Abstract
The brain cytoplasmic RNA, BC1, is a small non-coding RNA that is found in different RNP particles, some of which are involved in translational control. One component of BC1-containing RNP complexes is the fragile X mental retardation protein (FMRP) that is implicated in translational repression. Peptide mapping and computational simulations show that the tudor domain of FMRP makes specific contacts to BC1 RNA. Endogenous BC1 RNA is 2′-O-methylated in nucleotides that contact the FMRP interface, and methylation can affect this interaction. In the cell body BC1 2′-O-methylations are present in both the nucleus and the cytoplasm, but they are virtually absent at synapses where the FMRP–BC1–mRNA complex exerts its function. These results strongly suggest that subcellular region-specific modifications of BC1 affect the binding to FMRP and the interaction with its mRNA targets. We finally show that BC1 RNA has an important role in translation of certain mRNAs associated to FMRP. All together these findings provide further insights into the translational regulation by the FMRP–BC1 complex at synapses.
INTRODUCTION
Absence of the fragile X mental retardation protein (FMRP) causes the fragile X syndrome. FMRP is a multi-functional RNA-binding protein with roles in localization, translation (1,2) and stability of mRNAs (3,4). At synapses, two FMRP complexes have been identified: FMRP forms mRNPs with non-coding RNAs such as the brain cytoplasmic RNAs BC1/BC200 (5,6) and the miRNAs (7). FMRP interacts with BC1 RNA through its N-terminal region shown to contain a novel RNA-binding domain (8,9) containing two tudor motifs (10). The tudor domain has been described to typically recognize methylated amino acids (11). One activity of FMRP is to repress local translation (1,2), a process implicated in synapse maturation, learning and memory (12).
BC1 RNA is a small non-protein coding RNA (sncRNA) initially identified in rat brain (13,14) that is highly expressed in neurons (15,16) and enriched at synapses (17). BC1 RNA forms diverse ribonucleoprotein particles (RNPs) with different protein partners including FMRP (5,9,18), the Testis–Brain Protein (TBP) (19), Staufen (20), Pur alpha and beta (21), poly(A)-binding protein 1 (PABP1) (22,23), eIF4A (24) and hnRNPA2 (25). Some of the BC1 RNP particles are involved in neuronal translational control as well; in particular, the FMRP–BC1 complex represses translation of a defined subset of FMRP target mRNAs (5,6). In this context, BC1 RNA acts as a bridging molecule between FMRP and the substrate mRNAs (5,18) and helps recruiting additional factors that are responsible for translation inhibition (6).
We show here that BC1 RNA is 2′-O-methylated in the 5′-hairpin that is involved in mRNA translational regulation in vivo (5). These 2′-O-methylations are not detected at synapses where absence of BC1 RNA affects translation of some FMRP target mRNAs. Peptide mapping, molecular modelling and docking simulations showed that the second tudor domain of FMRP recognizes the modified region of BC1 RNA and surprisingly the 2′-O-methylations affect the interaction of BC1 RNA with FMRP.
We propose that the 2′-O-methyl modifications of BC1 RNA influence its activity in controlling translation at synapses.
MATERIALS AND METHODS
Details of general molecular procedures, RNA quantification by RT–qPCR and the primer sequences used in this study may be found in the Supplementary Data.
Animal care
Animal care was conducted conforming to the institutional guidelines that are in compliance with national and European laws and policies. Mouse strains that have been used in this study are: C57BL/6-129SV Wild-Type (WT) and C57BL/6-129SV BC1 Knock-Out (KO). All animals used in this study were 3 weeks old.
Detection of 2′-O-methylations
2′-O-methylation was analysed as previously reported (26–28). Fifteen microgram of total RNA from WT mouse brain or 2 µg of synaptosomal RNA (highly enriched for BC1 RNA, Supplementary Figure S1B) were used for each primer extension assay with low (4 and 2 µM) or high (1 mM) deoxyribonucleotide triphosphate (dNTP) concentration. 2′-O-methylation creates a primer extension stop when the dNTPs are limiting (4 and 2 µM, Supplementary Figure S2A). Avian Myeloblastosis Virus (AMV) reverse transcriptase (Q-biogen) and radiolabelled primers were used in the primer extension assays. Reverse transcription for RT–qPCR reactions used the Moloney Murine Leukemia Virus reverse transcriptase from Invitrogen. For the sequence of the oligos used, see Supplementary Data.
RNA sequencing
Two hundred nanogram of BC1 and U2 transcripts were sequenced using AMV reverse transcriptase (Q-biogen). The RNAs were denatured for 10 min at 70°C and renatured for 10 min on ice. The RT reactions were performed at 45°C for 35 min in the presence of 0.5 mM of each ddATP, ddGTP, ddCTP and ddTTP, and stopped with blue/formamide buffer. The RNA was denatured for 1 min at 90°C before being loaded onto an 8 M urea–10% polyacrylamide gel. The gel was then dried and exposed for 12 or 24 h using a PhosphoImager screen (GE Healthcare).
Expression of the FMRP-NT
Two separate sources of FMRP-N Terminus (FMRP-NT) were used. Independent experiments were performed using the FMRP protein domains prepared according to Zalfa et al. (9), produced in house, and the same protein domains produced by the Protein Expression and Purification Core Facility of the EMBL, Heidelberg.
Mutagenesis of the FMRP-N Terminus
In order to mutagenize the FMRP-NT, we used the Quick Change Lightning Multi Site-Directed Kit (from Quiagen) and the pET-M11-FMRP-NT plasmid which has the same insert as described in (9), cloned into NcoI-KpnI site of pET-M11 (EMBL protein expression group).
To generate the R70E-R11E double mutant, the DNA oligos TV1 and TV2 were used. Mutagenized triplet is shown in bold.
TV1: 5′-GGTTTATTCCGAAGCAAATGAAAAAG-3′
TV2: 5′-GTCACAATTGAGGAGCTACGATCTG-3′
To generate the Y103A-N104A double mutant, the DNA oligo TV5 was used. Mutagenized triplets are shown in bold.
TV5: 5′- GTGATGCTACGGCTGCTGAAATTGTCACAATTG 3′
The two mutagenized plasmids were sequenced before use.
Electrophoretic mobility shift assay
BC1 RNA was in vitro transcribed in the presence of α-[32P] UTP using T7 RNA polymerase, or reconstituted by ligation of [32P]-labelled oligos in the presence of a splint DNA oligo (29) (Supplementary Data). Directly before use, RNAs were denatured/renatured in electrophoretic mobility shift assay (EMSA) buffer in the presence of 1 µg of tRNAs. Then, 1 × 105–2 × 105 cpm of RNA (corresponding to 10 and 20 fmol, respectively) were incubated with 1–5 pmoles of FMRP N-terminus for 20 min at room temperature in the presence of 150 mM KCl, 5 mM MgCl2, 2 mM DTT, 5% glycerol, 20 mM HEPES, pH 7.6 and 1 µg yeast tRNA. The entire reaction (20 µl) was loaded on a native 1 × TBE/6% acrylamide gel and run in 0.5× TBE from 2 to 4 h at 170 V in a cold room. The autoradiogram was recorded with a PhosphorImager (GE Healthcare) and the bands were quantified. From the equation Kd = ([R][P]/[RP]) → [RP]/[R] = [P]/ Kd, the ratio of [bound RNA]/[free RNA] was plotted against [P], the slope giving 1/Kd.
Sucrose gradients to separate mRNPs and polysomes
Total mouse brain was homogenized in 3 ml of lysis buffer (10 mM Tris–HCl pH 7.5, 100 mM NaCl, 10 mM MgCl2, 1% Triton X-100, 1 mM dithiothreitol DTT, 30 U/ml RNasin). After 5 min of incubation on ice, the extract was centrifuged for 4 min at 9300 g at 4°C. The supernatant (up to 0.5 ml) was frozen in liquid N2 and either stored at −80°C or immediately loaded onto a 15–50% (w/v) sucrose gradient and centrifuged at 4°C for 1 h 50 min at 37 000 rpm in a Beckman SW41 rotor. Same procedure was used to isolate Polysomes and mRNPs from synaptoneurosomes. See Supplementary Data for Polysome-mRNP analysis.
Synaptoneurosome preparation and RNA extraction
Synaptoneurosomes are particles containing both pre- and postsynaptic compartments (30). Synaptoneurosomes were prepared as previously described (6). Details can be found in Supplementary Data.
Nuclear and cytoplasmic extracts
Cortices were weighed and resuspended in fractionation buffer (75 mg in 600 µl). Nuclear and cytoplasmic extracts were prepared using the PARIS kit according to manufacturer's recommendation (Ambion). RNA quality control was performed using the Experion system (Bio-Rad).
Immunoprecipitation followed by RT–PCR and RT–qPCR analysis
Total brain and synaptosomes were lysed in 100 mM NaCl, 10 mM MgCl2, 10 mM Tris–HCl pH 7.5, 1% Triton X-100, 1 mM DTT, 40 U/ml RNAse OUT (Invitrogen), 10 μl/µl Protease inhibitor cocktail (Sigma-Aldrich), 5 mM β-glycerophosphate, 0.5 mM Na3VO4. After 5 min of incubation on ice, lysates were centrifuged for 5 min at 12 000 g at 4°C. The protein concentration was determined and the equivalent of 500 μg protein of the supernatant was used for the IP using anti-FMRP antibodies (16) or purified rabbit IgGs as negative control, and protein A Dynabeads (Invitrogen). RT–PCR and RT–qPCR were performed as described in Supplementary Data.
Mass spectrometric analysis of UV cross-links
The FMRP–BC1 particles (5 nmol of FMRP-NT and 10 nmol of BC1 RNA) were incubated in EMSA-binding buffer for 15 min on ice and irradiated on glass dishes at 254 nm with four 8-Watt germicidal lamps (G8T5, Herolab, Wiesloch, Germany) in parallel at a distance of 4 cm for 2 min on ice.
The reactions were diluted with buffer containing 1 M of urea, and digested with trypsin, RNase A and RNase T1. Peptides cross-linked to an RNA moiety were enriched on TiO2, and analysed by Electro Spray Ionization Quadrupole Time Of Flight as described by (31–33). Cross-linked peptides exhibit the combined mass of the peptide and the RNA fragment; in the Q-TOF sequencing, the cross-linked amino acid is identified as the one where the sequencing ladder breaks off.
Computational procedures
The MC-Sym web server (http://www.major.iric.ca/MC-Sym/) has been used to generate the atomic structure of the BC1 5′-end RNA (34). As a template we used the BC1 5′-hairpin 2D structure experimentally determined by Rozhdestvensky and collaborators (35). This analysis, i.e. the secondary structure determined by chemical and enzymatic assays, revealed that BC1 RNA 5′-domain has an extended rod-like stem-loop structure. Details regarding the molecular modelling and the molecular docking procedures can be found in Supplementary Data.
RESULTS AND DISCUSSION
BC1 RNA represses translation of FMRP mRNA targets at synapses
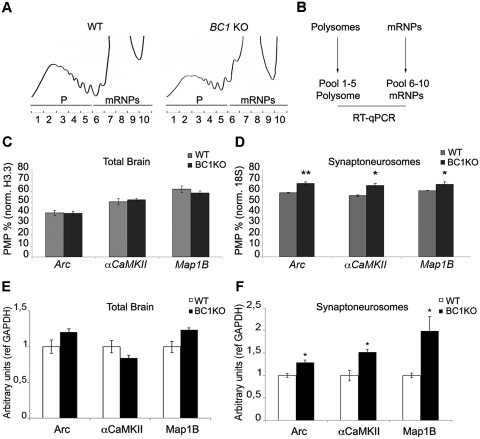
BC1 RNA is highly localized in dendrites and axons and it is thought to regulate local protein synthesis (5,36). Previous reports have shown a functional interaction between BC1 RNA and FMRP at synapses (5,9,37,38). To investigate the contribution of BC1 RNA to FMRP-mediated translational regulation, we performed polysome-mRNP gradient analysis. Cytoplasmic whole-brain extracts from 3 weeks old WT and BC1 KO mice (39) were fractionated on continuous sucrose gradients. Figure 1A shows the sedimentation profiles for the two genotypes. Fractions that contain actively translating polysomes (P) and translationally silent mRNPs (mRNPs), respectively, were pooled and the extracted RNAs were used to determine the translational efficiency by RT–qPCR (as percentage of mRNAs on polysomes, PMP, Figure 1B) of Arc, αCamKII and Map1B mRNAs that are translationally controlled by FMRP (5,40–44). When the polysomal-mRNP analysis was carried out from total brain, no increase in the PMP value was observed for Arc, αCamKII and Map1B mRNAs (Figure 1C). These data show that in total brain of BC1 KO animals, the translational efficiency of some FMRP target mRNAs is not altered compared to WT (Figure 1C).
Figure 1.
BC1 RNA represses mRNA translation at synapses. (A) Cytoplasmic brain extracts from WT and BC1 KO mice were centrifuged through a 15–50% sucrose gradient; absorbance at 254 nm was monitored continuously and plotted against the fraction numbers. (B) Fractions 1–5, corresponding to polysomes (P) and the fractions 6–10 containing mRNPs (mRNP) were pooled and further analysed by RT–qPCR. (C) Translational efficiency of Arc, αCaMKII and Map1B mRNAs from WT (grey histograms) and BC1 KO (black histograms) mice were quantified and expressed as PMP in the histograms. (D) Same as in panel (C) using synaptosomal preparation. Error bars represent SE: *P < 0.05 or **P < 0.01 for BC1 KO versus WT by Student's test, n = 4. (E) Protein levels of Arc, αCaMKII and Map1B from WT (in white) and BC1 KO (in black) from total brain. (F) Same as in (E) using synaptic protein extracts. Error bars represent SE: *P < 0.05 for KO versus WT by Student's test, n = 3.
We then investigated whether loss of BC1 RNA affects mRNA translation at synapses. Synaptoneurosomes (30) from WT and BC1 KO mice were prepared as previously described (6). The quality of the preparations was verified by the enrichment of synaptic proteins and RNAs (Supplementary Figure S1A and S1B). The PMP of Arc, αCamKII and Map1B mRNAs resulted higher in synaptoneurosomes from BC1 KO animals than in their WT littermates (Figure 1D), showing that these mRNAs are more efficiently translated in the absence of BC1 RNA at synapses similarly to what we observed in synaptoneurosomes from Fmr1 KO mice (5). Western blot analysis confirmed the increased level of proteins encoded by those mRNAs at synapses (Figure 1F), meanwhile no protein increase is observed in total brain (Figure 1E). These data suggest that BC1 RNA controls the translation of three FMRP mRNA targets mainly at synapses and this nicely coincides with the observation of a translation dysregulation in synaptoneurosomes from Fmr1 KO mice (5). These findings strongly suggest that the FMRP/BC1 RNP exerts its major function at synapses.
BC1 RNA is differentially 2′-O-methylated in neurons
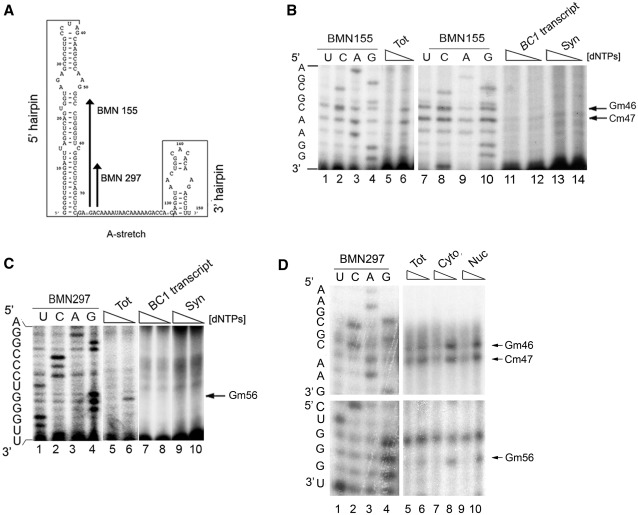
Post-transcriptional RNA modifications can have important roles in modulating its functions by influencing secondary and tertiary structure (45,46). Endogenous BC1 RNA was therefore isolated from mouse brain and examined for the presence of 2′-O-methylation, one of the most common RNA modifications. The analysis was performed on the BC1 5′-hairpin that is implicated in the interaction with FMRP and its target mRNAs (5,9).
We first verified the efficacy of the previously established methodology (26–28) by analysing, in mouse brain, the well-documented U2 snRNA 2′-O-methylations (47) (Supplementary Figure S2A). Using the BC1-specific primers BMN155 and BMN297 (Figure 2A) and total brain RNA, 2′-O-methylations were clearly detected in the 5′-stem-loop at positions G46 (Gm46), C47 (Cm47) and G56 (Gm56) (Figure 2B and C: compare lanes 5 and 6; 2′-O-methylations are detected by strong signals with less dNTPs, n = 5). No 2′-O-methylations were observed using in vitro-transcribed BC1 RNA (Figure 2B and C, lanes 11 and 12 and lanes 7 and 8, respectively), or in the region located between the two hairpins of the endogenous BC1 RNA (nucleotides 75–127, data not shown). Since the FMRP–BC1 complex controls mRNA translation at synapses (5,6) (Figure 1D and F) we prepared synaptoneurosomes in order to analyse the 2′-O-methylation status of BC1 RNA at synapses. In this case, no 2′-O-methylations were detected on the synaptic BC1 RNA (Figure 2B and C, compare lanes 13, 14 and 9 and 10, respectively, n = 4). The presence of intact BC1 RNA molecules was verified by the presence of a stop of elongation at nucleotide 1 of BC1 RNA (Supplementary Figure S2B). Because BC1 RNA was found differentially 2′-O-methylated in the cell body versus the synapses, we investigated its methylation status in the nucleus. Nuclear and cytoplasmic compartments, used for this analysis, were revealed by the presence and enrichment for dyskerin and Glyceraldehyde 3-phosphate dehydrogenase (Supplementary Figure S1C). As shown in Figure 2D, BC1 RNA from the cytoplasmic or nuclear fractions is 2′-O-methylated (compare lanes 7 and 8 and lanes 9 and 10 for cytoplasmic and nuclear fraction, respectively) underlining that absence of these 2′-O-methylations may have a function at synapses. These findings suggest that BC1 2′-O-methylations occur in nucleus.
Figure 2.
BC1 RNA is 2′-O-methylated. (A) Primers used to detect BC1 RNA 2′-O-methylations: BMN297, BMN155 are indicated on the BC1 secondary structure. (B) Low dNTP concentration primer extension analysis using the BMN155 primer on total brain RNA from WT mice (lanes 5 and 6), on the BC1 transcript (lanes 11 and 12), and on RNA from synaptoneurosomes (lanes 13 and 14). The arrows denote 2′-O-methylations at positions G46, C47. For each sample, the second lane shows the lower dNTP concentration; stops due to low dNTP indicate the presence of a 2′-O-methylation. Sequencing reactions are shown in lanes 1–4 and 7–10. (C) Same as in (B) using the BMN297 primer; sequencing (lanes 1–4), low dNTP concentration primer extension analysis on total brain (lanes 5 and 6), on BC1 transcript (lanes 7 and 8) and on synaptosomal preparation (lanes 9 and 10). (D) Same as in (C) using total brain (lanes 5 and 6), cytoplasmic (lanes 7 and 8) and nuclear extracts (lanes 9 and 10).
To our knowledge this is the first time that a non-coding RNA is shown to be differently modified according to its subcellular location.
BC1 RNA 2′-O-methylations affect FMRP binding
Since the unmodified BC1 RNA is present at synapses, where the FMRP–BC1 complex acts as a translational inhibitor, we studied this interaction further.
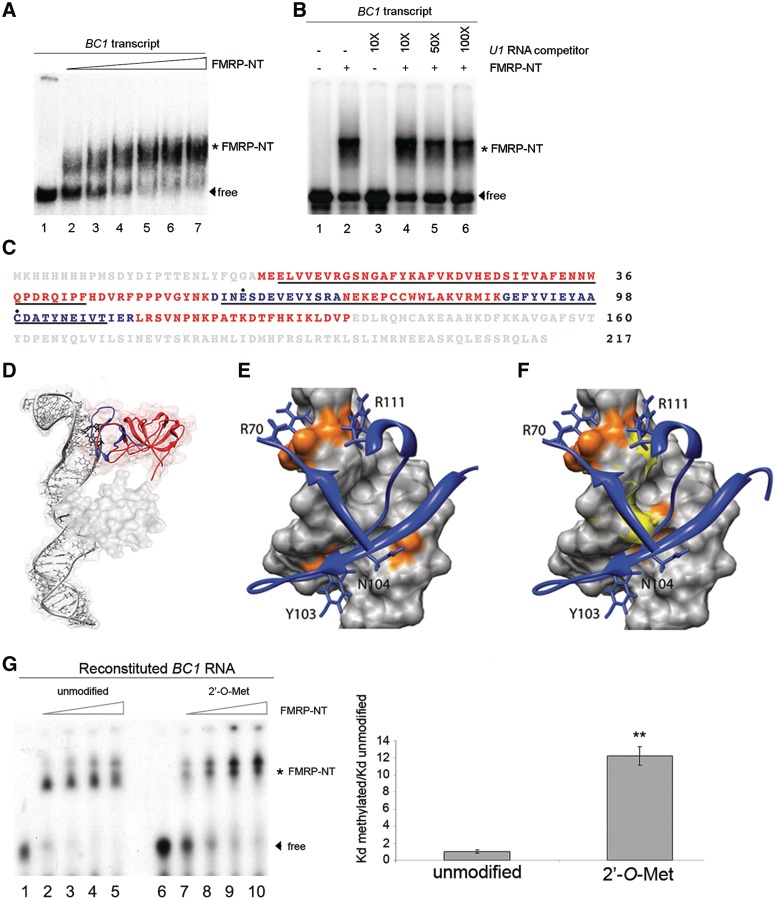
The N-terminus of FMRP (FMRP-NT; residues 1–217) contains two tudor domains (10), has RNA-binding properties (8,48), and interacts directly with the BC1 5′-hairpin (5,9). Because FMRP-NT tends to aggregate (48) (and data not shown) we tested its correct folding via the binding to the entire unmodified BC1 RNA (Figure 3A and B) as well as to poly riboG and poly riboC (data not shown). The FMRP-NT binds BC1 RNA with high affinity (apparent Kd = 128 ± 22.7 nM, the same within experimental error as in ref. (9); see Figure 3A for the gel and Supplementary Figure S3A for the Kd plot). Furthermore, a similarly structured RNA, U1 snRNA, was unable to compete with BC1 RNA for the FMRP binding (Figure 3B, compare lane 2 with lanes 4, 5 and 6).
Figure 3.
Functional mapping of the FMRP-NT/BC1 interaction. (A) EMSA using the entire BC1 RNA without (lane 1) or with increasing amounts of FMRP-NT (from 20 to 500 ng, lanes 2–7). The asterisk denotes the shifted FMRP–BC1 complex. Arrowhead denotes unbound BC1 (free). (B) Competition experiment using unlabelled U1 RNA. Lane 1 radiolabelled BC1 RNA, lane 2 radiolabelled BC1 and FMRP-NT (100 ng, asterisk). Lanes 4 and 6 show binding of FMRP-NT and BC1 RNA in the presence of 10–100-fold excess of unlabelled U1 RNA. Lanes 1 and 3 do not contain FMRP-NT. (C) Identification of FMRP residues that are in contact with BC1 RNA. The two tudor domains are underlined in black. The tryptic peptides containing the cross-linked amino acids (highly reactive: E61 and C99, black dots) are shown in blue. (D) Docking model of the interaction of FMRP-NT with the 5′-end of BC1. The backbone of the tudor domains is shown in red, with the two cross-linked peptides highlighted in blue. (E and F) Electrostatic and hydrogen-bond interactions of the FMRP-NT with BC1 RNA. FMRP is shown as a blue ribbon, and the lateral chains of the amino acids involved in the binding network are indicated as blue sticks. The RNA is shown as grey molecular surface with the nucleotides involved in the hydrogen bond and electrostatic network (orange spot), the 2′-O-methylated atoms are shown as a yellow surface. (G) EMSA using FMRP-NT (from 25 to 100 ng) and unmodified BC1 RNA (lanes 2–5) or 2′-O-methylated BC1 RNA (lanes 7–10). Lanes 1 and 6, unbound BC1 RNA. The asterisk indicates the FMRP-NT/BC1 RNA complex. The histogram shows the ratio of the Kd of 2′-O-methylated BC1 construct versus the Kd of unmodified BC1 construct. Error bars represent SE: **P < 0.01, Student's test, n = 3.
In order to gain insights into the FMRP-NT/BC1 interaction, we cross-linked BC1 RNA to FMRP-NT by UV exposure, enriched the tryptic peptides that were covalently bound to RNA, and analysed them by mass spectrometry using a recently described methodology (31–33) in which mass spectroscopy sequencing identifies the cross-linked amino acid. The two amino acids that cross-link to BC1 RNA are indicated in Figure 3C (E61 and C99, black dots) and are both situated in the second tudor domain (underlined). Next we carried out a molecular modelling of the BC1 RNA tertiary structure. Based on this structure, the 3D structure of the first 134 amino acids of FMRP-NT (10), and the cross-links identified by mass spectrometry (Figure 3C, blue peptides), we performed a molecular docking simulation to generate a protein–RNA complex (Figure 3D). The molecular docking simulation shows that the hypothesized recognition between FMRP-NT and 5′-end BC1 RNA is stabilized through both salt bridges and hydrogen-bond interactions. Specifically the FMRP residues R70, Y103, N104 and R111 interact through hydrogen bonds with the RNA nucleotides G31, C32, C45, G46, C47 and A48 (Figure 3E, orange nucleotides and Supplementary Table S1). Furthermore, salt bridge interactions occur between the two arginines R70 and R111 and the phosphate groups of the nucleotides C47 and A48 (Figure 3E and Supplementary Table S1). Importantly, two of the three 2′-O-methylated nucleotides (G46 and C47) are directly involved in FMRP-NT binding while the third (G56) is localized outside of this binding region. When two methyl groups are added, by molecular modelling, to the nucleotides G46 and C47 (Figure 3F, methyl groups in yellow), significant changes in the BC1 surface occur that affect the interaction with FMRP-NT. In particular, the addition of 2′-O-methylations increases the steric hindrance and makes access of FMRP more difficult (Figure 3F). In order to characterize the effect of BC1 2′-O-methylations on FMRP binding, the BC1 5′-hairpin was synthesized in an unmodified or 2′-O-methylated form, ligated to the BC1 3′-end (A-stretch plus 3′-hairpin) to constitute the entire BC1 RNA (Supplementary Figure S3B) and tested for its ability to associate to FMRP-NT. As shown in Figure 3G, the 2′-O-methylated BC1 RNA binds significantly less to FMRP-NT (right panel: the Kd of the 2′-O-methylated BC1 RNA oligonucleotide is ∼10-fold higher than the Kd of the unmodified one). 2′-O-methylation of these nucleotides in the minimal FMRP-binding domain of BC1 confirmed these data (Supplementary Figure S3C). We suggest that the 2′-O-methylation critically alters the BC1 surface shape and, reducing the electrostatic interactions, decreases the affinity between FMRP-NT and BC1.
While the tudor domains of FMRP are necessary for RNA binding, they are not sufficient: another 83 amino acids are needed to create the RNA-binding domain (9). We could not model this extra domain with high confidence, because there are no reference structures showing sufficient identity in the databases. However, molecular docking suggests that this domain might be able to form additional contacts to the basis of the 5′-hairpin. In particular, the helix-loop-helix (HLH) motif in this region (10) might insert itself into the major groove of the RNA region containing the third 2′-O-methylated nucleotide (G56), creating additional specific contacts (Figure 3D and data not shown). It is thus the second tudor domain of FMRP together with the adjacent residues that creates a novel RNA-binding surface. A crucial contribution to the binding comes from R70 and R111 that form salt bridges to the negative charges of the phosphates at positions 47 and 48. While these contacts are formed by the tudor domain, its canonical methyl binding site is not involved in the RNA binding. Instead, the tudor domain serves as a scaffold that positions the amino acids on one site of the beta barrel in such away that they can form specific interactions with the BC1 RNA.
Structural analysis of the FMRP-NT/BC1 interaction
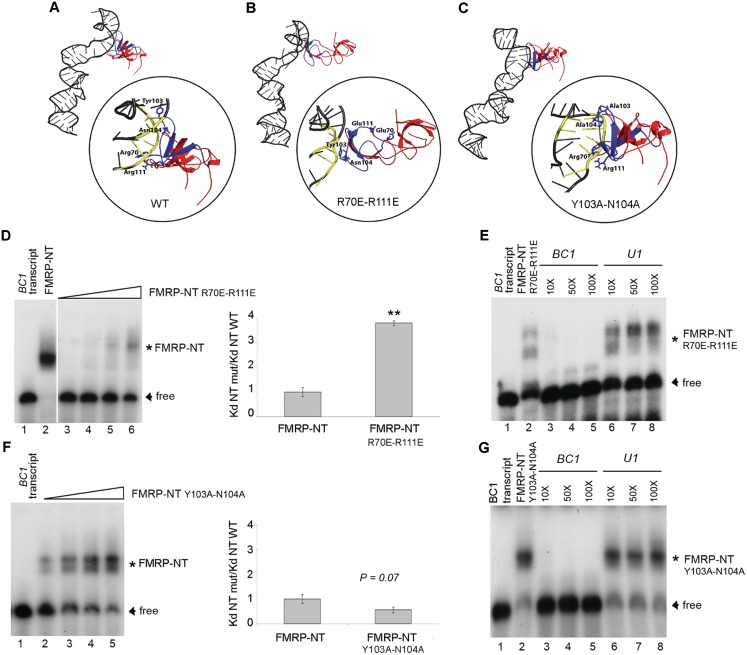
According to the molecular docking simulation and the experimental cross-linking data, the second tudor domain gives an important contribution to the stability of the FMRP-NT/BC1 complex. In order to verify this model, two different FMRP-NT double mutants were created. In the first mutant the positive charge, carried by the side chain of residues R70 and R111 (Figure 3E and F), was reversed by mutating both the residues into glutamic acid, while in the second mutant the hydrogen-bond interactions of residues Y103 and N104 (Figure 3E and F) were removed by mutating both residues into alanines. The effect of these mutations was predicted by docking simulation, comparing the HADDOCK score (49) where a lower value predicts stronger binding.
The introduction of two alanines at positions 103 and 104 actually removes the hydrogen bonds formed by the wild type residues (compare Figure 4A and C and see Supplementary Table S1). Due to the reduced steric hindrance of the alanine residues, however, the FMRP protein wraps more tightly around BC1 RNA improving other energy terms in the HADDOCK score (i.e. the van der Waals, the buried surface area, the binding and the desolvation energies). The total score therefore decreases from −38.0 in the WT to −49.0 in the mutant, predicting a higher binding affinity. The docking model was experimentally validated by EMSA experiments with the in vitro-transcribed unmodified BC1 RNA and the mutant protein (produced in Escherichia coli; Supplementary Figure S4A). As expected, the apparent Kd decreases by a factor of 2.7 (Figure 4F, strong tendency: P = 0.07, n = 5; apparent Kd FMRP NT WT = 128 ± 22.7 nM and Kd NT Mut = 51 ± 4.1 nM) showing the validity of the modelling. On the other hand the reversion of the positive charges (R70E-R111E, Figure 4A and B) repel the negatively charged phosphates of the RNA backbone, leading to a change in the FMRP orientation towards BC1 and thus to a considerable decrease in the area of interaction surface (Figure 4B). Therefore, the HADDOCK score increases from −38.0 to −25.0, showing a significantly lower affinity. Again the docking prediction was verified by EMSA using the mutant protein (Supplementary Figure S4B). The Kd increased significantly by a factor of 3.8 (Figure 4D, P < 0.01, n = 3. Apparent Kd FMRP NT WT = 128 ± 22.7 nM and Kd FMRP NT Mut = 488 ± 12 nM, respectively). Competition experiments, using both unlabelled BC1 and U1 RNAs, showed that the mutated FMRP-NT/BC1 interaction remains specific (Figure 4E and G). In conclusion the electrostatic interactions of the two arginines R70 and R111 guide the interaction of the second tudor domain with BC1 RNA. Interestingly the binding partners of the two arginines are the phosphates at the 3′-side of the 2′-O-methylated nucleotides G46 and C47.
Figure 4.
Effect of R70, R111, N104, Y103 to the binding of BC1 RNA. (A–C) Docking models of the FMRP-NT WT, FMRP-NT R70E-R111E and FMRP-NT Y103A-N104A interaction with BC1, respectively. The yellow RNA backbone represents the nucleotides involved in the interaction. The key amino acids detected by mass spectrometry analysis are depicted as blue sticks. (D) EMSA experiment using BC1 RNA, FMRP-NT WT and FMRP-NT R70E-R111E mutant (10–100 ng). Shifted and unbound RNA are indicated by asterisk and arrowhead, respectively; the histogram shows the ratio of the Kd of mutant FMRP-NT versus the Kd of WT FMRP-NT. Error bars represent SE: **P < 0.01, Student's test, n = 3. (E) Competition experiments using unlabelled BC1 (lanes 3–5) and U1 (lanes 6–8) transcripts. (F) and (G) The same as in (D) and (E) but using the Y103A-N104A mutant. P = 0.07 Student's test, n = 5.
The FMRP–BC1–mRNA inhibitory complex at synapses
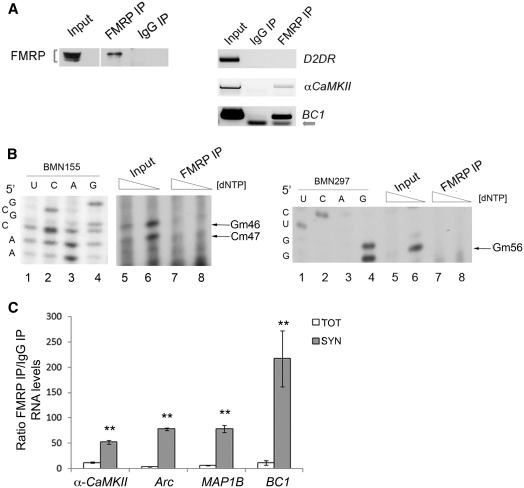
To investigate if BC1 2′-O-methylations have an effect on the affinity of FMRP for its mRNA targets, we immunoprecipitated FMRP complexed with BC1 RNA from total brain (Figure 5A, left panel). At first, we monitored that BC1 RNA was indeed associated to FMRP (Figure 5A, right panel), together with a well-known FMRP target (αCaMKII mRNA). The specificity of the interaction is revealed by the absence of D2DR mRNA in the FMRP complex (38). To address whether the BC1 bound to FMRP is 2′-O-methylated or not, we performed low dNTP primer extension assays from FMRP immunoprecipitated RNAs (Figure 5B). Using the BC1-specific primers, BMN155 and BMN297, the 2′-O-methylations were detected in the 5′-stem-loop only in the input RNA (Figure 5B, left and right panels: compare lanes 5 and 6) while the BC1 immunoprecipitated by FMRP was not 2′-O-methylated (Figure 5B: compare lanes 7 and 8). Since the in vitro data showed that FMRP binds less efficiently to 2′-O-methylated BC1 RNA (Figure 3G, Supplementary Figure S3C), we wanted to investigate if the presence or absence of 2′-O-methylations on BC1 RNA had also an effect on stabilizing FMRP target mRNAs in the complex. To address this point we immunoprecipitated FMRP from total brain and synaptoneurosomal extracts and detected the associated mRNAs in the complex. RT–qPCR assays showed a higher co-immunoprecipitation efficiency of FMRP target mRNAs/RNA (αCamKII, Arc, Map1B, BC1) in synaptoneurosome preparations versus total brain (Figure 5C, Supplementary Figure S5). Since at synapses BC1 is mainly detected in the non-methylated form, these findings suggest that absence of 2′-O-methylations favours the FMRP–BC1–mRNA association. Importantly, this complex has been previously shown in vivo to inhibit mRNA translation at synapses (5,6) and Figure 1D and F).
Figure 5.
At synapses, where BC1 is mainly unmethylated, FMRP binds its mRNA targets with higher affinity. (A) Immunoprecipitation of FMRP-associated mRNAs/RNA. Left panel, Western blot of FMRP immunoprecipitated from total brain extracts. Input (1/10), FMRP IP (1/3), the IgG IP (1/3). Right panel, RT–PCR from immunoprecipitated RNA derived from the input (1/10), IgG IP (2/3) and FMRP IP (2/3). The arrow points to the primers used to amplify BC1 RNA (B) Upper panel, low dNTP concentration primer extension analysis using the BMN155 primer on total brain RNA (Input, lanes 5 and 6) and immunoprecipitated BC1 RNA (lanes 7 and 8, n = 5). The arrows denote 2′-O-methylations at positions G46, C47. For each sample, the second lane shows the lower dNTP concentration. Sequencing reactions are shown in lanes 1–4. Lower panel as upper panel using the BMN297 primer able to detect the 2′-O-methylation on the G56, n = 3. (C) Enrichment of FMRP mRNA targets in immunoprecipitated FMRP complex from both total brain and at synapses. RT–qPCR of FMRP co-immunoprecipitated mRNAs/RNA. Shown is the ratio of the RNA precipitated using FMRP and rabbit unspecific IgGs from total (white histogram) and synaptoneurosomes (grey histogram) brain extracts, each normalized for the respective input. Error bars represent SE: **P < 0.01, Student's test, n = 3.
In conclusion, in this study, we show that (i) BC1 is differentially 2′-O-methylated according to its subcellular location. (ii) The tudor domain of FMRP, with its RNA-binding activity, binds FMRP recognizing the 2′-O-methylation status of BC1 RNA. (iii) At synapses, where BC1 RNA is not 2′-O-methylated, the FMRP–BC1–mRNA interaction is increased. (iv) At synapses BC1 regulates the translation of some FMRP target mRNAs.
At structural level, methylation of the sugar 2′-OH-group favour the 3′-endo conformation which stabilize helical conformations (46). It is possible to envision that the endo-conformation affects the binding to a given protein. In a highly complex cell like the neuron, which undergoes many changes during brain development, elaborate and fine-tuned mechanisms are required to regulate gene expression. We propose that changes in the 2′-O-methylation status of BC1 RNA contribute to the regulation of synaptic gene regulation and consequently neuronal plasticity.
These findings provide new insights into translational control at synapses and suggest that RNA modifications could have an important influence in genetic brain disease, a heretofore unknown relationship.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figures 1–5, Supplementary Methods and Supplementary References [3,6,16,29,49,50–54].
FUNDING
Telethon (GGP10150); Fonds Wetenschappelijk Onderzoek (G066709N); Vlaams Instituut Voor Biotechnologie and European Union Seventh Framework Programme under grant agreement n° (HEALTH-F4-2010-242167 “SynSys” project) (to C.B.); Methusalem (to B.D.S.); the National Autism Association Research (NAAR) (to C.L.) and European Molecular Biology Organization (ASTF 170-2008 to C.L.). Funding for open access charge: Telethon.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Tamas Kiss (CNRS, LBME, Toulouse, France) for plasmids expressing U1 and U2 snRNAs, Eliane Cherrette for assistance, Jonathan Royaert for technical help, Emanuela Pasciuto for sharing preliminary data and Hüseyin Besir, Giuseppe Novelli and Maria-Giulia Farace for access to facilities, Silvia De Rubeis and Esperanza Fernandez for comments on the article.
REFERENCES
- 1.Bagni C, Greenough WT. From mRNP trafficking to spine dysmorphogenesis: the roots of fragile X syndrome. Nat. Rev. Neurosci. 2005;6:376–387. doi: 10.1038/nrn1667. [DOI] [PubMed] [Google Scholar]
- 2.Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zalfa F, Eleuteri B, Dickson KS, Mercaldo V, De Rubeis S, di Penta A, Tabolacci E, Chiurazzi P, Neri G, Grant SG, et al. A new function for the fragile X mental retardation protein in regulation of PSD-95 mRNA stability. Nat. Neurosci. 2007;10:578–587. doi: 10.1038/nn1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang M, Wang Q, Huang Y. Fragile X mental retardation protein FMRP and the RNA export factor NXF2 associate with and destabilize Nxf1 mRNA in neuronal cells. Proc. Natl Acad. Sci. USA. 2007;104:10057–10062. doi: 10.1073/pnas.0700169104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zalfa F, Giorgi M, Primerano B, Moro A, Di Penta A, Reis S, Oostra B, Bagni C. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell. 2003;112:317–327. doi: 10.1016/s0092-8674(03)00079-5. [DOI] [PubMed] [Google Scholar]
- 6.Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, Di Marino D, Mohr E, Massimi M, Falconi M, et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134:1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 7.Edbauer D, Neilson JR, Foster KA, Wang CF, Seeburg DP, Batterton MN, Tada T, Dolan BM, Sharp PA, Sheng M. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65:373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adinolfi S, Ramos A, Martin SR, Dal Piaz F, Pucci P, Bardoni B, Mandel JL, Pastore A. The N-terminus of the fragile X mental retardation protein contains a novel domain involved in dimerization and RNA binding. Biochemistry. 2003;42:10437–10444. doi: 10.1021/bi034909g. [DOI] [PubMed] [Google Scholar]
- 9.Zalfa F, Adinolfi S, Napoli I, Kuhn-Holsken E, Urlaub H, Achsel T, Pastore A, Bagni C. Fragile X mental retardation protein (FMRP) binds specifically to the brain cytoplasmic RNAs BC1/BC200 via a novel RNA-binding motif. J. Biol. Chem. 2005;280:33403–33410. doi: 10.1074/jbc.M504286200. [DOI] [PubMed] [Google Scholar]
- 10.Ramos A, Hollingworth D, Adinolfi S, Castets M, Kelly G, Frenkiel TA, Bardoni B, Pastore A. The structure of the N-terminal domain of the fragile X mental retardation protein: a platform for protein-protein interaction. Structure. 2006;14:21–31. doi: 10.1016/j.str.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 11.Maurer-Stroh S, Dickens NJ, Hughes-Davies L, Kouzarides T, Eisenhaber F, Ponting CP. The Tudor domain ‘Royal Family': Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem. Sci. 2003;28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- 12.Costa-Mattioli M, Sossin WS, Klann E, Sonenberg N. Translational control of long-lasting synaptic plasticity and memory. Neuron. 2009;61:10–26. doi: 10.1016/j.neuron.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sutcliffe JG, Milner RJ, Bloom FE, Lerner RA. Common 82-nucleotide sequence unique to brain RNA. Proc. Natl Acad. Sci. USA. 1982;79:4942–4946. doi: 10.1073/pnas.79.16.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milner RJ, Bloom FE, Lai C, Lerner RA, Sutcliffe JG. Brain-specific genes have identifier sequences in their introns. Proc. Natl Acad. Sci. USA. 1984;81:713–717. doi: 10.1073/pnas.81.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muslimov IA, Lin Y, Heller M, Brosius J, Zakeri Z, Tiedge H. A small RNA in testis and brain: implications for male germ cell development. J. Cell Sci. 2002;115:1243–1250. doi: 10.1242/jcs.115.6.1243. [DOI] [PubMed] [Google Scholar]
- 16.Ferrari F, Mercaldo V, Piccoli G, Sala C, Cannata S, Achsel T, Bagni C. The fragile X mental retardation protein-RNP granules show an mGluR-dependent localization in the post-synaptic spines. Mol. Cell. Neurosci. 2007;34:343–354. doi: 10.1016/j.mcn.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 17.Chicurel ME, Terrian DM, Potter H. mRNA at the synapse: analysis of a synaptosomal preparation enriched in hippocampal dendritic spines. J. Neurosci. 1993;13:4054–4063. doi: 10.1523/JNEUROSCI.13-09-04054.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson EM, Kinoshita Y, Weinreb DB, Wortman MJ, Simon R, Khalili K, Winckler B, Gordon J. Role of Pur alpha in targeting mRNA to sites of translation in hippocampal neuronal dendrites. J. Neurosci. Res. 2006;83:929–943. doi: 10.1002/jnr.20806. [DOI] [PubMed] [Google Scholar]
- 19.Muramatsu T, Ohmae A, Anzai K. BC1 RNA protein particles in mouse brain contain two y-,h-element-binding proteins, translin and a 37 kDa protein. Biochem. Biophys. Res. Commun. 1998;247:7–11. doi: 10.1006/bbrc.1998.8657. [DOI] [PubMed] [Google Scholar]
- 20.Mallardo M, Deitinghoff A, Muller J, Goetze B, Macchi P, Peters C, Kiebler MA. Isolation and characterization of Staufen-containing ribonucleoprotein particles from rat brain. Proc. Natl Acad. Sci. USA. 2003;100:2100–2105. doi: 10.1073/pnas.0334355100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobayashi S, Agui K, Kamo S, Li Y, Anzai K. Neural BC1 RNA associates with pur alpha, a single-stranded DNA and RNA binding protein, which is involved in the transcription of the BC1 RNA gene. Biochem. Biophys. Res. Commun. 2000;277:341–347. doi: 10.1006/bbrc.2000.3683. [DOI] [PubMed] [Google Scholar]
- 22.Muddashetty R, Khanam T, Kondrashov A, Bundman M, Iacoangeli A, Kremerskothen J, Duning K, Barnekow A, Huttenhofer A, Tiedge H, et al. Poly(A)-binding protein is associated with neuronal BC1 and BC200 ribonucleoprotein particles. J. Mol. Biol. 2002;321:433–445. doi: 10.1016/s0022-2836(02)00655-1. [DOI] [PubMed] [Google Scholar]
- 23.Kondrashov AV, Kiefmann M, Ebnet K, Khanam T, Muddashetty RS, Brosius J. Inhibitory effect of naked neural BC1 RNA or BC200 RNA on eukaryotic in vitro translation systems is reversed by poly(A)-binding protein (PABP) J. Mol. Biol. 2005;353:88–103. doi: 10.1016/j.jmb.2005.07.049. [DOI] [PubMed] [Google Scholar]
- 24.Wang H, Iacoangeli A, Popp S, Muslimov IA, Imataka H, Sonenberg N, Lomakin IB, Tiedge H. Dendritic BC1 RNA: functional role in regulation of translation initiation. J. Neurosci. 2002;22:10232–10241. doi: 10.1523/JNEUROSCI.22-23-10232.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muslimov IA, Iacoangeli A, Brosius J, Tiedge H. Spatial codes in dendritic BC1 RNA. J. Cell Biol. 2006;175:427–439. doi: 10.1083/jcb.200607008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maden BE. Mapping 2′-O-methyl groups in ribosomal RNA. Methods. 2001;25:374–382. doi: 10.1006/meth.2001.1250. [DOI] [PubMed] [Google Scholar]
- 27.Motorin Y, Muller S, Behm-Ansmant I, Branlant C. Identification of modified residues in RNAs by reverse transcription-based methods. Methods Enzymol. 2007;425:21–53. doi: 10.1016/S0076-6879(07)25002-5. [DOI] [PubMed] [Google Scholar]
- 28.Yu YT, Shu MD, Steitz JA. A new method for detecting sites of 2′-O-methylation in RNA molecules. RNA. 1997;3:324–331. [PMC free article] [PubMed] [Google Scholar]
- 29.Stark MR, Pleiss JA, Deras M, Scaringe SA, Rader SD. An RNA ligase-mediated method for the efficient creation of large, synthetic RNAs. RNA. 2006;12:2014–2019. doi: 10.1261/rna.93506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Rubeis S, Bagni C. In: Encyclopedia of Neuroscience. Binder MD, Hirokawa N, Windhorst U, Hirsch MC, editors. Springer; 2009. Part 19. pp. 3982–3985. [Google Scholar]
- 31.Kuhn-Holsken E, Lenz C, Dickmanns A, Hsiao HH, Richter FM, Kastner B, Ficner R, Urlaub H. Mapping the binding site of snurportin 1 on native U1 snRNP by cross-linking and mass spectrometry. Nucleic Acids Res. 2010;38:5581–5593. doi: 10.1093/nar/gkq272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lenz C, Kuhn-Holsken E, Urlaub H. Detection of protein-RNA crosslinks by NanoLC-ESI-MS/MS using precursor ion scanning and multiple reaction monitoring (MRM) experiments. J. Am. Soc. Mass Spectrom. 2007;18:869–881. doi: 10.1016/j.jasms.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 33.Martin Richter F, Hsiao HH, Plessmann U, Urlaub H. Enrichment of protein-RNA crosslinks from crude UV-irradiated mixtures for MS analysis by on-line chromatography using titanium dioxide columns. Biopolymers. 2009;91:297–309. doi: 10.1002/bip.21139. [DOI] [PubMed] [Google Scholar]
- 34.Parisien M, Major F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature. 2008;452:51–55. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- 35.Rozhdestvensky TS, Kopylov AM, Brosius J, Huttenhofer A. Neuronal BC1 RNA structure: evolutionary conversion of a tRNA(Ala) domain into an extended stem-loop structure. RNA. 2001;7:722–730. doi: 10.1017/s1355838201002485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao X, Yeo G, Muotri AR, Kuwabara T, Gage FH. Noncoding RNAs in the mammalian central nervous system. Annu. Rev. Neurosci. 2006;29:77–103. doi: 10.1146/annurev.neuro.29.051605.112839. [DOI] [PubMed] [Google Scholar]
- 37.Gabus C, Mazroui R, Tremblay S, Khandjian EW, Darlix JL. The fragile X mental retardation protein has nucleic acid chaperone properties. Nucleic Acids Res. 2004;32:2129–2137. doi: 10.1093/nar/gkh535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Centonze D, Rossi S, Napoli I, Mercaldo V, Lacoux C, Ferrari F, Ciotti MT, De Chiara V, Prosperetti C, Maccarrone M, et al. The brain cytoplasmic RNA BC1 regulates dopamine D2 receptor-mediated transmission in the striatum. J. Neurosci. 2007;27:8885–8892. doi: 10.1523/JNEUROSCI.0548-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Skryabin BV, Sukonina V, Jordan U, Lewejohann L, Sachser N, Muslimov I, Tiedge H, Brosius J. Neuronal untranslated BC1 RNA: targeted gene elimination in mice. Mol. Cell. Biol. 2003;23:6435–6441. doi: 10.1128/MCB.23.18.6435-6441.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- 41.Zhang YQ, Bailey AM, Matthies HJ, Renden RB, Smith MA, Speese SD, Rubin GM, Broadie K. Drosophila fragile X-related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell. 2001;107:591–603. doi: 10.1016/s0092-8674(01)00589-x. [DOI] [PubMed] [Google Scholar]
- 42.Lu R, Wang H, Liang Z, Ku L, O'Donnell WT, Li W, Warren ST, Feng Y. The fragile X protein controls microtubule-associated protein 1B translation and microtubule stability in brain neuron development. Proc. Natl Acad. Sci. USA. 2004;101:15201–15206. doi: 10.1073/pnas.0404995101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park S, Park JM, Kim S, Kim JA, Shepherd JD, Smith-Hicks CL, Chowdhury S, Kaufmann W, Kuhl D, Ryazanov AG, et al. Elongation factor 2 and fragile X mental retardation protein control the dynamic translation of Arc/Arg3.1 essential for mGluR-LTD. Neuron. 2008;59:70–83. doi: 10.1016/j.neuron.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hou L, Antion MD, Hu D, Spencer CM, Paylor R, Klann E. Dynamic translational and proteasomal regulation of fragile X mental retardation protein controls mGluR-dependent long-term depression. Neuron. 2006;51:441–454. doi: 10.1016/j.neuron.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 45.Helm M. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res. 2006;34:721–733. doi: 10.1093/nar/gkj471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Motorin Y, Helm M. tRNA stabilization by modified nucleotides. Biochemistry. 2010;49:4934–4944. doi: 10.1021/bi100408z. [DOI] [PubMed] [Google Scholar]
- 47.Massenet S, Mougin A, Branlant C. In: Post-Transcriptional Modifications in the U Small Nuclear RNAs. Grosjean H, Benne R, editors. Washington DC: ASM Press; 1998. pp. 201–227. [Google Scholar]
- 48.Adinolfi S, Bagni C, Musco G, Gibson T, Mazzarella L, Pastore A. Dissecting FMR1, the protein responsible for fragile X syndrome, in its structural and functional domains. RNA. 1999;5:1248–1258. doi: 10.1017/s1355838299990647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 50.Hoe HS, Fu Z, Makarova A, Lee JY, Lu C, Feng L, Pajoohesh-Ganji A, Matsuoka Y, Hyman BT, Ehlers MD, et al. The effects of amyloid precursor protein on postsynaptic composition and activity. J. Biol. Chem. 2009;284:8495–8506. doi: 10.1074/jbc.M900141200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sheng M. Molecular organization of the postsynaptic specialization. Proc. Natl Acad. Sci. USA. 2001;98:7058–7061. doi: 10.1073/pnas.111146298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoareau-Aveilla C, Bonoli M, Caizergues-Ferrer M, Henry Y. hNaf1 is required for accumulation of human box H/ACA snoRNPs, scaRNPs, and telomerase. RNA. 2006;12:832–840. doi: 10.1261/rna.2344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vriend G. WHAT IF: a molecular modeling and drug design program. J. Mol. Graph. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. 29. [DOI] [PubMed] [Google Scholar]
- 54.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–38. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.