Abstract
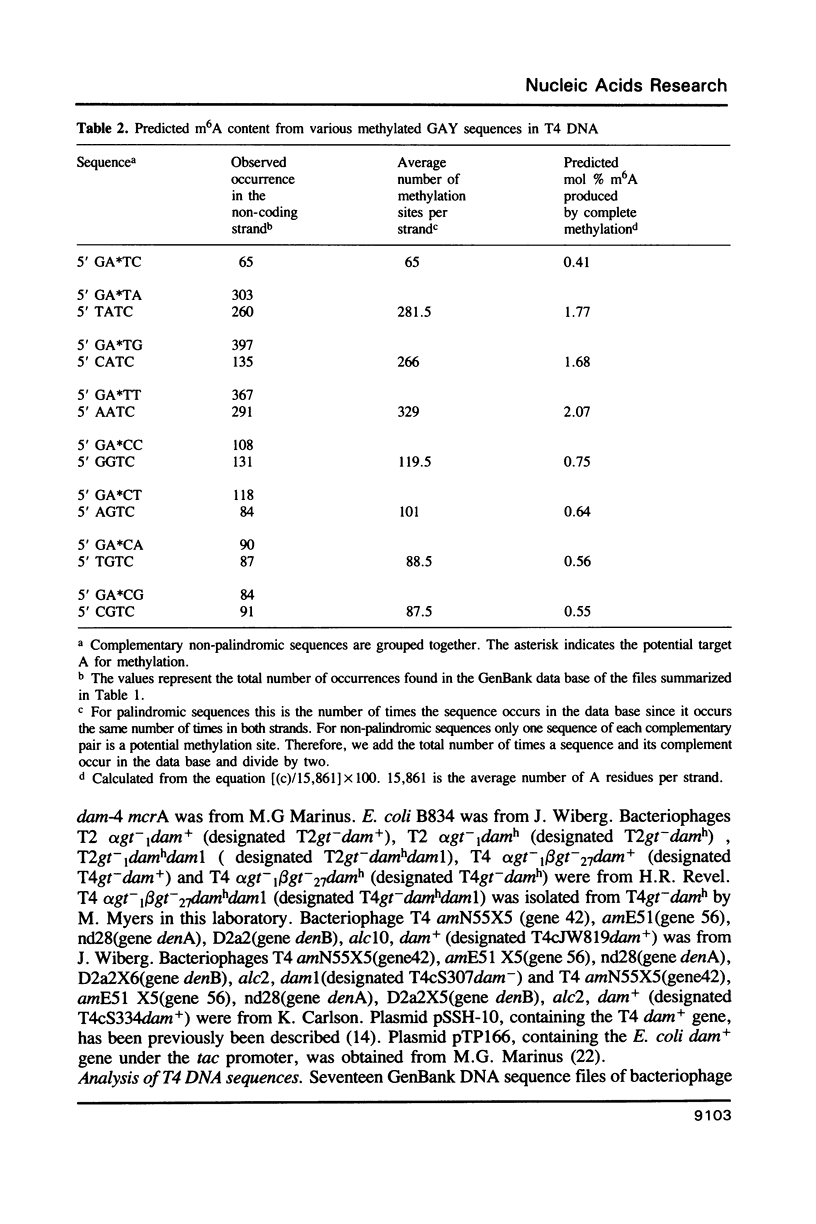
Bacteriophages T2 and T4 encode DNA-[N6-adenine] methyltransferases (Dam) which differ from each other by only three amino acids. The canonical recognition sequence for these enzymes in both cytosine and 5-hydroxymethylcytosine-containing DNA is GATC; at a lower efficiency they also recognize some non-canonical sites in sequences derived from GAY (where Y is cytosine or thymine). We found that T4 Dam fails to methylate certain GATA and GATT sequences which are methylated by T2 Dam. This indicates that T2 Dam and T4 Dam do not have identical sequence specificities. We analyzed DNA sequence data files obtained from GenBank, containing about 30% of the T4 genome, to estimate the overall frequency of occurrence of GATC, as well as non-canonical sites derived from GAY. The observed N6methyladenine (m6A) content of T4 DNA, methylated exclusively at GATC (by Escherichia coli Dam), was found to be in good agreement with this estimate. Although GATC is fully methylated in virion DNA, only a small percentage of the non-canonical sequences are methylated.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barras F., Marinus M. G. Arrangement of Dam methylation sites (GATC) in the Escherichia coli chromosome. Nucleic Acids Res. 1988 Oct 25;16(20):9821–9838. doi: 10.1093/nar/16.20.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks J. E., Hattman S. In vitro methylation of bacteriophage lambda DNA by wild type (dam+) and mutant (damh) forms of the phage T2 DNA adenine methylase. J Mol Biol. 1978 Dec 15;126(3):381–394. doi: 10.1016/0022-2836(78)90047-5. [DOI] [PubMed] [Google Scholar]
- Doolittle M. M., Sirotkin K. Bacteriophage T2 and T4, dam+ and damh and Eco dam+ methylation: preference at different sites. Biochim Biophys Acta. 1988 Feb 28;949(2):240–246. doi: 10.1016/0167-4781(88)90088-7. [DOI] [PubMed] [Google Scholar]
- Fujimoto D., Srinivasan P. R., Borek E. On the nature of the deoxyribonucleic acid methylases. Biological evidence for the multiple nature of the enzymes. Biochemistry. 1965 Dec;4(12):2849–2855. doi: 10.1021/bi00888a041. [DOI] [PubMed] [Google Scholar]
- GOLD M., HAUSMANN R., MAITRA U., HURWITZ J. THE ENZYMATIC METHYLATION OF RNA AND DNA. 8. EFFECTS OF BACTERIOPHAGE INFECTION ON THE ACTIVITY OF THE METHYLATING ENZYMES. Proc Natl Acad Sci U S A. 1964 Aug;52:292–297. doi: 10.1073/pnas.52.2.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gefter M., Hausmann R., Gold M., Hurwitz J. The enzymatic methylation of ribonucleic acid and deoxyribonucleic acid. X. Bacteriophage T3-induced S-adenosylmethionine cleavage. J Biol Chem. 1966 May 10;241(9):1995–2006. [PubMed] [Google Scholar]
- Geier G. E., Modrich P. Recognition sequence of the dam methylase of Escherichia coli K12 and mode of cleavage of Dpn I endonuclease. J Biol Chem. 1979 Feb 25;254(4):1408–1413. [PubMed] [Google Scholar]
- Hattman S., Brooks J. E., Masurekar M. Sequence specificity of the P1 modification methylase (M.Eco P1) and the DNA methylase (M.Eco dam) controlled by the Escherichia coli dam gene. J Mol Biol. 1978 Dec 15;126(3):367–380. doi: 10.1016/0022-2836(78)90046-3. [DOI] [PubMed] [Google Scholar]
- Hattman S. DNA methylation of T-even bacteriophages and of their nonglucosylated mutants: its role in P1-directed restriction. Virology. 1970 Oct;42(2):359–367. doi: 10.1016/0042-6822(70)90279-5. [DOI] [PubMed] [Google Scholar]
- Hattman S., Wilkinson J., Swinton D., Schlagman S., Macdonald P. M., Mosig G. Common evolutionary origin of the phage T4 dam and host Escherichia coli dam DNA-adenine methyltransferase genes. J Bacteriol. 1985 Nov;164(2):932–937. doi: 10.1128/jb.164.2.932-937.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S., van Ormondt H., de Waard A. Sequence specificity of the wild-type dam+) and mutant (damh) forms of bacteriophage T2 DNA adenine methylase. J Mol Biol. 1978 Mar 5;119(3):361–376. doi: 10.1016/0022-2836(78)90219-x. [DOI] [PubMed] [Google Scholar]
- Hausmann R., Gold M. The enzymatic methylation of ribonucleic acid and deoxyribonucleic acid. IX. Deoxyribonucleic acid methylase in bacteriophage-infected Escherichia coli. J Biol Chem. 1966 May 10;241(9):1985–1994. [PubMed] [Google Scholar]
- Hehlmann R., Hattman S. Mutants of bacteriophage T2 gt with altered DNA methylase activity. J Mol Biol. 1972 Jun 28;67(3):351–360. doi: 10.1016/0022-2836(72)90455-x. [DOI] [PubMed] [Google Scholar]
- Kholmina G. V., Rebentish B. A., Skoblov Iu S., Mironov A. A., Iankovskii N. K. Vydelenie i kharakteristika novoi sait-spetsificheskoi éndonukleazy Eco Rv. Dokl Akad Nauk SSSR. 1980;253(2):495–497. [PubMed] [Google Scholar]
- Kim J. S., Davidson N. Electron microscope heteroduplex study of sequence relations of T2, T4, and T6 bacteriophage DNAs. Virology. 1974 Jan;57(1):93–111. doi: 10.1016/0042-6822(74)90111-1. [DOI] [PubMed] [Google Scholar]
- Lacks S., Greenberg B. Complementary specificity of restriction endonucleases of Diplococcus pneumoniae with respect to DNA methylation. J Mol Biol. 1977 Jul;114(1):153–168. doi: 10.1016/0022-2836(77)90289-3. [DOI] [PubMed] [Google Scholar]
- Marinus M. G. DNA methylation in Escherichia coli. Annu Rev Genet. 1987;21:113–131. doi: 10.1146/annurev.ge.21.120187.000553. [DOI] [PubMed] [Google Scholar]
- Marinus M. G., Poteete A., Arraj J. A. Correlation of DNA adenine methylase activity with spontaneous mutability in Escherichia coli K-12. Gene. 1984 Apr;28(1):123–125. doi: 10.1016/0378-1119(84)90095-7. [DOI] [PubMed] [Google Scholar]
- McClelland M., Nelson M. The effect of site-specific DNA methylation on restriction endonucleases and DNA modification methyltransferases--a review. Gene. 1988 Dec 25;74(1):291–304. doi: 10.1016/0378-1119(88)90305-8. [DOI] [PubMed] [Google Scholar]
- Miner Z., Hattman S. Molecular cloning, sequencing, and mapping of the bacteriophage T2 dam gene. J Bacteriol. 1988 Nov;170(11):5177–5184. doi: 10.1128/jb.170.11.5177-5184.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miner Z., Schlagman S., Hattman S. Single amino acid changes which alter the sequence specificity of the T4 and T2 (Dam) DNA-adenine methyltransferases. Gene. 1988 Dec 25;74(1):275–276. doi: 10.1016/0378-1119(88)90302-2. [DOI] [PubMed] [Google Scholar]
- Nelson M., Christ C., Schildkraut I. Alteration of apparent restriction endonuclease recognition specificities by DNA methylases. Nucleic Acids Res. 1984 Jul 11;12(13):5165–5173. doi: 10.1093/nar/12.13.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revel H. R., Hattman S. M. Mutants of T2gt with altered DNA methylase activity: relation to restriction by prophage P1. Virology. 1971 Aug;45(2):484–495. doi: 10.1016/0042-6822(71)90348-5. [DOI] [PubMed] [Google Scholar]
- Scarbrough K., Hattman S., Nur U. Relationship of DNA methylation level to the presence of heterochromatin in mealybugs. Mol Cell Biol. 1984 Apr;4(4):599–603. doi: 10.1128/mcb.4.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schildkraut I., Banner C. D., Rhodes C. S., Parekh S. The cleavage site for the restriction endonuclease EcoRV is 5'-GAT/ATC-3'. Gene. 1984 Mar;27(3):327–329. doi: 10.1016/0378-1119(84)90078-7. [DOI] [PubMed] [Google Scholar]
- Schlagman S. L., Hattman S., Marinus M. G. Direct role of the Escherichia coli Dam DNA methyltransferase in methylation-directed mismatch repair. J Bacteriol. 1986 Mar;165(3):896–900. doi: 10.1128/jb.165.3.896-900.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlagman S. L., Hattman S. Molecular cloning of a functional dam+ gene coding for phage T4 DNA adenine methylase. Gene. 1983 May-Jun;22(2-3):139–156. doi: 10.1016/0378-1119(83)90098-7. [DOI] [PubMed] [Google Scholar]
- Schlagman S. L., Miner Z., Fehér Z., Hattman S. The DNA [adenine-N6]methyltransferase (Dam) of bacteriophage T4. Gene. 1988 Dec 20;73(2):517–530. doi: 10.1016/0378-1119(88)90516-1. [DOI] [PubMed] [Google Scholar]
- Sternberg N. Evidence that adenine methylation influences DNA-protein interactions in Escherichia coli. J Bacteriol. 1985 Oct;164(1):490–493. doi: 10.1128/jb.164.1.490-493.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ormondt H., Gorter J., Havelaar K. J., de Waard A. Specificity of deoxyribonucleic acid transmethylase induced by bacteriophage T2. I. Nucleotide sequences isolated from tmicrococcus luteus DNA methylated in vitro. Nucleic Acids Res. 1975 Aug;2(8):1391–1400. doi: 10.1093/nar/2.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]




