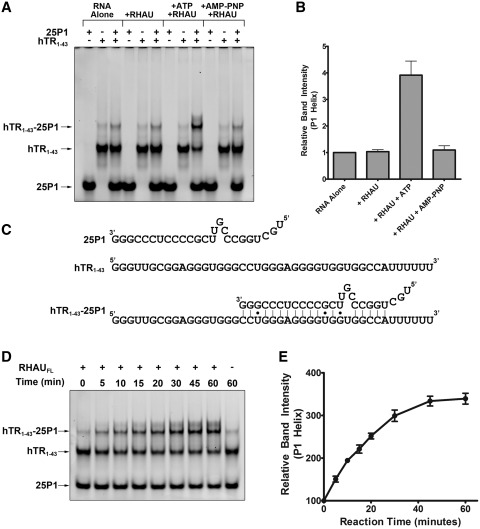
Quadruplex formation within the 5′ region of hTR (Figure 2F) was previously hypothesized to inhibit the base pairing necessary to form the upstream P1 helix (24). The P1 helix of hTR is a critical structural element for establishing the template boundary of reverse transcription (27). To study the function of RHAU, we purified the full-length protein from HEK293T cells (Supplementary Figure S2). Preliminary experiments demonstrated that the purified protein was capable of unwinding a synthetic intermolecular quadruplex but did not show a clear impact on hTR (Supplementary Figure S3). Studying the impact of RHAU on an intramolecular quadruplex is confounded by the likelihood that, without a method of structure stabilization, the RNA quickly refolds into a quadruplex upon dissociation of the enzyme. To circumvent this obstacle, we used a previously published method using a complementary 25 nt internal fragment of hTR, referred to as 25P1, to study the potential role for RHAU in resolution of the 5′ quadruplex to promote the formation of the P1 helix (24). 25P1 and hTR1–43 were incubated either alone or together in the presence and absence of RHAU and ATP to determine if the helicase activity of RHAU could promote hTR unwinding and annealing of 25P1 (Figure 4A and B). As is clearly shown by the electrophoretic mobility shift assay, 25P1 and hTR1–43 form a detectable level of complex when combined alone. Inclusion of RHAU does not enhance duplex formation; however, inclusion of RHAU in the presence of ATP results in a nearly 4-fold increase in formation of the P1 helix. To confirm the dependence of this observation on ATP hydrolysis, the experiment was repeated using a non-hydrolysable ATP analog AMP-PNP. Under these conditions there was no promotion of the P1 helix structure (Figure 4B). Time-course analysis of the helicase activity reveals a maximal conversion of the quadruplex RNA to the P1 duplex following 45 min incubation (Figure 4D and E). These data strongly suggest that the ATP-dependent helicase activity of RHAU renders quadruplex-prone regions of hTR accessible for base pairing with the 25P1 RNA.
Figure 4.
RHAU promotes the formation of the hTR P1 helix. (A) Native TBE gel electrophoresis of the RNAs 25P1 and hTR1–43 both alone and in complex. Duplex formation was assessed with 25P1 and hTR1–43 alone and together in the presence and absence of the full length recombinant RHAU protein and either 1 mM ATP or 1 mM AMP-PNP. Approximately 200 nM of hTR1–43 was combined with 400 nM 25P1 in a 25 µl reaction ± 50 nM RHAU and 1 mM ATP/1 mM AMP-PNP for 30 min at 30°C. RNAs were separated by native electrophoresis and stained with SYBR Gold. (B) Densitometry analysis of the hTR1–43-25P1 complex band intensity relative to the RNA alone lane. Data reveals an ATP-dependent 4-fold increase in P1 helix formation in the presence of RHAU. Data represent the mean of three independent experiments ± standard deviation. Additional gel images are provided in Supplementary Figures S4 and S5. (C) Schematic representing sequence details of the 25P1 RNA, hTR1–43 as well as the expected double stranded interaction product. (D) Time-course analysis of the hTR43-25P1 duplex formation in the presence of RHAU and 1 mM ATP. RHAU was added to the reaction mixture and the tubes were incubated at 30°C for the indicated time-points. RNAs were then separated by Native TBE gel electrophoresis. (E) Densitometry analysis of the hTR1–43 -25P1 complex band intensity relative to the 0 min time-point. Data represent the mean of three independent experiments ± standard deviation. Additional gel images are provided in Supplementary Figure S6.
An internal quadruplex is responsible for P1 helix inhibition
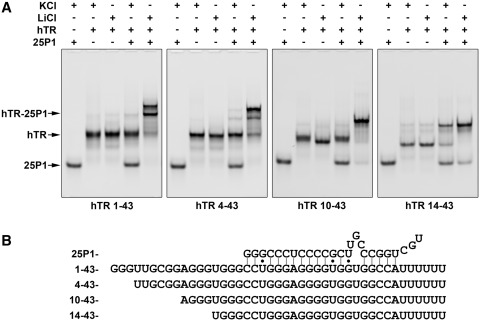
In Figure 2A, we demonstrate that hTR1–17 is the smallest fragment capable of forming a stable quadruplex. Analyzing the sequence involved in formation of the P1 helix reveals that nucleotides 1–17 correspond to the region that is not involved in interactions with 25P1 (Figure 5B). To assess whether the quadruplex consisting of the first four guanine tracts is responsible for inhibition of P1 helix formation, we prepared truncations from the 5′-end of hTR that disrupt formation of the 1–17 quadruplex but leave the region involved in P1 helix formation intact. Interactions of each of these truncations with 25P1 were assessed by electrophoretic mobility shift assays under buffer conditions containing KCl or LiCl. While KCl stabilizes the quadruplex structure, previous work has demonstrated that substitution of LiCl is sufficient to destabilize the quadruplex structure (24). Therefore, all truncations were expected to form an interaction with 25P1 in the presence of LiCl but not KCl unless the inhibitory quadruplex was abolished by truncation. As is shown in Figure 5A, hTR1–43, hTR4–43 and hTR10–43 do not interact with 25P1 in the presence of KCl, but show nearly complete interaction in the presence of LiCl. This suggests that a quadruplex exists in these RNAs that prevents interaction with 25P1. In addition, these results suggest that the inhibitory quadruplex forms independent of the first two guanine tracts and that deletion of the third guanine tract (hTR14–43) results in disruption of the inhibitory quadruplex. This is evident in that hTR14–43 forms a complex with 25P1 in the presence of KCl. Substitution of LiCl promotes complete interaction with 25P1, indicating that a quadruplex involving the remaining four guanine tracts may persist to some degree. In summary, while the first 17 nt are sufficient to form a stable quadruplex, the major structure inhibiting P1 helix formation in hTR1–43 appears to be a quadruplex between nucleotides 11 and 28.
Figure 5.
Deletion of the first 13 nt of the hTR RNA disrupts a quadruplex responsible for blocking P1 helix formation. (A) Electrophoretic mobility shift assays examining the formation of P1 helix in hTR RNAs containing successive truncations from the 5′-end. Each hTR truncation was incubated in the presence and absence of 25P1 in a buffer containing either 100 mM KCl or 100 mM LiCl. All RNAs with the exception of hTR14–43 failed to interact in the presence of KCl. Quadruplex disruption by LiCl resulted in nearly complete interaction of the hTR RNAs with 25P1. hTR14–43 demonstrated significant interaction in the presence of KCl that was enhanced in the presence of LiCl. The two upper bands present in the 25P1 duplex formed with hTR1–43 and hTR4–43 are likely due to alternative conformations of the single stranded RNA outside of the duplex. (B) Schematic detailing the sequence of each hTR truncation as well as the 25P1 RNA and the expected interaction site.
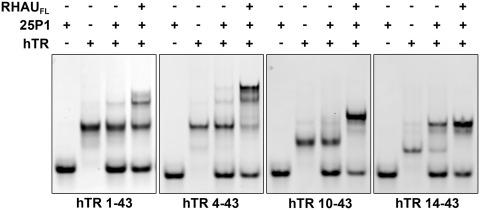
RHAU unwinds an internal quadruplex to promote P1 helix formation
To assess whether RHAU is capable of unwinding the internal inhibitory quadruplex present in the hTR 5′ truncations, we assessed RHAU helicase activity on each of the RNAs in the presence of the complementary strand 25P1. hTR1–43 demonstrated a similar enhancement of the interaction with 25P1 in the presence of RHAU and ATP (Figure 6). RHAU had a similar impact on hTR4–43, but demonstrated an enhanced activity. Activity appeared further enhanced when RHAU was incubated with hTR10–43 with complete conversion of the RNA to the double-stranded P1 helix. While the majority of hTR14–43 formed a duplex with 25P1 independent of RHAU, inclusion of the helicase enhanced the interaction and resulted in complete conversion of the free RNA to the 25P1 duplex. These data demonstrate that RHAU is capable of unwinding quadruplexes within all of the hTR 5′ truncations to promote an interaction with 25P1.
Figure 6.
RHAU promotes the formation of the hTR P1 helix in all hTR 5′ truncations. Binding reactions were performed for each hTR RNA truncation with a 2-fold excess of the 25P1 RNA in the presence and absence of the full length RHAU protein. RNAs were incubated at 30°C for 30 min and then resolved by native TBE polyacrylamide gel electrophoresis. RHAU strongly enhanced P1 helix formation for each of the RNAs with enhanced efficiency observed in the 5′ truncated forms.
The 1–17 quadruplex and P1 helix are not mutually exclusive
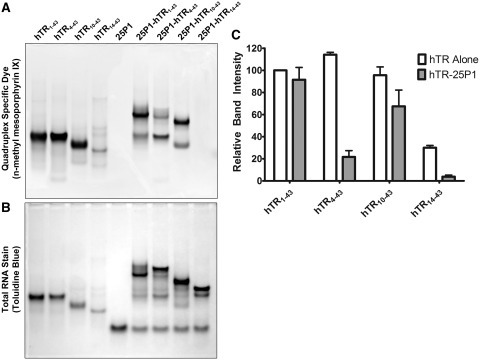
Previous reports assume that a quadruplex in the 5′ region of hTR and the P1 helix are mutually exclusive structures. The data presented suggests that the 1–17 quadruplex is not the structure responsible for P1 helix inhibition and that an internal quadruplex, formed with guanines 11–13, 15–17, 21–23 and 26–28, represents the primary inhibitory structure. As we previously demonstrated that hTR1–17 is capable of forming quadruplex, and as the first 17 nt do not base pair in the P1 helix, we set out to determine if a quadruplex in the first 17 nt could persist in the presence of the P1 helix. hTR1–43 as well as the 5′ truncations hTR4–43, hTR10–43 and hTR14–43 were heated to 95°C in the presence of a 2-fold molar excess of 25P1 to induce the formation of the P1 helix. The free RNAs as well as the hTR-25P1 duplexes were then separated by native TBE polyacrylamide gel electrophoresis and stained with the quadruplex specific dye n-methyl mesoporphyrin IX. As is shown in Figure 7A, the free RNAs hTR1–43, hTR4–43 and hTR10–43 all stain with similar intensity (quantified in Figure 7C) and a significant reduction in staining is observed for hTR14–43. This result is expected based both upon the primary sequence of the truncations (Figure 5B) and our data which demonstrates that hTR14–43 does not form a stable inhibitory quadruplex (Figure 6). As is expected, 25P1 alone (lane 5) demonstrates no detectable staining for the quadruplex specific dye. When hTR1–43-25P1 complex is formed (lane 6), the staining intensity is nearly identical to hTR1–43 alone (lane 1). The 25P1 complex formed with hTR4–43 demonstrates significantly reduced quadruplex-specific staining when compared with hTR1–43, which is expected as only three guanine runs remain single stranded in the hTR4–43-25P1 duplex. As expected, the complex formed with hTR14–43 is undetectable by staining with n-methyl mesoporphyrin IX. Unexpectedly, the duplex formed with hTR10–43 still shows ∼70% of the staining intensity of the free RNA, suggesting the possibility that the remaining sequence adopts a quadruplex structure. The same gel was subsequently stained with toluidine blue to reveal total RNA. As was expected, toluidine blue stained the smaller truncations with reduced intensity as they were present in equimolar quantities. These data suggest that the free nucleotides 1–17 in the hTR1–43-25P1 complex form a stable quadruplex that is abolished upon deletion of the first guanine tract.
Figure 7.
A switch between an internal and terminal quadruplex occurs upon P1 helix formation. (A) hTR1–43 and successive 5′ truncations were heated to 95°C either alone or in the presence of a 2-fold molar excess of 25P1 for 5 min and then allowed to cool to room temperature. Approximately 500 pmols of each RNA was separated by native TBE polyacrylamide gel electrophoresis and stained with the quadruplex-specific fluorescent dye n-methyl mesoporphyrin IX. A significant decrease in staining intensity was observed for hTR14–43 indicating a loss of quadruplex structure. The hTR1–43-25P1 complex stained with the quadruplex-specific dye; however, this staining was completely lost in the hTR14–43-25P1 complex. (B) Following fluorescent visualization of the gel it was further stained with the total RNA stain toluidine blue. (C) Densitometry quantification of the bands in (A). In the case of hTR-25P1 complexes, the dominant band, as observed in (B), was chosen for quantification. Data represents the mean of three independent experiments ± standard deviation. Additional gel images are provided in Supplementary Figure 7.
RHAU can interact with the terminal 1–17 quadruplex retained in the hTR1–43-25P1 duplex
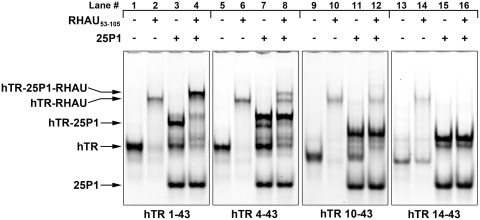
After confirming the presence of a quadruplex structure following P1 helix formation in hTR1–43 (Figure 7), we sought to determine whether this quadruplex retained affinity for RHAU. Electrophoretic mobility shift assays were performed with each of the hTR 5′ truncations alone or in complex with the complementary 25P1 RNA. As is clearly shown by the electrophoretic mobility shifts in Figure 8 (lanes 2, 6, 10 and 14), each of the free RNAs exhibit affinity for RHAU53–105; however hTR14–43 exhibited substantially reduced binding. Interestingly, P1 helix assembled with hTR1–43, forms a complex with RHAU53–105 (upper band, lane 4) as can be seen by the complete shift of the hTR-25P1 band seen in lane 3 to a higher molecular weight complex (lane 4). This indicates that the persisting quadruplex retains the ability to interact with RHAU. This interaction was disrupted upon truncation of the first 3 nt as hTR4–43 exhibited minimal interaction (lane 8). While all of the dominant hTR4–43-25P1 duplex observed in lane 7 remains in lane 8, a complex does form that appears to result from a subpopulation of hTR4–43-25P1 (second band from top, Figure 8, lane 7) that stained with the quadruplex-specific dye (Figure 7, lane 7), possibly due to incomplete base pairing with 25P1. In the case of hTR10–43 and hTR14–43, no interaction is evident for the hTR-25P1 complex with RHAU53–105 (lanes 12 and 16). These data mirror what was observed in Figure 7, in that RNAs or complexes demonstrating affinity for the quadruplex dye n-methyl mesoporphyrin IX also exhibit affinity for RHAU53–105.
Figure 8.
The RHAU RSM domain interacts with the terminal quadruplex of the hTR1–43-25P1 complex. hTR1–43 and successive 5′ truncations were heated to 95°C and allowed to cool either alone or in the presence of a 2-fold molar excess of 25P1. This process converted the majority of the hTR RNA into a complex with 25P1 (hTR-25P1, upper band in lanes 3, 7, 11 and 15). RHAU53–105 was added at a 3-fold molar excess and binding reactions were incubated for 15 min at room temperature. RNAs and RNA–protein complexes were separated by native TBE polyacrylamide gel electrophoresis and stained with SYBR Gold. RHAU53–105 demonstrated an interaction with all of the hTR truncations (lanes 2, 6, 10 and 14); however, significantly decreased affinity was observed for hTR14–43 (lane 14). RHAU53–105 demonstrated an interaction with the hTR1–43-25P1 complex (lane 4, upper band). This interaction is diminished in the case of the hTR4–43-25P1 (lane 8, upper band), and abolished in the case of both hTR10–43-25P1 (lane 12) and hTR14–43-25P1 (lane 16). Faint bands not identified by arrows represent residual alternative conformations of the RNA species.
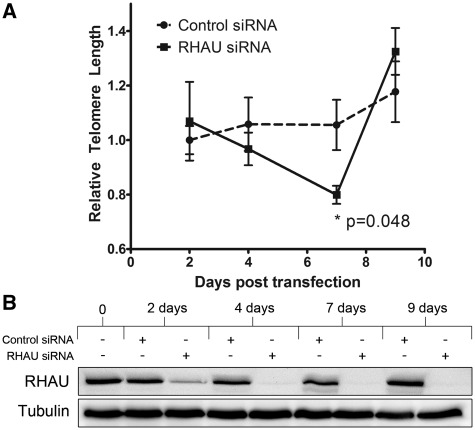
RHAU knockdown results in a reduction of average telomere length
To gain insight into the potential role for RHAU in regulating telomerase activity, we performed siRNA knockdowns of RHAU over the course of 9 days and assessed relative telomere lengths in the cells at various time points. This time course represents approximately 12–15 doublings of the initially transfected HEK293T cells. Telomere lengths were measure by a previously published monochrome, multiplex RT-PCR method, which was initially validated over a range of DNA concentrations (Supplementary Figure 8) (33). As is shown in Figure 9A, relative telomere length steadily decreased in the RHAU knockdown cells until 7 days, at which point it achieved a level that differed significantly (0.799 vs 1.06) from that of the cells transfected with a non-specific control siRNA (P = 0.048). Following 9 days of transfection, the decrease was reversed, indicating the possibility of a compensatory mechanism being used by the cells following prolonged RHAU siRNA knockdown. A similar reduction (13%) in telomere length following 7 days knockdown was observed in HeLa cells; however, the data were not statistically significant (P = 0.077 by paired one-tail t test; Supplementary Figure 9).
Figure 9.
RHAU knockdown by siRNA results in a reduction in average relative telomere length 7 days post-transfection that is recovered by 10 days. (A) Average relative telomere lengths were assessed by a quantitative RT-PCR-based assay using primers that generate a fixed length product and standardized to primers specific for the ALB gene (33). Data represent the average of three independent experiments measured in triplicate relative to the CT obtained for untransfected cells harvested at day zero. Asterisks indicates P = 0.048 at Day 7 as measured by paired one-tail t-test. (B) Western blot demonstrating efficiency of the RHAU siRNA knockdown. Cells harvested at each time-point were lysed in RIPA buffer and 40 µg from each time point was analyzed by SDS/PAGE followed by western blotting with antibodies specific for RHAU. Blots were reprobed with antibodies specific for β-tubulin as a loading control.
DISCUSSION
The biological relevance of G4-quadruplex structures in both DNA and RNA is only beginning to be unravelled. Indeed, while prediction algorithms suggest a widespread prevalence of G4-quadruplexes in both DNA and RNA, little in vivo evidence exists confirming the existence and significance of these unique structural motifs (35,36). The discovery of RHAU as a G4-specific helicase lends great support to the notion that G4-quadruplexes are not an in vitro artifact but are indeed a regulatable biological phenomenon (15). The enrichment of quadruplex structures in regulatory regions of the genome as well as the 3′ and 5′ regions of mRNAs suggests that quadruplexes play a significant role in the control of gene transcription and translation, a concept with far reaching implications (10,13). Further study of RHAU, the only known enzyme to modify these structural elements, can provide great insight into the potential array of functions that may be ascribed to the G4-quadruplex. Recent work by Lattmann et al. (31) presents greater than 100 mRNAs that have affinity for RHAU and therefore it can be expected that the role of RHAU in mRNA stability, processing and translational control will quickly expand.
The hTR contains as many as seven guanine tracts that are conserved in a wide variety of mammalian species (24). While certain homologs, such as the murine telomerase RNA, lack these ∼40-nt upstream of the template region, it does not preclude the possibility that they possess a regulatory function (37). In fact, while the P1 helix that forms 5′ of the template region has been ascribed a critical role in template boundary definition in human telomerase, other species appear to use alternative mechanisms (27). Furthermore, Li et al. (25) demonstrate interactions between the first 17 nt of hTR and two internal regions that appear to modulate activity of the enzyme. The same group also demonstrated that the truncation of the hTR at the 5′- end results in an enzyme with increased activity, implicating the region in a negative regulatory role (38). Herein, we sought to elucidate the regulatory function of the quadruplex prone 5′ region of hTR by investigating an interaction with the RNA helicase RHAU and studying the downstream consequences.
Supporting two other studies (30,31), we identified and confirmed an interaction of RHAU with the hTR. Through RNA coimmunoprecipitations and streptavidin pull-down assays, we demonstrate with two separate approaches an interaction between the endogenous RHAU protein and the hTR. Truncations of hTR from the 3′-end revealed that a quadruplex structure persists upon reduction of the first 43 nt of the RNA down to a small 17 nt fragment. While this fragment appears to form a stable quadruplex, the shortest RNA with affinity for the endogenous RHAU protein consists of the first 20 nt. Whether this is due to a lack of affinity for RHAU in the absence of the three non-guanine nucleotides 3′ of the quadruplex or if it is due to reduced accessibility of the 3′ biotin tag remains to be determined. While one previous study suggested the possibility of a quadruplex existing within the first 17 nt of the hTR RNA (25), the bulk of recent reports presume quadruplex formation occurs with guanine tracts that exist 3′ of this region (24,29,31). These data demonstrate quite conclusively that the first 17 nt are sufficient to form a stable quadruplex, but do not rule out the possibility of alternate quadruplex forms.
The RSM region of RHAU has been identified as a key component for the interaction of RHAU with G4-quadruplex DNA and RNA. A previously published study by Sexton et al. (30) determined that an N-terminal truncation consisting of the first 200 amino acids of RHAU had affinity for hTR. A recent article by Lattmann et al. (31) further demonstrated that a RHAU mutant lacking the RSM region was unable to interact with hTR. We show, through electromobility shift assays, that a 53 amino acid fragment containing the RSM domain is sufficient to interact with the 5′ region of hTR and fails to interact with the same RNA containing quadruplex disrupting G to C substitutions. Taken together, these data are quite convincing that RHAU interacts in a quadruplex-dependent manner with hTR through interactions with the RSM.
While the interaction between RHAU and hTR is now well defined, the precise functional consequences of this interaction remain elusive. Previous work by Sexton et al. suggests that RHAU interacts with a predominantly inactive pool of the telomerase enzyme and functions to promote RNA accumulation through unwinding the 5′ G4-quadruplex. They suggest the quadruplex acts as a protective cap for hTR before incorporation into the telomerase holoenzyme. This was determined by overexpression of hTR mutants containing G to C substitutions within the quadruplex forming region, which demonstrated reduced RNA incorporation into the telomerase holoenzyme as well as reductions in telomere length and telomerase activity. These results suggest that the formation of a 5′ quadruplex is important for the production of a functional telomerase enzyme (30). In contrast to the findings of Sexton et al. (30), a recent study by Lattmann et al. (31) demonstrated that RHAU associated with a functional telomerase enzyme and that RHAU depletion by IP from cell lysates removed ∼25% of telomerase activity. Lattmann et al. (31) also demonstrated that the presence of ATP and MgCl2 resulted in a reduction of RHAU association with telomerase, indicating that following ATP hydrolysis, RHAU is released from the RNA. While the work by Lattmann et al. (31) suggests a probable role in the resolution of a G4-quadruplex to promote P1 helix formation, no experimental data is provided to support this hypothesis. We have shown that the full length RHAU protein promotes duplex formation between the 5′ region of hTR and an RNA fragment consisting of the complementary internal strand of the P1 helix (25P1). Furthermore, this P1 helix formation is ATP dependent, indicating that the helicase activity of RHAU is critical for the quadruplex disruption.
While we initially identified the minimal sequence capable of forming a G4-quadruplex as the first 17 nt of hTR, the fact that these nucleotides do not overlap with the region that base pairs to form P1 helix leaves open the possibility that this quadruplex is not responsible for disrupting the interaction with 25P1. Truncations from the 5′-end of hTR1–43 confirm this to be true, as, in the absence of the first 9 nt, a quadruplex persists that strongly inhibits an interaction between hTR and 25P1. It is only when nucleotides 1–13 are removed that the secondary structure is relaxed sufficiently to allow for an interaction to take place under quadruplex favorable conditions. This supports the prevailing notion that the primary quadruplex structure in hTR makes use of guanine tracts beyond the first 17 nt (24,29,31).
We demonstrated that the full-length RHAU protein is able to promote the formation of the P1 helix in all of the hTR variants tested, and appears to have enhanced activity in the absence of the first three guanine nucleotides. The molecular mechanism that results in this increased activity remains unclear, but may be due to the possibility that a quadruplex persists in hTR1–43 that sequesters some of the enzyme in the reaction even after the P1 helix is formed. This quadruplex, in fact, does persist as we see that a quadruplex forms in the first 17 nt that are not base paired in the P1 helix in the hTR1–43-25P1 complex. This quadruplex is abolished upon deletion of the first three guanines at the 5′-end of the RNA. While it is not clear if this explains the increased resolvase activity of RHAU on hTR4–43 compared with hTR1–43, it raises the interesting possibility that RHAU may be able to interact with hTR even in the presence of the stable P1 helix. Our investigation of this possibility with RHAU53–105 confirms that an interaction is likely, as RHAU53–105 demonstrated similar affinity for hTR in complex with 25P1 when compared with hTR alone. Furthermore, we have found that while nearly 100% of the hTR1–43-25P1 complex is bound by RHAU53–105, a portion of the free hTR RNA remains unbound in both the condition of the RNA alone and the residual-free hTR in the hTR-25P1 complex. This raises several interesting possibilities such that RHAU may have a function at hTR beyond the formation of the P1 helix. Intriguingly, Li et al. (25) demonstrated affinity of the first 17 nt of hTR for 2 internal regions of the RNA and identified both positive and negative regulatory roles depending upon which region bound. Therefore, it appears while an internal quadruplex and P1 helix formation may be mutually exclusive, the same scenario could be true for the 1–17 quadruplex and its other potential interaction sites. This raises the possibility that RHAU induces a sliding of the quadruplex along the 5′ region of hTR and in so doing acts as a switch to regulate interactions between the 5′ region of hTR and other internal sites.
While the bulk of the study was focused upon interactions between RHAU, hTR and their impact on base pairing with 25P1, we also sought to gain insight into the broader impact RHAU may have on telomerase function. Through measurements of relative telomere lengths in cells in which RHAU was knocked down by siRNA, we found an expected trend of telomere reduction up until 7 days post-transfection when compared with control cells. This reduction was sharply reversed at later time points, an observation that may be explained by compensatory mechanisms within the cell. While a significant impact was observed in HEK293T cells, HeLa cells did not exhibit statistically significant deviation from control. As such, it remains to be seen whether this impact is consistent amongst cell lines using telomerase mediated telomere extension. Furthermore, investigation of RHAU in the context of recombination mediated (alternative lengthening of telomeres) telomere extension remains an area of future investigation (39). Measurements of telomere lengths and telomerase activity in cell isolates may be confounded by many potential indirect impacts of RHAU siRNA knockdown and we are currently developing refined methods for studying the impact of RHAU and its helicase function on the activity, fidelity and processivity of the telomerase enzyme. While the interaction between RHAU and hTR is now fairly well established, significant work likes ahead in precisely defining the impact of this helicase and the function of the telomerase holoenzyme.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online: Supplementary Figures 1–9.
FUNDING
An establishment grant from the Manitoba Health Research Council; MHRC post-doctoral fellowship (to E.B.); the Canada Research Chair program (to J.S.). NSERC undergraduate student research award (to S.N. and N.O.). Funding for open access charge: Manitoba Health Research Council Establishment Grant.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr Yoshikuni Nagamine for the monoclonal anti-RHAU 12F33 hybridoma. The authors would also like to thank Dr Thomas Klonisch and Dr Spencer Gibson for the provided cell lines.
REFERENCES
- 1.Fuller-Pace FV. DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 2006;34:4206–4215. doi: 10.1093/nar/gkl460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Linder P. Dead-box proteins: a family affair—active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34:4168–4180. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fu JJ, Li LY, Liu SF, Xing XW, Liu G, Lu GX. Expression research for human DDX36 and mouse Ddx36 gene in the adult testis. Yi. Chuan. Xue. Bao. 2003;30:201–208. [PubMed] [Google Scholar]
- 4.Fu JJ, Li LY, Lu GX. Molecular cloning and characterization of human DDX36 and mouse Ddx36 genes, new members of the DEAD/H box superfamily. Sheng. Wu. Hua. Xue. Yu. Sheng. Wu. Wu. Li. Xue. Bao. 2002;34:655–661. [PubMed] [Google Scholar]
- 5.Iwamoto F, Stadler M, Chalupnikova K, Oakeley E, Nagamine Y. Transcription-dependent nucleolar cap localization and possible nuclear function of DExH RNA helicase RHAU. Exp. Cell Res. 2008;314:1378–1391. doi: 10.1016/j.yexcr.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 6.Tran H, Schilling M, Wirbelauer C, Hess D, Nagamine Y. Facilitation of mRNA deadenylation and decay by the exosome-bound, DExH protein RHAU. Mol. Cell. 2004;13:101–111. doi: 10.1016/s1097-2765(03)00481-7. [DOI] [PubMed] [Google Scholar]
- 7.Vaughn JP, Creacy SD, Routh ED, Joyner-Butt C, Jenkins GS, Pauli S, Nagamine Y, Akman SA. The DEXH protein product of the DHX36 gene is the major source of tetramolecular quadruplex G4-DNA resolving activity in HeLa cell lysates. J. Biol. Chem. 2005;280:38117–38120. doi: 10.1074/jbc.C500348200. [DOI] [PubMed] [Google Scholar]
- 8.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lipps HJ, Rhodes D. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol. 2009;19:414–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 11.Chang CC, Kuo IC, Ling IF, Chen CT, Chen HC, Lou PJ, Lin JJ, Chang TC. Detection of quadruplex DNA structures in human telomeres by a fluorescent carbazole derivative. Anal. Chem. 2004;76:4490–4494. doi: 10.1021/ac049510s. [DOI] [PubMed] [Google Scholar]
- 12.Salazar M, Thompson BD, Kerwin SM, Hurley LH. Thermally induced DNA.RNA hybrid to G-quadruplex transitions: possible implications for telomere synthesis by telomerase. Biochemistry. 1996;35:16110–16115. doi: 10.1021/bi961442j. [DOI] [PubMed] [Google Scholar]
- 13.Verma A, Yadav VK, Basundra R, Kumar A, Chowdhury S. Evidence of genome-wide G4 DNA-mediated gene expression in human cancer cells. Nucleic Acids Res. 2009;37:4194–4204. doi: 10.1093/nar/gkn1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giri B, Smaldino PJ, Thys RG, Creacy SD, Routh ED, Hantgan RR, Lattmann S, Nagamine Y, Akman SA, Vaughn JP. G4 Resolvase 1 tightly binds and unwinds unimolecular G4-DNA. Nucleic Acids Res. 2011;39:7161–7178. doi: 10.1093/nar/gkr234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Creacy SD, Routh ED, Iwamoto F, Nagamine Y, Akman SA, Vaughn JP. G4 resolvase 1 binds both DNA and RNA tetramolecular quadruplex with high affinity and is the major source of tetramolecular quadruplex G4-DNA and G4-RNA resolving activity in HeLa cell lysates. J. Biol. Chem. 2008;283:34626–34634. doi: 10.1074/jbc.M806277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang W, Smaldino PJ, Zhang Q, Miller LD, Cao P, Stadelman K, Wan M, Giri B, Lei M, Nagamine Y, et al. Yin Yang 1 contains G-quadruplex structures in its promoter and 5′-UTR and its expression is modulated by G4 resolvase 1. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim HN, Lee JH, Bae SC, Ryoo HM, Kim HH, Ha H, Lee ZH. Histone deacetylase inhibitor MS-275 stimulates bone formation in part by enhancing Dhx36-mediated TNAP transcription. J Bone Miner. Res. 2011;26:2161–2173. doi: 10.1002/jbmr.426. [DOI] [PubMed] [Google Scholar]
- 18.Chalupnikova K, Lattmann S, Selak N, Iwamoto F, Fujiki Y, Nagamine Y. Recruitment of the RNA helicase RHAU to stress granules via a unique RNA-binding domain. J. Biol. Chem. 2008;283:35186–35198. doi: 10.1074/jbc.M804857200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lattmann S, Giri B, Vaughn JP, Akman SA, Nagamine Y. Role of the amino terminal RHAU-specific motif in the recognition and resolution of guanine quadruplex-RNA by the DEAH-box RNA helicase RHAU. Nucleic Acids Res. 2010;38:6219–6233. doi: 10.1093/nar/gkq372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Sullivan RJ, Karlseder J. Telomeres: protecting chromosomes against genome instability. Nat. Rev. Mol. Cell Biol. 2010;11:171–181. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wyatt HD, West SC, Beattie TL. InTERTpreting telomerase structure and function. Nucleic Acids Res. 2010;38:5609–5622. doi: 10.1093/nar/gkq370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neidle S. Human telomeric G-quadruplex: the current status of telomeric G-quadruplexes as therapeutic targets in human cancer. FEBS J. 2010;277:1118–1125. doi: 10.1111/j.1742-4658.2009.07463.x. [DOI] [PubMed] [Google Scholar]
- 23.Artandi SE, DePinho RA. Telomeres and telomerase in cancer. Carcinogenesis. 2010;31:9–18. doi: 10.1093/carcin/bgp268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gros J, Guedin A, Mergny JL, Lacroix L. G-Quadruplex formation interferes with P1 helix formation in the RNA component of telomerase hTERC. Chembio Chem. 2008;9:2075–2079. doi: 10.1002/cbic.200800300. [DOI] [PubMed] [Google Scholar]
- 25.Li X, Nishizuka H, Tsutsumi K, Imai Y, Kurihara Y, Uesugi S. Structure, interactions and effects on activity of the 5′-terminal region of human telomerase RNA. J. Biochem. 2007;141:755–765. doi: 10.1093/jb/mvm081. [DOI] [PubMed] [Google Scholar]
- 26.Chen JL, Blasco MA, Greider CW. Secondary structure of vertebrate telomerase RNA. Cell. 2000;100:503–514. doi: 10.1016/s0092-8674(00)80687-x. [DOI] [PubMed] [Google Scholar]
- 27.Chen JL, Greider CW. Template boundary definition in mammalian telomerase. Genes Dev. 2003;17:2747–2752. doi: 10.1101/gad.1140303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim MM, Rivera MA, Botchkina IL, Shalaby R, Thor AD, Blackburn EH. A low threshold level of expression of mutant-template telomerase RNA inhibits human tumor cell proliferation. Proc. Natl Acad. Sci. USA. 2001;98:7982–7987. doi: 10.1073/pnas.131211098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lacroix L, Seosse A, Mergny JL. Fluorescence-based duplex-quadruplex competition test to screen for telomerase RNA quadruplex ligands. Nucleic Acids Res. 2011;39:e21. doi: 10.1093/nar/gkq1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sexton AN, Collins K. The 5′ guanosine tracts of human telomerase RNA are recognized by the G-quadruplex binding domain of the RNA helicase DHX36 and function to increase RNA accumulation. Mol. Cell Biol. 2011;31:736–743. doi: 10.1128/MCB.01033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lattmann S, Stadler MB, Vaughn JP, Akman SA, Nagamine Y. The DEAH-box RNA helicase RHAU binds an intramolecular RNA G-quadruplex in TERC and associates with telomerase holoenzyme. Nucleic Acids Res. 2011;39:9390–9404. doi: 10.1093/nar/gkr630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arthanari H, Basu S, Kawano TL, Bolton PH. Fluorescent dyes specific for quadruplex DNA. Nucleic Acids Res. 1998;26:3724–3728. doi: 10.1093/nar/26.16.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cawthon RM. Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res. 2009;37:e21. doi: 10.1093/nar/gkn1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ryder SP, Recht MI, Williamson JR. Quantitative analysis of protein-RNA interactions by gel mobility shift. Meth. Mol.r Biol. 2008;488:99–115. doi: 10.1007/978-1-60327-475-3_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kikin O, D'Antonio L, Bagga PS. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006;34:W676–W682. doi: 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kikin O, Zappala Z, D'Antonio L, Bagga PS. GRSDB2 and GRS_UTRdb: databases of quadruplex forming G-rich sequences in pre-mRNAs and mRNAs. Nucleic Acids Res. 2008;36:D141–D148. doi: 10.1093/nar/gkm982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hinkley CS, Blasco MA, Funk WD, Feng J, Villeponteau B, Greider CW, Herr W. The mouse telomerase RNA 5″-end lies just upstream of the telomerase template sequence. Nucleic Acids Res. 1998;26:532–536. doi: 10.1093/nar/26.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li X, Nishizuka H, Tsutsumi K, Imai Y, Kurihara Y, Uesugi S. Effects on telomerase activity of the 5′-terminal region of human telomerase RNA. Nucleic Acids Symp. Ser. 2005;49:313–314. doi: 10.1093/nass/49.1.313. [DOI] [PubMed] [Google Scholar]
- 39.Cesare AJ, Reddel RR. Alternative lengthening of telomeres: models, mechanisms and implications. Nat. Rev. Genet. 2010;11:319–330. doi: 10.1038/nrg2763. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.