Abstract
We report that the cationic porphyrin TmPyP4, which is known mainly as a DNA G-quadruplex stabilizer, unfolds an unusually stable all purine RNA G-quadruplex (M3Q) that is located in the 5′-UTR of MT3-MMP mRNA. When the interaction between TmPyP4 and M3Q was monitored by UV spectroscopy a 22-nm bathochromic shift and 75% hypochromicity of the porphin major Soret band was observed indicating direct binding of the two molecules. TmPyP4 disrupts folded M3Q in a concentration-dependent fashion as was observed by circular dichroism (CD), 1D 1H NMR and native gel electrophoresis. Additionally, when TmPyP4 is present during the folding process it inhibits the M3Q RNA from adopting a G-quadruplex structure. Using a dual reporter gene construct that contained the M3Q sequence alone or the entire 5′-UTR of MT3-MMP mRNA, we report here that TmPyP4 can relieve the inhibitory effect of the M3Q G-quadruplex. However, the same concentrations of TmPyP4 failed to affect translation of a mutated construct. Thus, TmPyP4 has the ability to unfold an RNA G-quadruplex of extreme stability and modulate activity of a reporter gene presumably via the disruption of the G-quadruplex.
INTRODUCTION
Matrix metalloproteinases (MMPs) are zinc-dependent endopeptidases, which are capable of degrading extracellular matrix proteins (1,2). Recently, it was reported that a 20 nucleotide all purine sequence (M3Q) located in the 5′-untranslated region (5′-UTR) of the MT3-MMP mRNA forms an extremely stable intramolecular G-quadruplex that inhibits translation in eukaryotic cells (3). G-quadruplexes are nucleic acid secondary structures containing two or more planar stacked hydrogen bonded G-tetrads which are stabilized by the presence of certain monovalent cations (4). Guanine-rich sequences of both RNA and DNA can form G-quadruplex structures via hydrogen bonding between Watson–Crick and major-groove base edges (4). DNA G-rich sequences are prevalent throughout the entire human genome (5–9), especially in some of the key growth regulatory genes and oncogenes (10–14). However, a computational survey found G-quadruplex forming sequences to be enriched within mRNA processing sites (15) and in the 5′-UTRs of mRNAs of genes related to cancer (16). G-quadruplex motifs have been characterized in several naturally occurring RNAs (3,16–23) and have been shown to have an inhibitory effect on translation (3,16,20,23,24). In particular, it has been shown that an RNA quadruplex motif (M3Q) located in the 5′-UTR of the MT3-MMP mRNA forms an extremely stable G-quadruplex structure and inhibits translation in eukaryotic cells when analyzed alone as well as in the context of the entire 5′-UTR (3). Despite the growing number of RNA quadruplexes being discovered, reports on their interactions with small molecules are few (25–28) even though other types of RNA secondary structures have been extensively studied in this regard for the purpose of drug development (29). Several small-molecule ligands have been reported to interact with DNA quadruplexes, on which they exert either a stabilizing or a destabilizing effect (9,30). Many of these studies employed the cationic porphyrin, 5,10,15,20-tetra(N-methyl-4-pyridyl)porphyrin (TmPyP4), which was usually found to stabilize DNA quadruplexes in vitro as well as in vivo (10,13,31–34). Besides the quadruplex structures, TmPyP4 can also bind to several duplex DNA sequences with comparable affinities (35,36). It was also reported that this ligand can unfold a bimolecular DNA quadruplex (37) but its effect on the stability of RNA quadruplexes is poorly understood at this time. In this report, CD spectrophotometry, 1D 1H NMR spectroscopy and gel electrophoresis convincingly demonstrate that TmPyP4 unfolds the extremely stable, 20 nucleotide RNA G-quadruplex forming sequence located in the 5′-UTR of the MT3-MMP mRNA. We also report the in vivo effect of TmPyP4 on translation of the MT3-MMP mRNA.
MATERIALS AND METHODS
RNA Purification
The RNA sequences 5′-rGAGGGAGGGAGGGAGAGGGA-3′ (M3Q) and 5′-rGAGAUAGUGAGUGAGAGAGA-3′ (mut-M3Q) were purchased from Dharmacon, Inc. RNA products were purified via denaturing 17% polyacrylamide gel electrophoresis (PAGE). Full-length products were visualized by UV shadowing and excised from the gel. The RNA was harvested via the crush and soak method by tumbling the crushed gel slices overnight at 4°C in a solution of 300 mM NaCl, 10 mM Tris–HCl and 0.1 mM EDTA (pH 7.5). The RNA was isolated by ethanol precipitation followed by two 70% ethanol washes of the precipitate. The final RNA pellet was dissolved in 10 mM Tris–HCl and 0.1 mM EDTA (pH 7.5). RNA concentrations were determined on the basis of their absorbance value at 260 nm and appropriate extinction coefficients (38). The RNAs were folded in the presence of 100 mM KCl, 10 mM Tris–HCl and 0.1 mM EDTA (pH 7.5) by heating for 5 min at 95°C followed by cooling to room temperature over a 90-min period.
5′-Labeling of RNA oligonucleotides
The RNA was 5′-end labeled by treating with T4 polynucleotide kinase (Promega) and [γ-32P] ATP (Perkin Elmer). The reaction was stopped by the addition of an equal volume of stop buffer [7 M urea, 10 mM Tris–HCl and 0.1 mM EDTA (pH 7.5)]. The radiolabeled full-length RNA was isolated by 17% denaturing PAGE. The RNA was extracted from the gel via the crush and soak method as described above.
Native gel electrophoresis
5′-End-labeled M3Q RNA was folded as described above. After cooling, samples were incubated with various concentrations of TmPyP4 (Calbiochem) for 15 min at room temperature in the dark. The samples were then loaded on a native 10% gel and electrophoresed at 4°C, dried on Whatman paper and exposed to a phosphorimager screen. The gel images were visualized by scanning the screen on a Typhoon Phosphorimager 8600 (Molecular Dynamics).
Circular Dichroism
All measurements were recorded at room temperature using folded RNA samples (4 µM). Spectra were recorded using a Jasco J-810 spectrophotometer with a 0.1-cm cell at a scan speed of 50 nm/min with a response time of 1 s. The spectra were averaged over three scans and the spectrum from a blank sample containing only buffer was subtracted from the averaged data. After the collection of the initial spectrum, 1 µl aliquots of TmPyP4 in the same buffer as the RNA were added to the sample in the cuvette and was mixed several times before obtaining a new spectrum.
UV–visible spectroscopic analysis
A concentrated sample of either M3Q or mut-M3Q folded in the presence of 100 mM KCl 10 mM Tris–HCl and 0.1 mM EDTA (pH 7.5) was titrated into a 4 µM sample of TmPyP4 (100 µl). The absorption spectra at different sample addition points were collected with a Cary 300 scan UV–Vis spectrophotometer (Varian) in the wavelength range from 350 to 500 nm using a 1-cm path length microcuvette. The concentration of free TmPyP4 was determined using an extinction coefficient of 2.26 × 105 M−1 cm−1 and absorbance values at 424 nm (39). The titration was stopped when three successive additions of the M3Q RNA resulted in no further shift of the Soret band. All values were corrected for dilution effect. The fraction (α) of TmPyP4 bound was determined as follows:
where Abs TmPyP4free is the absorbance of the free TmPyP4 in the absence of any added RNA, Absmixture is absorbance at any point after the beginning of the addition of the RNA and Absbound is the absorbance of fully bound TmPyP4 measured at 424 nm (the Soret maxima for TmPyP4). Concentrations of free TmPyP4 ([TmPyP4]free) and the concentration of bound TmPyP4 ([TmPyP4]bound) were calculated as follows:
[TmPyP4]bound = ([TmPyP4]corrected−[TmPyP4]free), where [TmPyP4]corrected represents the concentration of TmPyP4 corrected for the change in volume that occurred due to the titrated RNA. The percentage of hypochromicity of the Soret band is calculated as follows:
NMR
1D 1H NMR spectra of M3Q were recorded at 37°C using a 750 MHz Varian spectrometer. A 300 -µl sample of 430 µM M3Q was folded in 20 mM potassium phosphate buffer, pH 7.0 and 8% D2O as described above. Aliquots of 55 mM TmPyP4 were added in 0.2 M equivalents and allowed to equilibrate for ∼1 min between acquisitions. Imino proton NMR spectra were collected at 37°C utilizing the Watergate 3-9-19 sequence (41) with sweep width of 20 ppm. Recycle delays of 2 s were employed between each of 128 scans. NMR spectra were processed and superimposed with iNMR (http://www.inmr.net) using the following parameters: a 40 Hz solvent filter was applied to clean up the residual water signal, the fid was apodized with a cosine2 function, linear predicted from 2 k to 3 k points, and zero filled to 4 k for Fourier transformation and automatic baseline correction in the frequency domain. During spectral comparison and plotting, the same scale was maintained for each spectrum to permit accurate comparisons of signal intensity.
Plasmid construction
Plasmids p-M3Q and wt-UTR were constructed as previously described (3). The plasmid del-M3Q was constructed by using a QuikChange site-directed mutagenesis kit (Stratagene) where nucleotides 213–229 were deleted from the wt-UTR plasmid.
Cell culture
HeLa cells were grown in 96-well plates in Dulbecco's modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and the antibiotics streptomycin and penicillin at 37°C in 5% CO2 in a humidified incubator.
Dual luciferase assays
HeLa cells were transfected with plasmids described above using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Following incubation at 37°C for 2 h, each plate was supplemented with a final concentration of 0, 50 or 100 µM TmPyP4. Twenty-four hours after transfection, Renilla (RL) and firefly (FL) luciferase activities were measured using a Dual-Glo Luciferase assay system (Promega) as per the manufacturer's supplied protocol using a Synergy 2 microplate reader (BioTek Instruments). The ratio of Renilla to firefly luciferase activities was calculated for each plasmid. In each case, the value of the ratio obtained in the absence of TmPyP4 was normalized to 100%.
RESULTS AND DISCUSSION
TmPyP4 unfolds an RNA G-quadruplex (M3Q) structure and inhibits its formation
It has been previously shown that a G-quadruplex located in the 5′-UTR of MT3-MMP (M3Q) mRNA forms an extremely stable quadruplex structure and inhibits translation of a reporter gene in eukaryotic cells (3). The physiological processes related to up-regulation of MT3-MMP make it a target to find small molecules that could selectively interact with the M3Q motif and affect translation. To determine the effect of TmPyP4 on the folded M3Q G-quadruplex, several biochemical and biophysical techniques were utilized. As shown in Figure 1A, the CD spectrum of folded M3Q in 100 mM KCl shows a peak at 263 nm and a trough at 240 nm, which is characteristic of a parallel RNA quadruplex (3,4). Titrating increasing concentrations of TmPyP4 resulted in a decrease in the CD signal at 263 nm, reflecting a disappearance of the quadruplex structure presumably due to its unfolding. According to previously published results, at this concentration of K+, M3Q was unable to be unfolded even at a temperature as high as 95°C (3). Despite the extreme stability of this quadruplex structure under these conditions, it was remarkable that TmPyP4 was able to unfold M3Q. The possibility that the reduction in the CD signal could result from non-specific associations of the added ligand to the RNA and not due to a destabilization of the quaduplex was considered. To ensure that this was not the case and to distinguish between the two possibilities, CD spectra were taken at different wavelengths to check if there was a negative induced circular dichoism (ICD), but there was no evidence of such phenomenon indicating that the reduction at 263 nm was a direct effect of the quadruplex being destabilized (Supplementary Figure S1) (42,43). The results from Figure 1A at different concentrations of TmPyP4 were normalized and graphed against CD signal by assuming that the signal at 263 nm for M3Q in the absence of TmPyP4 corresponds to 100% folded while that at 100 µM TmPyP4 represented 0% folded. As shown in Figure 1B, the data fit well with an exponential function which permitted interpolation for a value of TmPyP4 corresponding to that for 50% unfolding (11 μM). Additionally, the presence of 20 µM TmPyP4 prevents M3Q quadruplex formation when present during the folding of the G-quadruplex, as indicated by the complete lack of any structural features in the CD spectrum (Supplementary Figure S2) (42,43). A possible explanation for the lack of spectral features of a quadruplex structure when TmPyP4 is added before folding of the M3Q motif might be that TmPyP4 binds to the unfolded form of M3Q RNA (44) thus preventing the formation of the quadruplex structure.
Figure 1.
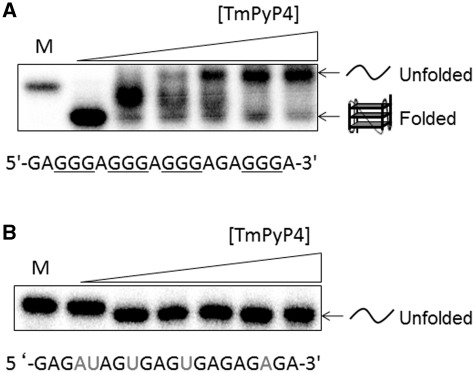
(A) CD spectra of 4 µM pre-folded (in 100 mM KCl) M3Q in the absence and presence of increasing concentrations of TmPyP4. The chemical structure of TmPyP4 is shown in the inset. The arrow defines the decrease in CD signal as a function of increasing TmPyP4 concentration. (B) Plot of calculated fraction folded versus TmPyP4 concentration.
To determine whether TmPyP4 can interact and influence the folding and subsequent migration patterns of M3Q RNA, native gel electrophoresis was performed. As shown in the first two lanes of Figure 2A, M3Q has a greater mobility than a mutated version of this sequence (mut-M3Q) which has been previously shown not to form any structure (3). Since the two oligonucleotides have the same charge, the greater mobility of M3Q can be ascribed to formation of a quadruplex structure (45). Therefore, if TmPyP4 unfolds the M3Q quadruplex, then a species with lower mobility should be observed. At total concentrations of TmPyP4 from 5 to 50 μM, an RNA species emerges that has a lower mobility than mut-M3Q which may represent a complex of TmPyP4 with unfolded M3Q. It has been previously shown by the Fry group that the mobility of an RNA sequence that presumably adopts a quadruplex structure can be retarded by TmPyP4 (45). The mut-M3Q sequence shows no change in mobility in the presence of up to 50 µM of TmPyP4 indicating the specificity of the interaction between TmPyP4 and the M3Q sequence. The lower mobility of the M3Q band resulting in the unfolding of the sequence can be corroborated with CD experiments (Figure 1), which indicate that the M3Q quadruplex is almost completely unfolded in the presence of 50 μM TmPyP4. In addition, the binding of TmPyP4 with unfolded M3Q was evidenced by spectral shifts of its Soret absorption band (see below). Interestingly, at concentrations of the ligand from 1 to 10 μM, a band with intermediate mobility was observed, which may reflect the existence of a transient, intermediate complex of TmPyP4 with partially unfolded M3Q. While this may be an intermediate structure, its exact identity cannot be established at this point. It should also be noted that the different levels of retardation in mobility of the M3Q band observed may also be due to binding of the cationic TmPyP4 molecule resulting in a diminished net negative charge on the RNA which would cause its lower mobility. Considering the prevailing dynamic, non-equilibrium experimental condition where RNA anions and TmPyP4 cations are being separated by electrophoresis, the cause of different lower mobilities cannot be determined unambiguously. Overall, the CD and native gel electrophoresis data suggest a change in nature of the M3Q quadruplex structure in the presence of TmPyP4 presumably due to unfolding of M3Q.
Figure 2.
(A) Native gel shift assay of M3Q (final concentration of ∼250 nM) in the absence and presence of increasing concentrations of TmPyP4. M denotes mut-M3Q. Concentrations used are 0, 1, 5, 10, 20 and 50 µM TmPyP4. (B) Native gel shift assay of mut-M3Q in the absence and presence of increasing concentrations of TmPyP4. M denotes the single stranded RNA marker mut-M3Q. Concentrations of TmPyP4 are the same as in A.
Proton NMR reveals unfolding of the M3Q RNA quadruplex
The disruption of the folded M3Q quadruplex structure in the presence of TmPyP4 was further established by 1H NMR. It has been shown that the imino protons from the Hoogsteen hydrogen bonding between the guanines of the tetrads in the quadruplex show distinct peaks from 10 to 12 ppm (4). Figure 3 shows the imino peaks from the tetrads in the M3Q quadruplex structure that are visible even at 37°C which still resulted in sharp imino peaks attesting to the extreme stability of the quadruplex structure; most RNA imino protons from bases engaged in Watson–Crick pairs, exchange broaden at about room temperature. Thus, the NMR data suggest the presence of a thermodynamically hyper-stable quadruplex structure at a biologically relevant temperature. TmPyP4 was titrated into the preformed M3Q RNA G-quadruplex in 0.2 equivalent portions and the spectra were taken immediately. Titration of this RNA with the TmPyP4 molecule resulted in accelerated imino exchange (Figure 3) consistent with G-quadruplex structure melting. As the concentration of TmPyP4 is increased there is a reduction in intensity of the imino peaks until eventually there is a complete lack of signal while new signals appear upfield of the main cluster of iminos at substoichiometric levels. The new iminos' appearance and disappearance of other imino signals is consistent with a ligand dissociation that is slower than the NMR time-scale. Such signal behavior is often seen with modest to higher affinity complexes with Kd's typically <20 µM (46,47). These new peaks probably represent ring-current shifted iminos resulting from bound but not fully unfolded molecules (48). Because these resonances are upfield shifted, we would hypothesize that this ligand bound and partially unfolded state represents stacking of the TmPyP4 upon the G-tetrad (49,50), although additional structure probing would be required to validate this notion. Several of the resonances in the anomeric proton region are present even at the highest concentration of TmPyP4. No visible precipitate was present at this highest concentration; this and the persistent signal from non-exchangeable protons suggest that the disappearance of imino resonances is not due to non-specific aggregation.
Figure 3.
NMR spectra of 0.42 mM M3Q titrated with TmPyP4. 1D 3-9-19 Watergate spectra were collected at 37°C. PAGE purified and dialyzed M3Q was pre-folded in NMR buffer (20 mM potassium phosphate, pH 7.0 and 8% D2O) and titrated with 0.2 M equivalent increments of 55 mM TmPyP4. Each experiment was collected for 5 min with 128 scans and 2-s recycle delays. The left portion of the spectrum shows the imino region, while the right shows a portion of the anomeric, i.e. H1′-region.
UV–Vis spectroscopy shows binding of M3Q to TmPyP4
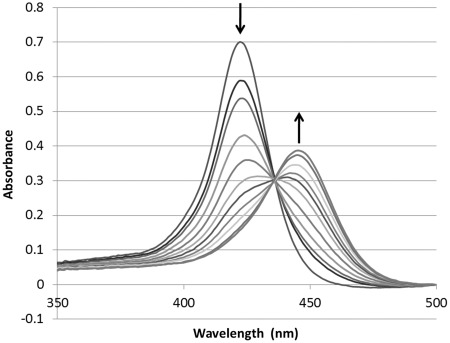
The binding of M3Q and TmPyP4 was also monitored via UV–Vis spectroscopy. Folded M3Q was titrated into a solution of TmPyP4 and the Soret band was monitored as a function of M3Q concentration. As the concentration of M3Q was increased there was substantial hypochromicity (maximum 75%) as well as a rather unusually large bathochromic shift of 22 nm (from λmax 424 to 446 nm), which is indicative of binding of TmPyP4 to the M3Q RNA G-quadruplex (Figure 4) (51). A sharp isosbestic point was observed at 436 nm. When mut-M3Q was titrated, no shift or hypochromicity was apparent even after addition of up to five equivalents of TmPyP4, which suggests specificity of TmPyP4 interaction with the M3Q RNA (Supplementary Figure S3). These results correlate well with the native gel electrophoresis data on the specificity of the interaction between M3Q and TmPyP4.
Figure 4.
Visible absorption spectra of TmPyP4 in the absence and presence of increasing concentration of pre-folded M3Q. The initial concentration of TmPyP4 was 4 µM (100 µl) to which 0.21 nmoles of the pre-folded M3Q RNA quadruplex was added in 0.5 µl increments. The experiment was performed at 100 mM KCl, 0.1 mM EDTA and 10 mM Tris–HCl (pH. 7.5). The arrow pointing downward indicates the decrease in the λmax 424 nm of TmPyP4 with the addition of M3Q and the arrow pointing upward indicates the shifted Soret band.
Hypochromicity and a bathochromic shift of the Soret band reported previously were observed in cases of stabilization of the G-quadruplex structure (51,52). An important aspect in case of destabilization is the fate of the Soret band after presumed disruption of the M3Q G-quadruplex structure. We observed that the Soret band shift remained unchanged even after destabilization of the M3Q quadruplex. Porphyrins are known to bind to single-stranded nucleic acids resulting in shift of the Soret band in the same amount and direction as the change when interacting with a nucleic acid secondary structure (44,53,54), which may explain the lack of change in shift of the Soret band even after the quadruplex structure was destabilized. Presumably the ability of TmPyP4 to prevent the M3Q quadruplex formation is due to the fact that it can bind to single-stranded RNA.
The mode of interaction between TmPyP4 and RNA quadruplexes is unknown, while the mechanism of interaction between DNA G-quadruplexes and the TmPyP4 is a point of debate in the literature (25,54,55). We observed a large bathochromic shift of the Soret band accompanied by a high percentage of hypochromicity by UV spectroscopy when the pre-folded M3Q RNA was titrated into a TmPyP4 solution. The large shift in the Soret band resulted from the transfer of the π-electrons from the purine bases to the pyrrole rings (56), suggesting stacking on the G-quadruplex or perhaps intercalation between the tetrads, both of which have been proposed in the literature (25,54,55).
It is important to note that the mechanisms of binding are for reported stabilization of the G-quadruplex structures due to TmPyP4 binding (25,54,55). However, here we report unfolding of an usually stable RNA quadruplex. Although the mode of recognition of this stable RNA quadruplex by TmPyP4 is unclear at this point, the unfolding detected by the various biophysical methods described above indicates that the interaction between TmPyP4 and unfolded and non-quadruplex form of M3Q is thermodynamically favorable. Previous reports have shown that TmPyP4 can bind to non-quadruplex nucleic acid forms. For example, it has been reported that TmPyP4 binds to various double-stranded DNA sequences with affinities that are similar to quadruplex DNA (35,36), which raises the question on the selectivity of its binding. Additionally, the TmPyP4 can bind to single-stranded DNA with preference for sequences with stretches of guanines; however the particular mechanism of such interaction is still unknown.
TmPyP4 enhances translation activity of a reporter gene in live eukaryotic cells
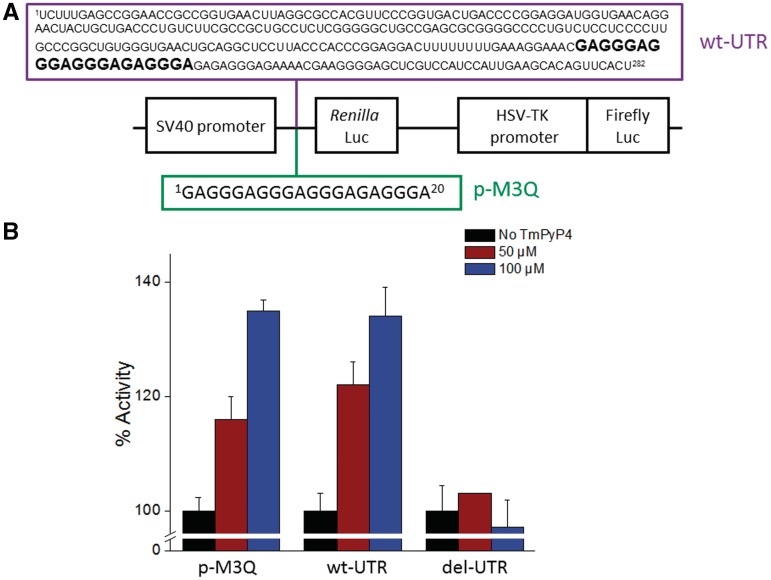
The possibility that TmPyP4 could unfold the M3Q motif in eukaryotic cells and affect translation efficiency was addressed by using a dual luciferase reporter construct in which the M3Q sequence was placed just before a Renilla luciferase gene, while the upstream firefly luciferase was under the control of a herpes simplex virus thymidine kinase promoter (p-M3Q, Figure 5A). It has been shown previously that the quadruplex has an inhibitory effect on the translation of the Renilla mRNA in this particular plasmid (3). This plasmid was transfected in HeLa cells and incubated with 0, 50 or 100 µM TmPyP4, respectively for 24 h. The cells where inspected prior to the luciferase assay and no detectable change in the cell number or morphology in any of the treatment groups was observed. As can be seen in Figure 5B, 50 µM TmPyP4 had a moderate effect on translation with an increase of 15 ± 4%. However, when the concentration of TmPyP4 was increased to 100 µM it had an increase of 35 ± 2% in translation. In order to increase translation of Renilla mRNA, the TmPyP4 would have to relieve the repressive effect of the quadruplex, presumably by destabilizing the quadruplex structure. It was also of interest to compare these results with those found from having the M3Q sequence in the context of the entire 282 nucleotide 5′-UTR of MT3-MMP mRNA (wt-UTR). A similar increase in activity would suggest that TmPyP4 is binding to the quadruplex embedded within the entire 5′-UTR and is modulating its activity. The entire 5′-UTR was placed in front of the Renilla gene and again assayed after the cells where incubated in the presence of 0, 50 or 100 µM TmPyP4, respectively for 24 h. As shown in Figure 5B, the data for the two constructs correlate well with wt-UTR increasing activity 22 ± 4% in the presence of 50 µM of TmPyP4 and 37 ± 5% when the cells were treated with 100 µM TmPyP4. While evaluating the enhancement of translation by TmPyP4 it should be taken into account that the translation was repressed by the G-quadruplex alone (no ligand) by 55%. We performed qRT-PCR experiments that showed no change in mRNA levels (Supplementary Figure S4) of the wt-UTR construct at 0 and 100 µM concentration of TmPyP4 which shows that the gain in activity was at the translational level. We then tested the effect of TmPyP4 on a mutant of wt-UTR in which the M3Q sequence was deleted (del-UTR). The strategy of using the deletion mutant as a control was based upon two previous independent reports (26,45) where the quadruplex forming segment deleted to test the functional consequence of small-molecule G-quadruplex interactions. Figure 5B shows that at concentrations up to 100 µM there is no increase in Renilla luciferase activity. The lack of any change in the level of translation of the deletion mutant can be explained by presuming that TmPyP4 is specifically disrupting the M3Q quadruplex in vivo to significantly mitigate its repressive effect on translation.
Figure 5.
(A) Schematic of dual luciferase bi-cistronic constructs (3). (B) Histogram showing percentage of activity of the translation of the Renilla gene as a function of TmPyP4 concentration.
The destabilization of the M3Q sequence by TmPyP4 is unprecedented
Until recently, the majority of reports have focused on the small-molecule interactions with DNA G-quadruplexes, however, the interactions between small molecules and RNA G-quadruplexes are barely understood. The studies on clearly established RNA G-quadruplexes interacting with small molecules reported stabilization of the quadruplex structure. Balasubramanian and coworkers have shown the stabilization and repression of translation in a eukaryotic cell-free system by targeting an RNA quadruplex located in the 5′-UTR of NRAS with a pyridine-2,6-bis-quinolino-dicarboxamide derivative (26). Neidle and coworkers have reported the interactions between tetrasubstituted naphthalene diimides among other ligands, and the telomeric RNA quadruplex forming sequence (27). Recently, Hartig and coworkers (28) also reported stabilization of designed RNA quadruplexes in the presence of bisquinolinium compounds.
In the current report, the interaction between the previously well-characterized quadruplex forming sequence, M3Q, and the known DNA stabilizing cationic porphyrin TmPyP4 is described. A variety of biochemical and biophysical data strongly suggest the destabilization of the very stable M3Q RNA G-quadruplex structure by TmPyP4, unlike most of the previous studies that reported stabilization of DNA quadruplexes in the presence of this small molecule (25). Interestingly, when we analyzed a DNA version of the M3Q sequence in the presence of TmPyP4 the quadruplex (Tm = 71 ± 1°C at 100 mM KCl, Supplementary Figure 5A) was also destabilized. However, it required a higher concentration to unfold the DNA quadruplex compared to the M3Q RNA version (Supplementary Figure 5B) although the RNA quadruplex could not be melted at 100 mM KCl (Tm = 72 ± 1 at 1 mM KCl,). Thus, the DNA version of M3Q is unfolded by TmPyP4 but not as efficiently as it unfolds the RNA version of M3Q.
Destabilization of G-quadruplex structures by small-molecules have been shown in only a few reports and they mostly involve DNA quadruplexes (43,45). The Balasubramanian group has described a triarylpyridine molecule that induced unfolding of the quadruplex formed by the DNA sequence found within the c-kit promoter, a structure that based upon the reported Tm is significantly less stable than M3Q (43,57). Destabilization of a d(CGG) repeat in presence of TmPyP4 was reported by the Fry group.(37) Additionally, Fry and coworkers (45) have shown that the interactions of an RNA quadruplex forming motif r(CGG)33 were destabilized by TmPyP4, however, according to the authors the nature of the secondary structure that this sequence adopts is still rather unclear.
CONCLUSION
The functional consequence of the M3Q G-quadruplex destabilization would result in increased translation of the reporter mRNA. In vitro biochemical and biophysical data presented in this report indicated destabilization of the M3Q G-quadruplex which may be valuable for other G-quadruplexes that regulate translation. The findings of this report can be useful in cases where up-regulation of genes may have a therapeutic benefit.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5.
FUNDING
The Kent State University (to S.B., start-up). Funding for open access charge: The University of Akron (to T.C.L., start-up).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENT
We thank Dr Fred Walz for his comments on the article.
REFERENCES
- 1.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 2.McQuibban GA, Gong JH, Tam EM, McCulloch CA, Clark-Lewis I, Overall CM. Inflammation dampened by gelatinase A cleavage of monocyte chemoattractant protein-3. Science. 2000;289:1202–1206. doi: 10.1126/science.289.5482.1202. [DOI] [PubMed] [Google Scholar]
- 3.Morris MJ, Basu S. An unusually stable G-quadruplex within the 5'-UTR of the MT3 Matrix Metalloproteinase mRNA represses translation in eukaryotic cells. Biochemistry. 2009;48:5313–5319. doi: 10.1021/bi900498z. [DOI] [PubMed] [Google Scholar]
- 4.Neidle S, Balasubramanian S. Cambridge, UK: Royal Society of Chemistry; 2006. Quadruplex Nucleic Acids. [Google Scholar]
- 5.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eddy J, Maizels N. Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes. Nucleic Acids Res. 2008;36:1321–1333. doi: 10.1093/nar/gkm1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mani P, Yadav VK, Das SK, Chowdhury S. Genome-wide analyses of recombination prone regions predict role of DNA structural motif in recombination. PLoS ONE. 2009;4:e4399. doi: 10.1371/journal.pone.0004399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stephen N. The structures of quadruplex nucleic acids and their drug complexes. Curr. Opin. Struct. Biol. 2009;19:239–250. doi: 10.1016/j.sbi.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simonsson T, Pecinka P, Kubista M. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 1998;26:1167–1172. doi: 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dexheimer TS, Sun D, Hurley LH. Deconvoluting the structural and drug-recognition complexity of the G-Quadruplex-forming region upstream of the bcl-2 P1 promoter. J. Am. Chem. Soc. 2006;128:5404–5415. doi: 10.1021/ja0563861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cogoi S, Xodo LE. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006;34:2536–2549. doi: 10.1093/nar/gkl286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun D, Guo K, Rusche JJ, Hurley LH. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005;33:6070–6080. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kostadinov R, Malhotra N, Viotti M, Shine R, D'Antonio L, Bagga P. GRSDB: a database of quadruplex forming G-rich sequences in alternatively processed mammalian pre-mRNA sequences. Nucleic Acids Res. 2006;34:D119–D124. doi: 10.1093/nar/gkj073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumari S, Bugaut A, Huppert JL, Balasubramanian S. An RNA G-quadruplex in the 5'-UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sundquist WI, Heaphy S. Evidence for interstrand quadruplex formation in the dimerization of human immunodeficiency virus 1 genomic RNA. Proc. Natl Acad. Sci. USA. 1993;90:3393–3397. doi: 10.1073/pnas.90.8.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schaeffer C, Bardoni B, Mandel JL, Ehresmann B, Ehresmann C, Moine H. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 2001;20:4803–4813. doi: 10.1093/emboj/20.17.4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonnal S, Schaeffer C, Creancier L, Clamens S, Moine H, Prats A-C, Vagner S. A single internal ribosome entry site containing a G quartet RNA structure drives fibroblast growth factor 2 gene expression at four alternative translation initiation codons. J. Biol. Chem. 2003;278:39330–39336. doi: 10.1074/jbc.M305580200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arora A, Dutkiewicz M, Scaria V, Hariharan M, Maiti S, Kurreck J. Inhibition of translation in living eukaryotic cells by an RNA G-quadruplex motif. RNA. 2008;14:1290–1296. doi: 10.1261/rna.1001708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christiansen J, Kofod M, Nielsen FC. A guanosine quadruplex and two stable hairpins flank a major cleavage site in insulin-like growth factor II mRNA. Nucleic Acids Res. 1994;22:5709–5716. doi: 10.1093/nar/22.25.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morris MJ, Negishi Y, Pazsint C, Schonhoft JD, Basu S. An RNA G-quadruplex is essential for cap-independent translation initiation in human VEGF IRES. J. Am. Chem. Soc. 2010;132:17831–17839. doi: 10.1021/ja106287x. [DOI] [PubMed] [Google Scholar]
- 23.Gomez D, Guedin A, Mergny J-L, Salles B, Riou J-Fo, Teulade-Fichou M-P, Calsou P. A G-quadruplex structure within the 5'-UTR of TRF2 mRNA represses translation in human cells. Nucleic Acids Res. 2010;38:7187–7198. doi: 10.1093/nar/gkq563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wieland M, Hartig JS. RNA quadruplex-based modulation of gene expression. Chem. Biol. 2007;14:757–763. doi: 10.1016/j.chembiol.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 25.Arora A, Maiti S. Differential biophysical behavior of human telomeric RNA and DNA quadruplex. J. Phys. Chem. B. 2009;113:10515–10520. doi: 10.1021/jp810638n. [DOI] [PubMed] [Google Scholar]
- 26.Bugaut A, Rodriguez R, Kumari S, Hsu S-T, Balasubramanian S. Small molecule-mediated inhibition of translation by targeting a native RNA G-quadruplex. Org. Biomol. Chem. 2010;21:2771–2776. doi: 10.1039/c002418j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collie G, Reszka AP, Haider SM, Gabelica V, Parkinson GN, Neidle S. Selectivity in small molecule binding to human telomeric RNA and DNA quadruplexes. Chem. Commun. 2009;48:7482–7484. doi: 10.1039/b901889a. [DOI] [PubMed] [Google Scholar]
- 28.Halder K, Largy E, Benzler M, Teulade-Fichou M-P, Hartig JS. Efficient suppression of gene expression by targeting 5′-UTR-Based RNA quadruplexes with bisquinolinium compounds. Chem. Bio. Chem. 2011;12:1663–1668. doi: 10.1002/cbic.201100228. [DOI] [PubMed] [Google Scholar]
- 29.Thomas JR, Hergenrother PJ. Targeting RNA with small molecules. Chem. Rev. 2008;108:1171–1224. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- 30.Monchaud D, Teulade-Fichou M-P. A hitchhiker's guide to G-quadruplex ligands. Org. Biomol. Chem. 2008;6:627–636. doi: 10.1039/b714772b. [DOI] [PubMed] [Google Scholar]
- 31.de Armond R, Wood S, Sun D, Hurley LH, Ebbinghaus SW. Evidence for the presence of a Guanine quadruplex forming region within a polypurine tract of the Hypoxia Inducible Factor 1 alpha promoter. Biochemistry. 2005;44:16341–16350. doi: 10.1021/bi051618u. [DOI] [PubMed] [Google Scholar]
- 32.Guo K, Pourpak A, Beetz-Rogers K, Gokhale V, Sun D, Hurley LH. Formation of pseudosymmetrical G-quadruplex and i-motif structures in the proximal promoter region of the RET oncogene. J. Am. Chem. Soc. 2007;129:10220–10228. doi: 10.1021/ja072185g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qin Y, Rezler EM, Gokhale V, Sun D, Hurley LH. Characterization of the G-quadruplexes in the duplex nuclease hypersensitive element of the PDGF-A promoter and modulation of PDGF-A promoter activity by TMPyP4. Nucleic Acids Res. 2007;35:7698–7713. doi: 10.1093/nar/gkm538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun D, Liu W-J, Guo K, Rusche JJ, Ebbinghaus S, Gokhale V, Hurley LH. The proximal promoter region of the human vascular endothelial growth factor gene has a G-quadruplex structure that can be targeted by G-quadruplex interactive agents. Mol. Cancer Ther. 2008;7:880–889. doi: 10.1158/1535-7163.MCT-07-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren J, Chaires JB. Sequence and structural selectivity of nucleic acid binding ligands. Biochemistry. 1999;38:16067–16075. doi: 10.1021/bi992070s. [DOI] [PubMed] [Google Scholar]
- 36.Luedtke NW. Targeting G-quadruplex DNA with small molecules. CHIMIA. 2009;63:134–139. [Google Scholar]
- 37.Weisman-Shomer P, Cohen E, Hershco I, Khateb S, Wolfovitz-Barchad O, Hurley LH, Fry M. The cationic porphyrin TMPyP4 destabilizes the tetraplex form of the fragile X syndrome expanded sequence d(CGG)n. Nucleic Acids Res. 2003;31:3963–3970. doi: 10.1093/nar/gkg453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Puglisi JD, Tinoco I. Methods in Enzymol. Vol. 180. Academic Press; 1989. pp. 304–325. [DOI] [PubMed] [Google Scholar]
- 39.Pasternack RF, Huber PR, Boyd P, Engasser G, Francesconi L, Gibbs E, Fasella P, Cerio Venturo G, Hinds Ld. Aggregation of meso-substituted water-soluble porphyrins. J. Am. Chem. Soc. 1972;94:4511–4517. doi: 10.1021/ja00768a016. [DOI] [PubMed] [Google Scholar]
- 40.Keating LR, Szalai VA. Parallel-stranded guanine quadruplex interactions with a copper cationic porphyrin. Biochemistry. 2004;43:15891–15900. doi: 10.1021/bi0483209. [DOI] [PubMed] [Google Scholar]
- 41.Piotto M, Saudek V, Sklenář V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 42.Dash J, Shirude PS, Hsu S-TD, Balasubramanian S. Diarylethynyl amides that recognize the parallel conformation of genomic promoter DNA G-quadruplexes. J. Am. Chem. Soc. 2008;130:15950–15956. doi: 10.1021/ja8046552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waller ZAE, Sewitz SA, Hsu S-TD, Balasubramanian S. A small molecule that disrupts G-quadruplex DNA structure and enhances gene expression. J. Am. Chem. Soc. 2009;131:12628–12633. doi: 10.1021/ja901892u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Flynn S, George S, White L, Devonish W, Takle G. Water-soluble, meso-substituted cationic porphyrins–a family of compounds for cellular delivery of oligonucleotides. Biotechniques. 1999;26:736–742. doi: 10.2144/99264rr03. [DOI] [PubMed] [Google Scholar]
- 45.Ofer N, Weisman-Shomer P, Shklover J, Fry M. The quadruplex r(CGG)n destabilizing cationic porphyrin TMPyP4 cooperates with hnRNPs to increase the translation efficiency of fragile X premutation mRNA. Nucleic Acids Res. 2009;37:2712–2722. doi: 10.1093/nar/gkp130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Millet O, Loria JP, Kroenke CD, Pons M, Palmer AG. The static magnetic field dependence of chemical exchange linebroadening defines the NMR chemical shift time scale. J. Am. Chem. Soc. 2000;122:2867–2877. [Google Scholar]
- 47.Fielding L. NMR methods for the determination of protein-ligand dissociation constants. Curr. Top. Med. Chem. 2003;3:39–53. doi: 10.2174/1568026033392705. [DOI] [PubMed] [Google Scholar]
- 48.Johnson CE, Bovey FA. Calculation of nuclear magnetic resonance spectra of aromatic hydrocarbons. J. Chem. Phys. 1958;29:1012–1014. [Google Scholar]
- 49.Perkins SJ, Wüthrich K. Ring current effects in the conformation dependent NMR chemical shifts of aliphatic protons in the basic pancreatic trypsin inhibitor. Biochim. Biophys. Acta. 1979;576:409–423. doi: 10.1016/0005-2795(79)90416-1. [DOI] [PubMed] [Google Scholar]
- 50.Case DA. Calibration of ring-current effects in proteins and nucleic acids. J. Biomol. NMR. 1995;6:341–346. doi: 10.1007/BF00197633. [DOI] [PubMed] [Google Scholar]
- 51.Wheelhouse RT, Sun D, Han H, Han FX, Hurley LH. Cationic porphyrins as telomerase inhibitors: the interaction of tetra-(N-methyl-4-pyridyl)porphine with quadruplex DNA. J. Am. Chem. Soc. 1998;120:3261–3262. [Google Scholar]
- 52.Yamashita T, Uno T, Ishikawa Y. Stabilization of guanine quadruplex DNA by the binding of porphyrins with cationic side arms. Bioorg. Med. Chem. 2005;13:2423–2430. doi: 10.1016/j.bmc.2005.01.041. [DOI] [PubMed] [Google Scholar]
- 53.Iverson BL, Shreder K, Král V, Smith DA, Smith J, Sessler JL. Interactions between expanded porphyrins and nucleic acids. Pure Appl. Chem. 1994;66:845–850. [Google Scholar]
- 54.Ghazaryan AA, Dalyan YB, Haroutiunian SG, Tikhomirova A, Taulier N, Wells JW, Chalikian TV. Thermodynamics of interactions of water-soluble porphyrins with RNA duplexes. J. Am. Chem. Soc. 2006;128:1914–1921. doi: 10.1021/ja054070n. [DOI] [PubMed] [Google Scholar]
- 55.Parkinson GN, Ghosh R, Neidle S. Structural basis for binding of porphyrin to human telomeres. Biochemistry. 2007;46:2390–2397. doi: 10.1021/bi062244n. [DOI] [PubMed] [Google Scholar]
- 56.Shkirman S, Solov'ev K, Kachura T, Arabei S, Skakovskii E. Interpretation of the soret band of porphyrins based on the polarization spectrum of N-methyltetraphenylporphin fluorescence. J. Appl. Spectros. 1999;66:68–75. [Google Scholar]
- 57.Fernando H, Reszka AP, Huppert J, Ladame S, Rankin S, Venkitaraman AR, Neidle S, Balasubramanian S. A Conserved quadruplex motif located in a transcription activation site of the human c-kit oncogene. Biochemistry. 2006;45:7854–7860. doi: 10.1021/bi0601510. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.