Abstract
Bacteriophage ϕ29 genome consists of a linear double-stranded DNA with a terminal protein (TP) covalently linked to each 5′ end (TP-DNA) that together with a specific sequence constitutes the replication origins. To initiate replication, the DNA polymerase forms a heterodimer with a free TP that recognizes the origins and initiates replication using as primer the hydroxyl group of TP residue Ser232. The 3D structure of the DNA polymerase/TP heterodimer allowed the identification of TP residues that could be responsible for interaction with the DNA polymerase. Here, we examined the role of TP residues Arg158, Arg169, Glu191, Asp198, Tyr250, Glu252, Gln253 and Arg256 by in vitro analyses of mutant derivatives. The results showed that substitution of these residues had an effect on either the stability of the TP/DNA polymerase complex (R158A) or in the functional interaction of the TP at the polymerization active site (R169A, E191A, Y250A, E252A, Q253A and R256A), affecting the first steps of ϕ29 TP-DNA replication. These results allow us to propose a role for these residues in the maintenance of the equilibrium between TP-priming domain stabilization and its gradual exit from the polymerization active site of the DNA polymerase as new DNA is being synthesized.
INTRODUCTION
DNA polymerases are unable to start de novo DNA synthesis, requiring the use of a 3′-OH group, provided by a short RNA or DNA molecule. This fact, together with the internal initiation of asymmetric DNA replication, creates the so-called end replication problem of linear chromosomes, as once the last primer molecule is removed, there will remain a short region of unreplicated ssDNA at the end of the chromosome that would lead to a continuous shortening of the daughter DNA molecule after successive rounds of DNA replication. Many organisms, as several prokaryotic and eukaryotic viruses, as well as linear plasmids from bacteria, fungi and higher plants, possess replication origins, constituted by inverted terminal repetitions together with a terminal protein (TP), placed at both ends of their linear chromosomes [TP-DNA; (1)] allowing both strands to be replicated continuously. Additionally, the TP provides the OH-group of a specific serine, threonine or tyrosine to prime initiation of DNA replication from the very ends of the linear chromosome, circumventing the end replication problem, the TP remaining covalently linked to such 5′-ends (parental TP) (1–3).
Bacteriophage ϕ29 infects Bacillus subtilis and has become the paradigm in the study of the protein-primed mechanism to understand the rules that govern the first steps of TP-DNA replication. The ϕ29 genome is a linear dsDNA molecule 19 285 bp long with a phage-encoded TP covalently attached at each 5′ DNA end (parental TP) that, together with the terminal 12 bp containing a 6 bp inverted terminal repeat (3′ TTTCAT 5′), assemble a minimal replication origin (4). The DNA polymerase and a free TP molecule (primer TP) form a heterodimer (5) that, on the one hand recognizes specifically the replication origins and on the other excludes initiation of replication at internal sites of the genome (6).
ϕ29 DNA polymerase consists of a N-terminal exonuclease domain (residues 1–190), containing the 3′-5′ exonuclease active site, and a C-terminal polymerization domain (residues 191–575) subdivided into the universally conserved subdomains: palm, fingers and thumb (the latter coloured in blue in Figure 1) (7). There are two insertions into the sequence of the polymerase domain called Terminal Protein Regions 1 and 2 (TPR1 and TPR2, coloured in pale yellow and dark blue in Figure 1, respectively) (8,9), specifically present in the protein-primed DNA polymerases subgroup. Altogether, the exonuclease domain and the palm, thumb, TPR1 and TPR2 subdomains of the polymerase forms an internal ring-like structure that encircles both DNA polymerase substrates, DNA during DNA-primed elongation and the priming domain of TP during the first steps of ϕ29 genome replication (8; Figure 1).
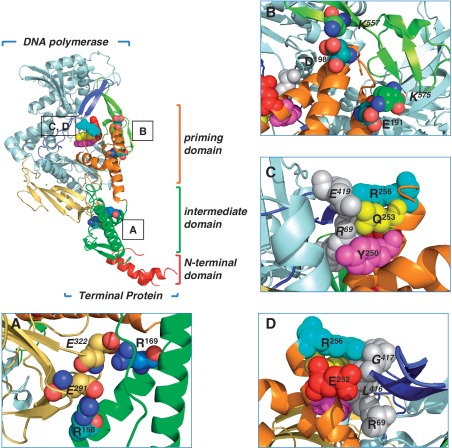
Figure 1.
Ribbon representation of ϕ29 TP/DNA polymerase heterodimer. DNA polymerase is coloured in blue except for TPR1, TPR2 and thumb subdomains that are in pale yellow, dark blue and light green, respectively. TP N-terminal, intermediate and priming domains are coloured in red, green and orange, respectively. (A) A detailed view of TP-intermediate domain residues R169 and R158 proposed to make salt bridges with DNA polymerase residues E322 and E291 of the TPR1 subdomain, respectively. (B) The proposed interactions of TP-priming domain residues E191 and D198 with the DNA polymerase thumb subdomain residues K575 and K557, respectively. In (C), the proposed stacking interactions between TP-priming subdomain residues R256, Q253 and Y250 with DNA polymerase residues R96 (exonuclease domain) and E419 (TPR2 subdomain) are indicated. (D) A detailed view of the proposed interaction of TP-priming subdomain residues R256 and E252 with DNA polymerase residues R96 (exonuclease domain) and L416 and G417 belonging to the TPR2 subdomain. View main text for details. Coordinates of the ϕ29 TP/DNA polymerase heterodimer were from PDB code 2EX3 (6). The figure was made with the Pymol software (http://www.pymol.org).
ϕ29 TP, encoded by the viral gene 3, has 266 amino acids (1). Figure 1 shows the high resolution crystal structures of the ϕ29 DNA polymerase/TP complex. TP has an elongated three-domain structure consisting of a disordered N-terminal domain (residues 1–73) that does not interact with the polymerase (10), an intermediate domain (residues 74–172) that confers specificity to the interaction with the DNA polymerase (10) and is connected through a hinge region to the C-terminal priming domain (residues 173–266) that contains the priming Ser232 laying in a loop at the end of this domain and placed at the active site of the DNA polymerase (6). Both, the intermediate and priming domains make extensive contacts with the polymerase accounting for the affinity and specificity between both proteins (10).
Once the DNA polymerase/TP heterodimer recognizes the replication origins, the polymerase catalyses the formation of a covalent bond between the initiator dAMP and the hydroxyl group of Ser232 of the primer TP in the initiation reaction (1). This reaction is directed by the second thymine (T) at the 3′-end of the template strand. Then, by a mechanism named ‘sliding-back’, the TP-dAMP initiation product translocates backwards one position to recover the template information corresponding to the terminal 3′ T, the penultimate T serving again as template to direct the incorporation of the second nucleotide (11). This mechanism requires a terminal repetition of two nucleotides at the template strand and could provide a way to prevent mutations at the ϕ29 DNA ends during initiation as the 3′–5′ exonuclease activity of the ϕ29 DNA polymerase cannot proofread the TP-linked nucleotide (12).
The DNA polymerase/TP heterodimer does not dissociate just after initiation. There is a stage during which the DNA polymerase synthesizes a five nucleotides long DNA molecule while complexed with the TP (initiation mode), undergoes some structural change during incorporation of nucleotides 6–9 (transition), and dissociates from the TP when the 10th nucleotide is incorporated into the nascent DNA chain (elongation mode) (13). Finally, the same DNA polymerase catalyses chain elongation via a strand displacement mechanism to fulfil TP-DNA replication (14).
The structure of the ϕ29 DNA polymerase/TP heterodimer has enabled the identification of the TP residues Arg158, Arg169, Glu191, Asp198, Tyr250, Glu252, Gln253 and Arg256 as potential DNA polymerase ligands (6). The results presented in this paper by using TP mutants allow us to infer a role for these residues in the first steps of TP-DNA replication, whereby the functional interaction with the DNA polymerase is guaranteed.
MATERIALS AND METHODS
Nucleotides and DNAs
Unlabelled nucleotides and [α-32P]dATP (3000 Ci/mmol) were supplied by Amersham Pharmacia. Oligonucleotides were obtained from Invitrogen.
Site-directed mutagenesis of ϕ29 TP
TP mutants were obtained using the QuikChange site-directed mutagenesis kit provided by Stratagene, using as template the plasmid pT7-3 that contains the viral gene 3 (15). The presence of the desired mutation, as well as the absence of additional ones was determined by sequencing the entire gene.
Proteins
TP mutants were expressed in Escherichia coli BL21(DE3) cells harbouring gene 3 cloned into plasmid pT7-3 and further purified, essentially as described for the wild-type TP (16). Both, the wild-type ϕ29 DNA polymerase and mutant N62D were purified essentially as described (17). ϕ29 double-stranded DNA binding protein (DBP) and ϕ29 single-stranded DNA binding protein (SSB) were purified as described (16,18).
Protein-primed initiation assay (TP-dAMP formation)
The ability to carry out the initiation step during TP-DNA replication was analysed as described (19). The incubation mixture contained, in 25 µl, 50 mM Tris–HCl, pH 7.5, 10 mM MgCl2, 20 mM ammonium sulphate, 1 mM dithiothreitol (DTT), 4% glycerol, 500 ng of ϕ29 TP-DNA as template, 0.1 mg/ml bovine serum albumin (BSA), 0.2 µM dATP (1µCi [α-32P]dATP), 10 ng of either wild-type or mutant TP and 20 ng of DNA polymerase. Samples were incubated for 4 min at 30°C. In the case of the template-independent TP-deoxyadenylylation reaction ϕ29 TP-DNA was omitted, 20 ng of either wild-type or mutant TP were incubated with 40 ng of DNA polymerase in the presence of 1 mM MnCl2, and the incubation was maintained for 90 min at 30°C. Reactions were stopped by adding 10 mM EDTA–0.1% SDS, and the samples were filtered through Sephadex G-50 spin columns and further analysed by SDS–PAGE in 12% polyacrylamide gels. Quantification was done by densitometric analysis of the labelled band corresponding to the TP-dAMP complex detected by autoradiography.
Interference assay for DNA polymerase binding
The assay was performed essentially as described (19). Reactions were carried out as described for the template-independent TP-dAMP formation, using a fixed amount of DNA polymerase and different proportions of wild-type and mutant TP. The amounts of proteins used were as follows: 40 ng of DNA polymerase, 20 ng of wild-type TP and 20, 40, 80 or 160 ng of the indicated mutant TP. In all cases, the incubation was for 90 min at 30°C. After incubation, reactions were stopped and analysed as indicated for the protein-primed initiation assay.
Analysis of the interaction between TP mutants and DNA polymerase by glycerol-gradient centrifugation
The assay was performed essentially as described (20). The incubation mixture contained, in 150 µl, 50 mM Tris–HCl, pH 7.5, 1 mM DTT, 0.1 mg/ml BSA, 20 mM ammonium sulphate, 6 µg of DNA polymerase and 3 µg of either wild-type or mutant TP. After incubation for 30 min at 4°C, samples were loaded on top of a continuous 15–30% (v/v) glycerol gradient (4 ml) in the presence of 50 mM Tris–HCl, pH 7.5, 20 mM ammonium sulphate, the indicated concentration of NaCl, 1 mM EDTA and 7 mM 2-mercaptoethanol, and centrifuged at 4°C for 24 h at 58 000 rpm in a Beckman TST 60.4 rotor. Gradients were fractionated and subjected to SDS–PAGE in 12% polyacrylamide gels. The proteins in the gel were stained with SYPRO to identify the peaks corresponding to the TP/DNA polymerase heterodimer (97 kDa) and the free monomers of TP (31 kDa) and DNA polymerase (66 kDa).
TP-DNA replication assay
The replication assay was performed as described (19). The incubation mixture contained, in 25 µl, 50 mM Tris–HCl, pH 7.5, 10 mM MgCl2, 20 mM ammonium sulphate, 1 mM DTT, 4% glycerol, 0.1 mg/ml BSA, 20 µM each dNTP and [α-32P] dATP (1 µCi), 10 ng of either wild-type or mutant TP, 20 ng of DNA polymerase, and 500 ng of ϕ29 TP-DNA. After incubation for the indicated times at 30°C, the reaction was stopped by adding 10 mM EDTA-0.1% SDS, and the samples were filtered through Sephadex G-50 spin columns. Quantitation of the DNA synthesized in vitro was carried out from the amount of radioactivity (Cerenkov radiation) corresponding to the excluded volume. The labelled DNA was denatured by treatment with 0.7 M NaOH and subjected to electrophoresis in alkaline 0.7% agarose gels, as described (21). After electrophoresis, the position of unit-length ϕ29 DNA (19 285 bases) was detected by ethidium bromide staining, and then the gels were dried and autoradiographed.
Transition assay
The assay was performed essentially as described for the TP-DNA replication assay. For the analysis of the transition products, 10 ng of TP wild-type or the indicated mutant, 500 ng of TP-DNA and 20 ng of DNA polymerase mutant N62D, which has a low 3′–5′ exonuclease activity but retains the strand displacement ability (22), were incubated in the presence of 5 µM dATP, dGTP and dTTP for 2 min at 30°C. The reaction was stopped by adding 10 mM EDTA-0.1% SDS, and the samples were filtered through Sephadex G-50 spin columns. The samples were analysed in SDS–12% polyacrylamide gels (360 × 280 × 0.5 mm) to obtain enough resolution to distinguish the TP bound to the first elongation products.
TP-DNA amplification assay
The assay was performed essentially as described (23) in the presence of 50 mM Tris–HCl, pH 7.5, 10 mM MgCl2, 20 mM ammonium sulphate, 1 mM DTT, 4% glycerol, 0.1 mg/ml BSA, 80 µM each dNTP and [α-32P]dATP (1 µCi), 5 ng of ϕ29 TP-DNA, 10 ng of DNA polymerase, 5 ng of TP wild-type or the indicated mutant, 10 µg of ϕ29 SSB and 10 µg of ϕ29 DBP in a final reaction volume of 25 µl. After incubation for the indicated times at 30°C, samples were processed and quantitated as described for the TP-DNA replication assay, and the amplified DNA analysed by electrophoresis in alkaline agarose gels as described (23). After electrophoresis, the position of unit length ϕ29 DNA was detected by ethidium bromide staining. To study the parental TP function of mutant TPs, amplification assays were carried out for 80 min under the same conditions. Next, the reaction was supplemented with the indicated TP/DNA polymerase heterodimer (5 and 10 ng of TP and DNA polymerase, respectively) and the reaction was continued for another 90 min. Aliquots of the reaction mixtures were withdrawn at appropriate times and processed as described above.
RESULTS
TP residues potentially involved in making interactions with the DNA polymerase
Specific recognition of the DNA polymerase by the TP is mainly contributed through the interaction between the TPR1 insertion of the former (coloured in pale yellow in Figure 1) and the intermediate domain of the latter (coloured in green in Figure 1) (10). ϕ29 TP has many positively charged residues and among them Arg158 and Arg169 stand out as they were predicted to form two salt bridges with residues Glu291 and Glu322 of the TPR1 insertion of the DNA polymerase (Figure 1A), respectively, (6). In contrast, the TP priming domain is highly electronegative, mimicking the DNA in its interaction with the DNA polymerase. In this sense, specific interactions of residues Glu191 and Asp198 of TP (Figure 1B) with the electropositive residues Lys575 and Lys557 of the thumb subdomain of the DNA polymerase were predicted (6). Similarly, residue Arg69 of the exonuclease domain of the polymerase would interact with the TP-priming domain residues Gln253 and Tyr250 through a hydrogen bond and a stacking interaction, respectively (Figure 1C). Finally, residues Glu252, Gln253 and Arg256 of the C-terminal helix of the TP priming domain would pack against residues Leu416, Gly417 and Glu419 of the TPR2 subdomain of the polymerase (Figure 1C and D) (6).
To analyse their functional role, all the above-mentioned TP residues were changed into alanine, removing the positive charge of arginines, the polar group of the glutamine, the negative charge of glutamates and aspartate, and the aromatic group of the tyrosine residue. The mutant derivatives were overexpressed and purified as described in ‘Materials and Methods’ section and their involvement in the interaction with the DNA polymerase was analysed by a variety of in vitro assays corresponding to the different stages of the TP-primed ϕ29 DNA replication process.
Protein-primed initiation of ϕ29 TP-DNA replication (TP-dAMP formation)
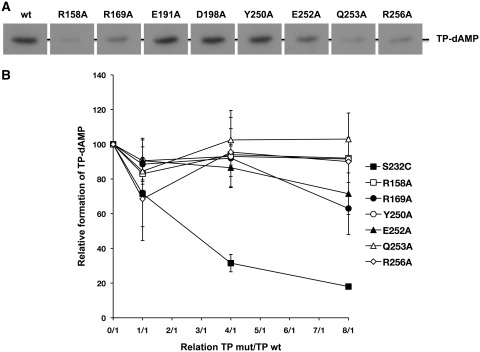
ϕ29 DNA replication starts from both origins placed at the termini of the linear ϕ29 TP-DNA molecule. The DNA polymerase forms a heterodimer with a free TP molecule and catalyses the template-directed insertion of 5′ dAMP onto the hydroxyl group of TP Ser232. To study the primer function of mutant TPs, the formation of the TP-dAMP complex (initiation reaction) was evaluated using as template ϕ29 TP-DNA. As shown in Figure 2A and Table 1, there were mutants severely (R158A) or moderately (R169A, Q253A and R256A) impaired in the formation of the TP-dAMP product. As the initiation of ϕ29 TP-DNA replication is a template-directed event, such deficiency could be a consequence of a faulty recognition/binding of the replication origins. To study this possibility, we made use of the ability exhibited by ϕ29 DNA polymerase to catalyse the deoxynucleotidylation of the TP in the absence of template, the formation of the TP-dAMP product relying exclusively on a functional DNA polymerase-TP interaction (24). As shown in Table 1, except for mutant D198A, the rest of the TP derivatives showed a drop in their priming ability. Thus, no activity was detected with mutants R158A, R169A, E252A, Q253A and R256A, and TP derivatives E191A and Y250A showed a decrease of 3- and 7-fold, respectively, in their priming activity relative to the template-directed reaction. Thus, the low TP-deoxynucleotidylation levels exhibited by TP mutants could be pointing to a defective interaction with the DNA polymerase.
Figure 2.
(A) In vitro protein-primed initiation with TP mutants. The initiation assay was performed as described in ‘Materials and Methods’ section, in the presence of 10 ng of either wild-type or the indicated mutant TP, 20 ng of DNA polymerase and 500 ng of ϕ29 TP-DNA. After 4 min incubation at 30°C, the reactions were stopped, processed and analysed by SDS–PAGE and autoradiography. The position of the TP–dAMP complex is indicated. (B) Competition for DNA polymerase between wild-type and mutant TPs. The assay of formation of TP-dAMP in the absence of TP-DNA by the wild-type ϕ29 TP/DNA polymerase heterodimer was performed in the presence of increasing amounts of TP mutants (the reaction conditions are described under ‘Materials and Methods’ section). Reactions were started by adding 1 mM MnCl2 and, after incubation for 90 min at 30°C, were stopped and analysed as indicated for the protein-primed initiation assay. The TP-dAMP formed in the different competition conditions relative to that formed in the absence of competition (100%) is indicated. As control of competition, the inactive TP mutant S232C was used. Data are represented as mean ± SD corresponding to three independent experiments.
Table 1.
Functions of wild-type and mutant ϕ29 TPs
| ϕ29 TP | TP-dAMP formationa |
TP-DNA replicationa | Transitiona,c | TP-DNA amplification |
||
|---|---|---|---|---|---|---|
| TP-DNAb | No template | (%)a Amplification factord | ||||
| wild-type | 100 | 100 | 100 | 1 | 100 | 50 ± 7 |
| R158A | 4 ± 1.5 | n.d. | 17 ± 3 | n.d. | 1 ± 0.2 | 1 ± 0.1 |
| R169A | 23 ± 6 | n.d. | 75 ± 13 | 1.7 ± 0.4 | 3 ± 0.3 | 3 ± 0.2 |
| E191A | 94 ± 11 | 30 ± 8 | 106 ± 17 | 2.2 ± 0.3 | 69 ± 2 | 35 ± 6 |
| D198A | 112 ± 1 | 102 ± 3 | 66 ± 8 | 0.3 ± 0.2 | 121 ± 10 | 61 ± 13 |
| Y250A | 74 ± 24 | 10 ± 2 | 100 ± 20 | 3.3 ± 0.4 | 130 ± 4 | 66 ± 10 |
| E252A | 70 ± 28 | n.d. | 103 ± 25 | 3.2 ± 0.8 | 52 ± 7 | 27 ± 7 |
| Q253A | 23 ± 5 | n.d. | 55 ± 12 | 4.7 ± 0.7 | 20 ± 6 | 10 ± 4 |
| R256A | 20 ± 8 | n.d. | 76 ± 19 | 2.6 ± 1.2 | 16 ± 5 | 10 ± 1 |
Data represent the mean value and the standard deviation obtained from at least three independent experiments.
aNumbers indicate the activity of mutant TPs with respect to the wild-type TP.
bTP-dAMP formation using ϕ29 TP-DNA as template of the reaction.
cTransition efficiency was calculated as the ratio of the truncated products TP-(dNMP)6-14/TP-(dNMP)1–2 considering such quotient 1 in the case of wild-type TP. See details in the main text.
dThe amplification factor was calculated as the ratio between the amount of DNA at the end of the reaction (input plus synthesized DNA) and the amount of input DNA.
n.d. Not detected.
The ability of mutant TPs to interact with the DNA polymerase was tested by carrying out interference assays in which wild-type and mutant TPs compete for the DNA polymerase (see ‘Materials and Methods’ section). As a control, the TP mutant S232C was used as it is essentially inactive as primer since it lacks the priming OH group, but conserves an intact capacity to interact with the DNA polymerase (25). As expected, the inhibition profile obtained with this mutant paralleled the theoretical one (Figure 2B). On the contrary, the wild-type TP was not competed by mutants R158A, Y250A, Q253A and R256A, and only slightly in the presence of a 8-fold excess of mutants R169A and E252A. These results indicate an impaired interaction of those TP mutants with the DNA polymerase.
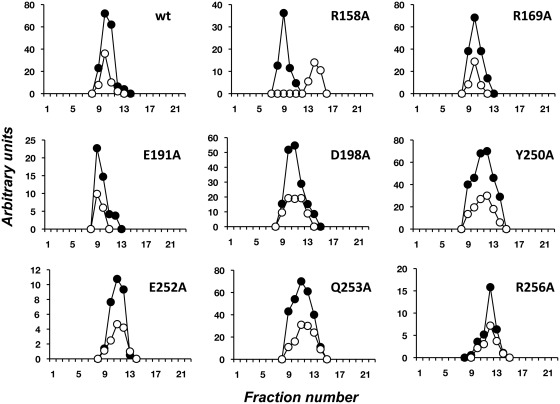
The interaction of mutant TPs with the DNA polymerase was also directly analysed by glycerol gradient ultracentrifugation in the presence of 0.2 M NaCl (see Materials and Methods). As it can be observed in Figure 3, the wild-type TP and the mutant derivatives, with the sole exception of mutant R158A, formed a heterodimer with the DNA polymerase. Thus, although it seemed that the stability of the heterodimer was not hindered by the mutations introduced, the poor deoxynucleotidylation activity exhibited by the mutant TPs in the absence of template, as well as the lack of competition with the wild-type TP (see above), would be reflecting a non-functional interaction with the DNA polymerase. To further analyse the stability of the DNA polymerase/TP complexes, glycerol gradient centrifugation analysis was carried out in the presence of 0.4 M NaCl (Supplementary Figure 1). Under these conditions, whereas the wild-type and mutant TPs E191A and D198A still formed a stable heterodimer with the DNA polymerase, mutants R158A, R169A, Y250A, E252A, Q253A and R256A eluted separately, showing that their interaction with the DNA polymerase is not as strong as that displayed by the wild-type DNA polymerase.
Figure 3.
Analysis of TP/DNA polymerase interaction by glycerol gradient ultracentrifugation. The assay was carried out as described under ‘Materials and Methods’ section, pre-incubating 3 µg of either wild-type or the indicated mutant TP with 6 µg of DNA polymerase. After incubation for 30 min at 4°C, samples were loaded on top of a continuous 15–30% glycerol gradient in the presence of 0.2 M NaCl. After centrifugation, the collected fractions were subjected to SDS–12% PAGE and further stained with SYPRO. Densitometric quantification, expressed in arbitrary units, of both DNA polymerase (full circles) and TP (open circles) are represented.
Protein-primed TP-DNA replication with TP mutants
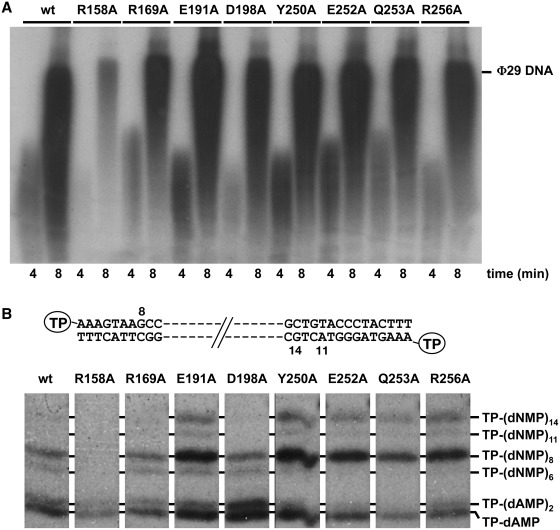
Once it catalyses the formation of the TP-dAMP product, the same DNA polymerase molecule elongates it via strand displacement to produce full-length ϕ29 TP-DNA. To ascertain to what extent the impaired DNA polymerase/mutant TP interaction affects the replication process, replication assays were carried out, using a minimal replication system based on ϕ29 TP-DNA, DNA polymerase and TP (14) (see ‘Materials and Methods’ section). As observed in Figure 4A and Table 1, when mutant R158A was used as primer TP, the efficiency of the reaction decreased 6-fold respect to the wild-type TP, in accordance with its poor priming proficiency. Nonetheless, when the rest of mutant TPs were used as primer, the efficiency of the reaction was either not affected (TP mutants E191A, Y250A, E252A) or only slightly reduced (TP mutants R169A, D198A, Q253A and R256A), regardless of their impaired interaction with the DNA polymerase. These results could suggest that formation of a catalytically competent heterodimer depends on contacts with the parental TP that could alleviate at least partially the defects caused by the mutations introduced in the TP.
Figure 4.
(A) TP-DNA replication by mutant TPs. The assays were carried as described in ‘Materials and Methods’ section in the presence of 20 ng of DNA polymerase and 10 ng of either wild-type or the indicated mutant TP. After incubation at the indicated times at 30°C, relative values were calculated (Table 1) and the length of the synthesized DNA was analysed by alkaline agarose gel electrophoresis. The migration position of unit-length ϕ29 DNA is indicated. (B) Analysis of the transition products of ϕ29 DNA replication. The assay was performed as described in ‘Materials and Methods’ section in the presence of 20 ng of DNA polymerase mutant N62D (22), 10 ng of either wild-type or the indicated mutant TP and 5 µM of each dATP, dGTP and dTTP. After incubation for 2 min at 30°C, the different transition products were detected and analysed by high resolution SDS–PAGE. The products expected during the first replication steps, starting from both ends of ϕ29 TP-DNA [(TP-dAMP, TP-(dAMP)2, TP-(dNMP)8 and TP-(dNMP11)], as well as TP-(dNMP)6 and TP-(dNMP)14, are indicated.
Transition from protein-priming to DNA-priming in ϕ29 DNA replication
After the initiation step has taken place, the ϕ29 DNA polymerase/TP heterodimer remains as a complex. Once the 10th nucleotide is inserted, the interaction between the two proteins is released to allow the same DNA polymerase molecule to continue TP-DNA replication (13). This stage between the TP-primed and DNA-primed modes is known as transition. Thus, considering the low initiation activity displayed by TP mutants R169A, Q253A and R256A, their relatively high level of replication could be pointing to an improvement in performing the transition phase by the DNA polymerase when it uses the mutant derivatives as primers, counteracting the reduced number of initiation events. To study this possibility the short reaction products obtained in the presence of three dNTPs (missing dCTP) were analysed by high resolution gel electrophoresis (see ‘Materials and Methods’ section). To better detect the amount of non-elongated initiation complexes and partially elongated products, the DNA polymerase variant N62D was used, as it has a low 3′–5′ exonuclease activity, preventing degradation of the replication intermediates, but retains a wild-type strand displacement proficiency (22). As shown in Figure 4B, when the DNA polymerase uses the wild-type TP as primer, it gives rise to the initiation products TP-(dAMP)1-2, TP-(dNMP)8, corresponding to the truncated elongation that started from the left origin, and a small amount of TP-(dNMP)6 that probably corresponds to molecules aborted during the transition from the initiation mode to the elongation one. As also observed in Figure 4B, with several of the mutant TPs, the DNA polymerase rendered the additional TP-(dNMP)14 product due to a misincorporation at position 12 from the right origin followed by its elongation, as previously described to occur with this exonuclease deficient DNA polymerase (26). Thus, when mutants R169A, E191A, Y250A, E252A, Q253A and R256A were used as primer TPs the ratio between the truncated products [TP-(dNMP)6-14] and the initiation complexes [TP-(dAMP)1-2] was higher than for the wild-type TP, possibly accounting for their improved TP-DNA replication/TP-dAMP formation ratio (Figure 4B and Table 1). This result could be explained on the basis that a weakened DNA polymerase/mutant TP interaction would favour the transition of ϕ29 DNA polymerase to the elongation stage. On the other hand, the lower proportion of the intermediate products with respect to the initiation ones obtained with TP mutant D198A is consistent with its reduced TP-DNA replication capacity relative to its wild-type initiation activity.
Protein-primed TP-DNA amplification with TP mutants
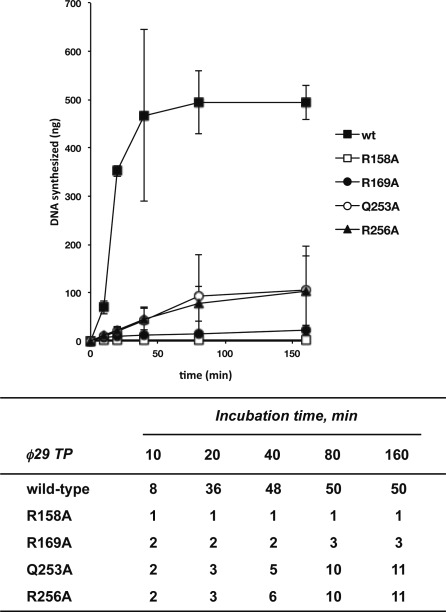
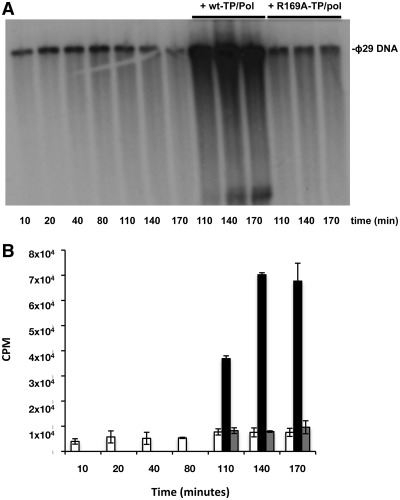
In contrast with in vitro replication, performed in the presence of the DNA polymerase and the TP, the addition to the reaction of DBP and SSB enables 103-fold amplification of limiting amounts (0.5 ng) of input ϕ29 TP-DNA (23). Figure 5 shows the amount of DNA amplified using wild-type or mutant TPs. Mutants E191A, D198A and Y250A led to amplification levels similar or even higher than those reached with the wild-type TP (Table 1), and the amplification yield reached with mutant E252A was only 2-fold lower than that obtained with the wild-type TP. Conversely, mutants R158A, R169A, Q253A and R256A were inefficient in supporting the amplification reaction most likely because of their deficient priming activity (Figure 5 and Table 1). In these latter cases, the amount of de novo synthesized DNA (in nanograms) was quantified from the amount of radioactivity incorporated into DNA as a function of time (Figure 6, upper panel), the amplification factor being calculated as the ratio between the amount of DNA at the end of the reaction (input DNA plus synthesized DNA) and the amount of input DNA (Figure 6, lower panel). As shown in Figure 6, with the wild-type TP, DNA synthesis reaches a plateau after 40 min giving rise to an amplification factor of 50. Whereas the DNA polymerase did not synthesize any detectable product when using TP mutant R158A, DNA synthesis with mutants R256A and Q253A did not reach a plateau at the longest time assayed (160 min), suggesting that these mutations slow down the starting of new replication rounds. Interestingly, when TP mutant R169A was used as primer, although synthesis reached a plateau after 80 min, the amount of de novo synthesized DNA corresponded to an amplification factor of 3 (Figure 6). This result suggests that DNA synthesis has stalled before completing a second round of replication, possibly due to the inactivation of the replication origin upon incorporation of the mutant TP that now would act as parental TP. If this were the case, only the wild-type TP-containing origins would be active in the second round of replication resulting in a maximum amplification factor of 3, as previously described for other TP mutants (27). Interestingly, renewed incorporation of dAMP was observed upon the addition of wild-type TP/DNA polymerase heterodimer after 80 min of reaction (Figure 7) ruling out the possibility that the mutation had affected the overall structure of the parental TP that might cause inactivation of the origins. As expected, addition of mutant TP/DNA polymerase heterodimer did not result in renewed dAMP incorporation. These results suggest that the parental TP mutant R169A could produce functional interactions with wild-type TP/DNA polymerase heterodimer but not with a heterodimer containing the mutant TP. Therefore, the mutation is critical when it is present in both the primer and parental TP, presumably affecting their proper interaction required for the initiation reaction.
Figure 5.
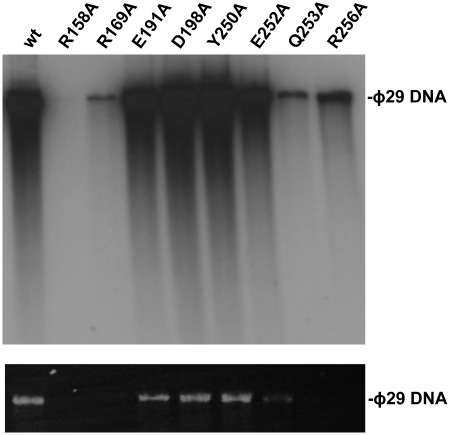
ϕ29 TP-DNA amplification with wild-type and mutant TPs. The assay was carried out as described under ‘Materials and Methods’ section, in the presence of 5 ng of ϕ29 TP-DNA, 10 ng of ϕ29 DNA polymerase, 5 ng of either wild-type or the indicated mutant TP, 10 µg of ϕ29 DBP and 10 µg of ϕ29 SSB. After 80 min of incubation at 30°C, the reaction was stopped with 10 mM EDTA. The relative activity values were calculated (Table 1), and the length and amount of the synthesized DNA was analysed by alkaline agarose gel electrophoresis followed by autoradiography (upper panel) and ethidium bromide staining (lower panel).
Figure 6.
Upper panel, kinetics of in vitro ϕ29 TP-DNA amplification with wild-type and mutant TPs. The amplification assays were carried out as described in ‘Materials and Methods’ section in the presence of 10 ng of DNA polymerase, 5 ng of either wild-type or the indicated mutant TP. After incubation for the indicated times at 30°C, the reaction was stopped and quantitated as total amount (in nanograms) of dNTP incorporated. Data are represented as mean ± SD corresponding to three independent experiments. In the lower panel, numbers show the amplification factor calculated as the ratio between the amount of DNA at the end of the reaction (input DNA plus synthesized DNA) and the amount of input DNA.
Figure 7.
In vitro ϕ29 TP-DNA amplification with TP mutant R169A is recovered upon addition of wild-type TP/DNA heterodimer. (A) Amplification assays were performed using the conditions described under ‘Materials and Methods’ section. After 80 min, TP/DNA polymerase heterodimers containing either wild-type or the TP mutant R169A were added to the reaction mixtures. The samples, withdrawn at the indicated times, were analysed as described in the legend of Figure 6. The amount of incorporated [α-32P]dAMP was measured at the indicated times and is represented in (B). White bars, amount of incorporated [α-32P]dAMP by mutant TP/DNA polymerase throughout the experiment. Black bars, amount of newly incorporated [α-32P]dAMP upon addition of wild-type TP/DNA polymerase heterodimers. Grey bars, amount of newly incorporated [α-32P]dAMP upon addition of mutant TP/DNA polymerase heterodimers after 80 min of reaction. Data are represented as mean ± SD corresponding to four independent experiments.
DISCUSSION
Extensive studies performed both in vitro and in vivo, mainly using bacteriophage ϕ29 and Adenovirus, provided the general insights about the mechanism of protein-primed DNA replication (1–3,28). First, the replicative DNA polymerase forms, with the corresponding TP, a heterodimer that recognizes the replication origins, located at both ends of the linear genome and comprising a 5′ covalently linked TP and a specific DNA sequence. The DNA polymerase then catalyses both, the formation of the covalent complex between a primer TP molecule and the initial dNMP, and its further elongation. Similarly, TP-DNA replication of bacteriophages PRD1 (29,30), Cp1 (31), and the ϕ29-related phages GA-1 (15,32) and Nf (15,33) was also shown to occur by a homologous protein-primed mechanism, involving its corresponding DNA polymerase and TP.
High-resolution crystal structures of the ϕ29 DNA polymerase/TP complex showed that the TP has a three domain folded structure (6). The positively charged intermediate domain is structurally complementary to the TPR1 subdomain of the DNA polymerase, whereas the electronegative charged C-terminal TP priming domain is accommodated at the polymerization domain of the polymerase being wrapped by the thumb and TPR2 subdomains of the latter (6).
Biochemical analyses of the DNA polymerase/TP heterodimers of phages ϕ29 and GA-1 have shown that the interaction between the TP intermediate domain and the DNA polymerase TPR1 subdomain confers the specificity between both proteins, dependent on their structural complementarity (10). The crystal structure of the TP/DNA polymerase heterodimer showed that residues Arg158 and Arg169 of the TP intermediate domain form salt bridges with DNA polymerase TPR1 subdomain residues Glu291 and Glu322, respectively (6). The fact that these four residues are highly conserved in other TPs and protein-primed DNA polymerases, together with the results presented here, agree with the hypothesis that not only the specificity, but also the stability of the heterodimer relies on these interactions, mainly the one contributed by TP Arg158 residue, as mutant R158A was very deficient in supporting a priming role in all the assays carried out, due to an impaired interaction with the DNA polymerase. It has been shown that interaction between the TP intermediate domain and the TPR1 subdomain of the DNA polymerase is required to allow the TP priming domain to be correctly positioned at the polymerase catalytic site (10). The results presented here with mutant R169A led us to propose a role for this residue in the proper interaction with the DNA polymerase in the heterodimer since, although the complex formed between mutant TP and the DNA polymerase is stable at 0.2 M NaCl, it displayed reduced initiation and amplification activities, and it was unable to compete the interaction between the wild-type TP and the DNA polymerase. It is also noteworthy that this mutant derivative cannot be acting simultaneously as parental and primer TP. This result is in agreement with the existence of a functional interaction between primer and parental TP during ϕ29 TP-DNA origins recognition, as it has been described for other TP intermediate domain residues (27,34).
The TP/DNA polymerase structure unveiled direct interactions between the TP priming domain and the TPR2 and thumb subdomains of the DNA polymerase. TP residues Glu252, Gln253 and Arg256 of the C-terminal helix of the TP priming domain would pack against residues Leu416, Gly417 and Glu419 of the TPR2 subdomain of the polymerase (6). In addition, Gln253 is also contacted by DNA polymerase residue Arg96 placed at the apex of a loop of the exonuclease domain that projects into the DNA binding region at the polymerization domain. Arg96 also interacts with TP priming domain residue Tyr250. The results presented here showed that TP mutants Y250A, E252A, Q253A and R256A are highly impaired in priming the TP-deoxyadenylylation reaction. As this reaction relies exclusively on a bona fide interaction between the DNA polymerase and the TP, the results point to the formation of faulty TP/DNA polymerase heterodimers. In agreement with this hypothesis, interference assays showed that these mutant TPs were not able to compete out the wild-type protein in its interaction with the DNA polymerase. Analysis of such interaction by glycerol gradient showed that these mutant TPs formed a stable TP/DNA polymerase heterodimer at 0.2 M NaCl but not at 0.4 M salt. Altogether, the results led us to suggest a role for these residues in the functional interaction with the DNA polymerase in the heterodimer to allow the TP priming loop to be correctly placed at the polymerization active site. Interestingly, when mutant TPs Y250A, E252A, Q253A and R256A were evaluated in their ability to support TP-DNA replication, they showed a nearly wild-type activity. As mentioned above, after incorporation of the initiating dAMP to the primer TP, the same ϕ29 DNA polymerase molecule elongates the initiation product going through a transition stage during which the DNA polymerase and the TP are still associated. Once both proteins dissociate, the ϕ29 DNA polymerase completes replication of the nascent DNA chain. Thus, for an efficient transition between protein-primed initiation and DNA elongation, dissociation of the TP/DNA polymerase heterodimer is required. Bearing in mind that interaction between the above mutant TPs and DNA polymerase is affected, this may favour the dissociation of the heterodimer during transition. As it has been shown, the amount of intermediate products detected with the mutant TPs during truncated replication assays was higher than with the wild-type protein, suggesting an enhanced transition efficiency that could account for the improvement exhibited by the mutant TPs in the replication reaction. This phenotype has been previously described for mutants at the RGD region of the TP, involved in making contacts with the DNA polymerase (35), as well as for ϕ29 DNA polymerase mutants defective in their interaction with the TP (19). Intriguingly, despite their efficient activity in TP-DNA replication, mutants Q253A and R256A were affected in amplifying the viral genome. These results suggest that a productive in vitro amplification of ϕ29 TP-DNA relies on a fine-tuned equilibrium among the different replication steps, initiation, transition and elongation. Thus, if such a balance is disturbed as in the case of a mutation in the TP (this work) or in the DNA polymerase (19), amplification efficiency is expected to drop.
Finally, other TP-priming domain residues as Glu191 and Asp198 were proposed to make contacts with the electropositive residues Lys575 and Lys557, respectively, of the DNA polymerase thumb subdomain (6). Both contacts seem also important to produce a functional heterodimer. Thus, mutant E191A displayed an impaired, although stable, interaction as deduced from its reduced TP-deoxynucleotidylation activity that was probably compensated in the replication and amplification assays by its improved initiation and transition efficiencies. Interestingly, a proper contact between TP residue Asp198 and the polymerase thumb subdomain seems to be critical to guarantee an efficient transition. Notwithstanding, the wild-type amplification activity showed by this mutant TP suggests that those defects are offset by its stronger priming capacity.
ϕ29 DNA polymerase/TP heterodimer identifies the replication origins at the genome termini, being the parental TP the main signal, as the use of terminal DNA fragments lacking parental TP makes the initiation reaction to fall 6- to 10-fold with respect to the activity obtained with TP-DNA (4,33). Recognition of parental TP is contributed by both the DNA polymerase and the primer TP (10,33–36). As it has been mentioned above, except for mutant D198A, the defects shown by the rest of mutant TPs in their interaction with the DNA polymerase were much more evident when TP-dAMP formation was tested without TP-DNA. These results suggest that formation of a catalytically competent heterodimer relies on contacts with parental TP that would be critical for a fine adjustment of the interaction between the DNA polymerase and the priming TP and that could relieve at some degree the defects caused in the heterodimer by the mutations introduced in the TP.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figure S1.
FUNDING
Spanish Ministry of Science and Innovation (grants BFU 2008-00215, and Consolider-Ingenio CSD2007-00015 to M.S.; Autonomous Community of Madrid (grant P-MAT-0283-0505 to M.S.); Fundación Ramón Areces to the Centro de Biología Molecular ‘Severo Ochoa’. Funding for open access charge: Research grant (Consolider-Ingenio CSD2007-00015) from the Spanish Ministry of Science and Innovation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to José M. Lázaro for his advice in protein purification. A.P. is a predoctoral fellow from Spanish Ministry of Education.
REFERENCES
- 1.Salas M. Protein-priming of DNA replication. Annu. Rev. Biochem. 1991;60:39–71. doi: 10.1146/annurev.bi.60.070191.000351. [DOI] [PubMed] [Google Scholar]
- 2.Salas M. Mechanisms of initiation of linear DNA replication in prokaryotes. Genet. Eng. 1999;21:159–171. doi: 10.1007/978-1-4615-4707-5_8. [DOI] [PubMed] [Google Scholar]
- 3.Salas M, Miller J, Leis J, DePamphilis M. Mechanisms for Priming DNA Synthesis. New York: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 4.Gutiérrez J, García JA, Blanco L, Salas M. Cloning and template activity of the origins of replication of phage ϕ29 DNA. Gene. 1986;43:1–11. doi: 10.1016/0378-1119(86)90002-8. [DOI] [PubMed] [Google Scholar]
- 5.Blanco L, Prieto I, Gutiérrez J, Bernad A, Lázaro JM, Hermoso JM, Salas M. Effect of NH4+ ions on ϕ29 DNA-protein p3 replication: formation of a complex between the terminal protein and the DNA polymerase. J. Virol. 1987;61:3983–3991. doi: 10.1128/jvi.61.12.3983-3991.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamtekar S, Berman AJ, Wang J, Lázaro JM, de Vega M, Blanco L, Salas M, Steitz TA. The phi29 DNA polymerase:protein-primer structure suggests a model for the initiation to elongation transition. EMBO J. 2006;25:1335–1343. doi: 10.1038/sj.emboj.7601027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamtekar S, Berman AJ, Wang J, Lázaro JM, de Vega M, Blanco L, Salas M, Steitz TA. Insights into strand displacement and processivity from the crystal structure of the protein-primed DNA polymerase of bacteriophage ϕ29. Mol.r Cell. 2004;16:609–618. doi: 10.1016/j.molcel.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 8.Blasco MA, Blanco L, Parés E, Salas M, Bernad A. Structural and functional analysis of temperature-sensitive mutants of the phage ϕ29 DNA polymerase. Nucleic Acids Res. 1990;18:4763–4770. [PMC free article] [PubMed] [Google Scholar]
- 9.Dufour E, Méndez J, Lázaro JM, de Vega M, Blanco L, Salas M. An aspartic acid residue in TPR-1, a specific region of protein-priming DNA polymerases, is required for the functional interaction with primer terminal protein. J. Mol. Biol. 2000;304:289–300. doi: 10.1006/jmbi.2000.4216. [DOI] [PubMed] [Google Scholar]
- 10.Pérez-Arnaiz P, Longás E, Villar L, Lázaro JM, Salas M, de Vega M. Involvement of phage φ29 DNA polymerase and terminal protein subdomains in conferring specificity during initiation of protein-primed DNA replication. Nucleic Acids Res. 2007;35:7061–7073. doi: 10.1093/nar/gkm749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Méndez J, Blanco L, Esteban JA, Bernad A, Salas M. Initiation of ϕ29 DNA replication occurs at the second 3′ nucleotide of the linear template: a sliding-back mechanism for protein-primed DNA replication. Proc. Natl Acad. Sci. USA. 1992;89:9579–9583. doi: 10.1073/pnas.89.20.9579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esteban JA, Salas M, Blanco L. Fidelity of ϕ29 DNA polymerase. Comparison between protein-primed initiation and DNA polymerization. J. Biol. Chem. 1993;268:2719–2726. [PubMed] [Google Scholar]
- 13.Méndez J, Blanco L, Salas M. Protein-primed DNA replication: a transition between two modes of priming by a unique DNA polymerase. EMBO J. 1997;16: 2519–2527. doi: 10.1093/emboj/16.9.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blanco L, Bernad A, Lázaro JM, Martín G, Garmendia C, Salas M. Highly efficient DNA synthesis by the phage ϕ29 DNA polymerase. Symmetrical mode of DNA replication. J. Biol. Chem. 1989;264:8935–8940. [PubMed] [Google Scholar]
- 15.Longás E, de Vega M, Lázaro JM, Salas M. Functional characterization of highly processive protein-primed DNA polymerases from phages Nf and GA-1, endowed with a potent strand displacement capacity. Nucleic Acids Res. 2006;34:6051–6063. doi: 10.1093/nar/gkl769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mencía M, Gella P, Camacho A, de Vega M, Salas M. Terminal protein-primed amplification of heterologous DNA with a minimal replication system based on phage ø29. Proc Natl Acad. Sci. USA. 2011;108:18655–18660. doi: 10.1073/pnas.1114397108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lázaro JM, Blanco L, Salas M. Purification of bacteriophage ϕ29 DNA polymerase. Methods Enzymol. 1995;262:42–49. doi: 10.1016/0076-6879(95)62007-9. [DOI] [PubMed] [Google Scholar]
- 18.Soengas MS, Gutiérrez C, Salas M. Helix-destabilizing activity of ϕ29 single-stranded DNA binding protein: effect on the elongation rate during strand displacement DNA replication. J. Mol. Biol. 1995;253: 517–529. doi: 10.1006/jmbi.1995.0570. [DOI] [PubMed] [Google Scholar]
- 19.de Vega M, Blanco L, Salas M. ϕ29 DNA polymerase residue Ser122, a single-stranded DNA ligand for 3′-5′ exonucleolysis, is required to interact with the terminal protein. J. Biol. Chem. 1998;273:28966–28977. doi: 10.1074/jbc.273.44.28966. [DOI] [PubMed] [Google Scholar]
- 20.Rodríguez I, Lázaro JM, Salas M, de Vega M. ϕ29 DNA polymerase-terminal protein interaction. Involvement of residues specifically conserved among protein-primed DNA polymerases. J. Mol. Biol. 2004;337: 829–841. doi: 10.1016/j.jmb.2004.02.018. [DOI] [PubMed] [Google Scholar]
- 21.McDonell MW, Simon MN, Studier FW. Analysis of restriction fragments of T7 DNA and determination of molecular weights by electrophoresis in neutral and alkaline gels. J. Mol. Biol. 1977;110:119–146. doi: 10.1016/s0022-2836(77)80102-2. [DOI] [PubMed] [Google Scholar]
- 22.de Vega M, Lázaro JM, Salas M, Blanco L. Primer-terminus stabilization at the 3′-5′ exonuclease active site of ϕ29 DNA polymerase. Involvement of two amino acid residues highly conserved in proofreading DNA polymerases. EMBO J. 1996;15:1182–1192. [PMC free article] [PubMed] [Google Scholar]
- 23.Blanco L, Lázaro JM, de Vega M, Bonnin A, Salas M. Terminal protein-primed DNA amplification. Proc. Natl Acad. Sci. USA. 1994;91:12198–12202. doi: 10.1073/pnas.91.25.12198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blanco L, Bernad A, Esteban JA, Salas M. DNA-independent deoxynucleotidylation of the ϕ29 terminal protein by the ϕ29 DNA polymerase. J. Biol. Chem. 1992;267:1225–1230. [PubMed] [Google Scholar]
- 25.Garmendia C, Hermoso JM, Salas M. Functional domain for priming activity in the phage phi 29 terminal protein. Gene. 1990;88:73–79. doi: 10.1016/0378-1119(90)90061-u. [DOI] [PubMed] [Google Scholar]
- 26.Dufour E, Rodríguez I, Lázaro JM, de Vega M, Salas M. A conserved insertion in protein-primed DNA polymerases is involved in primer terminus stabilisation. J. Mol. Biol. 2003;331:781–794. doi: 10.1016/s0022-2836(03)00788-5. [DOI] [PubMed] [Google Scholar]
- 27.Illana B, Lázaro JM, Gutiérrez C, Meijer WJ, Blanco L, Salas M. Phage ϕ29 terminal protein residues Asn80 and Tyr82 are recognition elements of the replication origins. J. Biol. Chem. 1999;274:15073–15079. doi: 10.1074/jbc.274.21.15073. [DOI] [PubMed] [Google Scholar]
- 28.King AJ, van der Vliet PC. A precursor terminal protein-trinucleotide intermediate during initiation of adenovirus DNA replication: regeneration of molecular ends in vitro by a jumping back mechanism. EMBO J. 1994;13:5786–5792. doi: 10.1002/j.1460-2075.1994.tb06917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caldentey J, Blanco L, Bamford DH, Salas M. In vitro replication of bacteriophage PRD1 DNA. Characterization of the protein-primed initiation site. Nucleic Acids Res. 1993;21:3725–3730. doi: 10.1093/nar/21.16.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caldentey J, Blanco L, Savilahti H, Bamford DH, Salas M. In vitro replication of bacteriophage PRD1 DNA. Metal activation of protein-primed initiation and DNA elongation. Nucleic Acids Res. 1992;20:3971–3976. doi: 10.1093/nar/20.15.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martín AC, Blanco L, García P, Salas M, Méndez J. In vitro protein-primed initiation of pneumococcal phage Cp-1 DNA replication occurs at the third 3′ nucleotide of the linear template: a stepwise sliding-back mechanism. J. Mol. Biol. 1996;260:369–377. doi: 10.1006/jmbi.1996.0407. [DOI] [PubMed] [Google Scholar]
- 32.Illana B, Blanco L, Salas M. Functional characterization of the genes coding for the terminal protein and DNA polymerase from bacteriophage GA-1. Evidence for a sliding-back mechanism during protein-primed GA-1 DNA replication. J. Mol. Biol. 1996;264:453–464. doi: 10.1006/jmbi.1996.0653. [DOI] [PubMed] [Google Scholar]
- 33.González-Huici V, Lázaro JM, Salas M, Hermoso JM. Specific recognition of parental terminal protein by DNA polymerase for initiation of protein-primed DNA replication. J. Biol. Chem. 2000;275:14678–14683. doi: 10.1074/jbc.m910058199. [DOI] [PubMed] [Google Scholar]
- 34.Serna-Rico A, Illana B, Salas M, Meijer WJ. The putative coiled coil domain of the ϕ29 terminal protein is a major determinant involved in recognition of the origin of replication. J. Biol. Chem. 2000;275: 40529–40538. doi: 10.1074/jbc.M007855200. [DOI] [PubMed] [Google Scholar]
- 35.Illana B, Zaballos A, Blanco L, Salas M. The RGD sequence in phage ϕ29 terminal protein is required for interaction with ϕ29 DNA polymerase. Virol. 1998;248:12–19. doi: 10.1006/viro.1998.9276. [DOI] [PubMed] [Google Scholar]
- 36.Freire R, Serrano M, Salas M, Hermoso JM. Activation of replication origins in ϕ29-related phages requires the recognition of initiation proteins to specific nucleoprotein complexes. J. Biol. Chem. 1996;271:31000–31007. doi: 10.1074/jbc.271.48.31000. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.