Abstract
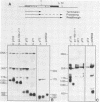
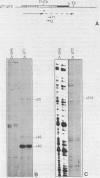
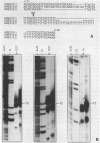
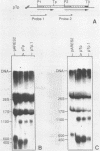
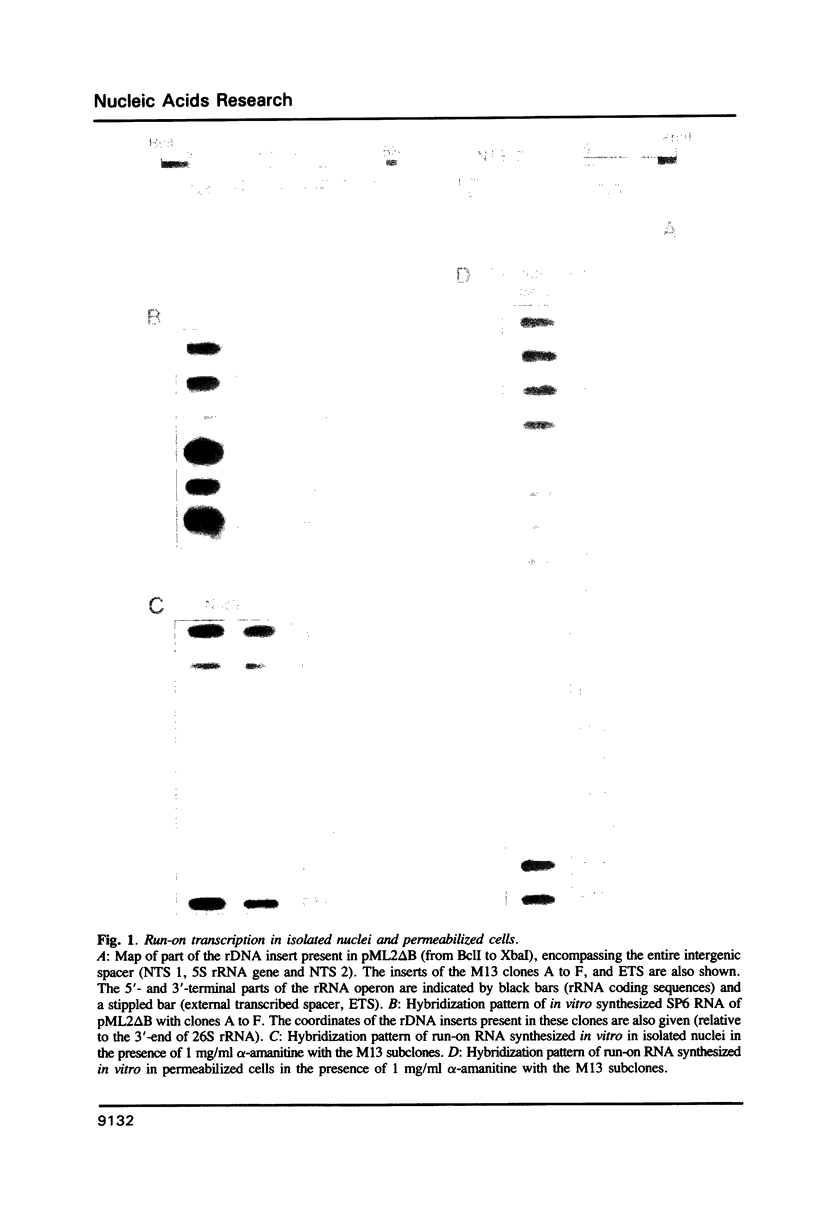
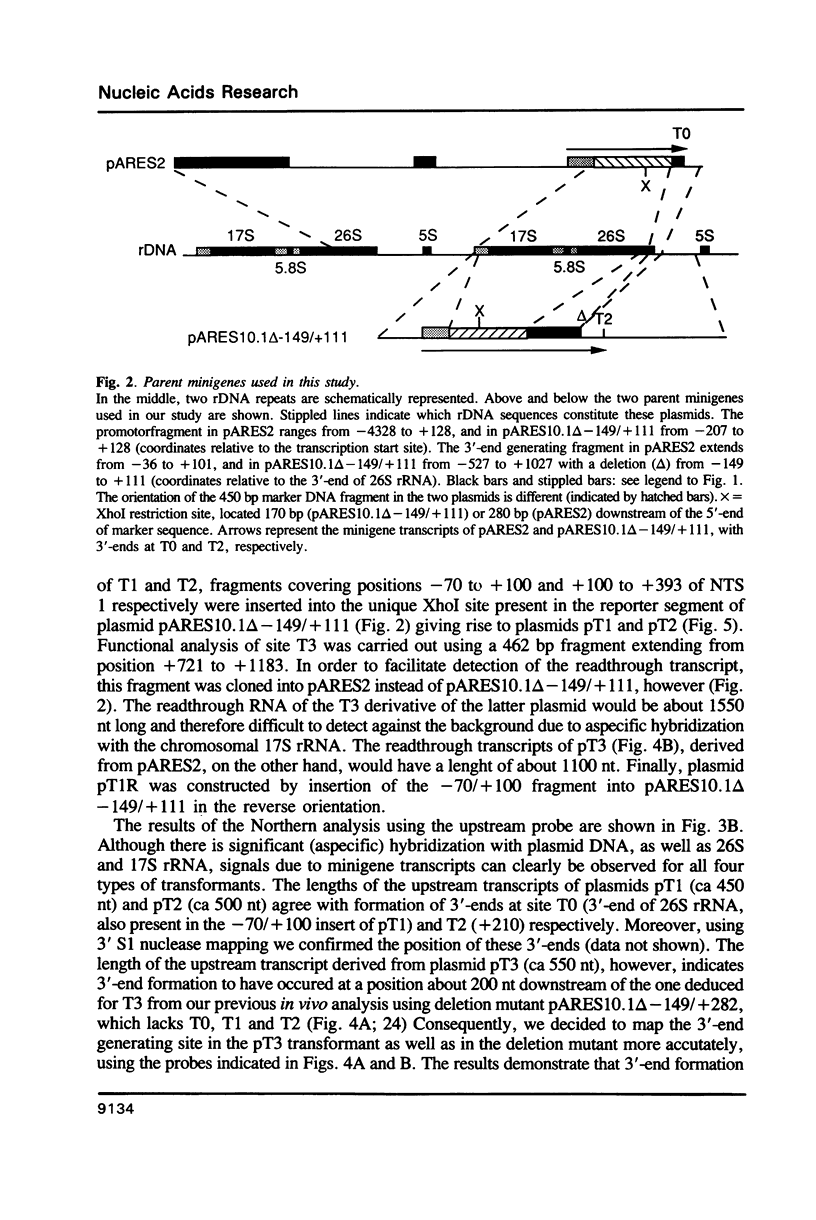
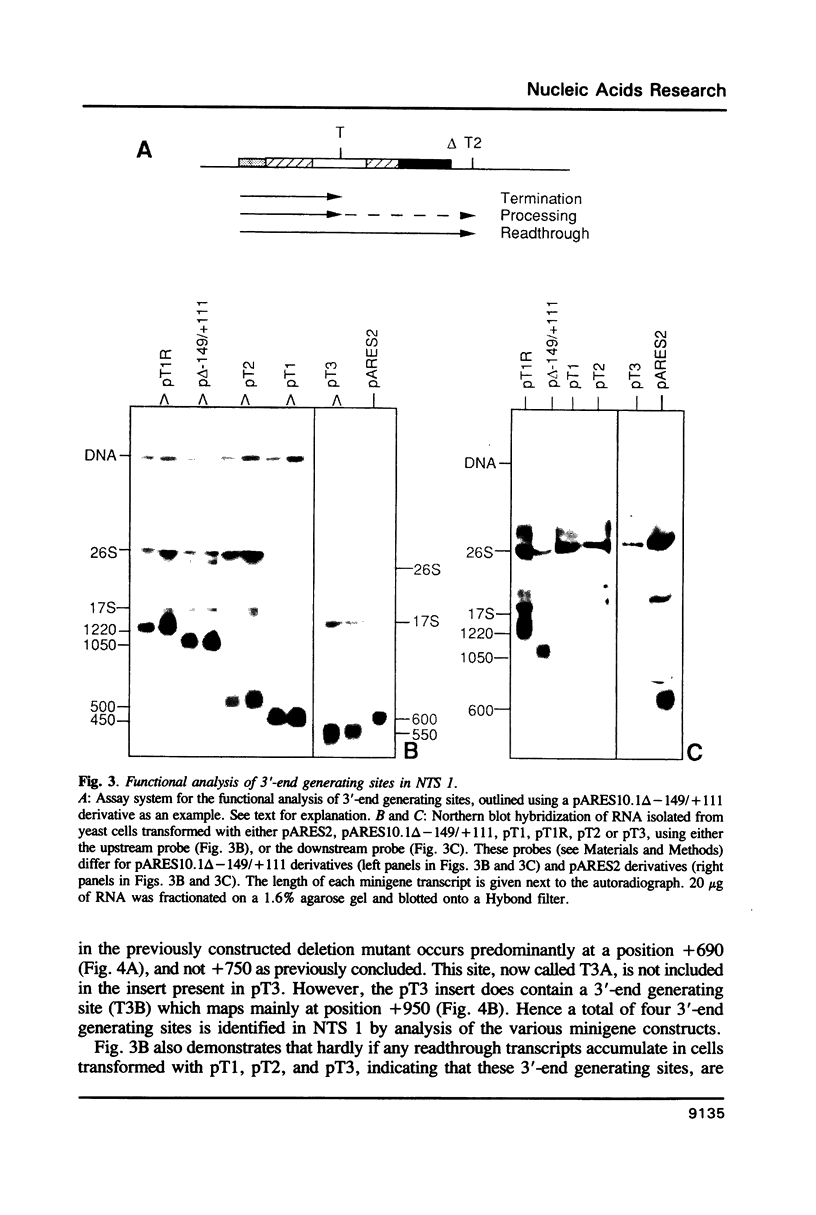
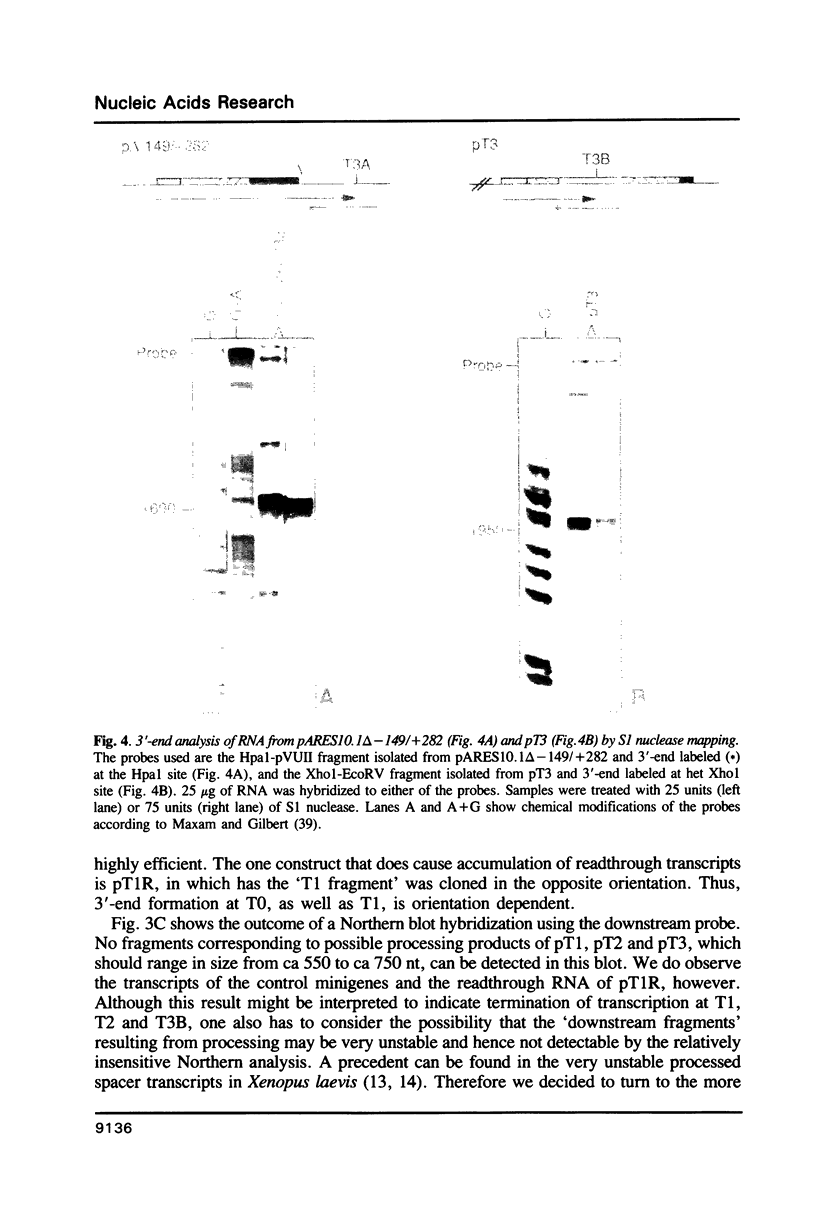
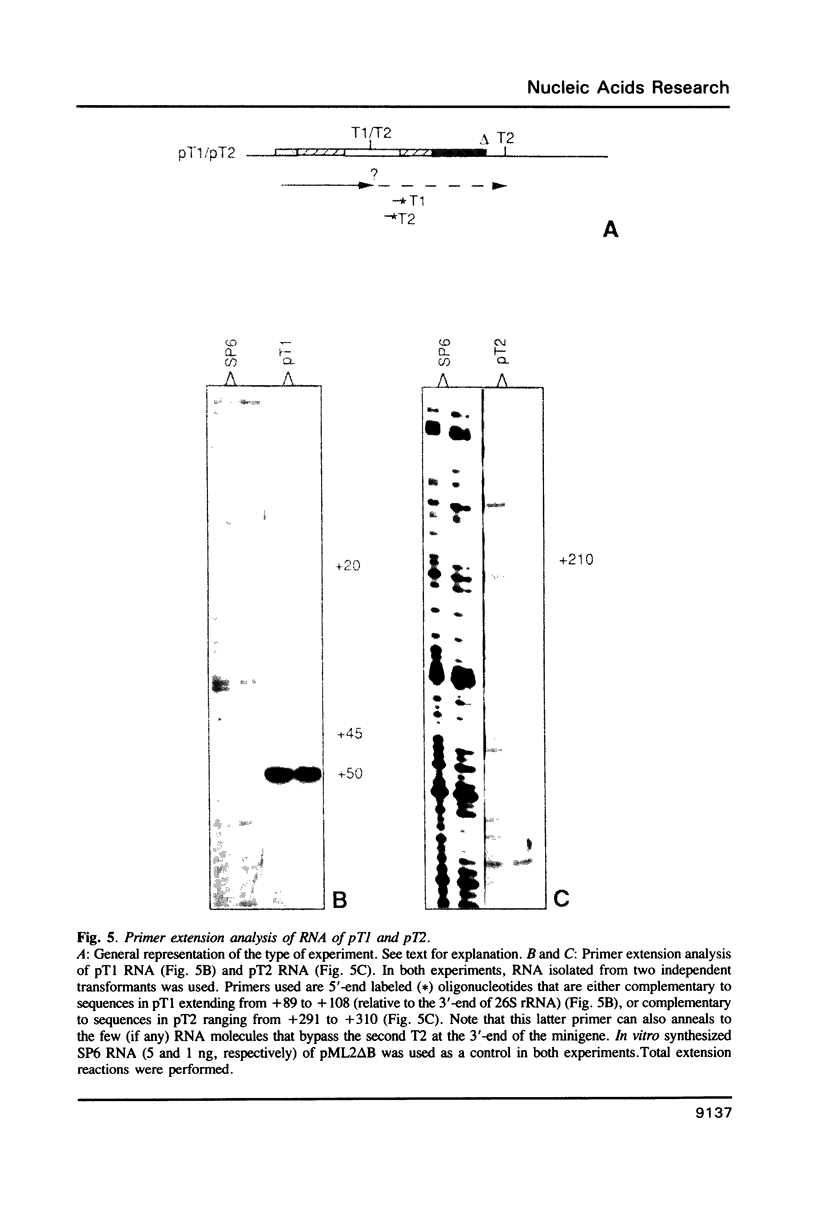
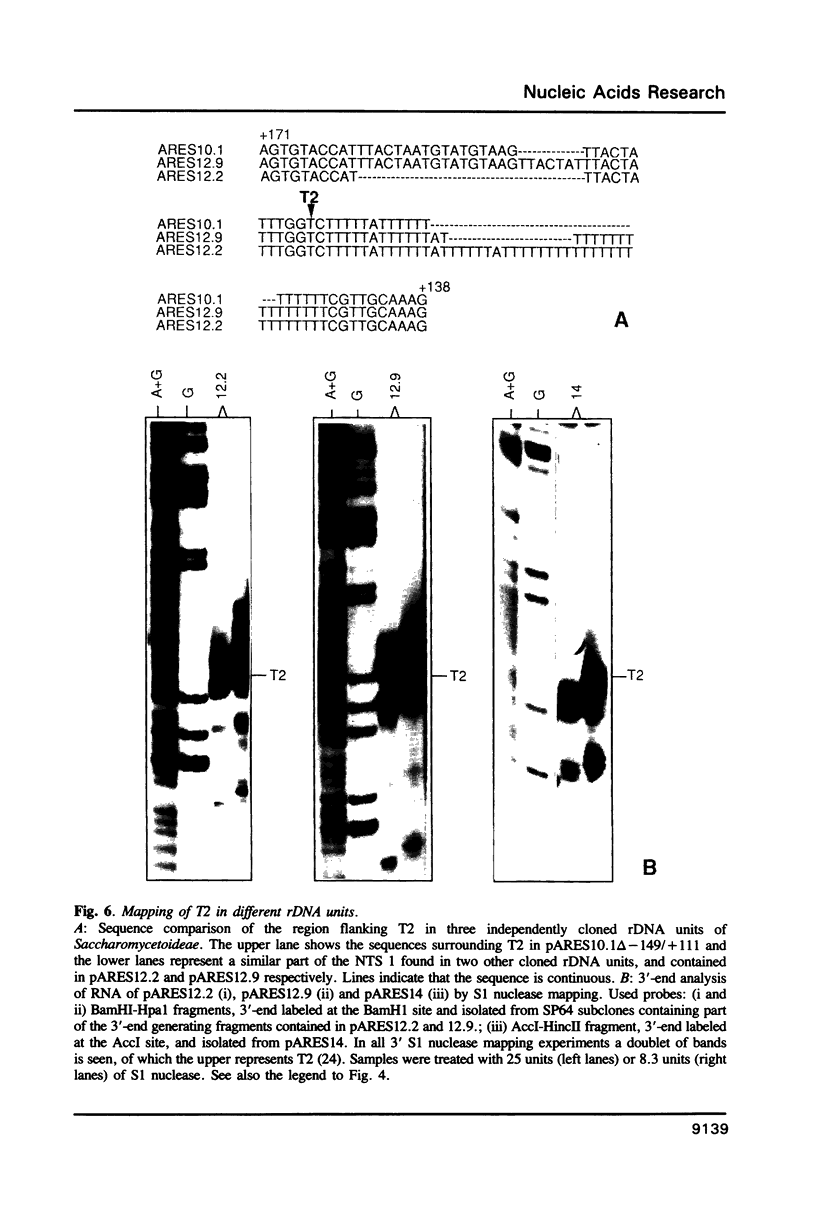
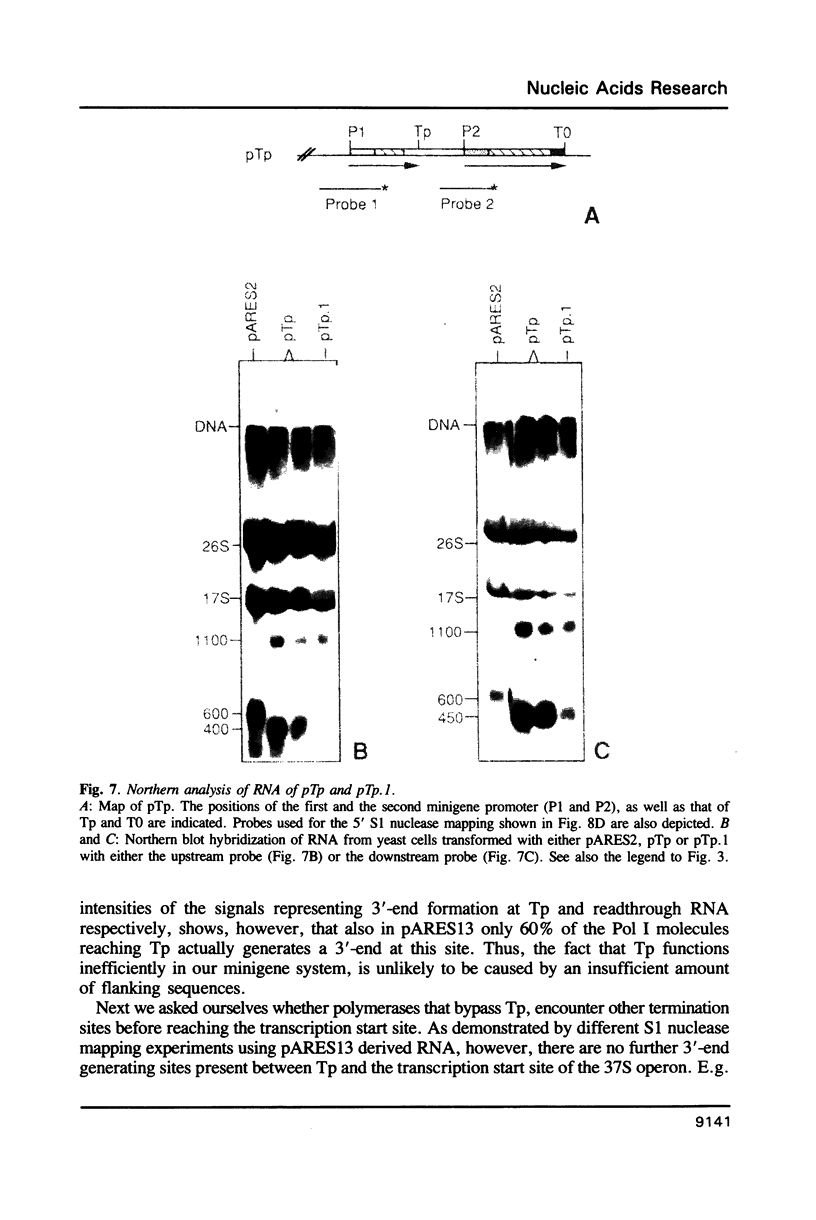
Analysis of the termination of transcription by yeast RNA polymerase I (Pol I) using in vitro run-on experiments in both isolated nuclei and permeabilized cells demonstrated that Pol I does not traverse the whole intergenic spacer separating consecutive 37S operons, but terminates transcription before reaching the 5S rRNA gene, that is within NTS 1. In order to discriminate between processing and termination at the 3'-end generating sites previously identified in vivo in NTS 1 (T1, T2 and T3), fragments containing these sites were inserted into the middle of the reporter DNA of an artificial rRNA minigene. RNA isolated from yeast cells transformed with these minigenes was analyzed for the presence of transcripts derived from sequences both up- and downstream of the insert by Northern blot hybridization, reverse transcription analysis and S1 nuclease mapping. In accordance with previously obtained results T1 (+15 to +50) was found to behave as a processing site. T2 (+210) however was concluded to be an efficient, genuine Pol I terminator. In addition to T2, two other terminators were identified in NTS 1: T3A (at +690) and T3B (at +950). Surprisingly, when the 3' terminal part of NTS 2 was tested for its capacity to generate 3'-ends, another terminator (Tp) was found to be present at a position 300 bp upstream of the transcription initiation site of the 37S-rRNA operon.
Full text
PDF







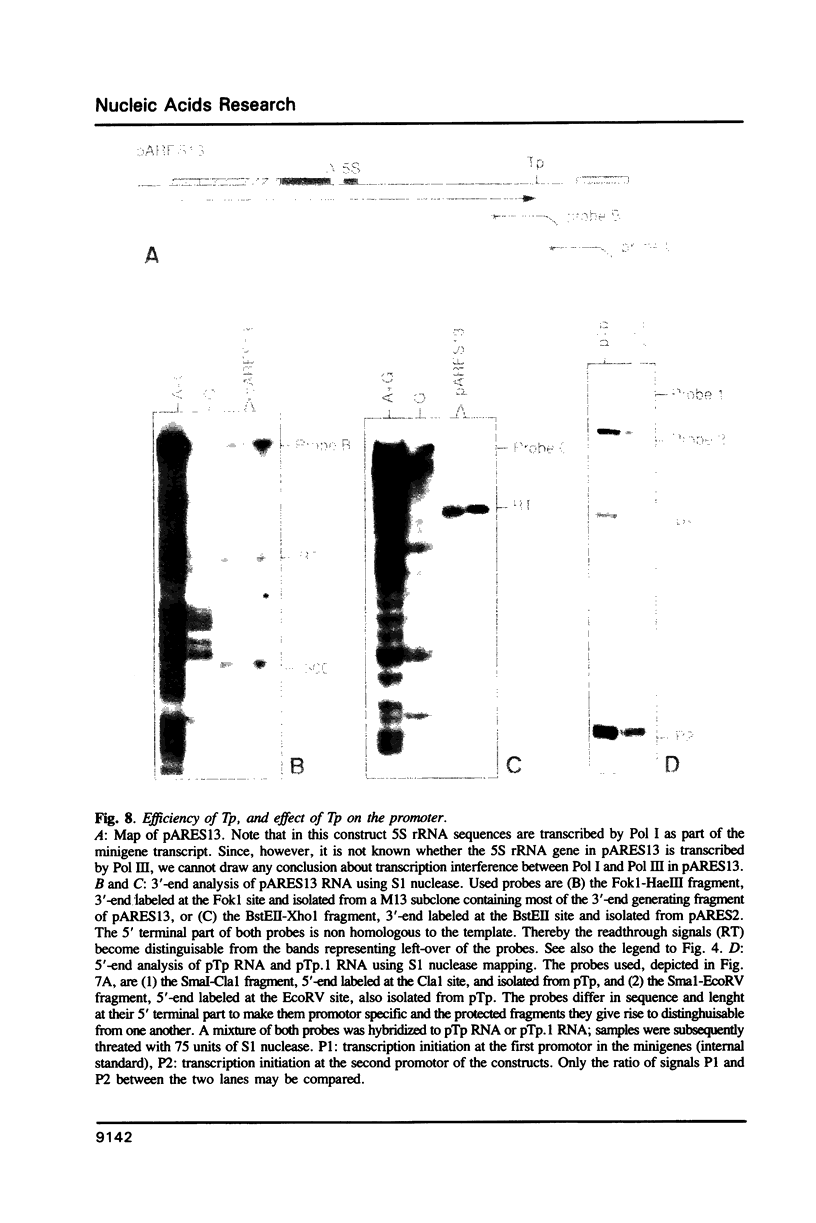




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker S. M., Platt T. Pol I transcription: which comes first, the end or the beginning? Cell. 1986 Dec 26;47(6):839–840. doi: 10.1016/0092-8674(86)90795-6. [DOI] [PubMed] [Google Scholar]
- Bartsch I., Schoneberg C., Grummt I. Evolutionary changes of sequences and factors that direct transcription termination of human and mouse ribsomal genes. Mol Cell Biol. 1987 Jul;7(7):2521–2529. doi: 10.1128/mcb.7.7.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch I., Schoneberg C., Grummt I. Purification and characterization of TTFI, a factor that mediates termination of mouse ribosomal DNA transcription. Mol Cell Biol. 1988 Sep;8(9):3891–3897. doi: 10.1128/mcb.8.9.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman E., Paule M. R. Promoter occlusion during ribosomal RNA transcription. Cell. 1988 Sep 23;54(7):985–992. doi: 10.1016/0092-8674(88)90113-4. [DOI] [PubMed] [Google Scholar]
- Beggs J. D. Transformation of yeast by a replicating hybrid plasmid. Nature. 1978 Sep 14;275(5676):104–109. doi: 10.1038/275104a0. [DOI] [PubMed] [Google Scholar]
- De Winter R. F., Moss T. A complex array of sequences enhances ribosomal transcription in Xenopus laevis. J Mol Biol. 1987 Aug 20;196(4):813–827. doi: 10.1016/0022-2836(87)90407-4. [DOI] [PubMed] [Google Scholar]
- De Winter R. F., Moss T. The ribosomal spacer in Xenopus laevis is transcribed as part of the primary ribosomal RNA. Nucleic Acids Res. 1986 Aug 11;14(15):6041–6051. doi: 10.1093/nar/14.15.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion E. A., Warner J. R. An RNA polymerase I enhancer in Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jun;6(6):2089–2097. doi: 10.1128/mcb.6.6.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion E. A., Warner J. R. The major promoter element of rRNA transcription in yeast lies 2 kb upstream. Cell. 1984 Dec;39(3 Pt 2):663–673. doi: 10.1016/0092-8674(84)90473-2. [DOI] [PubMed] [Google Scholar]
- Grummt I., Kuhn A., Bartsch I., Rosenbauer H. A transcription terminator located upstream of the mouse rDNA initiation site affects rRNA synthesis. Cell. 1986 Dec 26;47(6):901–911. doi: 10.1016/0092-8674(86)90805-6. [DOI] [PubMed] [Google Scholar]
- Grummt I., Maier U., Ohrlein A., Hassouna N., Bachellerie J. P. Transcription of mouse rDNA terminates downstream of the 3' end of 28S RNA and involves interaction of factors with repeated sequences in the 3' spacer. Cell. 1985 Dec;43(3 Pt 2):801–810. doi: 10.1016/0092-8674(85)90253-3. [DOI] [PubMed] [Google Scholar]
- Henderson S. L., Ryan K., Sollner-Webb B. The promoter-proximal rDNA terminator augments initiation by preventing disruption of the stable transcription complex caused by polymerase read-in. Genes Dev. 1989 Feb;3(2):212–223. doi: 10.1101/gad.3.2.212. [DOI] [PubMed] [Google Scholar]
- Henderson S., Sollner-Webb B. A transcriptional terminator is a novel element of the promoter of the mouse ribosomal RNA gene. Cell. 1986 Dec 26;47(6):891–900. doi: 10.1016/0092-8674(86)90804-4. [DOI] [PubMed] [Google Scholar]
- Jemtland R., Maehlum E., Gabrielsen O. S., Oyen T. B. Regular distribution of length heterogeneities within non-transcribed spacer regions of cloned and genomic rDNA of Saccharomyces cerevisiae. Nucleic Acids Res. 1986 Jul 11;14(13):5145–5158. doi: 10.1093/nar/14.13.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempers-Veenstra A. E., Musters W., Dekker A. F., Klootwijk J., Planta R. J. Deletion mapping of the yeast Pol I promoter. Curr Genet. 1985;10(4):253–260. doi: 10.1007/BF00365621. [DOI] [PubMed] [Google Scholar]
- Kempers-Veenstra A. E., Oliemans J., Offenberg H., Dekker A. F., Piper P. W., Planta R. J., Klootwijk J. 3'-End formation of transcripts from the yeast rRNA operon. EMBO J. 1986 Oct;5(10):2703–2710. doi: 10.1002/j.1460-2075.1986.tb04554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempers-Veenstra A. E., van Heerikhuizen H., Musters W., Klootwijk J., Planta R. J. Transcription of an artificial ribosomal RNA gene in yeast. EMBO J. 1984 Jun;3(6):1377–1382. doi: 10.1002/j.1460-2075.1984.tb01980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraig E., Haber J. E., Rosbash M. Sporulation and rna2 lower ribosomal protein mRNA levels by different mechanisms in Saccharomyces cerevisiae. Mol Cell Biol. 1982 Oct;2(10):1199–1204. doi: 10.1128/mcb.2.10.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A., Grummt I. 3'-end formation of mouse pre-rRNA involves both transcription termination and a specific processing reaction. Genes Dev. 1989 Feb;3(2):224–231. doi: 10.1101/gad.3.2.224. [DOI] [PubMed] [Google Scholar]
- Kuhn A., Grummt I. A novel promoter in the mouse rDNA spacer is active in vivo and in vitro. EMBO J. 1987 Nov;6(11):3487–3492. doi: 10.1002/j.1460-2075.1987.tb02673.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A., Normann A., Bartsch I., Grummt I. The mouse ribosomal gene terminator consists of three functionally separable sequence elements. EMBO J. 1988 May;7(5):1497–1502. doi: 10.1002/j.1460-2075.1988.tb02968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. Characterization of three sites of RNA 3' end formation in the Xenopus ribosomal gene spacer. Cell. 1986 May 9;45(3):431–443. doi: 10.1016/0092-8674(86)90329-6. [DOI] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. Ribosomal precursor 3' end formation requires a conserved element upstream of the promoter. Cell. 1987 Jul 3;50(1):51–57. doi: 10.1016/0092-8674(87)90661-1. [DOI] [PubMed] [Google Scholar]
- Lohr D., Ide G. I. In vitro initiation and termination of ribosomal RNA transcription in isolated yeast nuclei. J Biol Chem. 1983 Apr 25;258(8):4668–4671. [PubMed] [Google Scholar]
- Lucchini R., Reeder R. H. A test of 'polymerase handover' as a mechanism for stimulating initiation by RNA polymerase I. Nucleic Acids Res. 1989 Jan 11;17(1):373–387. doi: 10.1093/nar/17.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal R. K. The organization and transcription of eukaryotic ribosomal RNA genes. Prog Nucleic Acid Res Mol Biol. 1984;31:115–160. doi: 10.1016/s0079-6603(08)60376-1. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McStay B., Reeder R. H. A termination site for Xenopus RNA polymerase I also acts as an element of an adjacent promoter. Cell. 1986 Dec 26;47(6):913–920. doi: 10.1016/0092-8674(86)90806-8. [DOI] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestel R., Yip M., Holland J. P., Wang E., Kang J., Holland M. J. Sequences within the spacer region of yeast rRNA cistrons that stimulate 35S rRNA synthesis in vivo mediate RNA polymerase I-dependent promoter and terminator activities. Mol Cell Biol. 1989 Mar;9(3):1243–1254. doi: 10.1128/mcb.9.3.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchelson K., Moss T. The enhancement of ribosomal transcription by the recycling of RNA polymerase I. Nucleic Acids Res. 1987 Nov 25;15(22):9577–9596. doi: 10.1093/nar/15.22.9577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T., Mitchelson K., de Winter R. The promotion of ribosomal transcription in eukaryotes. Oxf Surv Eukaryot Genes. 1985;2:207–250. [PubMed] [Google Scholar]
- Murtif V. L., Rae P. M. In vivo transcription of rDNA spacers in Drosophila. Nucleic Acids Res. 1985 May 10;13(9):3221–3239. doi: 10.1093/nar/13.9.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musters W., Venema J., van der Linden G., van Heerikhuizen H., Klootwijk J., Planta R. J. A system for the analysis of yeast ribosomal DNA mutations. Mol Cell Biol. 1989 Feb;9(2):551–559. doi: 10.1128/mcb.9.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skryabin K. G., Eldarov M. A., Larionov V. L., Bayev A. A., Klootwijk J., de Regt V. C., Veldman G. M., Planta R. J., Georgiev O. I., Hadjiolov A. A. Structure and function of the nontranscribed spacer regions of yeast rDNA. Nucleic Acids Res. 1984 Mar 26;12(6):2955–2968. doi: 10.1093/nar/12.6.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Tower J. Transcription of cloned eukaryotic ribosomal RNA genes. Annu Rev Biochem. 1986;55:801–830. doi: 10.1146/annurev.bi.55.070186.004101. [DOI] [PubMed] [Google Scholar]
- Tautz D., Dover G. A. Transcription of the tandem array of ribosomal DNA in Drosophila melanogaster does not terminate at any fixed point. EMBO J. 1986 Jun;5(6):1267–1273. doi: 10.1002/j.1460-2075.1986.tb04356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbeet M. P., Klootwijk J., van Heerikhuizen H., Fontijn R., Vreugdenhil E., Planta R. J. Molecular cloning of the rDNA of Saccharomyces rosei and comparison of its transcription initiation region with that of Saccharomyces carlsbergensis. Gene. 1983 Jul;23(1):53–63. doi: 10.1016/0378-1119(83)90216-0. [DOI] [PubMed] [Google Scholar]
- Yip M. T., Holland M. J. In vitro RNA processing generates mature 3' termini of yeast 35 and 25 S ribosomal RNAs. J Biol Chem. 1989 Mar 5;264(7):4045–4051. [PubMed] [Google Scholar]