Abstract
While RNAi has traditionally relied on RNA duplexes, early evaluation of siRNAs demonstrated activity of the guide strand in the absence of the passenger strand. However, these single strands lacked the activity of duplex RNAs. Here, we report the systematic use of chemical modifications to optimize single-strand RNA (ssRNA)-mediated mRNA knockdown. We identify that 2′F ribose modifications coupled with 5′-end phosphorylation vastly improves ssRNA activity both in vitro and in vivo. The impact of specific chemical modifications on ssRNA activity implies an Ago-mediated mechanism but the hallmark mRNA cleavage sites were not observed which suggests ssRNA may operate through a mechanism beyond conventional Ago2 slicer activity. While currently less potent than duplex siRNAs, with additional chemical optimization and alternative routes of delivery, chemically modified ssRNAs could represent a powerful RNAi platform.
INTRODUCTION
RNA interference is a process inherently driven by RNA duplexes. The double-strand endonuclease Dicer processes long double-stranded RNAs (dsRNA) into small 21–23 nt RNA duplexes containing a 5′ phosphate (1–4). The resulting siRNAs and miRNAs load into the RNA-induced silencing complex (RISC) where the passenger strand (sense) is dissociated, leaving the active guide strand (antisense) loaded in Ago2 and available to base pair with target mRNAs and initiate degradation (5,6). Exogenous delivery of small interfering dsRNAs (siRNAs) to mammalian cells was shown to be an effective strategy to engage this endogenous mechanism of RNA-mediated regulation (7,8). This seminal discovery resulted in siRNA becoming an indispensible tool for studying gene function and now represents a new therapeutic modality (9,10).
Despite the central role of duplex RNAs in triggering RNAi, in the early days of the development of siRNAs there were reports of single-strand RNA (ssRNA) acting to initiate mRNA knockdown. Using extracts from human cell lines, Martinez et al. (11) reported that ssRNA could reconstitute RISC; however, compared to duplex siRNAs, 10- to 100-fold higher concentrations of ssRNA were required. They also transfected single strands and duplexes into cells, thus demonstrating that ssRNA knockdown was optimal with a size >17 nt and improved by the addition of a 5′ phosphate. Again, duplex siRNAs remained more potent. Schwarz et al. (12) evaluated both miRNA and siRNA, demonstrating that ssRNAs corresponding to the let-7 miRNA or luciferase could trigger target cleavage but required a 5′ phosphate. The observation that phosphorylation improves ssRNA activity indirectly argues for Ago-mediated activity. Phosphorylation of the 5′-end of the guide strand plays an important role in guide strand binding within the MID domain of Ago2 and the subsequent positioning of the guide strand for accurate cleavage (13,14).
Single-strand-mediated knockdown was shown to be dose-dependent but was ∼8-fold less effective than the corresponding duplex. Another study found that ssRNA targeting the human blood clotting initiator tissue factor (TF) was as effective in human cells as the duplex siRNA at high concentrations of 100–200 nM (15). In contrast to other reports, exogenous phosphorylation of the ssRNA was not found to improve knockdown activity. In a follow-up study, a dose–response comparison of the same single strand and duplex siRNAs found the dsRNA was 5- to 6-fold more potent than the corresponding ssRNA and analysis by northern blot demonstrated that ssRNA produced mRNA cleavage fragments (16).
Core components of RISC have been shown to bind to ssRNAs. In vitro binding studies with purified human Ago2 demonstrate significantly higher binding affinity for ssRNA which bound 70- to 100-fold tighter than dsRNA (17). Recombinant Dicer has also been shown to bind 21 nt ssRNAs in vitro with preferential affinity for ssRNAs containing a 5′ phosphate (18). This led the authors to hypothesize that Dicer may play a role in ssRNA knockdown by either facilitating loading of ssRNA into Ago2 or by binding to ssRNAs and thus protecting them from nucleases (18).
Chemical modifications have been employed to improve the pharmacokinetic properties of siRNAs including increasing nuclease stability, improving potency, reducing off target effects and abrogating immune stimulation (19–21). Generally, 2′ ribose modifications (2′-methoxy and 2′-fluoro in particular) are broadly tolerated within the guide strand of duplex siRNAs. Holen et al. reported the first evaluation of the impact of chemical modifications on ssRNA activity and compared them to corresponding duplex siRNAs (16). The positional chemical sensitivity seen for duplex siRNAs (15) was mirrored by the ssRNAs which the authors interpret as indicating ssRNA and dsRNA act through a common RNAi pathway (16). This hypothesis was supported by their observation that an excess of dsRNA can competitively block the activity of ssRNAs in vitro (16).
Hall et al. (22) reported that ssRNA potency can be significantly improved by the replacement of some of the phosphodiester backbone with boranophosphate modfications, where a non-bridging oxygen is replaced with an isoelectronic borane. Inclusion of 5–8 boranophosphate modifications allowed ssRNAs to achieve knockdown comparable to duplex siRNAs at ∼10–25 nM concentrations. Importantly, HeLa cells pre-treated with siRNAs targeting Ago2 had significantly reduced target knockdown when they were subsequently transfected with ssRNAs, suggesting that ssRNAs act through Ago2 (22). Despite their beneficial properties, boranophosphate modifications present a number of synthesis and chemistry challenges and have thus far not been widely adopted (21).
Recently, in unpublished work presented at the 7th Annual Oligonucleotide Therapeutics Society meeting (September 2011), Swayze et al. (23) described a modified ssRNA that included phosphorothioate backbone and 5′-end vinyl phosphonate modifications that were intended to improve the stability of the ssRNA molecule. This modification pattern was a refinement of an earlier published pattern of evenly spaced 2′F and 2′OMe modifications reported to improve the activity of duplex siRNAs (24). Swayze et al. (23) also reported that ssRNAs were inactive in Ago2 knockout cells indicating Ago2 mediates ssRNA activity. Importantly, subcutaneously administered unformulated ssRNAs were active in rodents and comparable to the potency of second generation RNase H antisense oligonucleotides.
As discussed above, the limited exploration of chemical modifications has shown promise for improving the activity of ssRNA. Therefore, we conducted a systematic analysis of the impact of 2′ ribose modifications on the activity of both duplex and ssRNA-mediated knockdown—screening six different RNAs to ensure the generality of our observations. Modifications of the 2′ ribose ring included methoxy (2′OMe), fluoro (2′F), and deoxy (2′H), which applied to either purines or pyrimidines present on the guide strand. Additionally, guide strands were synthesized with or without phosphorylation of the 5′-end. In vitro screening in tissue culture cells identified that 2′F content and 5′ phosphorylation are key for optimal ssRNA-mediated knockdown of target mRNAs. Optimized ssRNAs were tested in mice using a lipid nanoparticle delivery vehicle. Knockdown of target mRNA by ssRNA exceeded 90% at the highest dose tested; however, the duplex siRNAs in the same study had greater overall knockdown and much longer duration.
Modification of the guide strand 5′-end with abasic nucleotides negatively impacted the activity of both duplex and ssRNAs, consistent with an earlier report that an abasic nucleotide reduced Ago2 binding affinity and cleavage activity (17). Taken together with the role of 5′ phosphorylation, these observations offer indirect evidence that ssRNAs act through an Ago-mediated mechanism. However, analysis of cleavage products by 5′RACE and sequencing failed to identify distinct cleavage sites for ssRNAs despite the identification of hallmark Ago2 cleavage for duplex RNAs. This suggests ssRNA may act through a mechanism beyond canonical Ago2 slicer activity.
While 2′-fluoro content and 5′ phosphorylation were identified as greatly improving ssRNA activity, our side by side comparison with duplex siRNAs demonstrates that duplex RNAs are consistently more potent than their corresponding single strands both in vitro and in vivo. Importantly, as highlighted by preliminary results from Swayze et al. (23), with further improvements in chemical optimization and evaluation of alternative dosing strategies, such as sub-cutaneous delivery, ssRNA could represent a powerful alternative to duplex siRNAs.
MATERIALS AND METHODS
RNA oligo sequence and synthesis
A systematic screen of 2′ ribose modifications on six different target sites (Table 1) resulted in a total of 180 siRNAs which were then compared as duplex and single strands (360 in total). Individual sequences and modification patterns are listed in Supplementary Tables S3 and S4. Inverted abasic modifications present at the 5′ and 3′ of the passenger strand serve to block loading into Ago2 (25,26). Sequences for siRNAs were selected using a previously published algorithm developed to predict maximal knockdown while minimizing off-target hybridizations (27). siRNAs were synthesized using previously described methods (28,29). Individual oligonucleotide strands were synthesized separately using solid-phase methods, purified by ion-exchange chromatography and duplexed when appropriate. Knockdown studies were normalized using a non-targeting control siRNA sequence which contains ribose 2′OH (r), 2′F (f), 2′OMe (m) and 2′H (d) residues at the indicated positions as well as inverted abasic caps (iB) on the passenger strand: guide (5′–3′) (fC;fC;fU;mG;mA;mA;mG;mA;mG;mA;mG;fU;fU;mA;mA;mA;rA;rG;rA;mU;mU) and passenger (5′–3′) (iB;fU;fC;fU;fU;fU;fU;dA;dA;fC;fU;fC;fU;fC;fU;fU;fC;dA;dG;dG;dT;dT;iB).
Table 1.
Target gene, site, accession and sequence for siRNAs
In vitro mRNA knockdown
Mouse Hepa1–6 cells were cultured in Dulbecco's-modified Eagle Medium supplemented with 10% fetal bovine serum, 1% penicillin–steptomycin and 1% sodium bicarbonate. These cells were plated in a 96-well culture plates at a density of 3000 cells/well 24 h prior to transfection. Transfections were performed using Opti-MEM I Reduced Serum Media and Lipofectamine RNAiMAX as previously described (30). Final siRNA concentrations range from 100 to 1 nM for in vitro cell-based screens with concentrations varying for ssRNA (100 nM, 10 nM) and dsRNA (100 nM, 10 nM, and 1 nM) (see Supplementary Table S3). Final siRNA concentrations for the dose–response curves range from 40 to 0.002 nM along an eight-point, 4-fold titration curve. Twenty-four hours post-transfection cells were washed with phosphate-buffered saline and processed using the TaqMan Gene Expression Cells-to-CT (Invitrogen), per manufacturer's instructions, to extract RNA, synthesize cDNA and perform RT–qPCR using an Ssb (Mm00447374_m1) or ApoB (Mm01545154_g1) specific Taqman primer/probe set on an ABI Prism 7900HT Sequence Detector. Reverse transcription conditions were as follows: 60 min at 37°C followed by 5 min at 95°C. RT–qPCR conditions were as follows: 2 min at 50°C, 10 min at 95°C, followed by 40 cycles of 15 s at 95°C, and 1 min at 60°C. Gapdh mRNA levels were used for data normalization (Taqman part number 4308313). Knockdown of Ssb/ApoB was calculated as the percent knockdown in Ssb/ApoB cDNA measured in experimentally treated cells relative to the Ssb/ApoB cDNA levels measured in non-targeting, control-treated cells. The comparative Ct calculation method for knockdown has previously been described (31). Briefly, ΔCt = CtTarget mRNA − CtGapdh and ΔΔCt = ΔCt(siRNA) − ΔCt(non-targeting control) and relative expression level = 2−ΔΔCt and % knockdown = 100 × (1 − 2−ΔΔCt). Potency (EC50) was calculated using a four-parameter curve fit tool and Prism graphing software (GraphPad Software).
Lipid nanoparticle formulation
siRNA lipid nanoparticles (LNPs) were assembled by simultaneous mixing of a lipid mixture in ethanol with an aqueous solution of siRNA followed by diafiltration as previously described (30,32). The cationic lipid CLinDMA (2-{4-[(3b)-cholest-5-en-3-yloxy]-butoxy}-N,N-dimethyl-3-[(9Z,12Z)-octadeca-9,12-dien-1-yloxy]propan-1-amine), cholesterol and PEG-DMG (monomethoxy(polyethyleneglycol)-1,2-dimyristoylglycerol) were mixed together at a molar ratio of 50:44:6. PEG-DMG was purchased from NOF Corporation, cholesterol from Northern Lipids, and CLinDMA was synthesized by Merck & Co., Inc. Particle size was measured by dynamic light scattering using a Wyatt DynaPro plate reader and percent encapsulation was determined using a SYBR Gold fluorescence assay (Invitrogen) and were within pre-established quality metrics (32).
In vivo mRNA knockdown
All in vivo work was approved by an Institutional Animal Care and Use Committee and adhered to standards recommended by the Association for Assessment and Accreditation of Laboratory Animal Care, International. C57/BL6 mice (Jackson Labs) were dosed by intravenous injection with siRNA formulated in lipid nanoparticle at 3 or 6 mg/kg as previously described (30,33). Animals were euthanized by CO2 inhalation and immediately after euthanasia liver sections were excised, placed in RNA Later (Qiagen) and stored at 4°C until ready for analysis. Liver tissue was homogenized in Qiazol using stainless steel beads and a TissueLyser (Qiagen). Following homogenization, chloroform was added and samples were centrifuged. The aqueous layer was combined with an equal volume of 70% ethanol and samples were purified using an RNeasy purification kit per manufacturer's directions (Qiagen). The resulting RNA was then normalized, cDNA was synthesized, and RT-qPCR was performed using ApoB specific Taqman primer/probe sets on an ABI Prism 7900HT Sequence Detector. Reverse transcription conditions were as follows: 60 minutes at 37°C followed by 5 min at 95°C. RT–qPCR conditions were as follows: 2 min at 50°C, 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. Gapdh mRNA levels were used for data normalization. Knockdown of ApoB was calculated using the same method described for the in vitro experiments.
5′-RACE and sequencing of cleavage products
5′-RLM-RACE was performed using GeneRacer kit (Invitrogen) following the manufacturer’s instructions. Briefly, 100 ng untreated RNA was directly ligated to the RNA linker. After phenol/chloroform extraction and precipitation, the first-strand cDNA was synthesized using the Oligo-dT primer. For the first-round 5′RACE reaction, 1 µl of the first-strand cDNA was amplified using the GeneRacer 5′ primer and a target gene specific primer (Supplementary Table S5) with cycling as following: 1 cycle of 94°C for 2 min, then 5 cycles of 94°C for 30 s and 72°C for 1 min, and then 5 cycles of 94°C for 30 s, and 70°C for 1 min, followed by 20 cycles of 94°C for 30 s, 65°C for 30 s and 68°C for 1 min. After additional 10-min incubation at 68°C and cool down to 4°C, µl of the first PCR reaction was used for the second round 5′RACE (Nested PCR) using the GeneRacer 5′ Nested primer and gene-specific nested primer with the 1 cycle at 94°C for 2 min, and 25 cycles of 94°C for 30 s, 65°C for 30 s, and 68°C for 1 min. From this reaction, 10 µl was analyzed on a 2% agarose gel. See Supplementary Table S5 for sequence and other information for the primers used. To confirm the cleavage site of target mRNA, the gel bands corresponding to the size of the expected 5′RACE amplicon was excised and purified using QIAquick Gel Extraction kit (QIAGEN). The purified PCR products were sequenced by GENEWIZ (South Plainfield, NJ, USA).
RESULTS
Selection of siRNAs and modification patterns
ApoB has been a frequent target of siRNA knockdown because of its potential therapeutic value in lowering serum cholesterol. Furthermore, given its predominant expression in liver hepatocytes, ApoB transcripts can be readily targeted in vivo using currently available siRNA delivery technologies (34–36). The Ssb gene encodes the La antigen which is involved in transfer RNA maturation (37) and has been implicated in the autoimmune disease Sjogren's syndrome (38). Ssb was selected primarily for its ubiquitous cellular expression pattern. Four siRNAs targeting ApoB and two siRNAs targeting SSB were chosen to evaluate the generality of our findings across multiple siRNAs (Table 1).
Various combinations of ribose (2′OH), 2′-deoxy (2′H), 2′-fluoro (2′F), and 2′-methoxy (2′OMe) modifications were assigned based on the pyrimidine (Y) or purine (R) identity of the siRNA sequence (Table 2). Methoxy modification of position 14 of the guide strand was not permitted due to reported sensitivity of this position to modifications (17). These combinations were also evaluated in the presence or absence of 5′ phosphorylation of the guide strand (5′PO4). Placement of abasic residues at the 5′ of the guide strand have been reported to reduce Ago2 binding and cleavage activity (17). Therefore, as a negative control, a separate set of oligos were synthesized with three abasic residues (i.e. ribose sugar without nucleotide base) at positions 1–3. The chemical structures of the 2′ modifications are shown with adenosine as the representative nucleotide. All together, 30 different siRNA modification patterns were evaluated for each of the ApoB or SSB target sites (a total of 180 unique guide strands). Each of the different guide strands was evaluated as single strands or as duplexes with a passenger strand to create a conventional siRNA duplex. Passenger strands are largely ribonucleotides with 2′OMe uridine overhangs and inverted abasic residues on each end. Inverted abasics block the loading of these strands into Ago2, thus preventing passenger strand competition with the guide strand (25,26). Sequence information for each guide strand and passenger strand are provided in Supplementary Tables S3 and S4. For in vitro studies, all knockdown data were normalized to a chemically modified non-specific siRNA duplex which was designed to lack homology to nearly all mammalian transcripts (see ‘Materials and Methods’ section).
Table 2.
Summary of modification strategy used to evaluate structure–activity relationship for single strand and duplex siRNAs
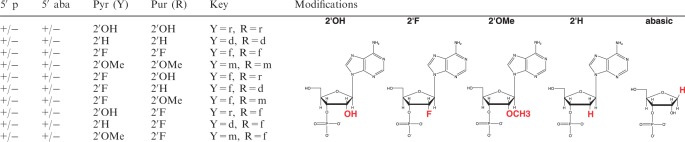 |
Oligos were prepared with or without 5′ phosphate (5′p). Additionally, oligos containing a 5′PO4 were tested and with or without three abasic residues at the 5′ end of the guide strand (5′aba). Unmodified ribose (2′OH) is compared to 2′-fluoro (2′F), 2′-deoxy (2′H), and 2′-methoxy (2′OMe) modifications assigned to pyrimidines (Pyr) or purines (Pur). Key column lists nomenclature used in subsequent figures. Structures are shown with adenosine as the representative nucleotide. A total of 30 different siRNAs were designed for each target site. Sequences and modification patterns are listed in Supplementary Table S3.
2′F modifications are critical for the activity of single strand RNA in vitro
The mouse hepatocyte derived cell line Hepa1-6 was transfected with single strand and duplex siRNAs at 100 and 10 nM concentrations. For dsRNAs, 10 nM is likely a saturating concentration as 100 nM did not result in significant improvements in observed target knockdown. Therefore, two siRNAs were also screened at 1 nM. Target knockdown was measured 24 h later and compared to unmodified controls. Figure 1 shows the results for ApoB (8786), though a similar trend was observed for the other five siRNAs tested (Supplementary Figure S1). Generally, dsRNAs were broadly tolerant of modifications though there was notable sensitivity to 2′-deoxy which is consistent with Ago2 preference for RNA (rather than DNA) substrates (13,14). High percentage methoxy modification of the guide strand was tolerated within dsRNAs though a loss in knockdown was seen at the submaximal 1 nM concentration. Such tolerance of methoxy modification is likely due to our inclusion of 2′OH at position 14 of the guide strand which has previously been reported as highly sensitive to methoxy modifications (17).
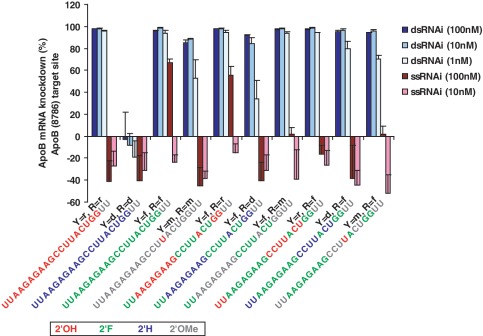
Figure 1.
Comparison of ssRNA and dsRNA oligos containing pyrimidine (Y) and purine (R) 2' modifications. ApoB (8786) sequence shown. In vitro cell-based evaluation of ApoB mRNA knockdown (error bars represent standard deviation of four replicates). Unmodified ribose (‘r’) is compared to 2'-fluoro (‘f’), 2'-methoxy (‘m’), and 2'-deoxy (‘d’) in all combinations on purines (R) and pyrimidines (Y). Oligos do not contain a 5' phosphate. Guide strand oligo sequence and color-coded modification patterns are depicted. Five other siRNAs were tested and a similar requirement of 2'F for ssRNA knockdown was observed (Supplementary Figure S1).
Strikingly, single strands containing 2′F content exhibit the most pronounced mRNA knockdown of the various single-strand modification patterns tested with 67% knockdown compared to the negligible activity seen for unmodified (Figure 1, Supplementary Table S3). Five other siRNA sequences were also evaluated and a similar 2′F dependence for ssRNA activity was observed, though the magnitude of response was variable (Supplementary Figure S1). This variance is likely attributable to the fact that these RNA oligos were not phosphorylated, and there is precedent in the literature that 5′ phosphorylation of the guide strand is key for ssRNA knockdown (11,12).
5′ phosphorylation improves the potency of ssRNA
The same series of RNA modifications evaluated in Figure 1 were then tested in the presence of 5′ phosphorylation of the guide strand. Figure 2 shows that addition of a 5′ phosphate significantly improves the potency of ssRNA, resulting in significant knockdown even at a lower concentration of 10 nM. The overall effectiveness of 2′F-modified ssRNA activity for the other five siRNAs is also more evident when the ssRNAs are phosphorylated (Supplementary Figure S2). In the presence of 5′ phosphorylation, the beneficial impact of 2′F content is significantly more obvious (compare to Supplementary Figure S1). While the ssRNA knockdown at this lower concentration still lags that of the comparable duplex siRNAs, the combination of 2′F modifications and 5′ phosphorylation are key to improving the potency of ssRNA knockdown: improving knockdown from negligible levels to 65–95% (Supplementary Table S3).
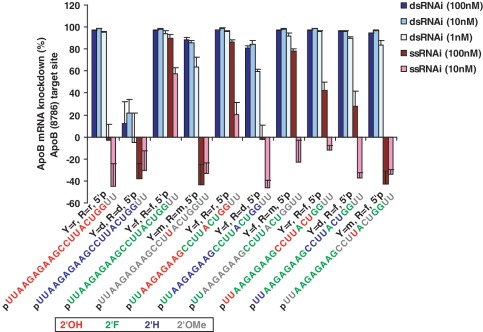
Figure 2.
Comparison of purine versus pyrimidine 2′ modifications for ApoB (8786) in the presence of 5′ phosphorylation of the guide strand. In vitro cell-based evaluation of ApoB mRNA knockdown (error bars represent standard deviation of four replicates). Unmodified ribose (‘r’) is compared to 2′-fluoro (‘f’), 2′-methoxy (‘m’), and 2′-deoxy (‘d’) in all combinations on purines (R) and pyrimidines (Y). All RNA oligos contain 5′ phosphate (5′p). Guide strand oligo sequence and color-coded modification patterns are depicted. Five other siRNAs with were also tested with 5′ phosphorylation (Supplementary Figure S2) and a similar requirement for 5′ phosphorylation for optimal in vitro ssRNA knockdown was observed (compare to Supplementary Figure S1).
Pyrimidines modified with 2′-fluoro appear to confer improved knockdown to ssRNAs than similarly modified purines. It does not seem that overall 2′F content of the RNA oligo can explain this difference as the ApoB siRNAs are either approximately evenly split between purine and pyrimidine content or are purine rich (58% purine). The Ssb sequences are predominantly pyrimidine rich (58–74%). The underlying mechanism for 2′F pyrimidine preference for ssRNA silencing activity remains unclear. Overall, phosphorylated ssRNAs containing 2′F modifications of both purines and pyrimidines exhibited the best knockdown activity in vitro.
ssRNA potency significantly underperforms duplex siRNAs
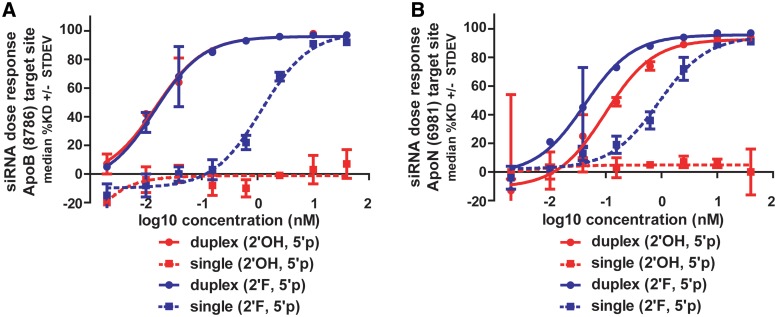
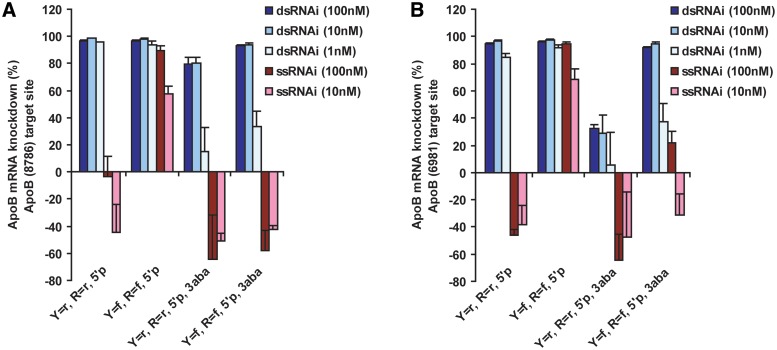
At the two concentrations tested, ssRNAs appear less potent than their corresponding duplex siRNAs. Two ApoB siRNAs were selected for dose response measurements and calculation of EC50 values (Figure 3). Unmodified and 2′F-modified RNAs were compared as single strands and duplexes over an eight-point dose–response ranging from 40 nM down to 0.002 nM (all oligos were phosphorylated). The value of 2′F modifications for ssRNA activity was immediately evident for both ApoB (8786) and (6981) sequences: unmodified ssRNAs essentially had no knockdown over the dose range while 2′F ssRNA exhibited a dose response. However, the duplex siRNAs were found to be 20- to 80-fold more potent than their corresponding single strands with picomolar potencies for dsRNAs compared to the nanomolar potencies measured for ssRNAs (Supplementary Table S1). ApoB (8786) duplexes possess 15 pM potencies (for both unmodified and 2′F) while the 2′F ssRNA EC50 is ∼1.2 nM. The ApoB (6981) 2′F duplex is ∼2-fold more potent than the unmodified duplex (2′OH); however, both are significantly more potent than the 2′F ssRNA (∼0.9 nM). Despite the lower overall potencies relative to duplexes, it is important to note that ssRNAs are still capable of maximal mRNA knockdown >90% and that this level of knockdown was enabled by chemical modifications.
Figure 3.
In vitro potency of single strands compared to duplex siRNAs. (A) ApoB (8786) and (B) ApoB (6981) compared as 2′OH and 2′F guide strands with 5′ phosphorylation. Single strands are ∼20- to 80-fold less potent than the corresponding duplex siRNAs containing identical guide strands. Potency and mRNA knockdown values listed in Supplementary Table S1.
ssRNA knockdown in vivo
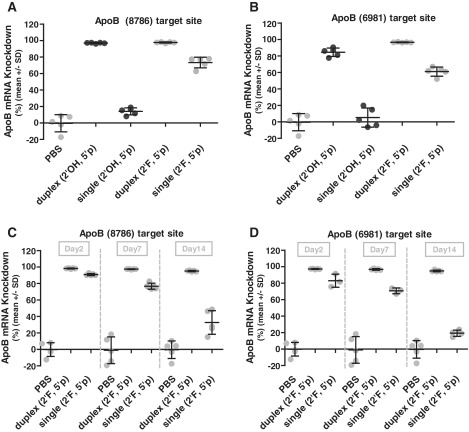
We extended our comparison of ssRNA and dsRNA into mice to identify whether ssRNAs were competent for knockdown in vivo. Single strand and duplex RNAs were formulated in lipid nanoparticles (LNP) which permitted side by side comparison of these RNAs while using a well-established delivery vehicle with proven capabilities (30,32). Since the nanoparticle encapsulates the siRNAs, it is likely shielded from serum nucleases which eliminated an additional round of siRNA chemical optimization for serum stability. The same ApoB siRNAs assayed for cell-based potency (Figure 3) were formulated in LNPs and intravenously dosed at 3 mg/kg into mice. Three days later the mice were sacrificed and ApoB mRNA knockdown was measured from harvested livers. Compared to buffer control, the duplex siRNAs reduced ApoB expression by 85–98% (Figure 4A and B and Supplementary Table S2). Unmodified ssRNAs had no significant knockdown which was consistent with in vitro observations. However 2′F-modified single strands were quite effective, reducing ApoB expression levels by 61–74% and demonstrating that ssRNA functions in vivo.
Figure 4.
In vivo knockdown of single strand versus duplex siRNA demonstrates ssRNAi activity in vivo. RNAs formulated in a lipid nanoparticle (LNP) delivery vehicle. Modified guide strands (2'F) are compared to unmodified (2'OH). All oligos contain 5' phosphorylation. Comparison of Day 3 ApoB mRNA knockdown after an LNP delivered 3 mg/kg dose of dsRNA or ssRNA for target sites (A) ApoB (8786) and (B) ApoB (6981). Knockdown of 60–75% was observed for phosphorylated single strands containing 2'F modifications. Duration of mRNA knockdown after an LNP delivered 6 mg/kg dose was measured over 2 weeks (Day 2, Day 7, Day 14 time points) for (C) ApoB (8786) and (D) ApoB (6981). Knockdown values shown in Supplementary Table S2.
To evaluate duration of knockdown, a separate in vivo study was then conducted with the 2′F-modified ssRNA and dsRNA by using a higher dose (6 mg/kg) and conducting a 2-week time-course. Groups of mice were sacrificed at Days 2, 7, and 14, and ApoB mRNA knockdown was measured (Figure 4C and D). At the higher dose, ssRNA knockdown increased to nearly 90% at Day 2, although the effect was transient—knockdown reduced to 71–77% at 7 days and returned to baseline after 14 days. In marked contrast, duplex siRNAs still had 95% knockdown two weeks after dosing. In summary, chemical modifications permit effective ssRNA knockdown in vivo and nearly ablate ApoB expression with an impressive 84–91% knockdown. However, maximal ssRNA knockdown is transient and duration activity is exceeded by duplex siRNAs.
Indirect evidence suggests ssRNA acts through an Ago-mediated mechanism
ssRNA is hypothesized to act through Ago-mediated mechanisms similar to duplex siRNAs (see ‘Introduction’ section). To address this, we tested a series of modified siRNAs where positions 1–3 of the 5′-end of the guide strand have been replaced with abasic residues. Abasic incorporation has previously been reported to adversely affect Ago2 binding and cleavage activity (17). Both double-strand siRNA and ssRNA are sensitive to the incorporation of abasic residues which dramatically reduce the potency of knockdown for two ApoB siRNAs when compared to RNA oligos without abasics (Figure 5A and B). Similar results are seen for all six siRNAs tested in this study across multiple modification patterns (Supplementary Figure S3). Duplex and ssRNAs also show a similar sensitivity to 2′-deoxy incorporation (Figure 1 and Supplementary Figure S1). Previously, similar types of chemical modification sensitivity have been used to argue that ssRNA and dsRNA act through a common pathway (16).
Figure 5.
Inclusion of three abasic residues at positions 1–3 (3aba) of the guide strand decreases the in vitro mRNA knockdown of both duplex and single strands of ApoB siRNAs. Ribose (2'OH, ‘r’) and fluoro (2'F, ‘f’) oligos with phosphate (5'p) are compared to those containing 3aba for (A) ApoB (8786) and (B) ApoB (6981). Abasic (3aba) data for additional ApoB and SSB siRNAs are shown in Supplementary Figure S3.
5′RACE identifies only dsRNA cleavage products
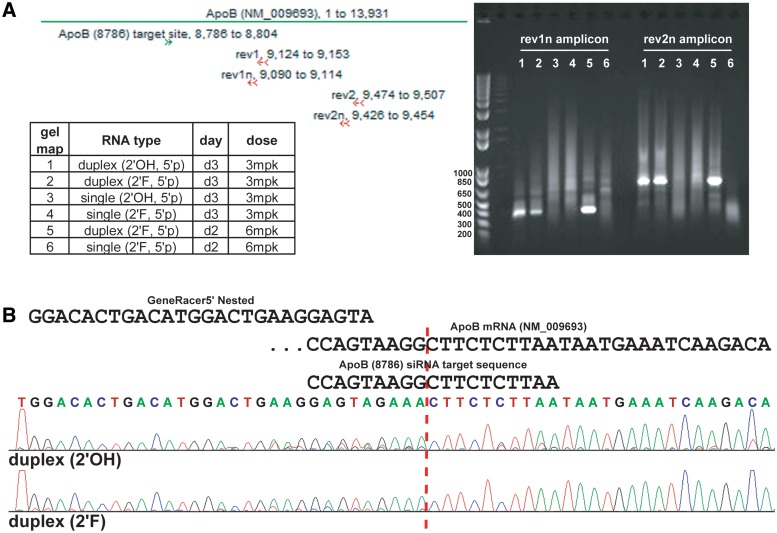
Conventional Ago2-mediated cleavage of target mRNA occurs between positions 9–10 of the region hybridizing to the guide strand. The PCR-based rapid amplification of cDNA ends (RACE) methodology is routinely applied to measure the 5′-end of the predicted siRNA cleavage product. PCR primers are designed to produce an amplicon of predicted size based on the site of siRNA cleavage of the target mRNA. Two primers are ‘nested’ together in subsequent PCR amplifications to confer specificity to the 5′RACE. Two separate PCR amplicons were designed (Supplementary Table S5) and used for 5′RACE from mouse liver mRNA isolated from animals treated with ApoB (8786) dsRNA and ssRNA (Figure 4). The rev1n amplicon should be 319 nt and the rev2n amplicon should be 659 nt in size based on the predicted site for ApoB (8786) cleavage. Both amplicons produce distinct bands of expected size for samples treated with the duplex siRNAs (Figure 6A). However, ssRNA treated samples did not produce a cleavage product despite yielding 74–91% ApoB knockdown in vivo. Since the ssRNA and dsRNA samples were amplified side by side, we are confident that the failure to isolate ssRNA cleavage products is not a technical artifact. Purification and sequencing of a subset of the rev1n amplicons from the duplex siRNA samples confirmed the predicted Ago2 cleavage site and the presence of the 5′RACE primer (Figure 6B).
Figure 6.
5'RACE demonstrates duplex RNA, but not ssRNA, cleaves ApoB mRNA producing a cleavage product size consistent with the ApoB (8786) cleavage site. (A) Agarose gel of two different 5' RACE products (rev1n and rev2n amplicons; see map of primers) from mRNA purified from mouse livers treated with duplex or ssRNA (Figure 4). ApoB (8786) cleavage compared from in vivo mouse livers treated with unmodified (2'OH) or 2'F-modified duplex and single strands. (B) Representative sequence data from duplex siRNA cleavage products showing cleavage at the hallmark site indicative of Ago2 slicer activity and the presence of 5' RACE primer.
DISCUSSION
Chemical modifications can improve the performance of ssRNA-mediated knockdown of target mRNAs in vitro and in vivo. In particular, 2′F ribose modification and inclusion of 5′ phosphorylation are required for optimal ssRNA activity (Figures 1 and 2). However, traditional duplex siRNAs containing identical guide strands are significantly more potent in vitro (Figure 3). Single strand RNA was compared to siRNA in vivo with lipid nanoparticle formulated materials dosed intravenously to mice. While duplex RNAs were consistently more potent, ssRNA did knockdown ApoB mRNA by up to 90% at a 6mpk dose (Figure 4). This level of ssRNA knockdown was dependent on chemical modifications, as unmodified ssRNAs possessed no significant activity in vivo despite that the same guide strand was performing well in the context of a duplex siRNA. Overall, while still less potent than duplexes, we demonstrate that chemically optimized ssRNAs are capable of robust mRNA knockdown in vivo. When coupled with additional chemical modifications and alternative dosing regiments, ssRNA could present significant advantages over traditional duplex siRNA (discussed below).
Multiple lines of indirect evidence suggest that ssRNAs engage an Ago-mediated mechanism for mRNA knockdown. First, 5′ phosphorylation is a requirement for optimal ssRNA-mediated mRNA knockdown—an observation consistent across all six siRNAs tested (Figures 1, 2, and Supplementary Figures S1, and S2). The role of 5′ phosphate in ssRNA activity is suggestive of an Ago-mediated mechanism given the well-characterized phosphate binding pocket within the MID domain of Ago2 (13,14). In vitro cleavage assays conducted with purified human Ago2 demonstrated that phosphorylated ssRNAs possessed significantly higher cleavage rates than corresponding ssRNAs without a phosphate (17). A single abasic at position 1 reduced the binding affinity and cleavage activity of Ago2 (17) and we found substituting positions 1–3 of the guide strand with abasic residues was deleterious to the activity of both duplex siRNAs and ssRNAs (Figures 5 and S3). Lastly, the length dependence of ssRNA requiring greater than 17 nt for optimal activity (11) is also similar to the well-characterized Ago2 preferences for the size of the RNA guide strand (39,40).
Fully modified 2′F RNA ssRNA does not constitute an RNase H substrate (41,42) and together with the impact of abasics on ssRNA activity argue against a RNase H cleavage role. Inclusion of three abasic residues at the 5′ of the antisense strand negatively impacts ssRNA-mediated knockdown despite 16 nt of intact bases which is a strong, but indirect, argument against the involvement of RNase H. Also, the antisense RNase H-mediated mechanism does not require 5′ phosphorylation for maximal activity (41). Lastly, while 2′-deoxy is the natural substrate for RNase H, 2′-deoxy modifications were deleterious for ssRNA (Figure 2) which argues against an RNase H mechanism being central to ssRNA activity. However, we cannot exclude the possibility that ssRNA acts through multiple pathways employing both RNase H and Ago-mediated mechanisms.
Despite the evidence for Ago involvement in ssRNA-mediated knockdown, we were unable to detect cleavage sites that are hallmarks of Ago2 slicer activity. Corresponding duplex siRNAs yielded robust cleavage products with two separate PCR amplicons, and sequence data support cleavage that occurs at the expected site (Figure 6). The detection of Ago2 cleavage products with duplex siRNAs argues that the failure to detect similar products with ssRNA is not a technical artifact but rather indicative of the absence of conventional Ago2 cleavage for the ssRNA tested. In vitro studies with Ago2 highlighted an RNase H activity (17), so it is intriguing to speculate that 2′F ssRNAs could be participating in an Ago2-mediated cleavage reaction beyond the hallmark cleavage of the nucleotide bond opposite positions 10–11 of the guide strand (although there is no evidence suggesting that DNA guide strands lead to alternative cleavage sites). Alternatively, ssRNAs may be complexing with other Ago family members which do not possess endonucleolytic cleavage activity but still can mediate knockdown by reducing the stability of the target mRNA (43). In fact, catalytically inactive mutant Ago2 has been shown to still maintain silencing activity (44). Taken together with the observation that siRNA-mediated target mRNA knockdown retains significant potency in the presence of 80% reduction of Ago2 by antisense oligonucleotides (45), multiple lines of evidence suggest the presence of a ‘slicer-independent’ mechanism in which ssRNA could be taking part.
The particular mechanism of how 2′F content improves the performance of ssRNA remains unclear. Incorporation of 2′F modifications into siRNAs is broadly tolerated and may confer beneficial potency and nuclease stability effects (24,28,46–49). Allerson et al. (24) identified an siRNA where the combination of 2′F and 2′OMe modifications resulted in greater than 500-fold improvement in potency relative to the unmodified siRNA. Manoharan et al. (49) found that 2′F incorporation improved siRNA potency approximately 2-fold in vivo. For the siRNAs tested in this study, 2′F content does not have a pronounced impact on the potency of the duplex siRNAs. Addition of 2′F to ApoB (6981) improves potency ∼2-fold (Figure 3), and there is a corresponding improvement in vivo (97% knockdown versus 85% for unmodified). However, there is no beneficial effect of 2′F incorporation in duplex ApoB (8786) either in vitro or in vivo (Figures 3 and 4). Additionally, 2′F content does not impact the in vitro knockdown of the four other duplexes tested (Supplementary Table S3). Therefore, the 2′F role appears specific to ssRNA, suggesting that 2′F content contributes to a process prior to Ago2 cleavage (e.g. RNA loading into RISC).
2′F pyrimidine modifications seem more favorable than corresponding purine modifications—even in sequences where pyrimidines are in the minority (Figure 2 and Supplementary Figure S2). This suggests that the role of 2′F in ssRNA is not simply a component of total 2′F content in the RNA molecule. Similar preference for 2′F pyrimidines was observed in a related study evaluating the activity of single stranded microRNA mimetics (Guillaume Chorn, Molly Klein-McDowell, Lihong Zhao, Matthew Saunders, Michael Flanagan, Aarron Willingham, Lee Lim, manuscript in preparation). In that study, positions 10 through 19 of miR-124 were sequence randomized while keeping the seed region constant and the single strand miR-124 oligos with the highest activity had elevated pyrimidine content. Enthalpy-driven enhanced base pairing affinity has been reported for 2′F-modified RNA (50); however, this would be expected to improve both duplex and ssRNA performance which was not broadly seen in this study.
Additional nuclease stability conferred by chemical modifications may contribute to the activity of ssRNA. However, 2′H and 2′OMe modifications would be expected to confer similar beneficial properties to ssRNA if stability were the sole rate-limiting factor. 2′OMe modifications are somewhat less tolerated in siRNAs, so the role of 2′F in ssRNA could be a combination of stability and broader tolerance. Overall the number of fully modified RNA oligos which are functional as dsRNA and not as ssRNA suggests that the 2′F improvement of ssRNA activity likely relies on properties beyond nuclease stability.
Further optimization and development of ssRNA with increased potency may result in a differentiating technology. Eliminating the passenger strand reduces synthesis cost by half and also halves the charge of the molecule which may aid in tissue biodistribution and cellular permeability. Intravenous or subcutaneous delivery of unformulated ssRNAs could benefit from the same pharmokinetic and toxicology properties of traditional antisense technology which itself has been successfully applied in late stage clinical trials in humans. Lastly, ssRNAs would not be susceptible to competition from non-specific double-strand binding proteins such as adenosine deaminases (ADAR) which have been reported to interfere with siRNA knockdown in specific tissues (51,52). The unpublished work of Swayze et al. (23) on the in vivo performance of an optimized ssRNA molecule is quite encouraging and highlights the potential value of ‘next generation’ ssRNA. However, it remains to be seen whether ssRNAs can confer beneficial drug-like properties beyond those currently available with either antisense oligonucleotides or duplex siRNAs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables S1–S5 and Supplementary Figures S1–S3.
FUNDING
Merck & Co., Inc. which employed the authors within the RNA Therapeutics department. Funding for open access charge: Merck Research Laboratories.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Lee Lim for invaluable collaboration and sharing early data on single stranded micro-RNA mimetics, Natalya Dubinina, Elmer Payson, Alex Freeman, Jing Kang, and Nelly Kuklin for their in vivo expertise, Enrique Vazquez, Mark Levorse, and Becky Arvary for their synthesis of oligonucleotides, Dipali Ruhela, Silvia Chang, Zhi Yu Hu, Radu Mihalia, and Alan Wei for their role in lipid nanoparticle formulation, and Jeremy Caldwell, Steve Colleti, and Peter Haeberli for valuable discussion and feedback.
REFERENCES
- 1.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 2.Billy E, Brondani V, Zhang H, Muller U, Filipowicz W. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc. Natl Acad. Sci. USA. 2001;98:14428–14433. doi: 10.1073/pnas.261562698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 5.Filipowicz W. RNAi: the nuts and bolts of the RISC machine. Cell. 2005;122:17–20. doi: 10.1016/j.cell.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 6.Ketting RF. The many faces of RNAi. Dev. Cell. 2011;20:148–161. doi: 10.1016/j.devcel.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 7.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 8.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davidson BL, McCray PB., Jr Current prospects for RNA interference-based therapies. Nat. Rev. 2011;12:329–340. doi: 10.1038/nrg2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dykxhoorn DM, Lieberman J. The silent revolution: RNA interference as basic biology, research tool, and therapeutic. Ann. Rev. Med. 2005;56:401–423. doi: 10.1146/annurev.med.56.082103.104606. [DOI] [PubMed] [Google Scholar]
- 11.Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–574. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 12.Schwarz DS, Hutvagner G, Haley B, Zamore PD. Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol. Cell. 2002;10:537–548. doi: 10.1016/s1097-2765(02)00651-2. [DOI] [PubMed] [Google Scholar]
- 13.Cenik ES, Zamore PD. Argonaute proteins. Curr. Biol. 2011;21:R446–R449. doi: 10.1016/j.cub.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 14.Joshua-Tor L, Hannon GJ. Ancestral roles of small RNAs: An ago-centric perspective. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a003772. doi:10.1101/cshperspect.a003772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amarzguioui M, Holen T, Babaie E, Prydz H. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res. 2003;31:589–595. doi: 10.1093/nar/gkg147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holen T, Amarzguioui M, Babaie E, Prydz H. Similar behaviour of single-strand and double-strand siRNAs suggests they act through a common RNAi pathway. Nucleic Acids Res. 2003;31:2401–2407. doi: 10.1093/nar/gkg338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lima WF, Wu H, Nichols JG, Sun H, Murray HM, Crooke ST. Binding and cleavage specificities of human Argonaute2. J. Biol. Chem. 2009;284:26017–26028. doi: 10.1074/jbc.M109.010835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kini HK, Walton SP. In vitro binding of single-stranded RNA by human Dicer. FEBS Lett. 2007;581:5611–5616. doi: 10.1016/j.febslet.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bramsen JB, Kjems J. Chemical modification of small interfering RNA. Method. Mol. Biol. 2011;721:77–103. doi: 10.1007/978-1-61779-037-9_5. [DOI] [PubMed] [Google Scholar]
- 20.Watts JK, Deleavey GF, Damha MJ. Chemically modified siRNA: tools and applications. Drug Discov. Today. 2008;13:842–855. doi: 10.1016/j.drudis.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 21.Behlke MA. Chemical modification of siRNAs for in vivo use. Oligonucleotides. 2008;18:305–319. doi: 10.1089/oli.2008.0164. [DOI] [PubMed] [Google Scholar]
- 22.Hall AH, Wan J, Spesock A, Sergueeva Z, Shaw BR, Alexander KA. High potency silencing by single-stranded boranophosphate siRNA. Nucleic Acids Res. 2006;34:2773–2781. doi: 10.1093/nar/gkl339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swayze EE, Prakash TP, Lima W, Crooke S. 2011. Activation of RNA interference in animals with single stranded oligonucleotides. In 7th Annual Meeting of the Oligonucleotide Therapeutics Society, September 8–10, 2011, Copenhagen, Denmark. [Google Scholar]
- 24.Allerson CR, Sioufi N, Jarres R, Prakash TP, Naik N, Berdeja A, Wanders L, Griffey RH, Swayze EE, Bhat B. Fully 2′-modified oligonucleotide duplexes with improved in vitro potency and stability compared to unmodified small interfering RNA. J. Med. Chem. 2005;48:901–904. doi: 10.1021/jm049167j. [DOI] [PubMed] [Google Scholar]
- 25.Campochiaro PA. Potential applications for RNAi to probe pathogenesis and develop new treatments for ocular disorders. Gene Ther. 2006;13:559–562. doi: 10.1038/sj.gt.3302653. [DOI] [PubMed] [Google Scholar]
- 26.Pei Y, Hancock PJ, Zhang H, Bartz R, Cherrin C, Innocent N, Pomerantz CJ, Seitzer J, Koser ML, Abrams MT, et al. Quantitative evaluation of siRNA delivery in vivo. RNA. 2010;16:2553–2563. doi: 10.1261/rna.2255810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 28.Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W, Hartsough K, Machemer L, Radka S, Jadhav V, et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat. Biotechnol. 2005;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- 29.Wincott F, DiRenzo A, Shaffer C, Grimm S, Tracz D, Workman C, Sweedler D, Gonzalez C, Scaringe S, Usman N. Synthesis, deprotection, analysis and purification of RNA and ribozymes. Nucleic Acids Res. 1995;23:2677–2684. doi: 10.1093/nar/23.14.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kenski DM, Cooper AJ, Li JJ, Willingham AT, Haringsma HJ, Young TA, Kuklin NA, Jones JJ, Cancilla MT, McMasters DR, et al. Analysis of acyclic nucleoside modifications in siRNAs finds sensitivity at position 1 that is restored by 5′-terminal phosphorylation both in vitro and in vivo. Nucleic Acids Res. 2010;38:660–671. doi: 10.1093/nar/gkp913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peacock H, Fucini RV, Jayalath P, Ibarra-Soza JM, Haringsma HJ, Flanagan WM, Willingham A, Beal PA. Nucleobase and ribose modifications control immunostimulation by a microRNA-122-mimetic RNA. J. Am. Chem. Soc. 2011;133:9200–9203. doi: 10.1021/ja202492e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abrams MT, Koser ML, Seitzer J, Williams SC, DiPietro MA, Wang W, Shaw AW, Mao X, Jadhav V, Davide JP, et al. Evaluation of efficacy, biodistribution, and inflammation for a potent siRNA nanoparticle: effect of dexamethasone co-treatment. Mol. Ther. 2010;18:171–180. doi: 10.1038/mt.2009.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ason B, Castro-Perez J, Tep S, Stefanni A, Tadin-Strapps M, Roddy T, Hankemeier T, Hubbard B, Sachs AB, Michael Flanagan W, et al. ApoB siRNA-induced liver steatosis is resistant to clearance by the loss of Fatty Acid Transport Protein 5 (Fatp5) Lipids. 2011;46:991–1003. doi: 10.1007/s11745-011-3596-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Judge AD, Bola G, Lee AC, MacLachlan I. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol. Ther. 2006;13:494–505. doi: 10.1016/j.ymthe.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 35.Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 36.Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, Harborth J, Heyes JA, Jeffs LB, John M, et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 37.Ben-Chetrit E. Target antigens of the SSA/Ro and SSB/La system. Am. J. Reprod. Immunol. 1992;28:256–258. doi: 10.1111/j.1600-0897.1992.tb00809.x. [DOI] [PubMed] [Google Scholar]
- 38.Mavragani CP, Tzioufas AG, Moutsopoulos HM. Sjogren's syndrome: autoantibodies to cellular antigens. Clinical and molecular aspects. Int. Arch. Allergy Immunol. 2000;123:46–57. doi: 10.1159/000024423. [DOI] [PubMed] [Google Scholar]
- 39.Czauderna F, Fechtner M, Dames S, Aygun H, Klippel A, Pronk GJ, Giese K, Kaufmann J. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 2003;31:2705–2716. doi: 10.1093/nar/gkg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sipa K, Sochacka E, Kazmierczak-Baranska J, Maszewska M, Janicka M, Nowak G, Nawrot B. Effect of base modifications on structure, thermodynamic stability, and gene silencing activity of short interfering RNA. RNA. 2007;13:1301–1316. doi: 10.1261/rna.538907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cerritelli SM, Crouch RJ. Ribonuclease H: the enzymes in eukaryotes. FEBS J. 2009;276:1494–1505. doi: 10.1111/j.1742-4658.2009.06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lima WF, Nichols JG, Wu H, Prakash TP, Migawa MT, Wyrzykiewicz TK, Bhat B, Crooke ST. Structural requirements at the catalytic site of the heteroduplex substrate for human RNase H1 catalysis. J. Biol. Chem. 2004;279:36317–36326. doi: 10.1074/jbc.M405035200. [DOI] [PubMed] [Google Scholar]
- 43.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Broderick JA, Salomon WE, Ryder SP, Aronin N, Zamore PD. Argonaute protein identity and pairing geometry determine cooperativity in mammalian RNA silencing. RNA. 2011;17:1858–1869. doi: 10.1261/rna.2778911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vickers TA, Lima WF, Wu H, Nichols JG, Linsley PS, Crooke ST. Off-target and a portion of target-specific siRNA mediated mRNA degradation is Ago2 ‘Slicer’ independent and can be mediated by Ago1. Nucleic Acids Res. 2009;37:6927–6941. doi: 10.1093/nar/gkp735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chiu YL, Rana TM. siRNA function in RNAi: a chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muhonen P, Tennila T, Azhayeva E, Parthasarathy RN, Janckila AJ, Vaananen HK, Azhayev A, Laitala-Leinonen T. RNA interference tolerates 2′-fluoro modifications at the Argonaute2 cleavage site. Chem. Biodivers. 2007;4:858–873. doi: 10.1002/cbdv.200790073. [DOI] [PubMed] [Google Scholar]
- 48.Prakash TP, Allerson CR, Dande P, Vickers TA, Sioufi N, Jarres R, Baker BF, Swayze EE, Griffey RH, Bhat B. Positional effect of chemical modifications on short interference RNA activity in mammalian cells. J. Med. Chem. 2005;48:4247–4253. doi: 10.1021/jm050044o. [DOI] [PubMed] [Google Scholar]
- 49.Manoharan M, Akinc A, Pandey RK, Qin J, Hadwiger P, John M, Mills K, Charisse K, Maier MA, Nechev L, et al. Unique gene-silencing and structural properties of 2′-fluoro-modified siRNAs. Angew. Chem. Int. Ed. 2011;50:2284–2288. doi: 10.1002/anie.201006519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pallan PS, Greene EM, Jicman PA, Pandey RK, Manoharan M, Rozners E, Egli M. Unexpected origins of the enhanced pairing affinity of 2′-fluoro-modified RNA. Nucleic Acids Res. 2011;39:3482–3495. doi: 10.1093/nar/gkq1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heale BS, Keegan LP, McGurk L, Michlewski G, Brindle J, Stanton CM, Caceres JF, O'Connell MA. Editing independent effects of ADARs on the miRNA/siRNA pathways. EMBO J. 2009;28:3145–3156. doi: 10.1038/emboj.2009.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hong J, Qian Z, Shen S, Min T, Tan C, Xu J, Zhao Y, Huang W. High doses of siRNAs induce eri-1 and adar-1 gene expression and reduce the efficiency of RNA interference in the mouse. Biochem. J. 2005;390:675–679. doi: 10.1042/BJ20050647. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.