Abstract
Non-canonical guanine quadruplex structures are not only predominant but also conserved among bacterial and mammalian promoters. Moreover recent findings directly implicate quadruplex structures in transcription. These argue for an intrinsic role of the structural motif and thereby posit that single nucleotide polymorphisms (SNP) that compromise the quadruplex architecture could influence function. To test this, we analysed SNPs within quadruplex motifs (Quad-SNP) and gene expression in 270 individuals across four populations (HapMap) representing more than 14 500 genotypes. Findings reveal significant association between quadruplex-SNPs and expression of the corresponding gene in individuals (P < 0.0001). Furthermore, analysis of Quad-SNPs obtained from population-scale sequencing of 1000 human genomes showed relative selection bias against alteration of the structural motif. To directly test the quadruplex-SNP-transcription connection, we constructed a reporter system using the RPS3 promoter—remarkable difference in promoter activity in the ‘quadruplex-destabilized’ versus ‘quadruplex-intact’ promoter was noticed. As a further test, we incorporated a quadruplex motif or its disrupted counterpart within a synthetic promoter reporter construct. The quadruplex motif, and not the disrupted-motif, enhanced transcription in human cell lines of different origin. Together, these findings build direct support for quadruplex-mediated transcription and suggest quadruplex-SNPs may play significant role in mechanistically understanding variations in gene expression among individuals.
INTRODUCTION
In addition to the canonical B DNA structure, DNA can adopt local secondary structure conformations. Role of non-canonical DNA structure has been implicated in important biological functions including replication, recombination and transcription (1). DNA secondary structures have also been associated with translocations and mutations that cause genome instability (1,2). This raises the intriguing possibility that locally formed DNA structure influences intrinsic cellular functions. In this context, it is interesting to consider the non-canonical secondary structure adopted by guanine-rich DNA sequences called the G-quadruplex or G4 DNA. Gathering evidence indicates involvement of G-quadruplex motifs in chromatin packaging (3–5), recombination (6) and CpG methylation (7) in addition to gene transcription, which is most studied.
G-quadruplex motifs are non-canonical Hoogsteen base-paired self-assembly of DNA strands in parallel/antiparallel orientation stabilized by charge coordination with monovalent cations (especially K+) (8–11). Initially observed to be enriched in bacterial promoters (12,13), potential G4 (PG4) motifs were subsequently found to be prevalent in human (14,15), chimpanzee (15), mouse (15), rat (15) and chicken (16) promoters. Furthermore, hundreds of PG4 motifs appear to be conserved among human, mouse and rat promoters (15). In vitro, c-MYC was the first case where a G-quadruplex-forming sequence in the nuclease hypersensitive element upstream of the P1 promoter was shown to affect transcription (17). Gene expression was also found to be influenced by G-quadruplex-forming sequence motifs within the core promoter of human c-KIT (18,19) and k-RAS (20) oncogenes. In addition, promoter G-quadruplex motifs have been reported for many genes, including VEGF, PDGF, HIF1α, BCL-2, RB, RET (21,22) and human telomerase hTERT (23,24). In case of thymidine kinase 1, we found a non-canonical G-quadruplex motif, formed by two-guanine repeats instead of three, to be functionally active (25).
More direct evidence in support of G-quadruplex-mediated transcription was obtained from chromatin immunoprecipitation (ChIP) experiments demonstrating that the non-metastatic factor NM23-H2 associates with the c-MYC promoter through a G-quadruplex motif (26). In addition, support for this mode of transcription was obtained from: interaction of recombinant hnRNP A1/Up1 with the KRAS promoter G-quadruplex (27); Myc-associated zinc finger protein (MAZ)/poly(ADP-ribose) polymerase 1 (PARP-1) binding to the G-quadruplex element in the murine KRAS promoter (28); and binding of nucleolin/hnRNP proteins to the G-quadruplex forming sequences of the VEGF promoter (29). Furthermore, similar motifs in the promoters of human sarcomeric mitochondrial creatine kinase, muscle creatine kinase and integrin alpha7 of mouse were shown to associate with the dimeric form of MyoD in vitro (30,31). Consistent with these findings, transcriptome profiling in presence of intracellular G-quadruplex binding ligands indicated genome-wide role of G-quadruplex motifs in transcription (32).
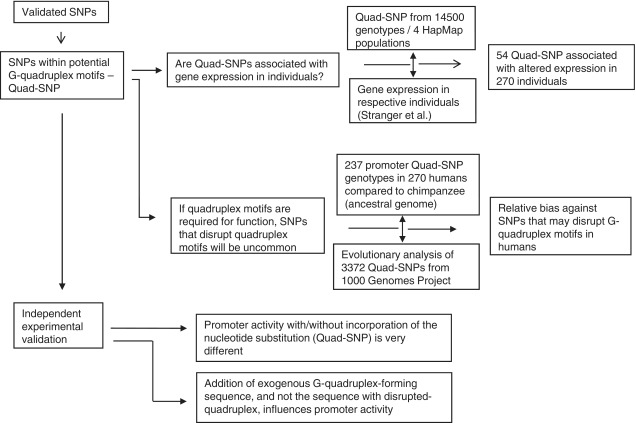
Taken together emerging computational/experimental evidence supporting G-quadruplex-mediated transcriptional functions raises an interesting question: can single nucleotide polymorphisms (SNP) that affect stability/formation of the quadruplex structure influence transcription, resulting in individual-specific gene expression change? This possibility has not been tested, although an independent line of study showed that SNPs that can potentially disrupt PG4 motifs were less frequent than expected (33). Using matched genotype information, SNP data from HapMap consortia (34) and gene expression profile of respective individuals (from lymphoblastoid-derived cell lines) (35), we asked whether difference in expression of a particular gene is associated with SNPs that disrupt PG4 motif(s) (Scheme 1). This was tested using human SNPs and the chimpanzee as an ancestral genome for comparison (36). Findings were verified using experimental gene expression reporter models that directly assayed the effect of G-quadruplex disruption on gene transcription in human cell lines.
Scheme 1.
Design of the study showing approaches adopted for computational and experimental tests.
METHODS
Analysis of HapMap data
Genotype information was extracted from HapMap data repository (HapMap Release 21). Data for significant correlation with gene expression (from lymphoblastoid cell lines of the HapMap individuals) was used as reported by Stranger et al. (35), who analysed 2.2 million SNPs with 13 643 distinct gene probes and found 1348 genes where the SNP was significantly associated with expression (cis-eQTL) of the respective gene in at least one of the four HapMap populations. We mapped the cis-eQTLs to PG4 motifs using an in-house PERL algorithm. For each Quad-SNP average gene expression [using data from http://www.ncbi.nlm.nih.gov/geo database (GSE6536)] of all individuals of a genotype was calculated and denoted as the expression value for that particular genotype, and this data was plotted as a heat map to accommodate all the SNPs and genotypes studied.
PG4 motif sequence retrieval and analysis
PG4 motifs were identified as described earlier (12). Briefly, we adopted a general pattern G3–L1–G3–L2–G3–L3–G3, where G is guanine; L is any nucleotide including G. The PG4 loops (L1, L2 and L3) could vary from one to seven bases. The program was rerun with cytosine instead of guanine to identify motifs on the complementary strand and was corrected for strand orientation with respect to positioning in the gene.
Sequence along with annotation of TSS for 18 056 unique human Refseq genes were retrieved from UCSC build hg18. PG4 motif sequence was identified as described earlier and mapped with validated SNPs using an in-house developed program. Validated SNPs were extracted from dbSNP (as per criteria: ‘by-frequency’, ‘byCluster’, ‘by2Hit2Allele’, ‘byOtherPop’ or by 1000 genomes). Random set of short regions for control analysis was made by extracting sequences of 15–33 nt from the same region that was used for extracting PG4 motifs, that is, 1 kb of TSS. For determining Quad-SNP found in stem/loop of PG4 motifs, we defined a PG4 motif stem position as any G residue which was: (i) flanked on both sides by G residues; (ii) preceded; or (iii) succeeded by at least two G residues. All other bases, including G residues were considered as loops.
Allele frequency data retrieval
Out of 1184 Quad-SNPs (vide infra) allele frequency data [Hapmart (34)] available for 356 Quad-SNPs in at least one population was used; 271 Quad-SNPs were found to be major alleles across all four populations. Of these, ancestral allele information [UCSC (hg18)] was available for 237, which were finally used for the comparative analysis. Population-wise derived allele frequency (DAF) analysis was done considering all the SNPs for which both allele frequency information and ancestral allele information was available: 291 (CEU), 279 (YRI), 282 (CHB) and 281 (JPT) Quad-SNPs. To confirm that the ancestral allele was invariant, we compared all human Quad-SNPs to their corresponding chimpanzee SNP (SNP125—http://hgdownload.cse.ucsc.edu/goldenPath/panTro1/database/snp125.txt.gz); human coordinates were converted to chimp (panTro1 or 2) using Liftover from UCSC. For all the 237 human Quad-SNPs used for DAF analysis, the corresponding chimpanzee position was found to be invariant. As a further test, we used the Genomic Evolutionary Rate Profiling (GERP) score (37), which was downloaded from conservation tract of UCSC table browser.
CD and melting experiments
CD spectra were recorded using a JASCO-810 instrument. Oligonucleotides were diluted to 3 µM final concentration in sodium cacodylate buffer (with 100 mM K+) prior to experiments, heated to 95°C and gradually cooled to ambient temperature overnight. CD scans were taken in a wavelength range of 220–320 nm at 20°C and scanning speed of 200 nm/min. For each oligo three scans were taken and spectrum of the buffer was subtracted. These samples were further used for melting experiments by first heating to a temperature of 95°C for 10 min and then slowly cooled to 25°C at a rate of 1°C/min. UV absorbance was measured at 295 nm.
Cell lines and culture conditions
Human fibrosarcoma HT1080 and lung adenocarcinoma A549 cell lines were obtained from National Centre for Cell Science, Pune and were maintained in MEM (HT 1080) or DMEM supplemented with 10% FBS (A549).
Cloning and reporter assays
Promoter of RPS3 gene was cloned in the promoter-less basic pGL3 vector (Promega) using XhoI and Hind III sites upstream of the luciferase gene following PCR amplification from normal genomic DNA using primers (FP—5′-AGAGCTCGAGAAAGAGAGAGGAAGGAAGGA-3′, RP—5′-AATAAGCTTGACCGACAAATGCTCACAAAC-3′). Clones were screened and sequenced for verification. Positive clones were subjected to site-directed mutagenesis using Quick Change Site-Directed mutagenesis Kit (Stratagene) to get desired single base change within the PG4 sequence (GGGCGG[G → C]CCCATGGGACCTTCTGGG). Prior to transfection, 12-well plates were seeded with 2.5 × 105 cells to achieve optimum confluency. Plasmid (1.5 µg) was transfected per well using lipofectamine 2000 (Invitrogen), according to manufacturer's protocol. For transfection control 5 ng of pGL4.73 was co-transfected. Cells were lysed after 24 h and luciferase assay was done using the dual luciferase assay kit from Promega, according to the manufacturer's protocol. Renilla counts were used for normalization. All experiments were done in triplicate.
Incorporation of the synthetic quadruplex motif
Synthetic quadruplex motif (GGGTGGGTGGGTGGG) and the sequence representing the corresponding disrupted motif (GAGTGAGTGAGTGAG) were cloned at the Bgl II site preceding the SV 40 promoter upstream of the luciferase gene (Renilla) in the psiCheck 2 vector (Promega). The firefly luciferase gene integrated within psiCheck 2 was used for normalization of transfection efficiency. Positive clones were confirmed by sequencing. A 12-well plate was seeded as mentioned earlier and 2 µg of plasmid was transfected using lipofectamine 2000 (Invitrogen), according to manufacturer's protocol. Cells were lysed after 48 h and luciferase assay was done as given in the previous section. All experiments were done in triplicate.
RESULTS
Presence of SNP within PG4 motifs is linked to altered gene expression in individuals
We hypothesized presence of SNPs that disrupt stability of PG4 motifs alter gene expression in individuals. To test this we sought to analyse SNP data (34) along with gene expression determined from lymphoblastoid cells of the respective individual where all SNPs that significantly correlate with altered expression of a gene have been reported (35).
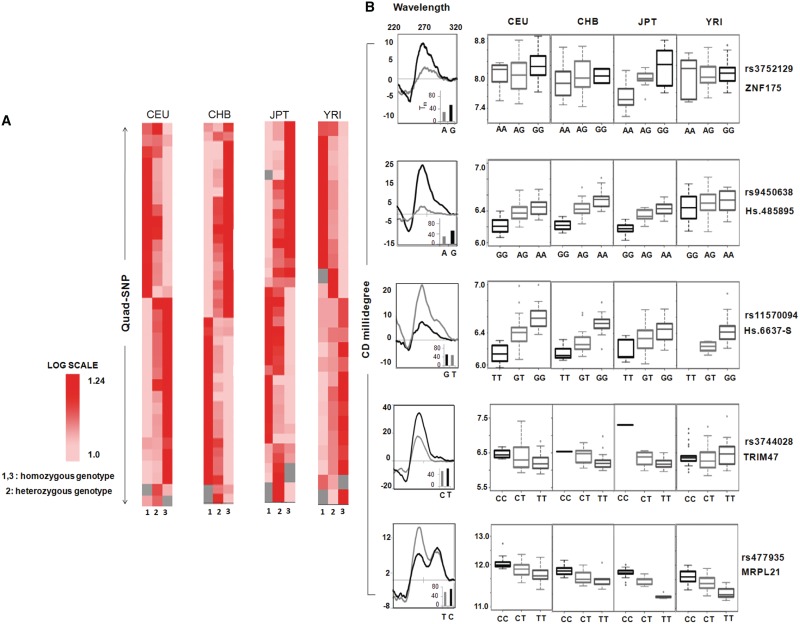
We found 54 SNPs lying within the potential quadruplex motif (Quad-SNP in following text) in 48 genes where change in genotype significantly correlated with altered gene expression in at least one population (18 genes harbouring 19 Quad-SNP were differentially expressed in all the four populations). This constituted 42 Quad-SNP in CHB (Chinese) population and 26, 33 and 41 in YRI (Yoruba from Ibadan), CEU (Caucasians of European origin) and JPT (Japanese), respectively. For every Quad-SNP, distinct change in expression of the corresponding gene associated with the genotypes across individuals was clearly observed and is shown population-wise in Figure 1A; each row represents a specific SNP, columns show respective genotypes (heterozygous in centre column flanked by homozygous). Difference in expression across the genotypes was statistically significant as reported earlier (P < 0.0001 in all cases, see Supplementary Table S1 for rs-id of SNPs). Interestingly, the heterozygous genotype always resulted in gene expression that was of an intermediate level with respect to the two homozygous groups—this is further illustrated using box plots for representative Quad-SNPs for genotypes in each population (Figure 1B, right panel; data for all Quad-SNP is given in Supplementary Table S1).
Figure 1.
Quadruplex-SNPs affect expression of corresponding genes across large number of individuals. (A) Heat map showing gene expression level in individuals representing the three genotypes within a population for 54 Quad-SNPs; each row represents a particular SNP (rs ID in Supplementary Data) within PG4 motifs. Average gene expression of all individuals representing a particular genotype within the population was used for 54 Quad-SNP in four populations; grey denotes cases when data was not available. (B) Right panel: Box plot of gene expression for individuals having a particular genotype resulting from the particular SNP is shown for five representative SNPs; gene name and rs ID as given on margin. Left panel: Quadruplex formation (in the five selected cases shown in right panel) and effect of the respective SNP on quadruplex structure as determined by CD spectroscopy; change in melting temperature (Tm) is given as inset.
Quad-SNP result in disruption of the G-quadruplex motif
Next, in order to test that the PG4 motifs detected above adopt the G-quadruplex motif and also to check whether the Quad-SNP results in altered stability of the motifs we randomly selected five sequences (with the SNP either in stem or loop, Supplementary Table S2). Oligonucleotides were synthesized with or without the variation and circular dichroism (CD) experiments were performed in the presence of K+ ion. As expected, all the five sequences showed distinct characteristic of the G-quadruplex motif comprising both parallel (260 nm) and antiparallel (∼290 nm) orientations (Figure 1B, left panel). Interestingly, we noted in all cases when a guanine base was disrupted in the stem of the PG4 motif, the characteristic peak at 260/290 nm was either disrupted or the peak height reduced indicating a general decrease in stability. Accordingly, the melting temperature of the disrupted sequence also decreased. In cases when the Quad-SNP was found within the loop, if the quadruplex showed reduced stability in the CD signature, a corresponding decrease was observed in the melting temperature. We noted one exception, rs11570094 (Figure 1B), where substitution of a guanine in the stem led to a CD spectrum which suggested increased stability, though the melting point was very similar.
Most Quad-SNP maintain the chimpanzee allele within humans
Next, using the 54 Quad-SNP found to be significantly associated with gene expression we asked whether the SNPs represented deviation or conservation in an evolutionary context. The chimpanzee genome was used to distinguish alleles into ancestral (when similar to chimpanzee) or derived (when different from chimpanzee) (38). In majority of cases (43 of 54), we found the ancestral allele was commonly present in all the four HapMap populations, in other words the derived form was found to be the minor allele. For 11 Quad-SNP, flipping was observed, that is, the major allele in human was different from the chimpanzee sequence. We noted with interest that only 4 out of the 11 flipped Quad-SNPs were found within the stem of the quadruplex and therefore were expected to directly affect stability of the quadruplex motif, while the remaining seven flipped bases were present within loop of the PG4 motif and therefore were not expected to significantly affect quadruplex stability.
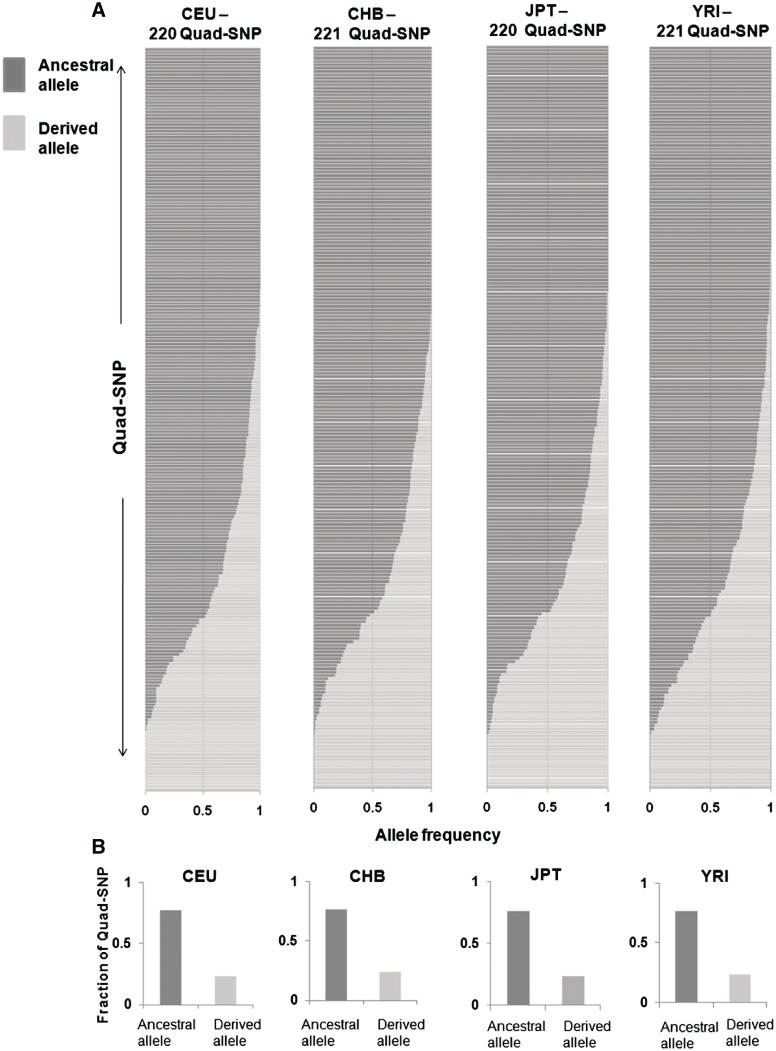
Given the prevalence of PG4 motifs near promoters in addition to multiple studies showing role of the quadruplex motif in gene expression (12,13,17,19,20) we next analysed the region within 1 kb of transcription start sites (TSS) in 18 056 unique human promoters. We found 1184 validated Quad-SNP (see ‘Methods’ section) in this region (note: only 54 Quad-SNP that were significantly associated with gene expression as reported in (35) were analysed in the previous sections). Out of 1184, we first used 237 Quad-SNP, where both allele frequency and ancestral allele data were available for all four populations, for further study (‘Methods’ section). Figure 2A depicts the genotype frequency of Quad-SNPs in each population where each row represents a bar graph showing ancestral/derived allele frequency for a given SNP. In line with our earlier observation using 54 Quad-SNP, here we found that in 195 out of 237 (82.2%) ancestral allele was the common or major allele whereas in only 42 (17.7%) flipping was observed. The fraction of ancestral versus derived alleles in each population further confirmed the finding that ancestral alleles were mostly maintained among Quad-SNP (Figure 2B).
Figure 2.
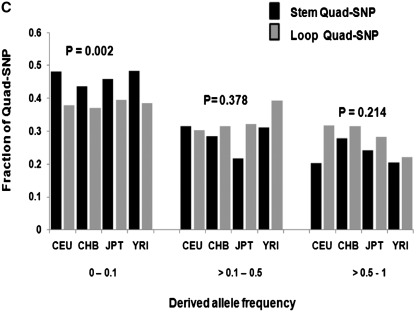
Promoter G-quadruplex motifs maintain the ancestral (chimpanzee) form. (A and B) Individual allele frequencies of Quad-SNP in the four HapMap populations—bar graph shows frequency of ancestral (chimpanzee) and derived allele for each SNP within a population (A) along with respective fractions of Quad-SNP that were either maintained with ancestral as major allele or flipped to the derived allele (B). (C) Categorization of stem/loop Quad-SNP with low (0–0.1), moderate (>0.1–0.5) or high (>0.5) derived allele frequencies shows stem SNP are significantly over-represented in the low category.
PG4 motif stems are maintained by evolutionarily restricting destabilizing substitutions
Since regions constituting the stem of the PG4 motifs are known to be relatively more important for structural stability we hypothesized that a Quad-SNP that potentially disrupts the structure could be under pressure to be conserved/promoted depending on the selective advantage that the PG4 motif may impart. In order to test this, we considered the Quad-SNPs that had low DAF (0–0.1), i.e. ones that appear to resist change from the ancestral form. Using these we asked whether there was any difference in number of SNP occurring within stems compared to loops for the population-specific Quad-SNPs [291 (CEU), 279 (YRI), 282 (CHB) and 281 (JPT)]. Interestingly, in all the four populations we found that the numbers of stem-Quad-SNP were significantly more than loop-Quad-SNP for the DAF category 0–0.1 (Figure 2C, two-tailed t-test, P = 0.002, Supplementary Table S3). In contrast, this difference between stem/loop Quad-SNPs was not significant in any of the other higher DAF categories. Together, this suggests the likelihood that stem SNPs that could potentially disrupt the structure are being disfavoured in an evolutionary sense.
Next we sought to check the selection constraint metric GERP (37) for Quad-SNPs. Based on the understanding that the rate of natural selection is compromised when purifying selection acts on a locus GERP estimates the ‘rejected substitution’ or RS score for given SNPs. Since the RS score is calculated by subtracting actual number of substitutions at a site from the expected number under neutral condition, sites under selective constrain attain positive scores (37,39,40). On analysing the population-specific Quad-SNPs, consistent with what was noted above, we found stem-Quad-SNPs had higher proportion of positive RS-scores compared to loop SNPs (two-tailed t-test, P < 0.001, Supplementary Figure S1). In addition, for further testing we used the recently released SNP data from population-scale sequencing of 1000 human genomes (41). We found 3372 Quad-SNP within 1 kb of 18 056 genes—2430 and 942 were within stem and loop of the G-quadruplex motif, respectively. Again, in line with our earlier observations, we found relatively higher proportion of stem-Quad-SNPs had positive RS scores (Supplementary Figure S1, P = 0.001).
Most promoter PG4 motifs are devoid of SNPs
Above studies indicated a possible bias against the presence of SNPs within PG4 motifs present in regulatory regions. This prompted us to ask whether SNPs were asymmetrically distributed in PG4 motifs, i.e. what proportion of PG4 motifs had any SNP at all. This was checked in 18 056 unique Refseq gene promoters (±1 kb of TSS) which had 72 263 validated SNPs (∼2 SNP/kb). Out of these, as mentioned earlier, we found 1184 SNPs within 32 716 PG4 motifs (comprising 820 903 bases, average 15- to 33-mers) found in this region resulting in a density of 1.4 SNP/kb indicating that PG4 motifs are depleted in SNPs (P < 0.0001; χ2 test) consistent with a previous study, which used a different computational program for detecting motifs (33). Furthermore, we analysed an equivalent number (32 000) of randomly picked short sequences of similar average GC% from ±1 kb of TSS—this gave a density of 2.1 SNP/kb (P < 0.0001; χ2 test). Interestingly out of the 32 716 PG4 motifs only 1113 had any SNP (P = 3.9e−149; χ2 test), i.e. >96% of the promoter PG4 motifs were devoid of any polymorphism. In order to check this further, we used the recently released SNP data from population-scale sequencing of 1000 human genomes (41). In this case, we found 3372 Quad-SNP, validated by the 1000 genome project, occurring within 2982 of the 32 716 PG4 motifs present within 1 kb of 18 056 genes. This again showed that only ∼9% of promoter PG4 motifs had one or more polymorphic sites indicating that the distribution of SNP within PG4 motifs was significantly skewed when compared to expected distribution (P = 8.2e−119; χ2 test). Together, these studies strongly indicated a possible bias against nucleotide substitutions that could lead to disruption of quadruplex units in the genome.
Quadruplex-disrupting SNP results in significantly altered promoter activity
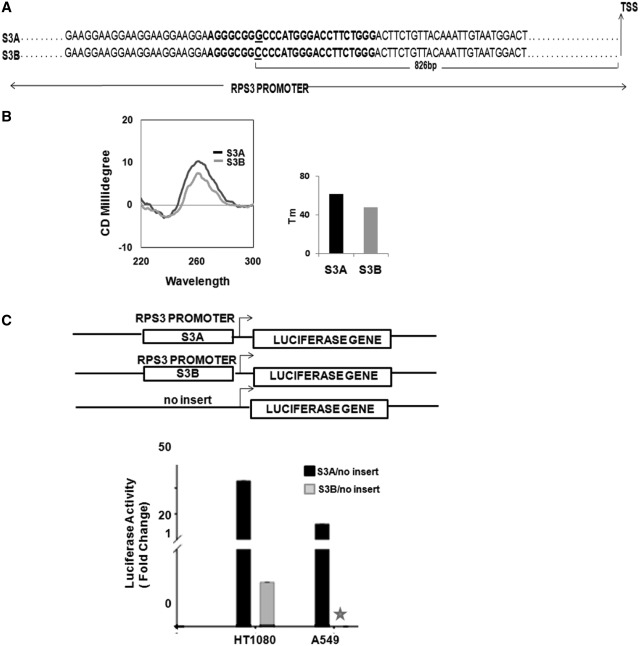
Next, to test above findings we sought to study a PG4 motif/SNP combination that was independent of the data sets analysed above and asked: (i) whether the specific nucleotide substitution resulted in disruption of the G-quadruplex structure and (ii) if the disruption caused any alteration in expression of the gene. For this the SNP (rs17880356, G to C) found in the promoter of the ribosomal protein S3 (RPS3) (Figure 3A), which plays a critical role in initiation of translation, was selected. In order to determine whether this sequence adopted the quadruplex motif, and if the substitution significantly disrupted the structure, we first synthesized two oligonucleotides, S3A and S3B comprising the PG4 motif representing both the alleles of the Quad-SNP found in RPS3, where S3A had the G-base while S3B had the substitution (G to C). G-quadruplex forming potential was determined using CD spectroscopy—S3A gave a well formed parallel quadruplex whereas S3B showed decrease in peak height at 260 nm suggesting loss of structural stability (Figure 3B). We also found that the Tm of the G-quadruplex motif was 62.1°C whereas that of the S3B motif was substantially decreased to 48°C, consistent with CD results (Figure 3B, right panel).
Figure 3.
Quad-SNP affects promoter activity of RPS3. (A) Scheme showing part of RPS3 promoter with sequence of the PG4 motif given in bold; Quad-SNP is underlined. (B) CD spectra of PG4 motif sequences S3A and S3B, melting temperature (Tm) in right frame. (C) Scheme showing promoter reporter systems inserted upstream of the firefly luciferase gene. Luciferase reporter activity of reporter clones with either S3A or S3B relative to no insert clone is shown below; activity in case of S3B in A549 cells was not detectable (asterisks). Experiments were done in triplicate; Renilla luciferase activity was used to normalize transfection efficiency.
Following this we sought to check the influence of the PG4 motif, and the substitution, on transcription of RPS3. To test promoter activity luciferase reporter systems were constructed using the 1.5-kb long putative promoter of human RPS3 harbouring the PG4 motif, which was cloned upstream of the firefly luciferase gene; expression of Renilla luciferase was used as control (Figure 3C). An additional construct was made to represent S3B after incorporating the specific SNP (G to C) within the PG4 motif. We first checked promoter activity in human fibrosarcoma cells. Remarkably, activity of S3A was found to be substantially high, which decreased by >80-fold on G to C substitution within the PG4 motif (S3B), supporting gene expression that is linked to presence of the quadruplex motif. Considering the extent of difference observed we sought to check this in a second cell line. On using the human adenocarcinoma cell line (A549), we found >25-fold increase in expression of S3A relative to the empty vector. In line with the earlier observation, here also in case of S3B expression was very low and could not be detected. Together, these experiments support our earlier findings and demonstrate that disruption of a promoter-PG4 motif could lead to significant change in gene expression.
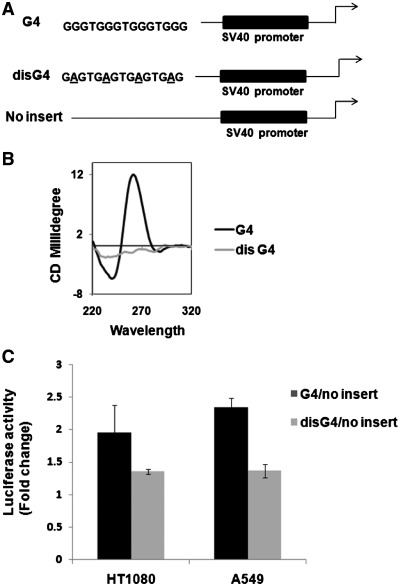
Insertion of synthetic G-quadruplex motif affects promoter activity of reporter construct inside cells
To test quadruplex-mediated transcription in a more direct fashion we made a synthetic G-quadruplex motif and incorporated this upstream of an exogenous promoter reporter system constituting the SV40 promoter upstream of the firefly luciferase gene (Figure 4A). An analogous system was made by introducing a similar sequence wherein the quadruplex motif was disrupted by specific nucleotide changes to constitute a negative control that did not adopt the quadruplex form. We confirmed that the substitutions led to disruption of the structure using CD (Figure 4B) and DNA melting experiments (data not shown). Following this luciferase activity was checked in two cell lines and reporter activity from firefly luciferase was normalized using Renilla luciferase counts to control for transfection efficiency. Promoter activity increased on quadruplex insertion by ∼1.9- and 2.3-folds in HT1080 and A549 cells, respectively (Figure 4C). In contrast, reporter activity when the quadruplex motif was disrupted was similar to the inherent SV40 promoter activity. Together these results showed that incorporation of the quadruplex motif results in altered promoter activity due to the presence of the structural motif and is lost when the structure is specifically disrupted.
Figure 4.
Incorporation of the G-quadruplex motif and not sequence per se induces promoter activity. (A) Scheme showing the constructs made to insert either a G-quadruplex-forming (G4) or disrupted G4 (disG4) as control sequence upstream of SV40 promoter in a luciferase reporter vector. (B) CD spectra of oligonucleotide used for G4 motif and disG4 showing disruption of the quadruplex motif in case of disG4. (C) Luciferase reporter activity of clones harbouring G4 or disG4 in human cell lines with respect to the no-insert construct. All experiments were done in triplicate using Renilla luciferase activity as transfection control.
DISCUSSION
Taken together results reported here show that integrity of the G-quadruplex secondary structure form is necessary for transcription. This is supported by multiple lines of findings demonstrating that any change in the quadruplex structure influences transcription. We found promoter quadruplex sequences not only harbour low number of polymorphic sites but are mostly devoid of any SNP that could potentially disrupt the structure. Interestingly, even in the small fraction of PG4 motifs with SNPs it was found that nucleotide changes, with respect to chimpanzee, occurred in a minor percentage of human populations. Moreover, this resistance to change with respect to chimpanzee was distinctly noted in SNPs that could potentially disrupt the quadruplex structure and not in ones that are expected to have limited effect on structure (e.g. SNPs within loop region of the quadruplex motif), supporting the notion that integrity of the structure was critical for function. These findings were further supported by results obtained from exogenous addition of a synthetic quadruplex motif: reporter gene expression was noted to be directly influenced by incorporation of the quadruplex structure, which was lost when nucleotide substitutions that specifically compromised the quadruplex secondary architecture was introduced.
At a genome-wide level, we found many SNPs within PG4 motifs where the individual genotypes were strongly correlated to gene expression (Figure 1). It was also evident from Figure 1 that largely individuals having heterozygous genotype had gene expression levels that were of an intermediate level relative to the corresponding homozygous genotypes. Thus, Quad-SNPs fitted well in an additive genetic model of inheritance, where the allelic change modulates the phenotype in a dose dependent manner (42). Though correlative, considered with other findings, this implicates quadruplex motifs in a broader sense suggesting that gene expression of individuals could be influenced by base changes that either form or disrupt a secondary DNA structure.
Recent data from population-scale sequencing in the 1000 genomes project (41) gives a much enhanced coverage of SNPs than obtained by HapMap. Indeed using this data set also we noted that SNPs occur in only a small proportion (<10%) of promoter PG4 motifs, in line with our observation from analysis of HapMap data. Further analysis of association with gene expression using SNPs from 1000 genomes data would be interesting. However, this was not possible as gene expression data of only a limited number of individuals are publicly available at this time, and many of the variants being rare would require expression data from large number of individuals to ascertain association with significance. Furthermore, we also noted that a recent study that compared HapMap3 and 1000 genomes genotypes for eQTLs using the CEU and YRI expression data sets found similar numbers of eQTLs between the two projects. Therefore, while resequencing gives many novel associations it is possible that most common effects have been captured with previous genotyping-based approaches (43). On the other hand, and perhaps more importantly, still in order to test causal link between G-quadruplex and Quad-SNP one would need to resort to experimental approaches. Keeping this in mind, we focused on evidence from transcriptional results that were caused by directed base changes that specifically disrupt G-quadruplex forms in order to build support for the G-quadruplex-gene expression connection among individuals.
Several of the 54 Quad-SNPs that significantly associated with gene expression across individuals were found at a relatively long distance from TSS (Supplementary Table S1). Influence on transcription for such instances is difficult to reason without direct evidence, though in case of eukaryotes optimum distance for regulatory control varies considerably and many cases of long-range regulation have been reported (44–46). On the other hand, using chromatin immunoprecipitation (ChIP) followed by sequencing, association of transcription factors have been noted that are in regions distant, and both upstream/downstream, of TSS (47). Therefore, the likelihood that SNPs that are far and both upstream/downstream of TSS can affect transcription cannot be ruled out.
Throughout this study we have considered loop sizes that were restricted to seven bases based on earlier reports (12,48) whereas more recent findings suggest that loops of stable quadruplex can be 10 bases or more (49,50). Therefore the number of PG4 motifs and SNPs detected in this study is perhaps a conservative set of possibilities.
In an earlier study it was reported that the G-tracts critical for stability of the G-quadruplex motif show low polymorphism (33). Authors further detected that any given short G-tract in the human genome had relatively low polymorphism irrespective of whether it was a part of G-quadruplex structure. These observations suggested a role of the G-tract that may not be related to the G-quadruplex motif. Our experiments using the RPS3 promoter and, particularly, the synthetic quadruplex reporter system show deformation of the quadruplex motif has distinct and remarkable effect on promoter activity. These experiments show in a relatively directly way that nucleotide base changes in integral positions of the quadruplex, namely the G-tract, are likely to have important functional consequence. On the other hand, though CD spectroscopy confirms G-quadruplex structure formation by oligonucleotides, it does not completely rule out constitution of other structural forms. Therefore contribution from non-G-quadruplex secondary structures is difficult to fully negate. Nonetheless, base substitutions in our experimental study were designed so that they perturb specifically G-quadruplexes and therefore are likely to support changes due to G-quadruplex structure formation/deformation.
In a recent genome-wide study, we found G-quadruplex motifs are closely associated with several DNA binding proteins in human, chimpanzee, mouse and rat (51). G-quadruplex association with SP1, hnRNP A1, MAZ and nucleolin has also been noted (27–29,52). Therefore, destabilization of G-quadruplex forms are likely to disrupt association with factors leading to impairment of enhancer/repressor functions. This could be a likely reason for the substantial change (in case of fibrosarcoma cells, Figure 3C) noted in transcription, given the single base change that was incorporated. On similar lines, we noted that moderate changes in G-quadruplex stability or even alteration in bases within potential loop regions (Figure 1), at times, resulted in significant change in gene expression among individuals. This again suggests the possibility that subtle changes in the G-quadruplex form/stability leads to relatively pronounced gene expression changes due to altered DNA binding of transcription factor(s).
Another interesting aspect of the findings stems from the fact that a distinct difference was noted in the frequency of polymorphisms within populations with respect to their occurrence in stems/loops of the G-quadruplex motifs. Stem SNPs maintained a bias towards the ancestral form (predominant in low DAF category, Figure 2C). This suggests an interesting evolutionary perspective. It is widely understood that natural selection acts to conserve/disrupt functional elements within a genome and thereby drives evolution and population differentiation (53). Therefore, if a particular locus is not diversifying then the ancestral allele would be expected to remain unchanged or conserved, whereas any change generally signifies selective advantage. Based on this, it is tempting to speculate that perhaps the structural form of a quadruplex is being maintained, whereas loop SNPs that are largely not expected to affect structure are relatively more amenable to change.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3 and Supplementary Figure 1.
FUNDING
Council of Scientific and Industrial Research (CSIR) (senior research fellowship to A.B. and V.K.Y.; project assistantships to P.M. and A.S. (Task Force Project SIP 006)); Indian Council of Medical Research (senior research fellowship to P.K. and R.H.); and Department of Science and Technology, Government of India (fellowship LS-03/2006-07 to S.C.). SKD acknowledges NIH/NIDDK (R01 DK039311). Funding for open access charge: CSIR (Task Force Project SIP 0006).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Authors acknowledge Mitali Mukerji, Arijit Mukhopadhyay, Amit Mandal and Munia Ganguli from IGIB for helpful discussions and careful reading of the manuscript.
REFERENCES
- 1.Bacolla A, Wells RD. Non-B DNA conformations, genomic rearrangements, and human disease. J. Biol. Chem. 2004;279:47411–47414. doi: 10.1074/jbc.R400028200. [DOI] [PubMed] [Google Scholar]
- 2.Zhao J, Bacolla A, Wang G, Vasquez KM. Non-B DNA structure-induced genetic instability and evolution. Cell Mol. Life Sci. 2010;67:43–62. doi: 10.1007/s00018-009-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Halder K, Halder R, Chowdhury S. Genome-wide analysis predicts DNA structural motifs as nucleosome exclusion signals. Mol. Biosyst. 2009;5:1703–1712. doi: 10.1039/b905132e. [DOI] [PubMed] [Google Scholar]
- 4.Hershman SG, Chen Q, Lee JY, Kozak ML, Yue P, Wang LS, Johnson FB. Genomic distribution and functional analyses of potential G-quadruplex-forming sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 2008;36:144–156. doi: 10.1093/nar/gkm986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong HM, Huppert JL. Stable G-quadruplexes are found outside nucleosome-bound regions. Mol. Biosyst. 2009;5:1713–1719. doi: 10.1039/b905848f. [DOI] [PubMed] [Google Scholar]
- 6.Mani P, Yadav VK, Das SK, Chowdhury S. Genome-wide analyses of recombination prone regions predict role of DNA structural motif in recombination. PLoS One. 2009;4:e4399. doi: 10.1371/journal.pone.0004399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Halder R, Halder K, Sharma P, Garg G, Sengupta S, Chowdhury S. Guanine quadruplex DNA structure restricts methylation of CpG dinucleotides genome-wide. Mol. Biosyst. 2010;6:2439–2447. doi: 10.1039/c0mb00009d. [DOI] [PubMed] [Google Scholar]
- 8.Balagurumoorthy P, Brahmachari SK. Structure and stability of human telomeric sequence. J. Biol. Chem. 1994;269:21858–21869. [PubMed] [Google Scholar]
- 9.GELLERT M, LIPSETT MN, DAVIES DR. Helix formation by guanylic acid. Proc. Natl Acad. Sci. USA. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 11.Sundquist WI, Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989;342:825–829. doi: 10.1038/342825a0. [DOI] [PubMed] [Google Scholar]
- 12.Rawal P, Kummarasetti VB, Ravindran J, Kumar N, Halder K, Sharma R, Mukerji M, Das SK, Chowdhury S. Genome-wide prediction of G4 DNA as regulatory motifs: role in Escherichia coli global regulation. Genome Res. 2006;16:644–655. doi: 10.1101/gr.4508806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yadav VK, Abraham JK, Mani P, Kulshrestha R, Chowdhury S. QuadBase: genome-wide database of G4 DNA–occurrence and conservation in human, chimpanzee, mouse and rat promoters and 146 microbes. Nucleic Acids Res. 2008;36:D381–D385. doi: 10.1093/nar/gkm781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verma A, Halder K, Halder R, Yadav VK, Rawal P, Thakur RK, Mohd F, Sharma A, Chowdhury S. Genome-wide computational and expression analyses reveal G-quadruplex DNA motifs as conserved cis-regulatory elements in human and related species. J. Med. Chem. 2008;51:5641–5649. doi: 10.1021/jm800448a. [DOI] [PubMed] [Google Scholar]
- 16.Du Z, Kong P, Gao Y, Li N. Enrichment of G4 DNA motif in transcriptional regulatory region of chicken genome. Biochem. Biophys. Res. Commun. 2007;354:1067–1070. doi: 10.1016/j.bbrc.2007.01.093. [DOI] [PubMed] [Google Scholar]
- 17.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rankin S, Reszka AP, Huppert J, Zloh M, Parkinson GN, Todd AK, Ladame S, Balasubramanian S, Neidle S. Putative DNA quadruplex formation within the human c-kit oncogene. J. Am. Chem. Soc. 2005;127:10584–10589. doi: 10.1021/ja050823u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernando H, Reszka AP, Huppert J, Ladame S, Rankin S, Venkitaraman AR, Neidle S, Balasubramanian S. A conserved quadruplex motif located in a transcription activation site of the human c-kit oncogene. Biochemistry. 2006;45:7854–7860. doi: 10.1021/bi0601510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cogoi S, Xodo LE. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006;34:2536–2549. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balasubramanian S, Hurley LH, Neidle S. Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat. Rev. Drug Discov. 2011;10:261–275. doi: 10.1038/nrd3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim KW, Lacroix L, Yue DJ, Lim JK, Lim JM, Phan AT. Coexistence of two distinct G-quadruplex conformations in the hTERT promoter. J. Am. Chem. Soc. 2010;132:12331–12342. doi: 10.1021/ja101252n. [DOI] [PubMed] [Google Scholar]
- 24.Palumbo SL, Ebbinghaus SW, Hurley LH. Formation of a unique end-to-end stacked pair of G-quadruplexes in the hTERT core promoter with implications for inhibition of telomerase by G-quadruplex-interactive ligands. J. Am. Chem. Soc. 2009;131:10878–10891. doi: 10.1021/ja902281d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Basundra R, Kumar A, Amrane S, Verma A, Phan AT, Chowdhury S. A novel G-quadruplex motif modulates promoter activity of human thymidine kinase 1. FEBS J. 2010;277:4254–4264. doi: 10.1111/j.1742-4658.2010.07814.x. [DOI] [PubMed] [Google Scholar]
- 26.Thakur RK, Kumar P, Halder K, Verma A, Kar A, Parent JL, Basundra R, Kumar A, Chowdhury S. Metastases suppressor NM23-H2 interaction with G-quadruplex DNA within c-MYC promoter nuclease hypersensitive element induces c-MYC expression. Nucleic Acids Res. 2009;37:172–183. doi: 10.1093/nar/gkn919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paramasivam M, Membrino A, Cogoi S, Fukuda H, Nakagama H, Xodo LE. Protein hnRNP A1 and its derivative Up1 unfold quadruplex DNA in the human KRAS promoter: implications for transcription. Nucleic Acids Res. 2009;37:2841–2853. doi: 10.1093/nar/gkp138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cogoi S, Paramasivam M, Membrino A, Yokoyama KK, Xodo LE. The KRAS promoter responds to Myc-associated zinc finger and poly(ADP-ribose) polymerase 1 proteins, which recognize a critical quadruplex-forming GA-element. J. Biol. Chem. 2010;285:22003–22016. doi: 10.1074/jbc.M110.101923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uribe DJ, Guo K, Shin YJ, Sun D. Heterogeneous nuclear ribonucleoprotein K and nucleolin as transcriptional activators of the vascular endothelial growth factor promoter through interaction with secondary DNA structures. Biochemistry. 2011;50:3796–3806. doi: 10.1021/bi101633b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yafe A, Shklover J, Weisman-Shomer P, Bengal E, Fry M. Differential binding of quadruplex structures of muscle-specific genes regulatory sequences by MyoD, MRF4 and myogenin. Nucleic Acids Res. 2008;36:3916–3925. doi: 10.1093/nar/gkn340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shklover J, Weisman-Shomer P, Yafe A, Fry M. Quadruplex structures of muscle gene promoter sequences enhance in vivo MyoD-dependent gene expression. Nucleic Acids Res. 2010;38:2369–2377. doi: 10.1093/nar/gkp1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verma A, Yadav VK, Basundra R, Kumar A, Chowdhury S. Evidence of genome-wide G4 DNA-mediated gene expression in human cancer cells. Nucleic Acids Res. 2009;37:4194–4204. doi: 10.1093/nar/gkn1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakken S, Rognes T, Hovig E. The disruptive positions in human G-quadruplex motifs are less polymorphic and more conserved than their neutral counterparts. Nucleic Acids Res. 2009;37:5749–5756. doi: 10.1093/nar/gkp590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 35.Stranger BE, Nica AC, Forrest MS, Dimas A, Bird CP, Beazley C, Ingle CE, Dunning M, Flicek P, Koller D, et al. Population genomics of human gene expression. Nat. Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hacia JG, Fan JB, Ryder O, Jin L, Edgemon K, Ghandour G, Mayer RA, Sun B, Hsie L, Robbins CM, et al. Determination of ancestral alleles for human single-nucleotide polymorphisms using high-density oligonucleotide arrays. Nat. Genet. 1999;22:164–167. doi: 10.1038/9674. [DOI] [PubMed] [Google Scholar]
- 37.Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S, Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fry AE, Trafford CJ, Kimber MA, Chan MS, Rockett KA, Kwiatkowski DP. Haplotype homozygosity and derived alleles in the human genome. Am. J. Hum. Genet. 2006;78:1053–1059. doi: 10.1086/504160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goode DL, Cooper GM, Schmutz J, Dickson M, Gonzales E, Tsai M, Karra K, Davydov E, Batzoglou S, Myers RM, et al. Evolutionary constraint facilitates interpretation of genetic variation in resequenced human genomes. Genome Res. 2010;20:301–310. doi: 10.1101/gr.102210.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cooper GM, Goode DL, Ng SB, Sidow A, Bamshad MJ, Shendure J, Nickerson DA. Single-nucleotide evolutionary constraint scores highlight disease-causing mutations. Nat. Methods. 2010;7:250–251. doi: 10.1038/nmeth0410-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Altshuler D, Durbin RM, Abecasis GR, Bentley DR, Chakravarti A, Clark AG, Collins FS, De La Vega FM, Donnelly P, Egholm M, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewis CM. Genetic association studies: design, analysis and interpretation. Brief. Bioinform. 2002;3:146–153. doi: 10.1093/bib/3.2.146. [DOI] [PubMed] [Google Scholar]
- 43.Montgomery SB, Lappalainen T, Gutierrez-Arcelus M, Dermitzakis ET. Rare and common regulatory variation in population-scale sequenced human genomes. PLoS Genet. 2011;7:e1002144. doi: 10.1371/journal.pgen.1002144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kleinjan DA, van HV. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am. J. Hum. Genet. 2005;76:8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- 46.Lettice LA, Heaney SJ, Purdie LA, Li L, de BP, Oostra BA, Goode D, Elgar G, Hill RE, de GE. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 2003;12:1725–1735. doi: 10.1093/hmg/ddg180. [DOI] [PubMed] [Google Scholar]
- 47.Kouwenhoven EN, van Heeringen SJ, Tena JJ, Oti M, Dutilh BE, Alonso ME, de IC-M, Smeenk L, Rinne T, Parsaulian L, et al. Genome-wide profiling of p63 DNA-binding sites identifies an element that regulates gene expression during limb development in the 7q21 SHFM1 locus. PLoS Genet. 2010;6:e1001065. doi: 10.1371/journal.pgen.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guedin A, Gros J, Alberti P, Mergny JL. How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Res. 2010;38:7858–7868. doi: 10.1093/nar/gkq639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yue DJ, Lim KW, Phan AT. Formation of (3+1) G-Quadruplexes with a Long Loop by Human Telomeric DNA Spanning Five or More Repeats. J. Am. Chem. Soc. 2011;133:11462–11465. doi: 10.1021/ja204197d. [DOI] [PubMed] [Google Scholar]
- 51.Kumar P, Yadav VK, Baral A, Kumar P, Saha D, Chowdhury S. Zinc-finger transcription factors are associated with guanine quadruplex motifs in human, chimpanzee, mouse and rat promoters genome-wide. Nucleic Acids Res. 2011;39:8005–8016. doi: 10.1093/nar/gkr536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raiber EA, Kranaster R, Lam E, Nikan M, Balasubramanian S. A non-canonical DNA structure is a binding motif for the transcription factor SP1 in vitro. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr882. (doi:10.1093/nar/gkr882; epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barreiro LB, Laval G, Quach H, Patin E, Quintana-Murci L. Natural selection has driven population differentiation in modern humans. Nat. Genet. 2008;40:340–345. doi: 10.1038/ng.78. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.