Abstract
The regulation of gene expression is central to developmental programs and largely depends on the binding of sequence-specific transcription factors with cis-regulatory elements in the genome. Hox transcription factors specify the spatial coordinates of the body axis in all animals with bilateral symmetry, but a detailed knowledge of their molecular function in instructing cell fates is lacking. Here, we used chromatin immunoprecipitation with massively parallel sequencing (ChIP-seq) to identify Hoxa2 genomic locations in a time and space when it is actively instructing embryonic development in mouse. Our data reveals that Hoxa2 has large genome coverage and potentially regulates thousands of genes. Sequence analysis of Hoxa2-bound regions identifies high occurrence of two main classes of motifs, corresponding to Hox and Pbx–Hox recognition sequences. Examination of the binding targets of Hoxa2 faithfully captures the processes regulated by Hoxa2 during embryonic development; in addition, it uncovers a large cluster of potential targets involved in the Wnt-signaling pathway. In vivo examination of canonical Wnt–β-catenin signaling reveals activity specifically in Hoxa2 domain of expression, and this is undetectable in Hoxa2 mutant embryos. The comprehensive mapping of Hoxa2-binding sites provides a framework to study Hox regulatory networks in vertebrate developmental processes.
INTRODUCTION
Differential gene transcription instructs the development of multicellular organisms. A central mechanism to control gene expression is the binding of sequence-specific transcription factors to the genome; DNA–protein interaction is mediated by short nucleotide sequences, known as cis-acting regulatory elements.
Hox transcription factors are sequence-specific DNA-binding proteins, encoded by 39 genes in mouse and human. The organization of Hox genes in clusters (four clusters in mammals) generates accurate spatio-temporal patterns of proteins expression across the developing embryo (1). Throughout the animal kingdom, Hox transcription factors specify the spatial coordinates of the body axis, to instruct whether a segment of the embryo will become head, thorax or abdomen (2). Mutations in single Hox genes can cause spectacular body transformations in Drosophila; and these effects can also occur in higher organisms, such as mice, but requires inactivation of multiple Hox genes (3,4).
Despite the recognized role of Hox proteins in embryonic development, and increasing evidence supporting their role in adult homeostasis and disease (5,6), a detailed knowledge of Hox molecular function in instructing cell fates is lacking. Few Hox target genes have been discovered in vertebrates, and the vast majority of Hox-binding sites in vivo are unknown (7). Hoxa2, a member of the Hox paralog group 2, controls the fate of the cranial neural crest that migrates from rhombomere 4 to the second branchial arch (IIBA) in the developing mouse embryo. Disrupting Hoxa2 function causes loss of IIBA typical elements, which are instead replaced by a duplicated set of first branchial arch elements (8–10). The knowledge of the entire genome sequence together with next generation sequencing provide an unprecedented opportunity to understand how binding of Hox transcription factors direct embryonic development. Here, we have used Hoxa2 chromatin immunoprecipitation coupled to massively parallel sequencing (ChIP-seq) to identify genome-wide binding locations of Hoxa2 at a defined step of IIBA development (Embryonic Day 11.5) in mouse. Our data reveals that Hoxa2 has large genome coverage and potentially regulates thousands of genes. Hoxa2 binding is detected on a considerable fraction of the genes dysregulated in Hoxa2 mutant embryos at the same developmental stage (E11.5). We identify a significant amount of genes involved in the Wnt-signaling pathway associated with Hoxa2-bound regions. We further show tissue- and stage-specific activation of the Wnt–β-catenin pathway in the IIBA, which is undetectable in Hoxa2 mutant embryos. Our data describe where Hoxa2 localizes in the genome while functioning to instruct embryonic development. As such, this study presents one of the first global maps of the in vivo interactions between Hox and chromatin during vertebrate embryogenesis.
MATERIALS AND METHODS
Mouse husbandry
Hoxa2 mutant mice were described previously (8). BAT-Gal transgenic mice express nuclear β-galactosidase under control of multimerized LEF/TCF-binding sites (11). CD1 mice were time-mated to obtain branchial arches. Whole mount in situ hybridization and LacZ staining were carried out as described (12,13), using Fzd4, Wnt5a (gifts from Lorenz Neidhardt and Rolf Kemler, respectively), and Hoxa2 probe (14). Animal experiments were carried out under ASPA 1986.
ChIP-seq assays
ChIP assays were carried out as described (15), with the following modifications for ChIP-seq. The crosslinked material was sonicated to 300-bp fragments (Vibracell sonicator, Sonics: eight times for 10 s at 50% output). Immunoprecipitation was performed starting from 12 pairs of second branchial arches from E11.5 embryos and incubating overnight at 4°C with 3 µg of anti-Hoxa2 antibody (15) or control IgG, followed by Dynabeads protein A (Invitrogen). A pool of immunoprecipitated DNA from eight individual immunoprecipitation (0.5 ng) was sent for sequencing using the SOLiD 4 platform according to the manufacturer's protocols (Life Technologies). Libraries were prepared according to the SOLiD ChIP-Seq Kit Guide (MAN0002594) with modifications outlined in the SOLiD ChIP-Seq Library Preparation with Barcodes (MAN0003616). Prior to library preparation the samples were sheared using a Covaris S2 (Covaris, Inc.) with the protocol recommended in the Library Preparation Guide (4445673 Rev. B 04/2010). The volumes for the end repair step were then doubled to accommodate the increase in volume of the samples.
Bioinformatics analysis
For ChIP-seq analysis, 50-bp sequences from two biological replicates of the Hoxa2 ChIP and matched input DNA controls (exp1 and exp2) were used. For exp1 only, in response to the observation of poor end of read quality scores, the ChIP and input sequences were truncated to the first 32 bp, using SOLiD_preprocess_filter_v1.pl (16) (http://hts.rutgers.edu/filter/index.html). Sequence reads were mapped to the NCBI37 (mm9/July 2007) release of the mouse (Mus musculus) genome (including the mitochondrial genome of mouse) using Corona-Lite version 4.2.2 (Life Technologies). Uniquely mapped reads with a maximum of five mismatches where converted into BED format for downstream analysis.
Identification of binding regions
The uniquely mapped reads were analysed using the MACS version 1.4 β software (17) (http://liulab.dfci.harvard.edu/MACS/) to identify binding regions (excluding reads mapping to the mitochondrial genome of mouse), using the matched input DNA reads as a control. The summit of each binding region is defined as the location with the highest read pileup. The bandwidth, mfold parameters were set to 250 and 10–30 respectively. The threshold P-value was set to P < 1 × 10−5. Among the 18 226 regions from ‘exp2’, 8245 with false discovery rate (FDR) <10 were selected for downstream analyses.
To associate Hoxa2-binding regions with potential target genes, RefSeq transcript coordinates (Release 41) were downloaded from the UCSC table browser and associated, via GALAXY (18) (http://galaxy.psu.edu/), with these regions. An association was made with one or more genes if the summit of the binding regions overlapped RefSeq transcripts or their promoter regions (defined as −10 to +2.5 kb of the TSS). If an overlap was not observed then the closest gene (5′ or 3′ of the gene) was selected.
The location of Hoxa2-binding regions, defined by their summit region coordinates (200 bp centred upon the MACS defined summit) relative to gene structure was determined using CEAS version 0.9.9.8 (19) (http://liulab.dfci.harvard.edu/CEAS/). The analysis was also performed using a dataset containing the same number of random genome coordinates (also 200 bp) selected from mappable regions of the mouse genome. The ‘CRG Align 50’ data (containing all 50-mer with no more than five mismatches relative to the mouse genome) was downloaded from the test version of the UCSC Genome Browser (http://genome-test.cse.ucsc.edu). The ChIP-seq data has been submitted to ArrayExpress under accession number E-MTAB-711.
Motif analysis
For motif discovery, over-represented sequence motifs were identified in 200-bp regions centered upon the summit of each binding region using Weeder version 1.4.2 (20) (http://159.149.109.9/modtools/). Matrices from the ‘Best Occurrences’ output were compared against all TRANSFAC v11.3 matrices using the webtool STAMP (21) (http://benoslab.pitt.edu/stamp/). Weblogos of the discovered matrices were created using STAMP. Weeder was run using the following parameters: S = scan both strands of the supplied sequences; M = assume that there may be more than one discovered motif per sequence; ‘medium’ = search for motifs of length 6 (one degenerate position), 8 (two degenerate positions) and 10 (three degenerate positions).
A PERL script was used to scan for motifs corresponding to the Hox and Pbx consensus binding sequences within 200 bp of Hoxa2-binding region summits and in background sequences. The script identified matches to a supplied IUPAC consensus sequence in the forward and reverse strands of unmasked sequences. Two background datasets were created by a PYTHON script (generate_background_sequences.py) part of the GimmeMotifs software package (22) (http://www.ncmls.eu/bioinfo/gimmemotifs/). For each summit region sequence a random sequence of the same length was created using a first order Markov Chain (modeling dinucleotide frequencies). A second set of sequences were created containing sequence from a matched genomic regions (relative to TSS).
Gene annotation analysis
The analysis of gene annotation enrichment was performed using GREAT (23) (http://great.stanford.edu/) using the ‘basal plus extension’ association rules and the whole mouse genome as background.
Statistical analysis
Fisher's Exact chi-square test for 2 × 2 contingency tables (using two-tailed P-values; http://www.quantitativeskills.com/sisa/) was applied in a number of instances to determine whether the occurrence of consensus sequences were significantly different from background sequences.
ChIP–qPCR
ChIP–qPCR was performed on a Bio-Rad Chromo4 Real-time PCR system using SYBR green (Dynamo) according to the manufacturer's instructions (50 µl PCR reaction for each primer set was performed using the following program: Step 1: 95°C 15 min; Step 2: 95°C 30 s, 55°C 1 min, 72°C 30 s, repeat 49 times; Step 3: melting curve from 55°C to 95°C). Results were analyzed using Bio-Rad Opticon Monitor 3.1.32. Fold enrichment of each bound region was calculated over a negative antibody control relative to input. In Figure 5A, values are expressed as percent input for each bound region and the corresponding negative antibody control. Primer sequences are listed in Supplementary Table S7.
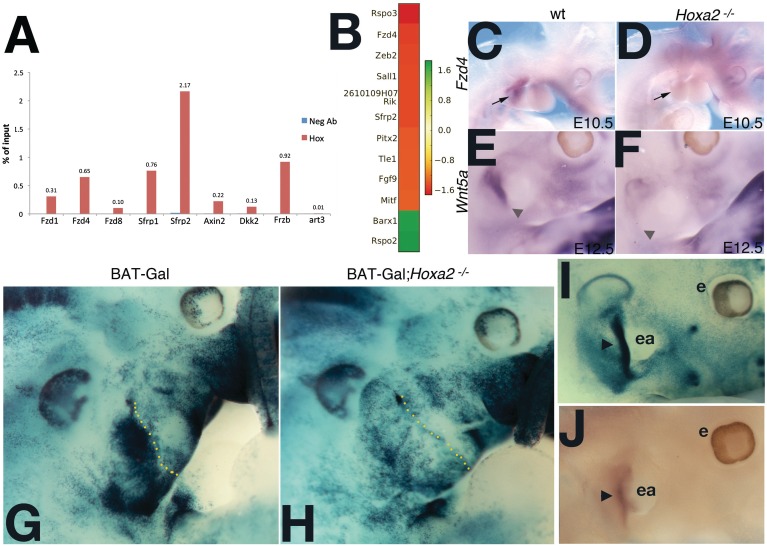
Figure 5.
Hoxa2 is upstream of Wnt–β-catenin signaling. (A) Validation of binding regions linked to genes in the Wnt-signaling pathway by ChIP–qPCR. Percent Input is shown for each Hoxa2-bound region and the corresponding negative antibody control (Neg Ab). Values represent the average of duplicate samples. Art3 is a negative control gene (unbound region). (B) The differentially expressed genes (from the list in Supplementary Table S6) linked to Wnt GO categories are shown together with their corresponding fold changes. (C–F) Expression of Fzd4 and Wnt5a in wild-type (C, E) and mutant (D, F) embryos. In situ hybridization on whole mount E10.5 wild-type and Hoxa2 mutant (C, D) and E12.5 wild-type and Hoxa2 mutant (E, F) using Fzd4 (C, D) and Wnt5a (E, F) probes. Arrow in (C) and (D), and arrowhead in (E) and (F), indicate the embryonic area where Fzd4 and Wnt5a are downregulated in the mutant, respectively. (G and H). Whole mount lacZ staining of E11.5 BAT-Gal (G) and BAT-Gal; Hoxa2−/− (H) embryos. (G) High Wnt canonical activity is detected in the IIBA of E11.0 BAT-Gal transgenic embryos. (H) Wnt canonical activity is lost in the IIBA (arrow) in the absence of Hoxa2. Dotted line separates IIBA from IBA. (I and J) BAT-Gal (I) and wild-type (J) E13.0 embryos. (I) Wnt canonical activity is mostly confined to the pinna of the outer ear (arrowhead). (J) In situ hybridization using Hoxa2 probe shows the pinna remains Hoxa2 positive at later stages (arrowhead). Ea, ear; e, eye.
Expression microarray
Second branchial arches of E11.0 and E11.5 embryos from Hoxa2+/− intercrosses were dissected out and snap-frozen in dry ice. After genotyping the embryos, pools were made with the wild-type and Hoxa2−/− branchial arches and total RNA was extracted using Trizol. RNA quality was checked using the RNA 6000 Nano Assay, and analyzed on an Agilent 2100 Bioanalyser (Agilent Technologies). RNA was quantified using a Nanodrop ultra-low-volume spectrophotometer (Nanodrop Technologies). Labeled targets were generated from total RNA (100 ng) using the 3′ IVT Express Kit and hybridized to GeneChip Mouse Genome 430 2.0 arrays. The arrays were then washed and stained using Fluidics Protocol FS450_0001 and images acquired using a GCS 3000HR scanner. The intensity files (.cel) generated by GeneChip Command Console (AGCC) software were then used for differential expression analyses. Technical quality control was performed with dChip (V2005) (www.dchip.org) (24) using the default settings. Background correction, quantile normalization and gene-expression analysis were performed using RMA in Bioconductor (25). Principal component analysis (PCA) was performed with Partek Genomics Solution (version 6.5, Copyright 2010, Partek Inc., St. Charles, MO, USA). Differential expression analysis was performed using Limma using the functions lmFit and eBayes (26). Gene lists of differentially expressed genes were controlled for FDR errors using the method of QVALUE (27). Microarray expression data has been submitted to ArrayExpress under accession number E-MEXP-3261.
RESULTS
Genome-wide mapping of Hoxa2-bound regions in the developing second branchial arch
The second branchial arch (IIBA) is the embryonic area most affected by inactivation of Hoxa2 in mouse. In the absence of Hoxa2, the IIBA generates skeletal elements typical of the more anterior first branchial arch (IBA). This abnormal skeletal development disrupts formation of the middle ear. Hoxa2 expression appears in the IIBA at E8.5 (28), following migration of Hoxa2-positive cranial neural crest cells from the hindbrain, and it is maintained throughout development of the IIBA. Hoxa2-positive cells are still detected in IIBA derivatives at later stages (E13.0). To define the genomic targets of Hoxa2, we performed Hoxa2 ChIP-seq on IIBAs collected at E11.5. Wild-type and Hoxa2 mutant embryos are visually indistinguishable at this stage; shortly after E11.5, the wild-type IIBA undergoes profound morphological changes to produce its final derivatives. Most importantly, Hoxa2 is strongly expressed in E11.5 IIBAs and it is still required for IIBA development at this stage (29) (Figure 1A).
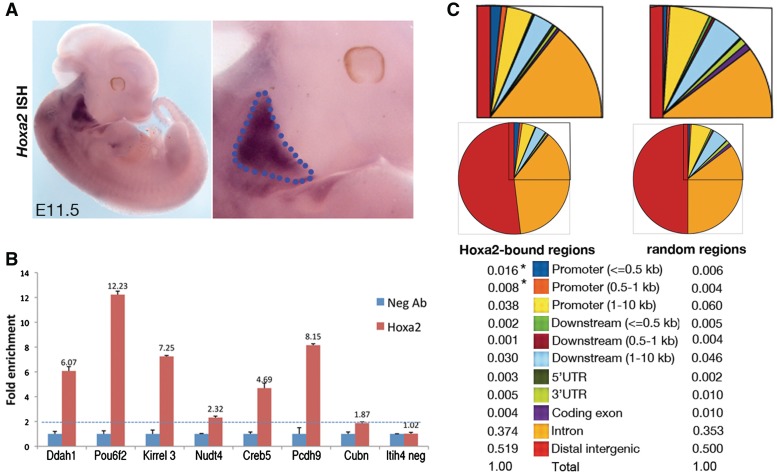
Figure 1.
Detection of Hoxa2-ChIP-seq peaks in the IIBA. (A) Whole-mount ISH on E11.5 wild-type embryos, using Hoxa2 probe (left). Magnification of the same picture shows the branchial area (right); the dotted line delimitates the IIBA. (B) ChIP–qPCR validation of FDR < 10 Hoxa2-bound regions. Fold enrichment of Hoxa2 over IgG negative control antibody (Neg Ab) is shown for each Hoxa2-bound region. Values represent the average of duplicate samples and are adjusted to the average enrichment of Pou6f2 from two independent experiments. Itih4 is a negative control (unbound region). For each sample the standard error of the mean is indicated by error bars. The dotted line represents a threshold of 2-fold or higher significant enrichment. (C) Distribution of Hoxa2 summit regions relative to Reference Sequence (RefSeq) gene structures. The pie charts indicate the proportion of reads for each gene structure; the corresponding numeric values are included. For each pie chart, an enlargement of the boxed area is shown above. CEAS detects significant enrichment of proximal promoter in Hoxa2-bound regions compared to random regions (asterisks).
DNA recovered from two independent Hoxa2 ChIP-seq experiments was used to construct fragment libraries and sequenced with the SOLID next generation sequencing platform (Life Technologies). In the second of the two experiments (exp2), we generated 12 242 621 and 16 973 356 uniquely mapping reads for Hoxa2 ChIP-seq and input library respectively. MACS peak calling software (17) identified 18 226 regions that are bound by Hoxa2 in vivo (P < 1e–5), of which 8245 with FDR < 10. The first experiment (exp1) resulted in fewer binding regions (less than one thousand) that almost entirely overlapped with exp2, with 89.4% binding regions of exp1 matching the entire set of binding regions detected in exp2 and 85% matching the subset of exp2-binding regions with FDR < 10.
To establish the consistency of Hoxa2-bound regions, we randomly selected 15 regions from the list of Hoxa2-bound regions with FDR < 10 in exp2 and tested these by ChIP–qPCR. We detected enrichment in the majority of the regions tested (12/15) (Figure 1B, Supplementary Figures S1 and S2). We therefore decided to concentrate on the list of 8245 binding regions with FDR < 10 detected in exp2, henceforth referred to as Hoxa2-bound regions (Supplementary Table S3).
By submitting Hoxa2-bound regions to the Cis-regulatory Elements Analysis System (CEAS) software (19), we found that a small fraction of Hoxa2 binding (2.4%) occur in the vicinity (within 1 kb) of transcriptional start sites (TSS); the occurrence of proximal promoters was 2-fold enriched (P = 2.46e–12) in Hoxa2-bound regions with respect to background sequences. The majority of Hoxa2-binding events (51.3%) occur at distances >10 kb from any known TSS, and a large fraction (37.4%) are located in introns (Figure 1C); these distributions indicate that Hoxa2 binding is widely distributed across the genome and slightly enriched at proximal promoters.
Hoxa2 ChIP-seq reveals abundance of Hox and Pbx–Hox-binding sites
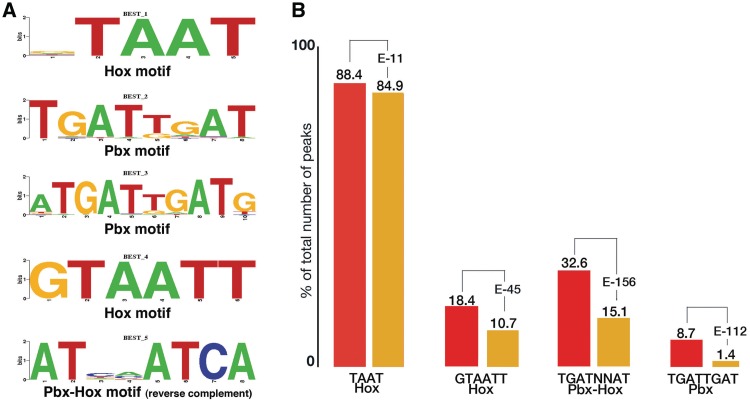
Hox proteins bind short AT-rich DNA sequences, commonly containing a TAAT core (7,30,31). It is difficult to understand how Hox proteins find their functional binding sites in the genome using four- to six-letter recognition sequences. Alternatively Hox short binding sites may simply reflect the lack of a better-defined recognition sequence, owing to the relatively small number of in vivo targets identified to date, and to the use of the homeodomain (rather than the entire protein) in most of the DNA-binding studies (7,32,33). Hoxa2 binds TAAT in vitro and in vivo (34,35). Analysis of Hox functional targets in vivo has shown that Hox can bind DNA cooperatively with TALE homeodomain proteins, Pbx and Meis (Exd and Hht in Drosphila) (7,36). To determine Hoxa2-binding preferences, we analyzed the sequence of Hoxa2-bound regions to search for over-represented motifs. Using the de novo motif discovery software (Weeder) (20) on 200-bp sequences centred upon each binding region summit, we found that the five top most highly ranked motifs could be categorized into three main classes (Figure 2A). The sequence TAAT was the most overrepresented motif; a longer version, flanked by G and T at 5′ and 3′ respectively, was also included in the five top motifs identified by Weeder. The second most overrepresented motif was TGATTGAT, which corresponds to Pbx-binding site (37). A longer version of the Pbx site also occurred at high frequency. Finally, included in the five top ranked motifs, we found the sequence TGATNNAT (where ‘N’ represents any nucleotide; sequence is inverted in Figure 2A), previously reported as a functional binding site for Pbx and Hox (7). We analyzed the distribution of these sequences in Hoxa2-bound regions. When compared to random sequences, Hoxa2-summit regions were found to be significantly enriched in all the top five motifs identified by Weeder (Figure 2B). The motif TAAT was present in the majority of Hoxa2-summit regions (88.4%) and a fraction of the Hoxa2-summit regions (18.4%) contained the extended version, GTAATT. We detected combined Pbx–Hox-binding sites in approximately one-third (32.6%) of Hoxa2-bound regions. Finally, a fraction of the Hoxa2-summit regions (7.9%) did not have any recognizable Hox or Pbx–Hox-binding site. The results above show that Hoxa2-bound regions mainly contain Hox and Pbx–Hox-binding sites. One third of Hoxa2-bound regions can potentially host binding of Hoxa2 together with its cofactor Pbx, but the largest fraction (60%) contains single Hox-binding motifs. The identification of TAAT as the most overrepresented motif by de novo discovery, and the significant enrichment observed in Hoxa2-bound regions, confirms that albeit short, this DNA sequence may indeed represent a functional recognition site in vivo.
Figure 2.
Sequence analysis of Hoxa2-bound regions. (A) Sequence logo of the top five motifs identified using de novo motif discovery. (B) Analysis of the distribution of motifs in Hoxa2-summit regions compared to background sequences. Red and yellow columns represent the occurrence of each motif (indicated below each column pair) in Hoxa2-summit regions (red) and in background sequences (yellow). The numbers on top of each column refer to the percentage of the peaks containing the motif. For all motifs shown, their occurrence in Hoxa2-bound regions was significantly higher than expected (only the exponent of the P-value is shown).
To identify additional overrepresented motifs, we masked Hox (TAAT) and Pbx (TGAT) motifs in Hoxa2-summit regions and performed again de novo motif discovery. We found additional AT rich motifs; among those only the sequence ATAAA was significantly overrepresented in Hoxa2-summit regions when compared to random sequences (55.5 and 48% in Hoxa2-bound regions and background sequences, respectively; P = 1.50e–20).
A well-established way in which Hox proteins achieve specificity in vivo is to bind DNA cooperatively with other DNA-binding factors. To identify factors interacting with Hoxa2, we performed de novo motif discovery within the 100-nt flanking each TAAT (50-nt upstream and downstream). We identified the sequence AATTA as the most prominent motif, (21.8% occurrence; P = 0.028). We found a distance of 6–10 nt between the two motifs (as defined by their motif midpoints) in 19.5% of the TAAT/ATTA pairs, suggesting the occurrence of palindromic binding sites in Hoxa2-summit regions. By scanning 100-nt sequences flanking each TGATNNAT motif, we found over-representation of the sequence TGACAG (occurrence in 8.4% of Hoxa2-summit regions containing Pbx–Hox-binding sites; P = 1.8e–25), which corresponds to the recognition sequence of the Hox cofactors Meis (Supplementary Figure S4). This finding suggests that Meis factors preferentially interact with Hoxa2 in the presence of Pbx. The over-representation of recognition sites for well-known Hox cofactors (Pbx and Meis) confirms the quality of Hoxa2 ChIP-seq regions. This result does not preclude that other factors function in important combinations with Hoxa2, but it suggests that no single transcription factor motif is commonly used in the entire Hoxa2-occupied set. A large fraction of Hoxa2-bound regions contains single Hox-binding sites; finding specific combinations will require focus on subsets of these regions selected by expression pattern of nearby genes.
Functional annotation links Hoxa2-peaks to the development of the branchial arches
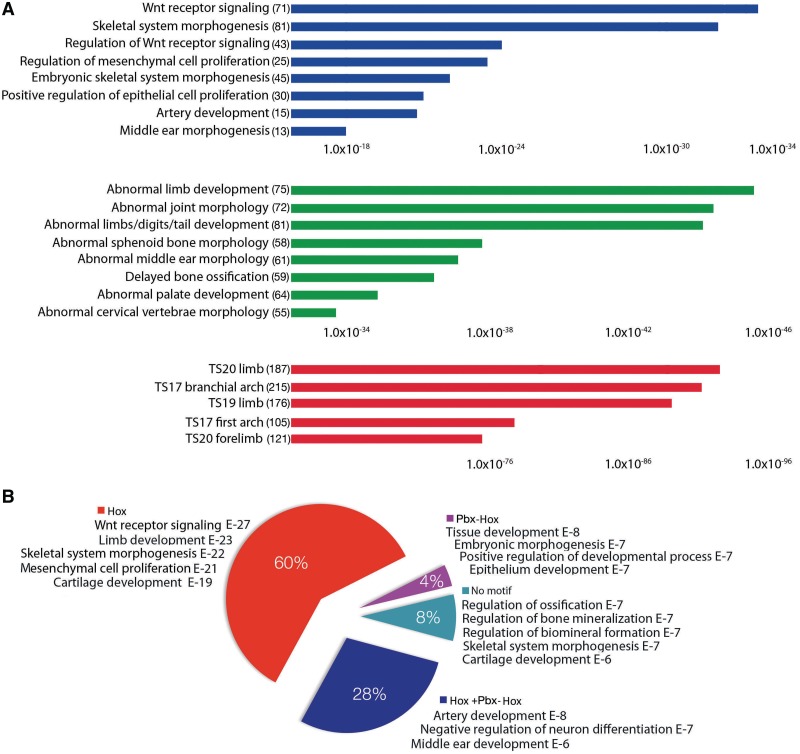
We used the web tool Genomic Regions Enrichment of Annotations Tool (GREAT) (23) to identify terms enriched in genes associated with Hoxa2-bound regions. Consistent with a functional significance of Hoxa2-binding events, GREAT analysis specifically identified enrichment of Hoxa2-bound regions near genes involved in biological processes regulated by Hoxa2 in embryonic development (Figure 3A). Hoxa2 loss-of-function affects formation of the middle ear, and ectopic expression disrupts development of the cranial skeleton (8–10,12,38–40). Middle ear morphogenesis and embryonic skeletal morphogenesis were identified in the top enriched biological processes. GREAT detected over-representation of genes expressed in the branchial arch in the gene-expression ontology, and genes whose mutations generate phenotypes such as middle ear abnormalities, delayed bone ossification and palatal development, another process controlled by Hoxa2 (41).
Figure 3.
Functional annotation of Hoxa2-bound regions. (A) Top over-represented categories in the Gene Ontology (GO) (blue), Mouse phenotypes (green) and Mouse expression (red). GO describes the biological processes associated with gene function; mouse phenotypes ontology contains data about mouse genotype–phenotype associations; Mouse Genome Informatics (MGI) expression contains information on tissue- and developmental-stage-specific expression in mouse. The number of genes contained in each category is indicated in brackets. The length of the bars corresponds to values on the x-axis, which are binomial raw (uncorrected) P- values. TS: Theiler Stage. (B) Top overrepresented categories in the Gene Ontology (GO) in Hoxa2-bound regions containing Hox (red), Hox-Pbx (purple), Hox and Hox-Pbx (blue), and no motif (turquoise). The proportion of peaks containing the different motifs with respect to the total of Hoxa2-bound regions is shown in the pie chart (together with percentage values for each fraction); for each class, the top five categories with P < 10−6 are shown.
Most interestingly, GREAT detected a strong enrichment of Hoxa2-regions near genes involved in the ‘Wnt-signaling pathway’, ranked as the top category. A closely related category, ‘regulation of Wnt-signaling pathway’, was also included in the top enriched biological processes. This novel association uncovers a potential role for Wnt signaling to instruct IIBA development downstream of Hoxa2.
Next, we classified regions according to the presence of Hox, Pbx–Hox, both Hox and Pbx–Hox, or no motif in their summit, and performed GREAT analysis on each class (Figure 3B). Consistent with each binding site (or their combination) having different functional outputs, we found that peaks containing Hox, Pbx–Hox or both binding sites were associated with genes involved in different biological processes. Binding regions containing a Hox-binding site had a similar profile to the one in Figure 3A, possibly because they constitute the largest class. We found the second largest class of peaks (28.7%; containing both Hox and Pbx–Hox-binding sites) to be enriched in genes involved in fewer specific processes, artery and middle ear development and neuron differentiation. Most notably ‘Wnt-signaling pathway’, the most overrepresented category in the previous class (and in the totality of Hoxa2 ChIP-seq peaks) was not included in the top overrepresented biological processes associated with peaks containing Hox and Pbx–Hox-binding sites. Regions containing only Pbx–Hox-binding sites were linked to genes involved in embryogenesis. Strikingly, peaks containing no Hox or Pbx–Hox-binding sites were exclusively and specifically associated with genes involved in bone formation. A simple explanation to this observation is that Hoxa2 binds these genomic regions via an additional transcription factor, possibly involved in regulation of bone. However, performing de novo motif discovery on this class of Hoxa2-peaks did not identify any known overrepresented motif. Finally, GREAT did not detect enrichment of gene ontology terms in the class of Hoxa2-peaks containing Pbx-binding site (i.e. the subset of the Pbx–Hox peaks containing Pbx recognition sequence TGATTGAT).
Identification of functional locus occupancy by Hoxa2 in E11.5 developing embryos
Next, we assigned Hoxa2-bound regions to genes. Using the nearest gene approach, we assigned 8245 Hoxa2-bound regions to a total of 3579 genes. A survey of the genes previously identified as regulated by Hoxa2 during embryonic development showed that 8 of 15 genes had at least one Hoxa2-bound region associated with them (Supplementary Table S5).
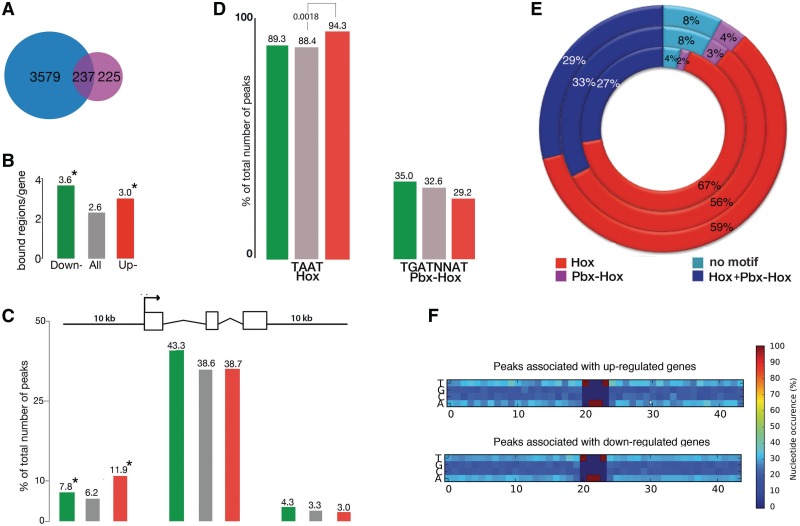
To assess the effect of Hoxa2 DNA binding on gene expression, we used microarrays in the same embryonic populations as those interrogated by ChIP-seq. By comparing E11.5 wild-type and Hoxa2-null mutant branchial arches, we identified 489 differentially expressed genes (fold difference > 1.3; P < 0.05) of which 359 and 130 were down and upregulated in mutant embryos, respectively. If Hoxa2-bound regions are functionally active, we should expect a marked enrichment of ChIP-seq genes in the genes dysregulated in Hoxa2 mutant branchial arches. We found that 48% (237/489) of Hoxa2-regulated genes had at least one Hoxa2-bound region assigned to them, which represents a highly significant enrichment compared with all genes (P = 1.1e–106) (Figure 4A). Upon separating Hoxa2-regulated genes into down and upregulated, we found that 50% and 42% of the genes down and upregulated in the Hoxa2 mutant, respectively, had at least one Hoxa2-bound region assigned to them (P = 8.9e–87 and 4.2e–23, respectively) (Supplementary Table S6). These results suggest that nearly half of the genes that are dysregulated in the Hoxa2 mutant are directly regulated (either positively or negatively) by Hoxa2.
Figure 4.
Intersection between ChIP-seq data and gene-expression analysis. (A) Venn diagram showing the overlap between the genes associated with Hoxa2-bound regions (blue) and those revealed by expression arrays. (B) Average number of Hoxa2-bound regions per gene in the entire ChIP-seq dataset (grey), Hoxa2-bound regions associated with down- (green) and up- (red) regulated genes. On average down- and upregulated genes are associated with a higher number of Hoxa2-bound regions (P = 2.2e–16 and 0.001, respectively). (C) Distribution of Hoxa2 summit regions relative to Reference Sequence (RefSeq) gene structures: promoter (10-kb upstream of the TSS), transcript, and downstream (10-kb downstream of the TTS). Hoxa2-summit regions associated with down and upregulated genes occur more frequently in promoters compared to the entire ChIP-seq dataset (P = 0.05 and 0.005, respectively); the color code is as in (B). (D) Analysis of the distribution of Hox and Pbx–Hox motifs in Hoxa2-summit regions associated with down- and upregulated genes [color code as in (B)]. TAAT is significantly enriched in Hoxa2-bound regions associated with upregulated genes. The numbers on top of each column refer to the percentage of Hoxa2-summit regions containing the motif. (E) Distribution of Hoxa2-bound regions containing Hox (red), Hox-Pbx (purple), Hox and Hox-Pbx (blue), and no motif (turquoise) in regulated genes. Numbers indicate the contribution of each class to the total of Hoxa2-bound regions associated to upregulated genes (inner ring), downregulated genes (middle ring) and the entire Hoxa2 data set (outer ring). (F) Extension of the TAAT motif in down- and upregulated genes. Plots of the density of each nucleotide around each TAAT motif contained in Hoxa2-summit regions associated with up and downregulated genes.
The list of bound and regulated genes included known functional targets of Hoxa2 (Robo2 and Six2) (15,34,42), genes whose expression is affected in the Hoxa2 mutant (Runx2 and Msx1) (12,29) and Hox genes themselves, consistent with Hox cross-regulation (43–47). Most interestingly, previously unknown targets included Meis1 and Meis2, whose protein products function as Hox cofactors (7,36), suggesting a positive regulatory loop between Hox and Meis proteins.
Further characterization of these putative Hoxa2 direct targets revealed 654 and 168 Hoxa2-bound regions associated with down and upregulated genes, respectively. The average number of Hoxa2-bound regions per gene was 3.6 and 3 for down and upregulated genes, respectively, which is significantly higher than the average of the ChIP-seq genes (Figure 4B). By analyzing the genomic distribution of Hoxa2-bound regions, we found a higher instance of promoters in Hoxa2-bound regions associated with regulated genes, compared to the totality of Hoxa2-bound regions. In contrast, Hoxa2-bound regions associated with down and upregulated genes were not preferentially localized to other genomic features (downstream, introns, intergenic) (Figure 4C).
We then asked if the occurrence of TAAT and TGATNNAT, identified as top enriched motifs in Hoxa2-peaks, was similar in Hoxa2-bound regions associated with down and upregulated genes. We detected a higher occurrence of TAAT in Hoxa2-bound regions associated with upregulated genes (P = 0.0018) compared to the totality of Hoxa2-bound regions (Figure 4D). To assess the effects of each binding site (or their combination) on gene expression, we classified Hoxa2-bound regions associated to upregulated and downregulated genes according to the presence of Hox, Pbx–Hox, both Hox and Pbx–Hox, or no motif. We found that ‘Hox’ regions are significantly over-represented in upregulated genes (P = 0.016) compared to downregulated genes (Figure 4E), indicating that TAAT preferentially occurs alone in sequences associated to repressed genes. The distributions of the remaining classes of Hoxa2-bound regions did not significantly differ in upregulated and downregulated genes; however, it is interesting to notice the lower frequency of ‘no motif’ sequences in upregulated genes compared to downregulated genes (P = 0.077).
In summary, Hoxa2 binds more frequently in the surrounding of regulated genes. Repressed genes display a significantly higher occurrence of Hox-binding sites alone in their binding regions, than activated genes. This result may reflect different effects of Hoxa2 on the transcriptional output when alone or in a complex with Pbx. Finally, we used Hoxa2-bound regions associated with down and upregulated genes to refine the Hoxa2 recognition site identified by de novo motif discovery (Figure 2A). Our search did not identify any obvious sequence restriction beyond the TAAT core motif, only a mild preference for T to follow, and the tendency to be embedded in stretches of AT-rich sequences (Figure 4F).
Hoxa2 controls Wnt–β-catenin-signaling pathway in the IIBA
The finding that Hoxa2-bound regions are associated to genes in the ‘Wnt-signaling pathway’ and ‘regulation of Wnt-signaling pathway’ categories, suggests that Hoxa2 may act upstream of the Wnt-signaling pathway in the IIBA. Verification of Hoxa2 binding on a subset of the regions linked to genes in the Wnt-signaling pathway by conventional ChIP–qPCR, confirmed that eight out of eight binding regions were bound by Hoxa2 in vivo (Figure 5A). Twelve of the 81 genes linked to ‘Wnt-signaling pathway’ and ‘regulation of Wnt-signaling pathway’ categories and associated with Hoxa2-bound regions, were differently regulated in microarrays comparison of E11.5 wild-type and mutant IIBA (Figure 5B). Among these 81 genes we found Wnt5a, whose inactivation in mouse affects outer ear development and Fzd4, previously identified as differentially expressed in a screen performed on E10.5 Hoxa2 mutant (our unpublished observation). ISH hybridization revealed expression of Fzd4 and Wnt5a in the IIBA of E10.5, and in the developing outer ear of E12.0 wild-type embryos, respectively (Figure 5C and E). Both expression domains were absent in the Hoxa2 mutant (Figure 5D and F). Next, we used BAT-GAL mice (11) to investigate the state of canonical Wnt–β-catenin signaling in the IIBA. At E11.5, the IIBA exhibits strong, localized β-galactosidase activity (Figure 5G). Wnt–β-catenin-signaling activity is greatly reduced in the IIBA of Hoxa2 mutant embryos, where the staining appears comparable to the one in the IBA (Figure 5H). Low β-galactosidase activity was detected in the IIBA of younger embryos (E10.5); this activity was comparable in wild-type and mutant embryos (not shown). At E13.0 β-galactosidase -positive cells labeled the pinna of the outer ear, which is Hoxa2-positive at the same stage (Figure 5I and J).
These findings indicate that Hoxa2 activates the canonical Wnt–β-catenin-signaling pathway in the IIBA. This Hoxa2-dependent Wnt–β-catenin activity is first observed in E11.5 IIBA. It is maintained at least until E13.0, when it is detected in the pinna, a IIBA-derivative that fails to form in the Hoxa2 mutant. In addition, the observed downregulation of Wnt5a in the Hoxa2 mutant suggests Hoxa2 may also regulate Wnt non-canonical, β-catenin independent pathways in the IIBA.
DISCUSSION
Hox proteins are key transcription factors in the antero-posterior (AP) patterning of the embryo, but the molecular basis of Hox function is proving difficult to understand. This analysis of Hoxa2 in vivo occupancy provides the basis to learn how Hox proteins binding to the genome informs embryonic development, and the structure of the body plan.
In view of their accurate snapshot of the biological processes downstream of Hoxa2, Hoxa2 ChIP-seq data provide real opportunities to disclose additional processes controlled by Hoxa2 (and possibly Hox proteins) in development and disease. We show here that Hoxa2 regulates the Wnt-signaling pathway, a finding uncovered solely on the basis of gene association to Hoxa2-bound regions, and we provide the molecular coordinates through which this control is executed. Hoxa2-peaks are frequently detected in the surroundings of genes associated to ‘Wnt-signaling pathway’ ontology definitions. Further investigation has revealed a stage- and tissue-specific activation of canonical Wnt–β-catenin signaling in Hoxa2 main domain of expression, which manifests exclusively in the presence of Hoxa2. Later in development, canonical Wnt–β-catenin signaling remains highly active in the outer ear outgrowth, which fails to form in the Hoxa2 mutant. Interestingly, the outer ear also develops abnormally following mesenchymal inactivation of β-catenin in the branchial arches (48). Collectively, these observations suggest that canonical Wnt–β-catenin signaling controls morphogenesis of the outer ear downstream of Hoxa2. The full understanding of Hoxa2 control of Wnt signaling requires characterizing the role and the contribution of Hoxa2-regulated events to Wnt–β-catenin activity in the IIBA. A first and essential step in this direction will be to correlate Wnt–β-catenin activity, which is regionally restricted in the IIBA, to the spatial expression of the individual Hoxa2 targets. Hoxa2 activation of Fzd4 is predicted to have a positive effect on Wnt–β-catenin signaling: Fzd4 serves as a receptor in Wnt–β-catenin and in non-canonical Wnt signaling (49–51), it is largely expressed in the IIBA, and loss of Fzd4 expression and of Wnt–β-catenin signaling is simultaneously observed in the Hoxa2 mutant. We also show here that Hoxa2 regulates Wnt5a, which is required for outer ear formation (52). Although Wnt5a preferentially signals via non-canonical pathways, it can both inhibit and activate Wnt–β-catenin signaling (and the latter requires the presence of Fzd4) (51,53).
Wnt is one of the few signaling pathways utilized to pattern organs and specify cell fate during embryonic development, and Hox proteins are key regulators of embryonic development, but surprisingly few instances of Hox regulation of Wnt have been described in vertebrate development (54,55). The ability to modify the activity of signaling pathways can change the local morphogenetic programs and generate segment-specific structures (56), and, in few cases, Hox proteins have been shown to modify the activity of the signaling pathways the cells are exposed to (57,58). From this perspective, Hoxa2 control of the Wnt-signaling pathway represents a perfectly suitable mechanism to generate second branchial arch-specific positional information.
Hox proteins bind AT-rich DNA sequences, centered on a TAAT core (7,30,31). The use of short and very similar recognition sequences does not explain the high specificity displayed by Hox proteins in vivo. We find that TAAT is the top enriched motif in Hoxa2-bound regions, and is significantly more enriched in Hoxa2-bound regions associated to repressed genes. GTAAT and TGANNAT, the additional motifs identified by sequence analysis, display a higher over representation in Hoxa2-bound regions (compared to background sequences) with respect to TAAT. The functional significance of GTAAT is unclear, as effort to extend Hoxa2 recognition sequence using ‘confident’ Hoxa2-bound regions (i.e. the ones linked to regulated genes) did not identify any strict sequence restriction beyond the TAAT core motif. Together, GTAATT and TGATNNAT could determine the specificity of about half Hoxa2-binding events (they are contained in 45% of Hoxa2-bound regions). The remaining half of Hoxa2 summit regions contains only TAAT, or no recognizable Hox or Hox-Pbx-binding site (8%), suggesting that additional unknown mechanisms are in place to determine specificity. Their discovery may require focus on subsets of regions selected by similar functions or by expression pattern of nearby genes.
Binding short, highly frequent motifs obviously does not help to discriminate functional targets, but provides large genome coverage, which may be important for additional functions. In a developmental context, Hox proteins endow cells with positional identity, to inform cells of the appropriate type of structure to be built in that specific body position. A possibility is that Hoxa2, in addition to regulating a specific set of target genes (possibly selected with the help of cofactors), acts as a ‘pioneer’ transcription factor and binds early chromatin to prepare for the subsequent steps of cell differentiation, i.e. recruitment of tissue-specific transcriptional regulators. Widespread genome coverage may also provide a built-in redundancy to Hoxa2 regulation, for the potential to bind in the vicinity of many genes, and to control downstream molecular mechanisms via multiple components. Indeed, a survey of the genes associated to Hoxa2-bound regions reveals many instances of homologous genes, a feature already evident within the restricted group of genes reported as Hoxa2 direct and functional targets (15,35,42). Hoxa2 ChIP-seq reveals binding in or around genes that are closely related to Six2 (Six1 and Six4), Robo2 (Robo1) and Meox1 (Meox2).
Mapping of binding sites across the genome reveals that Hoxa2 has large genome coverage, but we find that the majority of the regions bound by Hoxa2 at E11.5 are associated to genes with no evidence of dysregulation in Hoxa2 mutant IIBA. The binding profile of Hoxa2 reflects a mixture of binding sites from many different cell types that are sampled simultaneously from the entire IIBA. Simple biological possibilities are that many binding sites may have functional relevance at developmental stages different from the one assayed, in different tissues where Hoxa2 is active, or in unusual circumstances, such as the absence of other transcription factors. Furthermore binding sites that reflect fine-tuning regulation of gene expression are likely to be associated to expression changes below the cutoff applied to identify genes differentially expressed in the Hoxa2 mutant. An alternative possibility is that Hoxa2 binds promiscuously to many regions within portions of the genome that are physically accessible to it and much of this occupancy may not be associated with any regulatory activity. Relating the global Hoxa2 occupancy pattern to functional cis-regulatory modules (CRM) activity will be crucial to understand how many of these binding events are functionally relevant and to construct a reliable Hoxa2 regulatory network.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 3–7, Supplementary Figures 1–4.
FUNDING
Biotechnology and Biological Sciences Research Council (BB/E017355/1 to N.B., BB/H018123/2 to M.R., N.B and N.L.). The Bobola group is supported by the Manchester Academic Health Science Centre and the Manchester National Institute for Health Research Biomedical Research Centre. Funding for open access charge: School of Dentistry, University of Manchester.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Andy Sharrocks for help and critical reading of the manuscript, the members of the Genomic Technologies and Bioinformatics Core Facilities at the University of Manchester, Rolf Kemler for the BAT-GAL mice and the Wnt5a probe, Lorenz Neidhardt for the Fzd4 probe and Bettina Engist for technical assistance.
REFERENCES
- 1.Duboule D, Dolle P. The structural and functional organization of the murine HOX gene family resembles that of Drosophila homeotic genes. EMBO J. 1989;8:1497–1505. doi: 10.1002/j.1460-2075.1989.tb03534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carroll SB. Homeotic genes and the evolution of arthropods and chordates. Nature. 1995;376:479–485. doi: 10.1038/376479a0. [DOI] [PubMed] [Google Scholar]
- 3.Wellik DM. Hox patterning of the vertebrate axial skeleton. Dev. Dyn. 2007;236:2454–2463. doi: 10.1002/dvdy.21286. [DOI] [PubMed] [Google Scholar]
- 4.Wellik DM, Capecchi MR. Hox10 and Hox11 genes are required to globally pattern the mammalian skeleton. Science. 2003;301:363–367. doi: 10.1126/science.1085672. [DOI] [PubMed] [Google Scholar]
- 5.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat. Rev. Cancer. 2010;10:361–371. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- 6.Gouti M, Gavalas A. In: Hox Gene Expression. Papageorgiou S, editor. Boston, MA, USA: Landes Bioscience and Springer Science + Business Media; 2007. [Google Scholar]
- 7.Mann RS, Lelli KM, Joshi R. Hox specificity unique roles for cofactors and collaborators. Curr. Top. Dev. Biol. 2009;88:63–101. doi: 10.1016/S0070-2153(09)88003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gendron-Maguire M, Mallo M, Zhang M, Gridley T. Hoxa-2 mutant mice exhibit homeotic transformation of skeletal elements derived from cranial neural crest. Cell. 1993;75:1317–1331. doi: 10.1016/0092-8674(93)90619-2. [DOI] [PubMed] [Google Scholar]
- 9.Rijli FM, Mark M, Lakkaraju S, Dierich A, Dolle P, Chambon P. A homeotic transformation is generated in the rostral branchial region of the head by disruption of Hoxa-2, which acts as a selector gene. Cell. 1993;75:1333–1349. doi: 10.1016/0092-8674(93)90620-6. [DOI] [PubMed] [Google Scholar]
- 10.Barrow JR, Capecchi MR. Compensatory defects associated with mutations in Hoxa1 restore normal palatogenesis to Hoxa2 mutants. Development. 1999;126:5011–5026. doi: 10.1242/dev.126.22.5011. [DOI] [PubMed] [Google Scholar]
- 11.Maretto S, Cordenonsi M, Dupont S, Braghetta P, Broccoli V, Hassan AB, Volpin D, Bressan GM, Piccolo S. Mapping Wnt/beta-catenin signaling during mouse development and in colorectal tumors. Proc. Natl Acad. Sci. USA. 2003;100:3299–3304. doi: 10.1073/pnas.0434590100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanzler B, Kuschert SJ, Liu YH, Mallo M. Hoxa-2 restricts the chondrogenic domain and inhibits bone formation during development of the branchial area. Development. 1998;125:2587–2597. doi: 10.1242/dev.125.14.2587. [DOI] [PubMed] [Google Scholar]
- 13.Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo. 2nd edn. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 14.Mallo M. Retinoic acid disturbs mouse middle ear development in a stage-dependent fashion. Dev. Biol. 1997;184:175–186. doi: 10.1006/dbio.1997.8519. [DOI] [PubMed] [Google Scholar]
- 15.Kutejova E, Engist B, Self M, Oliver G, Kirilenko P, Bobola N. Six2 functions redundantly immediately downstream of Hoxa2. Development. 2008;135:1463–1470. doi: 10.1242/dev.017624. [DOI] [PubMed] [Google Scholar]
- 16.Sasson A, Michael TP. Filtering error from SOLiD Output. Bioinformatics. 2010;26:849–850. doi: 10.1093/bioinformatics/btq045. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goecks J, Nekrutenko A, Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shin H, Liu T, Manrai AK, Liu XS. CEAS: cis-regulatory element annotation system. Bioinformatics. 2009;25:2605–2606. doi: 10.1093/bioinformatics/btp479. [DOI] [PubMed] [Google Scholar]
- 20.Pavesi G, Mereghetti P, Mauri G, Pesole G. Weeder Web: discovery of transcription factor binding sites in a set of sequences from co-regulated genes. Nucleic Acids Res. 2004;32:W199–W203. doi: 10.1093/nar/gkh465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahony S, Benos PV. STAMP: a web tool for exploring DNA-binding motif similarities. Nucleic Acids Res. 2007;35:W253–W258. doi: 10.1093/nar/gkm272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Heeringen SJ, Veenstra GJ. GimmeMotifs: a de novo motif prediction pipeline for ChIP-sequencing experiments. Bioinformatics. 2011;27:270–271. doi: 10.1093/bioinformatics/btq636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl Acad. Sci. USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 26.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 27.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc. Natl Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nonchev S, Vesque C, Maconochie M, Seitanidou T, Ariza-McNaughton L, Frain M, Marshall H, Sham MH, Krumlauf R, Charnay P. Segmental expression of Hoxa-2 in the hindbrain is directly regulated by Krox-20. Development. 1996;122:543–554. doi: 10.1242/dev.122.2.543. [DOI] [PubMed] [Google Scholar]
- 29.Santagati F, Minoux M, Ren SY, Rijli FM. Temporal requirement of Hoxa2 in cranial neural crest skeletal morphogenesis. Development. 2005;132:4927–4936. doi: 10.1242/dev.02078. [DOI] [PubMed] [Google Scholar]
- 30.Noyes MB, Christensen RG, Wakabayashi A, Stormo GD, Brodsky MH, Wolfe SA. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell. 2008;133:1277–1289. doi: 10.1016/j.cell.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Pena-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Affolter M, Slattery M, Mann RS. A lexicon for homeodomain-DNA recognition. Cell. 2008;133:1133–1135. doi: 10.1016/j.cell.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 33.Svingen T, Tonissen KF. Hox transcription factors and their elusive mammalian gene targets. Heredity. 2006;97:88–96. doi: 10.1038/sj.hdy.6800847. [DOI] [PubMed] [Google Scholar]
- 34.Kutejova E, Engist B, Mallo M, Kanzler B, Bobola N. Hoxa2 downregulates Six2 in the neural crest-derived mesenchyme. Development. 2005;132:469–478. doi: 10.1242/dev.01536. [DOI] [PubMed] [Google Scholar]
- 35.Kirilenko P, He G, Mankoo BS, Mallo M, Jones R, Bobola N. Transient activation of meox1 is an early component of the gene regulatory network downstream of hoxa2. Mol. Cell. Biol. 2011;31:1301–1308. doi: 10.1128/MCB.00705-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moens CB, Selleri L. Hox cofactors in vertebrate development. Dev. Biol. 2006;291:193–206. doi: 10.1016/j.ydbio.2005.10.032. [DOI] [PubMed] [Google Scholar]
- 37.Bryne JC, Valen E, Tang MH, Marstrand T, Winther O, da Piedade I, Krogh A, Lenhard B, Sandelin A. JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 2008;36:D102–D106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tavella S, Bobola N. Expressing Hoxa2 across the entire endochondral skeleton alters the shape of the skeletal template in a spatially restricted fashion. Differentiation. 2009;79:194–202. doi: 10.1016/j.diff.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 39.Creuzet S, Couly G, Vincent C, Le Douarin NM. Negative effect of Hox gene expression on the development of the neural crest-derived facial skeleton. Development. 2002;129:4301–4313. doi: 10.1242/dev.129.18.4301. [DOI] [PubMed] [Google Scholar]
- 40.Massip L, Ectors F, Deprez P, Maleki M, Behets C, Lengele B, Delahaut P, Picard J, Rezsohazy R. Expression of Hoxa2 in cells entering chondrogenesis impairs overall cartilage development. Differentiation. 2007;75:256–267. doi: 10.1111/j.1432-0436.2006.00132.x. [DOI] [PubMed] [Google Scholar]
- 41.Smith TM, Wang X, Zhang W, Kulyk W, Nazarali AJ. Hoxa2 plays a direct role in murine palate development. Dev. Dyn. 2009;238:2364–2373. doi: 10.1002/dvdy.22040. [DOI] [PubMed] [Google Scholar]
- 42.Geisen MJ, Di Meglio T, Pasqualetti M, Ducret S, Brunet JF, Chedotal A, Rijli FM. Hox paralog group 2 genes control the migration of mouse pontine neurons through slit-robo signaling. PLoS Biol. 2008;6:e142. doi: 10.1371/journal.pbio.0060142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lampe X, Samad OA, Guiguen A, Matis C, Remacle S, Picard JJ, Rijli FM, Rezsohazy R. An ultraconserved Hox-Pbx responsive element resides in the coding sequence of Hoxa2 and is active in rhombomere 4. Nucleic Acids Res. 2008;36:3214–3225. doi: 10.1093/nar/gkn148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Popperl H, Bienz M, Studer M, Chan SK, Aparicio S, Brenner S, Mann RS, Krumlauf R. Segmental expression of Hoxb-1 is controlled by a highly conserved autoregulatory loop dependent upon EXD/Pbx. Cell. 1995;81:12. doi: 10.1016/s0092-8674(05)80008-x. [DOI] [PubMed] [Google Scholar]
- 45.Ferretti E, Marshall H, Popperl H, Maconochie M, Krumlauf R, Blasi F. Segmental expression of Hoxb2 in r4 requires two separate sites that integrate cooperative interactions between Prep1, Pbx and Hox proteins. Development. 2000;127:155–166. doi: 10.1242/dev.127.1.155. [DOI] [PubMed] [Google Scholar]
- 46.Jacobs Y, Schnabel CA, Cleary ML. Trimeric association of Hox and TALE homeodomain proteins mediates Hoxb2 hindbrain enhancer activity. Mol. Cell. Biol. 1999;19:5134–5142. doi: 10.1128/mcb.19.7.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lampe X, Picard JJ, Rezsohazy R. The Hoxa2 enhancer 2 contains a critical Hoxa2 responsive regulatory element. Biochem. Biophys. Res. Commun. 2004;316:898–902. doi: 10.1016/j.bbrc.2004.02.138. [DOI] [PubMed] [Google Scholar]
- 48.Huh SH, Ornitz DM. Beta-catenin deficiency causes DiGeorge syndrome-like phenotypes through regulation of Tbx1. Development. 2010;137:1137–1147. doi: 10.1242/dev.045534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu Q, Wang Y, Dabdoub A, Smallwood PM, Williams J, Woods C, Kelley MW, Jiang L, Tasman W, Zhang K, et al. Vascular development in the retina and inner ear: control by Norrin and Frizzled-4, a high-affinity ligand-receptor pair. Cell. 2004;116:883–895. doi: 10.1016/s0092-8674(04)00216-8. [DOI] [PubMed] [Google Scholar]
- 50.Ye X, Wang Y, Rattner A, Nathans J. Genetic mosaic analysis reveals a major role for frizzled 4 and frizzled 8 in controlling ureteric growth in the developing kidney. Development. 2011;138:1161–1172. doi: 10.1242/dev.057620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grumolato L, Liu G, Mong P, Mudbhary R, Biswas R, Arroyave R, Vijayakumar S, Economides AN, Aaronson SA. Canonical and noncanonical Wnts use a common mechanism to activate completely unrelated coreceptors. Genes Dev. 2010;24:2517–2530. doi: 10.1101/gad.1957710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamaguchi TP, Bradley A, McMahon AP, Jones S. A Wnt5a pathway underlies outgrowth of multiple structures in the vertebrate embryo. Development. 1999;126:1211–1223. doi: 10.1242/dev.126.6.1211. [DOI] [PubMed] [Google Scholar]
- 53.Mikels AJ, Nusse R. Purified Wnt5a protein activates or inhibits beta-catenin-TCF signaling depending on receptor context. PLoS Biol. 2006;4:e115. doi: 10.1371/journal.pbio.0040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Young T, Rowland JE, van de Ven C, Bialecka M, Novoa A, Carapuco M, van Nes J, de Graaff W, Duluc I, Freund JN, et al. Cdx and Hox genes differentially regulate posterior axial growth in mammalian embryos. Dev. Cell. 2009;17:516–526. doi: 10.1016/j.devcel.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 55.Rinn JL, Wang JK, Allen N, Brugmann SA, Mikels AJ, Liu H, Ridky TW, Stadler HS, Nusse R, Helms JA, et al. A dermal HOX transcriptional program regulates site-specific epidermal fate. Genes Dev. 2008;22:303–307. doi: 10.1101/gad.1610508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mallo M, Vinagre T, Carapuco M. The road to the vertebral formula. Int. J. Dev. Biol. 2009;53:1469–1481. doi: 10.1387/ijdb.072276mm. [DOI] [PubMed] [Google Scholar]
- 57.Bobola N, Carapuco M, Ohnemus S, Kanzler B, Leibbrandt A, Neubuser A, Drouin J, Mallo M. Mesenchymal patterning by Hoxa2 requires blocking Fgf-dependent activation of Ptx1. Development. 2003;130:3403–3414. doi: 10.1242/dev.00554. [DOI] [PubMed] [Google Scholar]
- 58.Mallo M, Magli MC. A look at life from the homeodomain. Workshop on homeodomain proteins, hematopoietic development and leukemias. EMBO Rep. 2006;7:976–980. doi: 10.1038/sj.embor.7400804. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.